ABSTRACT
Aptamers are high affinity single-stranded nucleic acid or protein ligands which exhibit specificity and avidity comparable to, or exceeding that of antibodies and can be generated against most targets. The functionality of aptamers is based on their unique tertiary structure, complexity and their ability to attain unique binding pockets by folding. Aptamers are selected in vitro by a process called Systematic Evolution of Ligands by Exponential enrichment (SELEX). The Kd values for the selected aptamer are often in the picomolar to low nanomolar range. Stable and nontoxic aptamers could be selected for a wide range of ligands including small molecules to large proteins. Aptamers have shown tremendous potential and have found multipurpose application in the field of therapeutic, diagnostic, biosensor and bio-imaging. While their mechanism of action can be similar to that of monoclonal antibodies, aptamers provide additional advantages in terms of production cost, simpler regulatory approval and lower immunogenicity as they are synthesized chemically. Human immunodeficiency virus (HIV) is the primary cause of acquired immune deficiency syndrome (AIDS), which causes significant morbidity and mortality with a significant consequent decrease in the quality of patient's lives. While cART has led to good viral control, people living with HIV now suffer from non-HIV comorbidities due to viral protein expression that cannot be controlled by cART. Hence pathophysiological mechanisms that govern these comorbidities with a focus on therapies that neutralize these HIV effects gained increased attention. Recent advances in HIV/AIDS research have identified several molecular targets and for the development of therapeutic and diagnostic using aptamers against HIV/AIDS. This review presents recent advances in aptamers technology for potential application in HIV diagnostics and therapeutics towards improving the quality of life of people living with HIV.
Introduction
Oligonucleotide aptamers are single stranded, short RNA or DNA molecules of 20–80 nucleotide length, capable of attaining tertiary complex structures to bind specifically to targets against which they are selected. These aptamers are selected by an iterative process called SELEX [Citation1,Citation2]. With increased rounds of selection, possible complex tertiary structures present in the random library are enriched to obtain increased binding efficacy against varied target molecules ranging from small molecules, peptides or protein targets. Aptamers can be considered functional analogs of antibodies. Binding of aptamers to its specific ligand is governed by specific structures, shape complementarity and conformation, involving binding pockets and bond formation such as hydrogen bond, electrostatic interaction and van der waals forces. Aptamers can be used as antagonists or agonists to modulate cell signaling [Citation3,Citation4] or as targeting ligands for delivery of drugs and other cargo in a tissue specific or cell type specific manner [Citation5,Citation6]. Recent advances in aptamer technology and automation of SELEX has increased the potential for aptamer use in therapeutics and diagnostics [Citation7–10]. Aptamers can be used in an extensive array of applications in diverse fields of diagnostics, delivery, bio-imaging, bio sensing and therapeutics. [Citation11–15] Unlike antibodies, aptamers, can be synthesized chemically and hence offer significant advantages in terms of production cost, simpler regulatory approval and lower immunogenicity when administered in preclinical doses 1000-fold greater than those used for animal and human therapeutic application [Citation9,Citation13]. Aptamers are highly specific and can discriminate between related proteins that share common sets of structural domains [Citation16,Citation17]. Aptamers are already in clinical use with Macugen™ approved by the FDA in 2004 for macular degeneration [Citation18-20]. Clinicaltrials.gov currently lists 19 trials with aptamers (completed, in progress or expected to begin).
Combination antiretroviral therapy (cART) has made HIV a treatable but a chronic disease. However, HIV patients continue to die of non-AIDS comorbidities almost a decade earlier that their non-HIV counterparts [Citation21]. Despite this progress, some comorbidities continue to remain highly prevalent among HIV-infected individuals. Most of these comorbidities are associated with underlying chronic inflammation or the actions of HIV proteins expressed by infected cells [Citation21,Citation22]. For instance, Tat is an immediate early gene of HIV and its expression cannot be suppressed by antiretrovirals [Citation23,Citation24]. Its protein transduction domain allows its secretion by infected cells and uptake by bystander cells where it mediates pleotropic effects [Citation25–28]. While aptamers hold great promise for HIV research, diagnostics and therapy, their application remains limited. Several aptamers for HIV-1 proteins as well as host proteins that interact with HIV-1 have been generated and characterized with functional role like neutralization of virions or its proteins [Citation29–32]. This review focuses on the prospects of aptamers as therapeutic leads and diagnostic tools for HIV/AIDS.
Systematic evolution of ligands by exponential enrichment (SELEX)
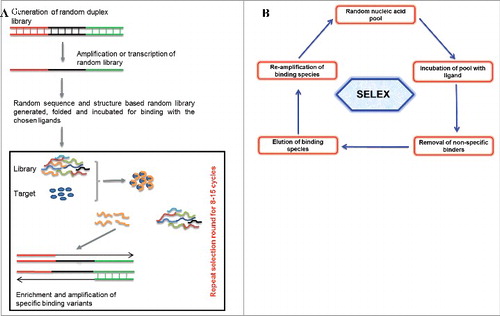
Aptamers are isolated and characterized for their binding affinity and specificity against the chosen ligands through an iterative in vitro selection process called SELEX [(), Citation1,Citation2]. A randomized oligonucleotide library folds into complex structural pool. Considering the susceptibility of oligonucleotides to serum nucleases, modified oligonucleotide are widely used for generating nuclease resistant aptamers [Citation33–35]. A starting aptamer library mostly consists of a central random region ranging from 20–60 nucleotide long, flanked by known sequences to allow primer binding for amplification, enrichment, cloning and characterization. Briefly, a usual SELEX protocol requires preclearing to remove library members that bind to the support, matrix or a ligand attached to your target of interest to facilitate separation. This is followed by incubation of the precleared library with the chosen ligand for their binding, nonspecific binders are removed by employing washes with increasing stringency and specific binding variants are amplified. In order to effectively obtain bound species with higher affinity, the selection stringency is cautiously controlled by adjusting conditions and adding negative or counter completive selections. The minimization of selection round is possible using advanced or automated SELEX protocols such as RAPID SELEX [Citation7,Citation8,Citation36]. Specificity check and counter negative selection are usually perform to get rid of weak binders and nonspecific variants. Aptamers selected from SELEX further need structural and binding characterization and based on such analysis best aptamer candidates are selected for biochemical, functional and biological application. The final selected and characterized aptamers could be used for several applications [Citation13,Citation37–42]. From the discovery of the first SELEX protocol till now, different types of SELEX have been established and merged with other advance technology based on their application in diverge fields [Citation10,Citation15,Citation43,Citation44].
Figure 1. (1A) Representation of aptamer selection. Briefly random oligonucleotide libraries with diverse structural complexities are generated and incubated for ligand binding. Non-specific binders are removed and high affinity binders are amplified. The selection cycle are usually repeated around 5-20 cycles. The sequences and structural characterization from chosen binders are done to achieve the best binding aptamer. (1B) Basic strategy of SELEX protocol showing incubation of random library with chosen ligands, their incubation for binding, removal of non-specific binder and finally the enrichment of the best binders for chosen targets.
Functional aptamers and its applications
Aptamers have found diverse application in the field of biomedical research. Aptamers have been used to target small molecules, large complex molecules, agonists and antagonists and function as inhibitors or activators of cell signaling in specific diseases [Citation41,Citation45–48]. Naturally occurring nucleic acid aptamers are present in the genome and also as a constituent of riboswitches which can be used to elucidate the role of nucleic acids in signal transduction [Citation49–52]. Aptamers are functionally comparable to antibodies and possess a wide repertoire of charge and structure combinations for extensive use in various biomedical, diagnostic, in vitro or in vivo bioimaging, and therapeutic applications. These are also broadly used as research tools in diverse areas like bioassays, imaging, pathogen detection, tissue staining, targeted delivery vehicles, aptazyme, biosensor and nanomedicine [Citation43,Citation44,Citation53–55]. Aptamers also have ability to recognize toxin molecules and hazardous chemicals which makes them suitable candidates for use in food safety and environment monitoring [Citation56–58]. Aptamers can be minimally truncated so as to retain only the biologically active portion from the initial SELEX without losing the functionality. In vitro modification of aptamers without loss of functionality (for eg; enhancing stability)[Citation34,35,59] combined with cost effective bulk synthesis makes them ideal candidates for therapeutic leads or for targeted drug delivery. For instance a number of reports have demonstrated the use of aptamers for cell type specific delivery of siRNAs [Citation60–62]. Aptamers have also found application in biomarker discovery during HIV infection and host interaction [Citation63–65]. Few aptamers have also been reported with ability to differentiate between cancerous and non-cancerous receptor that could be used as specific drug delivery agent and for biomarkers identification [Citation66–69].
Stages of the HIV life cycle
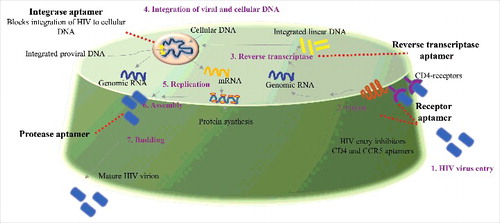
HIV infection promotes immunodeficiency by selectively targeting CD4 T-cells. CD4 cells are involved in crucial immune functioning. The HIV infection cycle can broadly be divided into seven steps or stages (binding, fusion, reverse transcription, integration, replication, assembly and budding) [(), Citation70]. An insight into the mechanism of HIV infection cycle is important to develop efficient anti HIV therapeutics and diagnostics. Therapeutic agents against HIV are designed to target one or more of the several stages of the HIV infection cycle. Currently six classes of anti-HIV drugs target four steps of HIV's lifecycle and are often used in combination called combination antiretroviral therapy. However, a number of these drugs can have side effects and in some cases also contribute to HIV related comorbidities. Aptamer-based targeting of HIV at multiple stages can be more specific, cheaper to produce synthetically and can be similarly designed to target multiple stages of the HIV infection cycle. shows the different stages in the HIV infection cycle that can be targeted by aptamers.
Figure 2. General layout of potential HIV aptamers: The whole cycle of HIV infection and replication consist of seven steps or stages (binding, fusion, reverse transcription, integration, replication, assembly and budding) called as “the HIV life cycle”. Aptamer targeted for different stages of HIV and its potential use for development of anti HIV medicines to treat HIV infection hold great significance.
Aptamers for human immunodeficiency virus (HIV)
Human immunodeficiency virus is the primary cause of AIDS and AIDS related comorbidities . The primary targets of HIV are CD4+ T cells, macrophages, and dendritic cells. Depletion of immune cells and immune dysregulation result in the AIDS. Since the advent of antiretroviral therapy, HIV infected patients have lived longer than ever before. However, viral replication persists even in presence of cART and leading to expression and secretion of viral proteins [Citation21,23,24] that can have pleotropic effects on bystander cells [Citation25–27]. Several aptamers have been selected based on their ability to neutralize HIV and/or its proteins with a goal towards developing them for therapeutic and diagnostic purposes. Aptamers could be further established and tested in preclinical and clinical studies as a potential new entry inhibitor drug against HIV/AIDS.
Potential HIV aptamer for diagnostic application
While most of the work with anti-HIV aptamers has been done with the intention of generating therapeutic aptamers, some studies have also identified aptamers that can be used for diagnostics. In the subsequent sections we will discuss the aptamers that have found application in HIV either as diagnostics or therapeutics. Initial diagnosis of HIV in human are mostly done by ELISA and real time based an antigen/antibody combination immunoassay that generally identifies HIV-1 and HIV-2 antibodies and HIV-1 p24 antigen HIV-1 infection. Aptamers that diagnose HIV proteins such as Tat, gp120 and Rev, have been primarily used in several biosensor systems and hold great significance as diagnostic tool development for HIV. Tat is an immediate early gene of HIV and its expression is suppressed by antiretrovirals [Citation73–76]. Its protein transduction domain allows secretion by infected cells and uptake by bystander cells where it mediates pleotropic effects [Citation24–26] leading to several HIV associated comorbidities in the cART era. Hence determining amounts of Tat in the serum can be a useful biomarker for subclinical HIV associated comorbidities before manisfestation of clinical disease. Tombelli et al., have designed and developed aptamer-based biosensors for the detection of HIV-1 Tat protein [Citation77]. High affinity aptamers selected to specifically bind HIV-1 Tat protein were immobilized on gold surface of piezoelectric quartz crystals SPR chips. A quartz crystal microbalance (QCM)-based and SPR-based biosensor were constructed using biotinylated aptamer. Binding characterization and specificity checks were performed. Rev proteins was used as negative control during for the specificity assays. They have shown that two biosensors were reproducible in terms of immobilization and binding [Citation77]. In another study by Minunni et al., [Citation78] RNA aptamer against HIV-1 Tat was used for the development of aptasensor. Development of aptamer was based on aptamer region with bio-recognition element. This designed biosensor was standardized using piezoelectric quartz-crystals. Concisely, biotinylated aptamer was immobilized on the gold electrode of the crystal using biotin-streptavidin interaction. The authors also investigated aptasensor specificity, sensistivity and reproducibility in the detail. The developed aptasensor was compared with an immunosenor where Tat specific monoclonal anti-Tat antibody was immobilized on carboxylated dextran. The sensitivity of the Tat aptasensor was found to be comparable to that of the immunosensor as an effective diagnostic tool. Similar to Tat, anti-Rev aptamers have also been used in several biosensor systems using methods such as based in SPR, QCM and the diamond field-effect transistor (FET) technique [Citation79]. Yamamoto et al 2000 have demonstrated molecular beacon aptamer fluorescence in the presence of Tat protein of HIV-1 [Citation80]. They have described Tat specific RNA aptamers with higher binding affinity than its canonical TAR RNA binding partner on HIV-1 RNA. They also demonstrated inhibition of Tat dependent transactivation in vitro and in vivo [Citation81]. They have suggested the use of aptamer derived oligomers to evaluate the HIV Tat and the potential applications of such tools as biosensors.
CD4 is the primary HIV receptor on target cells. Presence of CD4 receptor on target cells is critical for productive HIV infection [Citation82–84]. Blocking CD4 using monoclonal antibodies results in inhibition of viral entry [Citation85–87]. Kenneth et al., used recombinant human CD4 on bead based affinity matrix to screen 2-F modified random RNA library to select high affinity CD4 aptamers [Citation88]. Aptamers were conjugated with different fluorophores like phycoerytherin and fluorescein. Fluorophore conjugated aptamers were used for cell surface CD4 staining followed by flow cytometry analysis demonstrating that aptamers could be developed for diagnostic applications. Aptamers can be easily modified for diverse applications for instance Zhou et al., generated aptamer coated surface to capture the cells expressing CD4 antigen [Citation89]. Briefly, glass or silicon surfaces were modified with amine-terminated silanes followed by coating with thiolated anti-CD4 RNA aptamers. Data showed aptamer-functionalized surfaces have comparable capture efficiency to substrates containing anti-CD4 antibody. These aptamers have potential scope in diagnosis and in the generation of aptamer based monitoring and diagnostic kits for HIV/AIDS. Jing et al., developed two families of G-quartet oligonucleotides named as T40217-T40222, with possible formation of a tail to tail G-quartet dimer and T40224-T40227, with phosphorothioate (PT) linkages in the guanine loops [Citation90]. The authors suggested that G-quartet motif is the crucial prerequisite for the nuclease resistance. Such stable and resistant aptamer hold great promise in development as effective and diagnostic tool.
Potential therapeutic aptamers against proteins involved in HIV infection
Aptamers against Gp120
Entry of HIV-1 into host cells requires the virion surface envelope (Env) glycoproteins to interact with specific cell surface receptors on target cells [Citation91]. Hence targeting Env proteins for inhibition of viral entry has great therapeutic potential. HIV Gp120 is plays a critical role by interacting with its primary receptor CD4 in combination of either CCR5 or CXCR4 to effect viral entry [Citation92,Citation93]. A number of aptamers selected against gp120 have demonstrated neutralization of HIV infectivity by blocking vital entry. Mufhandu et al[30] have demonstrated a family of gp120 binding RNA aptamers that can block viral entry by binding to and neutralizing HIV gp120. Briefly, a synthetic derivative of aptamer, called UCLA1 was used against a large panel of HIV-1 subtype C viruses. The authors have previously isolated 2′-F-pyrimidine substituted RNA aptamers against HIV-1BaLgp120 and shown that they neutralized infectivity in cell-based assays [Citation94]. The binding of UCLA1 to HIV-1 subtype C gp120 resulted in neutralization of the same subtype. The chosen aptamers had been tested both in cell lines and primary cells for their toxicity and their binding sites on gp120 were characterized. Furthermore UCLA1 demonstrated high efficacy at low doses, no toxicity, while also demonstrating a synergistic effect with entry inhibitors [Citation30,Citation94]. Overall, these studies suggest that UCLA1 has potential for further development as a HIV-1 entry inhibitor. The fluoro-modification of the aptamer backbone also resulted in the UCLA1 aptamer being more stable in biological fluids. An additional derivative, named UCLA005 with a further chemical modification (UCLA005v11) lead to a 4-fold increase in aptamer stability, consequently suggesting that modified aptamers are highly stable. [Citation95] Zhou et al 2011 used a nitrocellulaose based SELEX approach to isolate 2'-F modified aptamers against gp120 with nanomolar (nM) affinity . The selected aptamers could be rapidly internalized into cells expressing HIV-1 envelope protein and neutralize HIV-1 infectivity. With aptamer A-1, the authors generated an exclusive dual inhibitory anti-gp120 aptamer-siRNA chimera with effective anti-HIV activities. They have also used this chimeras for cell-type specific delivery of the siRNA into HIV-1 infected cells and proposed the combinatorial of nucleic-acid based therapeutic agents such as aptamer, miRNA and siRNA for effective suppression of HIV. The aptamer-siRNA chimeras demonstrated by them can be developed as therapeutic leads for patients failing other current antiretroviral therapy . Likewise, Khati et al. in 2003 and Dey et al 2005 showed the isolation of 2-fluoro-modified RNA aptamers that bind specifically to the surface gp 120 [Citation94,Citation97]. Later, Dey et al 2005 has shown an anti-gp120 RNA aptamer neutralizes R5 strains of HIV-1 [Citation32]. They have also reported a comprehensive structural characterization of the selected aptamer. Binding and structural characterization were done by enzymatic, mutagenesis, secondary-structure analysis, surface plasmon resonance (SPR) and chemical probing methods. The study provide information to understand the molecular interactions between the virus and its host cell, besides this the selected aptamer possess properties to be developed as clinically relevant anti-viral HIV therapeutic. Cohen et al., have developed an aptamer that neutralizes R5 strains of HIV-1 by binding to the core residues of gp120 in the CCR5 binding site. They have earlier reported aptamers that bind the surface envelope gp120, of HIV-1, which have shown to neutralize infection of diverse sub-types of virus. Later truncated and chemically modified derivatives were generated and studied by the solid-phase synthesis. The structural understanding from their data may deliver a scope for development of possible anti-viral agents [Citation98]. Dey et al 2005 have reported an aptamer that neutralizes R5 strains of HIV-1 blocks via disturbance of gp120-CCR5 interaction. They isolated and characterized 2′-F-pyrimidine -substituted RNA aptamers that bind to gp120 of R5 strains of HIV-1. This modified aptamers were able to neutralize the infectivity of phylogenetically diverse R5 strains. They also elucidated the mechanism involved in such neutralization by demonstrating that the aptamer binds to the CCR5-binding site on gp120 in a relatively CD4-independent manner [Citation97].
Aptamers against Gag protein
Gag polyprotein can serve as another therapeutic target against HIV infection. A few studies have generated and characterized aptamers against Gag protein [Citation99,Citation100]. Gag polyprotein is a structural protein of HIV-1. Ramalingam et al., investigated the prospective of anti-Gag RNA aptamers to inhibit HIV-1 replication. They have generated and characterized RNA aptamers against full-length Gag protein of HIV-1. The selected aptamers were further established for their efficacy and functionality in terms of blocking the HIV replication in cultured cells. Interestingly the selected aptamers showed several fold inhibition in the extracellular capsid levels. Reduced cellular levels of mRNA for Gag was observed, suggesting a molecular structural control mechanism mediated via specific Gag-genomic RNA interactions [Citation99]. The authors also suggested hematopoietic stem cell (HSC)-based gene therapy, where HSCs are engineered to express anti-HIV molecules such as aptamers. Lochrie et al., isolated aptamers to HIV-1 Gag which bound to nucleocapsid (NC) region interfered with binding of psi (ψ) packaging signal [Citation100]. Their results suggest that RNA aptamers may offer an innovative way for inhibiting HIV replication.
Aptamers against Rev protein
The Rev protein of HIV-1 acts post transcriptionally to mediate export of unspliced and partially spliced viral RNA thereby allowing expression of late genes [Citation101]. HIV Rev specifically binds to to Rev responsive element (RRE) located on unspliced and partially spliced viral RNA to mediate RNA export. Inhibition of rev axis using siRNA or trans-dominant negative mutants have demonstrated significant suppression of viral replication [Citation102,Citation103]. However it is not known if the binding is “structure-specific” or sequence-specific. RNA constitutes an imperative structural component of numerous ribonucleoprotein (RNP) complexes often functioning as a scaffold to allow oligomerization of proteins, in ribosome and spliceosome. HIV viral replication requires assembly of RNP, consisting of Rev protein and RRE as an essential prelude to nuclear export of unspliced viral mRNAs. The specific mechanism necessitates protein-protein and protein-RNA interactions [Citation104]. RNA aptamers for HIV-1 Rev Protein have previously been isolated with high affinity and specificity using in vitro selection. In vitro studies has found that selected sequences also compete with the wild-type Rev-binding element (RBE). Symensma et al., did biological functional characterization of selected aptamer to determine whether the chosen aptamers could functionally substitute for RRE in vivo [Citation105]. Reporter system based hybrid RREs were generated and analyzed for their ability to facilitate Rev function. Remarkably, the data suggests that aptamers were functionally comparable to the wild-type element. Konopka et al., had shown that HIV-1 gene expression could be inhibited by the anti-HIV Rev-binding aptamer [Citation106]. They further improved the antiviral effect of the selected aptamers using a ribozyme directed, with receptor ligand-facilitated cationic liposome delivery. Their data concludes strong favor for the use of in vitro isolated and enriched ligands as possible anti-HIV agent.
Aptamers for nucleocapsid protein
Nucleocapsid (NC) protein of HIV-1 is involved in the encapsidation of viral RNA and assembly of viral particle [Citation107–109]. Inhibition of nucleocapsid synthesis inhibits viral replication, thereby making them attractive as potential targets for anti-HIV therapy [Citation110–113]. Kim et al 2002 isolated RNA aptamers that recognize the mature form of the NC protein [Citation114]. RNA aptamers selected in vitro was able to bind NC with high affinity, specificity and was able to compete for the packaging element (psi) RNA binding to the NC protein, suggesting that the aptamer can be used to inhibit viral packaging [Citation114]. Kim and Jeong, also screened two independent RNA libraries one of which had additional viral psi-sequences to isolate RNA aptamers that bound to the mature form of the NC protein of HIV-1. RNase foot printing was performed to characterize the RNA structures and to map the protein binding sites and data revealed that RNA aptamers that bind NC protein contain pseudoknots in addition to the characteristic stem-loop structure to facilitate RNA and NC (HIV-1) protein interaction [Citation115]. Later Kim and Jeong, further used one of such RNA aptamer to the NC protein, N70-13, to investigate its effect on NC protein both in vitro and in cell culture [Citation116]. They found complete abolishment of NC binding to the stable “transactivation response hairpin” and “psi RNA stem-loops of HIV-1 RNA”. The same aptamer were also expressed as an intramer that resulted in the inhibition of packaging of viral genomic RNA, underlying importance of targeting NC via aptamer tools for the optional development of clinical agent.
Potential therapeutic aptamers against receptors involved in HIV infection
Aptamers for CD4
Earlier, Zhang et al., have used RNA aptamers as probes specific for human CD4 for possible use in cell phenotyping [Citation117]. They found the synthetic CD4 aptamer showed comparable cell-binding specificity as standard CD4 antibody. Additionally, the aptamer can be combined with antibodies for use in multicolor flow cytometry analysis with significant repercussions for clinical application. Wheeler et al., reported aptamer-siRNA chimeras in which the CD4 aptamer was coupled with siRNA targeting HIV Gag and Vif or host CCR5 for targeted delivery to HIV susceptible cells. The chimeras were specifically taken up by CD4+ cells, which subsequently resulted in vitro and in vivo inhibition of HIV infection in primary CD4+ T cells and macrophages. They further demonstrated that CD4 aptamer/siRNA chimeras with hydroxyl ethylcellulose gel formulation resulted in complete transmission block in polarized human cervicovaginal explants and humanized mice [Citation118,Citation119]. Likewise, Guo et al., have also used CD4 aptamer and specific chimeric RNA scaffolds for targeted delivery [Citation120]. Blocking or modulating the interaction of viral gp120 and CD4-expressing T cells could be a clinically significant anti-viral strategy for suppressing HIV. Zhao et al., isolated ssDNA anti-CD4 aptamers that could interfere with HIV gp120 CD4 interaction suggesting strong potential of such molecules in as anti HIV therapy [Citation29].
Aptamers for CCR5
CCR5 is a G-protein coupled seven transmembrane receptor expressed by T cells and macrophages that act as co-receptor for macrophage-tropic HIV-1 [Citation121,Citation122]. Many reports have demonstrated selection of anti-CCR5 aptamers to inhibit HIV entry. Zhou et al., used live cell based SELEX combined with high-throughput sequencing technology to select cell-specific RNA aptamers against human CCR5 [Citation31]. These aptamers targeted HIV-1 susceptible cells and inhibited HIV-1 infectivity. Anti-CCR5 aptamer was able to inhibit R5 virus infection in primary peripheral blood mononuclear cells. Their data also suggest that CCR5-targeted aptamers and aptamer-siRNA conjugates offer a potential anti HIV/AIDS agent with targeted delivery of several therapeutic candidates. Scheideman et al., has reported peptide aptamers that bind the transmembrane region of CCR5 and inhibit CCR5 cell surface expression and HIV co-receptor function [Citation123]. Briefly, screening of library of randomized transmembrane domains was performed and theses variants were named as termed “traptamers,” for transmembrane aptamers. Six 44- or 45-amino-acid proteins with completely different transmembrane sequences were isolated and characterized. Traptamers were able to inhibit both, cell surface, and total expression of the HIV co-receptor CCR5. Truncation and standardization of two traptamers augmented its activity and resulted in higher activity in terms of inhibition of R5-tropic reporter virus. These active traptamers specifically recognized the transmembrane domains of CCR5. The authors have also constructed novel multiple proteins not found in nature that interfere with CCR5 expression and inhibit HIV infection [Citation123]. This novel strategy provides valuable insight to target organization of the transmembrane domains of CCR5 and to target biological cellular pathways mediated by them. These molecules could be developed as an innovative diagnostic approach or anti-HIV therapeutic.
Aptamers against enzymes involved in HIV infection
Aptamers for reverse transcriptase (RT)
HIV-1 RT has been targeted for the development of anti-HIV therapeutics drug. Aeksiri et al in 2014 have reported HIV-1 (RT) reverse transcriptase-aptamer interaction using molecular dynamics simulations [Citation124]. Authors have examined the conformational dynamics of aptamer bound HIV-1 RT complex with an aptamer (). Briefly, they have studied apo HIV-1 RT, HIV-1 RT bound RNA aptamer, and also HIV-1 RT bound DNA substrate. Their data establish that HIV-1 RT complex with an aptamer was more stable than DNA substrate complex. Aptamer and HIV-RT binding interaction showed fingers-and-thumb-closed conformation and key residues involved in interaction were identifies. Their data gives insight about HIV-RT and aptamer interaction at molecular level that will help in the development of efficient anti-HIV therapeutics. Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure was first demonstrated by Ferris et al in 1990 [Citation125]. They reported HIV-1 reverse transcriptase from HIV type 1 has two subunits, which have respective molecular weights of 66 and 51 kDa. Importance of HIV-1 reverse transcriptase in HIV replication and infection are well established, which signify its obvious targeting for AIDS therapeutic claim [Citation126,Citation127]. A number of antiretroviral are designed to target and inhibit HIV reverse transcriptase. DeStefano and Nair have reported DNA aptamer (37NT) for the reverse transcriptase of HIV HXB2 strain [Citation128]. They have developed novel small single-stranded loop-back DNA aptamer inhibitors of HIV reverse transcriptase. From the selected variants, a modified and characterized 37-nt aptamer was able to act as an effective inhibitor of HIV reverse transcriptase. The selected aptamer also competed with the natural template for the binding site in the enzyme. Nicken et al., showed simple model systems for screening RNA protein interactions within an intracellular context. They demonstrated inhibition of HIV-1 reverse transcriptase by RNA aptamers in Escherichia coli [Citation129]. Michalowski et al., recognized three DNA aptamers named RT5, RT6 and RT47 that inhibit HIV-1 reverse transcriptase [Citation130]. The binding motif responsible for inhibition consists of a bimodular structure comprising a 5′-stem-loop module linked to a 3′-G-quadruplex. Moreover, they also mentioned that chosen DNA aptamers inhibited reverse transcriptase from diverse primate lentiviruses with low nM IC50 values. These results provide reverse transcriptase inhibition by oligo nucleic acid based therapeutics. Joshi and Prasad reported an effective aptamer based inhibitor of HIV Type 1 replication by template analog reverse transcriptase [Citation131]. The inhibitors were generated using SELEX. The author also demonstrated that virion-encapsidated TRTIs can predispose virions for inhibition rapidly upon entry. In another study Ferreira-Bravo et al., selected 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with high affinity [Citation132]. They showed strong and specific inhibition of HIV-1 reverse transcriptase in extension assays with no effect on avian myeloblastosis or moloney murine leukemia reverse transcriptase. Lai and DeStefano reported and characterized DNA Aptamers to HIV reverse transcriptase selected by a primer-free SELEX method with other aptamers [Citation133].
Figure 3. (a) The interaction model between HIV-1 RT (reverse transcriptase) and the aptamer. The secondary structure of aptamer is the -HIV-1 RT complex. The p66 subdomains: fingers, palm, thumb, and RNase H and p51 interactions are shown using a surface representation (With permission of Springer) [Citationref 124]. (b) The dimeric 93del aptamer is positioned into the channel formed by the HIV1-IN (integrase) tetramer to block the HIV1-IN catalytic site (reproduced with permission from Proceedings of the National Academy of Sciences USA) [Citationref 134].
![Figure 3. (a) The interaction model between HIV-1 RT (reverse transcriptase) and the aptamer. The secondary structure of aptamer is the -HIV-1 RT complex. The p66 subdomains: fingers, palm, thumb, and RNase H and p51 interactions are shown using a surface representation (With permission of Springer) [Citationref 124]. (b) The dimeric 93del aptamer is positioned into the channel formed by the HIV1-IN (integrase) tetramer to block the HIV1-IN catalytic site (reproduced with permission from Proceedings of the National Academy of Sciences USA) [Citationref 134].](/cms/asset/cc5c8e2a-7e9e-43ee-925f-c80a4776ac4e/krnb_a_1414131_f0003_c.jpg)
Aptamers for integrase
Phan et al 2005 have reported NMR-based solution structure of the 93del (GGGGTGGGAGGAGGGT) HIV-1 integrase aptamer () [Citation134]. They have shown stable interlocked dimeric parallel-stranded DNA quadruplex which act as inhibitor of HIV-1 integrase. Guanine-rich DNA structure attains unusual stable dimeric quadruplex structures. The details NMR structural insight were presented, additionally integrase inhibition assays were performed (ref). Interestingly, they have proposed a model for positioning the 93del dimeric DNA quadruplex and HIV-1 integrase which can help in designing novel strategy for HIV/AIDS. Retroviral enzyme integrase is vital for viral replication which facilitates the integration of proviral DNA in the host chromosome [Citation135,Citation136]. Chiu and Davies first reported the structure and function of HIV-1 integrase in 2004. HIV-1 integrase is a multidomain enzyme, a potential target for therapeutic targeting [Citation137]. Ojwang et al., demonstrated an aptamer, named T30177, able to inhibit HIV integrase activity [Citation138]. T30177 represents a class of integrase inhibitors with promising properties for the development as therapeutic agent against HIV-1 in humans [Citation138]. T30177 is a novel integrase inhibitor and is being tested for its efficacy. T30177 was further derivatized to increase its stability and retained the efficient inhibition capability for HIV-1. T30177 inhibited replication of several strains of HIV-1 in human T-cell lines, peripheral blood lymphocytes, and macrophages. T30177 mediated inhibition was also reported in multiple clinical isolates of HIV-1 [Citation138]. Metifiot et al., 2005 targeted HIV-1 integrase with aptamers using purified RNase H domain of HIV-1 RT. Aptamers containing G-rich sequences were isolated and investigated. Additionally, a second in vitro selection was performed with an isolated RNase H domain of HIV-1 reverse transriptase (p15) as a target using a new DNA library. Their data suggested that prototype structures can be exploited to develop inhibitors of two related enzymes [Citation139]. As novel anti-HIV agents, the G-tetrad-forming oligonucleotides have been explored for their structure activity in relation with inhibition of integrase [Citation140].
Aptamers for proteases (PR)
Proteases are important therapeutic targets in various pathological condition. Dupont et al., has presented detailed review on biochemical mechanisms of aptamer selection, protein-aptamer recognition, protease inhibition, and advantages of aptamers for pharmacological intervention with pathophysiological functions of proteases [Citation141]. Several studies have reported protease-inhibiting aptamers directed against blood coagulation factors, which are currently undergoing clinical trials as anticoagulant drugs [Citation141–143]. Duclair et al., isolated high-affinity RNA Aptamers against the HIV-1 protease to inhibit protease activity [Citation144]. The authors also suggested that the anti-protease RNA aptamers have the prospective to be utilized in such an approach using intracellular expression to confer resistance to viral replication. Anti-HIV protease aptamers have been shown to inhibit both in vitro protease activity and late events of viral replication.
Conclusions and future perspective
HIV is pandemic in many areas of the world. In spite of the advances in combating HIV over many decades, there is no cure or an effective vaccine against HIV and it continues to affect many patients worldwide. Limited resources for HIV prevention is one of the challenge and this issue has more impact on developing countries. Given the precincts of existing HIV diagnostic, treatment and prevention methods and the financial load, an alternative second line potential treatment agents, new formulations, better pills and simple diagnostic tools, in low and middle income countries are desirable. Aptamers hold enormous potential as diagnostic agents and therapeutics for HIV/AIDS. Macugen an RNA aptamer for vascular endothelial growth factor (VEGF)-165 was the first FDA approved aptamer drug targeting VEGF for use against macular degeneration disease and many aptamers are under clinical trial [Citation18]. There is currently no cure or effective HIV vaccine, it is of the highest implication to discover specific molecular probes targeting HIV for early diagnosis and therapies using aptamer and SELEX technology can play a pivotal role in this. Several aptamer-based potential anti-HIV candidates have been generated using SELEX technology. Aptamer research over past years has found relevance as valuable diagnostic and therapeutic leads tools that could cure and diagnose HIV [Citation30,Citation31,Citation78]. Aptamers offer distinct advantages over ART in terms of cost and safety as alternative anti-HIV therapeutics. Moreover, aptamers specifically targeted to different HIV proteins can help neutralize these proteins and mitigate the non-AIDS comorbidities observed in people living with HIV. In the cART era the emphasis is on viral control. However, cART itself has its own side-effects can itself contribute to co-morbidities observed in HIV patients. Hence, it is of the highest implication to discover specific molecular probes targeting HIV for early diagnosis and therapies. The development of aptamer based biosensors can have profound impact in diagnostics use of aptamers for the detection of receptor proteins involved HIV infection. Aptamers to cellular proteins can also play a role in identifying unknown cellular pathways involved in HIV replication by functional proteomics approach [Citation145,Citation146]. Remarkably, targeted delivery of other nucleic acid based therapeutics like siRNA has been accomplished using aptamers to cell-surface receptors. Further advancement in the fields of SELEX technology, nanotechnology, array-based platforms, microfluidics, and others techniques can facilitate rapid selection and application of aptamers against HIV. [Citation147,Citation148] Aptamer loaded nano-formulation with aptamer would be considerable smaller facilitating tissue penetration and increasing efficacy. One of the major limitations of aptamers for application in therapy remains their stability in serum. This is because current aptamers have a nucleic acid backbone which is prone to degradation. However, recent advances in nucleic acid biology have now developed aptamers with base modifications that are more resistant to this degradation. [Citation148] For future therapeutic application aptamers can be delivered as nanoformulations to provide sustained delivery or even for transport across the blood brain barrier to mitigate the neurological complications of HIV [Citation18–20,Citation149,Citation150].
Conflict of interest
The authors have declared no conflict of interest.
Acknowledgment
Financial support from National Institute of Health grant R21-HL128141-01A1 are greatly acknowledged.
Additional information
Funding
References
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818. doi:10.1038/346818a0
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi:10.1126/science.2200121
- Nouvion A, Thibaut J, Lohez O, et al. Modulation of Nr-13 antideath activity by peptide aptamers. Oncogene. 2007;26:701–10. doi:10.1038/sj.onc.1209832
- Kahsai AW, Wisler JW, Lee J, et al. Conformationally selective RNA aptamers allosterically modulate the [beta] 2-adrenoceptor. Nat Chem Biol. 2016;12:709–16. doi:10.1038/nchembio.2126
- Zhang Y, Hong H, Cai W. Tumor-targeted drug delivery with aptamers. Curr Med Chem. 2011;18:4185–94. doi:10.2174/092986711797189547
- Zhou J, Rossi JJ. Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol Ther Nucleic Acids. 2014;3:e169. doi:10.1038/mtna.2014.21
- Cox JC, Ellington AD. Automated selection of anti-protein aptamers. Bioorg Med Chem. 2001;9:2525–31. doi:10.1016/S0968-0896(01)00028-1
- Cox JC, Hayhurst A, Hesselberth J, et al. Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res. 2002;30:e108–e. doi:10.1093/nar/gnf107
- Lee JH, Yigit MV, Mazumdar D, et al. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv Drug Deliv Rev. 2010;62:592–605. doi:10.1016/j.addr.2010.03.003
- Bunka DH, Platonova O, Stockley PG. Development of aptamer therapeutics. Curr Opin Pharmacol. 2010;10:557–62. doi:10.1016/j.coph.2010.06.009
- Zhang H, Ma Y, Xie Y, et al. A controllable aptamer-based self-assembled DNA dendrimer for high affinity targeting, bioimaging and drug delivery. Sci Rep. 2015;5:10099. doi:10.1038/srep10099
- Hamula CL, Guthrie JW, Zhang H, et al. Selection and analytical applications of aptamers. TrAC Trends Anal Chem. 2006;25:681–91. doi:10.1016/j.trac.2006.05.007
- Germer K, Leonard M, Zhang X. RNA aptamers and their therapeutic and diagnostic applications. Int J Biochem Mol Biol. 2013;4:27–40.
- Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–50. doi:10.1038/nrd3141
- Levy-Nissenbaum E, Radovic-Moreno AF, Wang AZ, et al. Nanotechnology and aptamers: applications in drug delivery. Trends Biotechnol. 2008;26:442–9. doi:10.1016/j.tibtech.2008.04.006
- Macedo B, Cordeiro Y. Unraveling Prion Protein Interactions with Aptamers and Other PrP-Binding Nucleic Acids. Int J Mol Sci. 2017;18:1023. doi:10.3390/ijms18051023
- Takahashi T, Tada K, Mihara H. RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of Aβ. Mol Biosyst. 2009;5:986–91. doi:10.1039/b903391b
- Ng EW, Shima DT, Calias P, et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–32. doi:10.1038/nrd1955
- Zhou B, Wang B. Pegaptanib for the treatment of age-related macular degeneration. Exp Eye Res. 2006;83:615–9. doi:10.1016/j.exer.2006.02.010
- Sundaram P, Kurniawan H, Byrne ME, et al. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013;48:259–71. doi:10.1016/j.ejps.2012.10.014
- Wu Y, Marsh JW. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J Virol. 2003;77:10376–82. doi:10.1128/JVI.77.19.10376-10382.2003
- Kelly J, Beddall MH, Yu D, et al. Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology. 2008;372:300–12. doi:10.1016/j.virol.2007.11.007
- Wu Y, Marsh JW. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science. 2001;293:1503–6. doi:10.1126/science.1061548
- Ensoli B, Bellino S, Tripiciano A, et al. Therapeutic immunization with HIV-1 Tat reduces immune activation and loss of regulatory T-cells and improves immune function in subjects on HAART. PLoS One. 2010;5:e13540. doi:10.1371/journal.pone.0013540
- Ensoli B, Barillari G, Salahuddin SZ, et al. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature. 1990;345:84. doi:10.1038/345084a0
- Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–93. doi:10.1016/0092-8674(88)90263-2
- Ensoli B, Buonaguro L, Barillari G, et al. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol. 1993;67:277–87.
- Chang HC, Samaniego F, Nair BC, et al. HIV‐1 Tat protein exits from cells via a leaderless secretory pathway and binds to extracellular matrix‐associated heparan sulfate proteoglycans through its basic region. AIDS. 1997;11:1421–31. doi:10.1097/00002030-199712000-00006
- Zhao N, Pei S-n, Parekh P, et al. Blocking interaction of viral gp120 and CD4-expressing T cells by single-stranded DNA aptamers. Int J Biochem Cell Biol. 2014;51:10–8. doi:10.1016/j.biocel.2014.03.008
- Mufhandu HT, Gray ES, Madiga MC, et al. UCLA1, a Synthetic Derivative of a gp120 RNA Aptamer, Inhibits Entry of Human Immunodeficiency Virus Type-1 Subtype C. J Virol. 2012;86:4989–99. doi:10.1128/JVI.06893-11.
- Zhou J, Satheesan S, Li H, et al. Cell-specific RNA aptamer against human CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity. Chem Biol. 2015;22:379–90. doi:10.1016/j.chembiol.2015.01.005
- DEY AK, GRIFFITHS C, LEA SM, et al. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA. 2005;11:873–84. doi:10.1261/rna.7205405
- Matsunaga K, Kimoto M, Hanson C, et al. Architecture of high-affinity unnatural-base DNA aptamers toward pharmaceutical applications. Sci Rep. 2014;5:18478. doi:10.1038/srep18478
- Chung CH, Kim JH, Jung J, et al. Nuclease-resistant DNA aptamer on gold nanoparticles for the simultaneous detection of Pb 2+ and Hg 2+ in human serum. Biosens Bioelectron. 2013;41:827–32. doi:10.1016/j.bios.2012.10.026
- Crouzier L, Dubois C, Edwards SL, et al. Efficient reverse transcription using locked nucleic acid nucleotides towards the evolution of nuclease resistant RNA aptamers. PLoS One. 2012;7:e35990. doi:10.1371/journal.pone.0035990
- Szeto K, Latulippe DR, Ozer A, et al. Rapid-SELEX for RNA aptamers. PLoS One. 2013;8:e82667. doi:10.1371/journal.pone.0082667
- Sun H, Zu Y. A highlight of recent advances in aptamer technology and its application. Molecules. 2015;20:11959–80. doi:10.3390/molecules200711959
- Mairal T, Özalp VC, Sánchez PL, et al. Aptamers: molecular tools for analytical applications. Anal Bioanal Chem. 2008;390:989–1007. doi:10.1007/s00216-007-1346-4
- Wang X-L, Li F, Su Y-H, et al. Ultrasensitive detection of protein using an aptamer-based exonuclease protection assay. Anal Chem. 2004;76:5605–10. doi:10.1021/ac0494228
- Hwang J, Nishikawa S. Novel approach to analyzing RNA aptamer-protein interactions: toward further applications of aptamers. J Biomol Screen. 2006;11:599–605. doi:10.1177/1087057106288491
- Bala J, Bhaskar A, Varshney A, et al. In vitro selected RNA aptamer recognizing glutathione induces ROS mediated apoptosis in the human breast cancer cell line MCF 7. RNA Biol. 2011;8:101–11. doi:10.4161/rna.8.1.14116
- Varshney A, Bala J, Santosh B, et al. Identification of an RNA aptamer binding hTERT-derived peptide and inhibiting telomerase activity in MCF7 cells. Mol Cell Biochem. 2017;427:157–67. doi:10.1007/s11010-016-2907-7
- Han K, Liang Z, Zhou N. Design strategies for aptamer-based biosensors. Sensors. 2010;10:4541–57. doi:10.3390/s100504541
- Lao Y-H, Phua KK, Leong KW. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–54. doi:10.1021/nn507494p
- Mondragón E, Maher LJ. RNA aptamer inhibitors of a restriction endonuclease. Nucleic Acids Res. 2015;43:7544–55. doi:10.1093/nar/gkv702
- Chi-hong BC, Chernis GA, Hoang VQ, et al. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc Natl Acad Sci. 2003;100:9226–31. doi:10.1073/pnas.1332660100
- Chang EK, Eckert MA, Ali MM, et al. Facile supermolecular aptamer inhibitors of L-selectin. PLoS One. 2015;10:e0123034. doi:10.1371/journal.pone.0123034
- Berezhnoy A, Castro I, Levay A, et al. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J Clin Invest. 2014;124:188–97. doi:10.1172/JCI69856
- Mandal M, Lee M, Barrick JE, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–9. doi:10.1126/science.1100829
- Gold L, Brody E, Heilig J, et al. One, two, infinity: genomes filled with aptamers. Chem Biol. 2002;9:1259–64. doi:10.1016/S1074-5521(02)00286-7
- Zimmermann B, Bilusic I, Lorenz C, et al. Genomic SELEX: a discovery tool for genomic aptamers. Methods. 2010;52:125–32. doi:10.1016/j.ymeth.2010.06.004
- Meyer MM, Roth A, Chervin SM, et al. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA. 2008;14:685–95. doi:10.1261/rna.937308
- Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107:3715–43. doi:10.1021/cr0306743
- Vinkenborg JL, Karnowski N, Famulok M. Aptamers for allosteric regulation. Nat Chem Biol. 2011;7:519–27. doi:10.1038/nchembio.609
- Zhou J, Rossi J. Aptamers as targeted therapeutics: Current potential and challenges. Nat Rev Drug Discov. 2017;16:181–202. doi:10.1038/nrd.2016.199.
- Parashar A, Rajput YS, Sharma R. Aptamer-Based Sensing of β-Casomorphin-7. J Agric Food Chem. 2015;63:2647–53. doi:10.1021/acs.jafc.5b00007
- Dong Y, Xu Y, Yong W, et al. Aptamer and its potential applications for food safety. Crit Rev Food Sci Nutr. 2014;54:1548–61. doi:10.1080/10408398.2011.642905
- Tang J, Yu T, Guo L, et al. In vitro selection of DNA aptamer against abrin toxin and aptamer-based abrin direct detection. Biosens Bioelectron. 2007;22:2456–63. doi:10.1016/j.bios.2006.09.015
- Lakhin A, Tarantul V, Gening L. Aptamers: problems, solutions and prospects. Acta Naturae (англоязычная версия). 2013;5:34–43.
- McNamara JO, Andrechek ER, Wang Y, et al. Cell type–specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–15. doi:10.1038/nbt1223
- Zhou J, Li H, Li S, et al. Novel dual inhibitory function aptamer–siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–9. doi:10.1038/mt.2008.92
- Chu TC, Twu KY, Ellington AD, et al. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73–e. doi:10.1093/nar/gkl388
- González VM, Martín ME, Fernández G, et al. Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses. Pharmaceuticals. 2016;9:78. doi:10.3390/ph9040078
- Wandtke T, Woźniak J, Kopiński P. Aptamers in diagnostics and treatment of viral infections. Viruses. 2015;7:751–80. doi:10.3390/v7020751
- Ulrich H, Wrenger C. Disease‐specific biomarker discovery by aptamers. Cytometry Part A. 2009;75:727–33. doi:10.1002/cyto.a.20766
- Tang Z, Shangguan D, Wang K, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem. 2007;79:4900–7. doi:10.1021/ac070189y
- Ostroff RM, Bigbee WL, Franklin W, et al. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS One. 2010;5:e15003. doi:10.1371/journal.pone.0015003
- Shangguan D, Meng L, Cao ZC, et al. Identification of liver cancer-specific aptamers using whole live cells. Anal Chem. 2008;80:721–8. doi:10.1021/ac701962v
- Chang YM, Donovan MJ, Tan W. Using aptamers for cancer biomarker discovery. J Nucleic Acids. 2013;2013:817350. doi:10.1155/2013/817350.
- Barré-Sinoussi F, Ross AL, Delfraissy J-F. Past, present and future: 30 years of HIV research. Nat Rev Microbiol. 2013;11:877–83. doi:10.1038/nrmicro3132
- Pivel L, Di Piramo S, Betancor E. Human acquired immunodeficiency syndrome (AIDS). Odontol Postgrado. 1987;1:51–80.
- Lloyd A. HIV infection and AIDS. P N G Med J. 1996;39:174–80.
- Gibellini D, Zauli G, Re MC, et al. Recombinant human immunodeficiency virus type‐1 (HIV‐1) Tat protein sequentially up‐regulates IL‐6 and TFG‐β1 mRNA expression and protein synthesis in peripheral blood monocytes. Br J Haematol. 1994;88:261–7. doi:10.1111/j.1365-2141.1994.tb05016.x
- Reinhold D, Wrenger S, Kähne T, et al. HIV-1 Tat: immunosuppression via TGF-β1 induction. Immunol Today. 1999;20:384. doi:10.1016/S0167-5699(99)01497-8
- Thatikunta P, Sawaya BE, Denisova L, et al. Identification of a cellular protein that binds to tat‐responsive element of TGFβ‐1 promoter in glial cells. J Cell Biochem. 1997;67:466–77. doi:10.1002/(SICI)1097-4644(19971215)67:4%3c466::AID-JCB5%3e3.0.CO;2-Q
- Cupp C, Taylor J, Khalili K, et al. Evidence for stimulation of the transforming growth factor beta 1 promoter by HIV-1 Tat in cells derived from CNS. Oncogene. 1993;8:2231–6.
- Tombelli S, Minunni M, Luzi E, et al. Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry. 2005;67:135–41. doi:10.1016/j.bioelechem.2004.04.011
- Minunni M, Tombelli S, Gullotto A, et al. Development of biosensors with aptamers as bio-recognition element: the case of HIV-1 Tat protein. Biosens Bioelectron. 2004;20:1149–56. doi:10.1016/j.bios.2004.03.037
- Schlecht U, Gronewold T, Malave A, et al. Detection of receptor-ligand interactions with an GHz impedance biosensor system. IEEE Sens J. 2007;7:1680–4. doi:10.1109/JSEN.2007.909230
- Yamamoto R, Kumar PK. Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV‐1. Genes Cells. 2000;5:389–96. doi:10.1046/j.1365-2443.2000.00331.x
- Yamamoto R, Katahira M, Nishikawa S, et al. A novel RNA motif that binds efficiently and specifically to the Tat protein of HIV and inhibits the trans‐activation by Tat of transcription in vitro and in vivo. Genes Cells. 2000;5:371–88. doi:10.1046/j.1365-2443.2000.00330.x
- Février M, Dorgham K, Rebollo A. CD4+ T cell depletion in human immunodeficiency virus (HIV) infection: role of apoptosis. Viruses. 2011;3:586–612. doi:10.3390/v3050586
- Cicala C, Martinelli E, McNally JP, et al. The integrin α4β7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci. 2009;106:20877–82. doi:10.1073/pnas.0911796106
- Park I-W, Koziel H, Hatch W, et al. CD4 receptor–dependent entry of human immunodeficiency virus type–1 env-pseudotypes into CCR5-, CCR3-, and CXCR4-expressing human alveolar macrophages is preferentially mediated by the CCR5 coreceptor. Am J Respir Cell Mol Biol. 1999;20:864–71. doi:10.1165/ajrcmb.20.5.3547
- Ugolini S, Mondor I, Parren PW, et al. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line–adapted HIV-1 neutralization. J Exp Med. 1997;186:1287–98. doi:10.1084/jem.186.8.1287
- Dalgleish AG, Beverley PC, Clapham PR, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312(5996):763–7.
- Smith DH, Byrn RA, Marsters SA, et al. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science. 1987;238:1704–8. doi:10.1126/science.3500514
- Davis KA, Abrams B, Lin Y, et al. Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res. 1998;26:3915–24. doi:10.1093/nar/26.17.3915
- Zhou Q, Liu Y, Shin D-S, et al. Aptamer-containing surfaces for selective capture of CD4 expressing cells. Langmuir. 2012;28:12544–9. doi:10.1021/la2050338
- Jing N, Marchand C, Guan Y, et al. Structure–activity of inhibition of HIV-1 integrase and virus replication by G-quartet oligonucleotides. DNA Cell Biol. 2001;20:499–508. doi:10.1089/104454901316976136
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. doi:10.1126/science.280.5371.1884
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi:10.1146/annurev.immunol.17.1.657
- Wilen CB, Tilton JC, Doms RW. Molecular mechanisms of HIV entry. Viral Molecular Machines: Springer. 2012;726:223–42. doi:10.1007/978-1-4614-0980-9_10.
- Khati M, Schüman M, Ibrahim J, et al. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2′ F-RNA aptamers. J Virol. 2003;77:12692–8. doi:10.1128/JVI.77.23.12692-12698.2003
- Moore MD, Cookson J, Coventry VK, et al. Protection of HIV neutralizing aptamers against rectal and vaginal nucleases implications for RNA-based therapeutics. J Biol Chem. 2011;286:2526–35. doi:10.1074/jbc.M110.178426
- Zhou J, Li H, Zhang J, et al. Development of cell-type specific anti-HIV gp120 aptamers for siRNA delivery. J Vis Exp. 2011;52:2954. doi:10.3791/2954
- Dey AK, Khati M, Tang M, et al. An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J Virol. 2005;79:13806–10. doi:10.1128/JVI.79.21.13806-13810.2005
- Cohen C, Forzan M, Sproat B, et al. An aptamer that neutralizes R5 strains of HIV-1 binds to core residues of gp120 in the CCR5 binding site. Virology. 2008;381:46–54. doi:10.1016/j.virol.2008.08.025
- Ramalingam D, Duclair S, Datta SA, et al. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J Virol. 2011;85:305–14. doi:10.1128/JVI.02626-09
- Lochrie MA, Waugh S, Pratt DG, et al. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997;25:2902–10. doi:10.1093/nar/25.14.2902
- Arrigo SJ, Chen I. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991;5:808–19. doi:10.1101/gad.5.5.808
- Unwalla HJ, Li H-T, Bahner I, et al. Novel Pol II fusion promoter directs human immunodeficiency virus type 1-inducible coexpression of a short hairpin RNA and protein. J Virol. 2006;80:1863–73. doi:10.1128/JVI.80.4.1863-1873.2006
- Unwalla HJ, Li M-J, Kim JD, et al. Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA. Nat Biotechnol. 2004;22:1573–8. doi:10.1038/nbt1040
- Daugherty MD, Booth DS, Jayaraman B, et al. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc Natl Acad Sci. 2010;107:12481–6. doi:10.1073/pnas.1007022107
- Symensma TL, Giver L, Zapp M, et al. RNA aptamers selected to bind human immunodeficiency virus type 1 Rev in vitro are Rev responsive in vivo. J Virol. 1996;70:179–87.
- Konopka K, Düzgünş N, Rossf J, et al. Receptor ligand-facilitated cationic liposome delivery of anti-HIV-1 Rev-binding aptamer and ribozyme DNAs. J Drug Target. 1998;5:247–59. doi:10.3109/10611869808995879
- Lapadat-Tapolsky M, Rocquigny HD, Gent DV, et al. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–9. doi:10.1093/nar/21.4.831
- Levin JG, Mitra M, Mascarenhas A, et al. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–74. doi:10.4161/rna.7.6.14115
- Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi:10.1016/j.virusres.2007.12.006
- Rice WG, Supko JG, Malspeis L, et al. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270:1194–8. doi:10.1126/science.270.5239.1194
- Breuer S, Chang MW, Yuan J, et al. Identification of HIV-1 inhibitors targeting the nucleocapsid protein. J Med Chem. 2012;55:4968–77. doi:10.1021/jm201442t
- Goudreau N, Hucke O, Faucher A-M, et al. Discovery and structural characterization of a new inhibitor series of HIV-1 nucleocapsid function: NMR solution structure determination of a ternary complex involving a 2: 1 inhibitor/NC stoichiometry. J Mol Biol. 2013;425:1982–98. doi:10.1016/j.jmb.2013.02.022
- Warui DM, Baranger AM. Identification of small molecule inhibitors of the HIV-1 nucleocapsid–stem-loop 3 RNA complex. J Med Chem. 2012;55:4132–41. doi:10.1021/jm2007694
- Kim SJ, Kim MY, Lee JH, et al. Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem Biophys Res Commun. 2002;291:925–31. doi:10.1006/bbrc.2002.6521
- Kim MY, Jeong S. RNA aptamers that bind the nucleocapsid protein contain pseudoknots. Mol Cells. 2003;16:413–7.
- Kim MY, Jeong S. Inhibition of the functions of the nucleocapsid protein of human immunodeficiency virus-1 by an RNA aptamer. Biochem Biophys Res Commun. 2004;320:1181–6. doi:10.1016/j.bbrc.2004.06.077
- Zhang P, Zhao N, Zeng Z, et al. Combination of an aptamer probe to CD4 and antibodies for multicolored cell phenotyping. Am J Clin Pathol. 2010;134:586–93. doi:10.1309/AJCP55KQYWSGZRKC
- Wheeler LA, Trifonova R, Vrbanac V, et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J Clin Invest. 2011;121:2401–12. doi:10.1172/JCI45876
- Wheeler LA, Vrbanac V, Trifonova R, et al. Durable knockdown and protection from HIV transmission in humanized mice treated with gel-formulated CD4 aptamer-siRNA chimeras. Mol Ther. 2013;21:1378–89. doi:10.1038/mt.2013.77
- Guo S, Tschammer N, Mohammed S, et al. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther. 2005;16:1097–110. doi:10.1089/hum.2005.16.1097
- Yang X, Jiao Y-M, Wang R, et al. High CCR5 density on central memory CD4+ T cells in acute HIV-1 infection is mostly associated with rapid disease progression. PLoS One. 2012;7:e49526. doi:10.1371/journal.pone.0049526
- Lopalco L. CCR5: from natural resistance to a new anti-HIV strategy. Viruses. 2010;2:574–600. doi:10.3390/v2020574
- Scheideman EH, Marlatt SA, Xie Y, et al. Transmembrane protein aptamers that inhibit CCR5 expression and HIV coreceptor function. J Virol. 2012;86:10281–92. doi:10.1128/JVI.00910-12
- Aeksiri N, Songtawee N, Gleeson MP, et al. Insight into HIV-1 reverse transcriptase–aptamer interaction from molecular dynamics simulations. J Mol Model. 2014;20:2380. doi:10.1007/s00894-014-2380-8
- Ferris AL, Hizi A, Showalter SD, et al. Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure. Virology. 1990;175:456–64. doi:10.1016/0042-6822(90)90430-Y
- Jonckheere H, Anné J, De Clercq E. The HIV-1 reverse transcription (RT) process as target for RT inhibitors. Med Res Rev. 2000;20:129–54. doi:10.1002/(SICI)1098-1128(200003)20:2%3c129::AID-MED2%3e3.0.CO;2-A
- De Clercq E. The role of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Antiviral Res. 1998;38:153–79. doi:10.1016/S0166-3542(98)00025-4
- DeStefano JJ, Nair GR. Novel aptamer inhibitors of human immunodeficiency virus reverse transcriptase. Oligonucleotides. 2008;18:133–44. doi:10.1089/oli.2008.0103
- Nickens DG, Patterson JT, Burke DH. Inhibition of HIV-1 reverse transcriptase by RNA aptamers in Escherichia coli. RNA. 2003;9:1029–33. doi:10.1261/rna.5550103
- Michalowski D, Chitima-Matsiga R, Held DM, et al. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008;36:7124–35. doi:10.1093/nar/gkn891
- Joshi P, Prasad VR. Potent inhibition of human immunodeficiency virus type 1 replication by template analog reverse transcriptase inhibitors derived by SELEX (systematic evolution of ligands by exponential enrichment). J Virol. 2002;76:6545–57. doi:10.1128/JVI.76.13.6545-6557.2002
- Ferreira-Bravo IA, Cozens C, Holliger P, et al. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015;43:9587–99.
- Lai Y-T, DeStefano JJ. DNA aptamers to human immunodeficiency virus reverse transcriptase selected by a primer-free SELEX method: characterization and comparison with other aptamers. Nucleic Acid Therapeutics. 2012;22:162–76.
- Phan AT, Kuryavyi V, Ma J-B, et al. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. PNAS. 2005;102:634–9. doi:10.1073/pnas.0406278102
- Craigie R. The molecular biology of HIV integrase. Future Virol. 2012;7:679–86. doi:10.2217/fvl.12.56
- Krishnan L, Engelman A. Retroviral integrase proteins and HIV-1 DNA integration. J Biol Chem. 2012;287:40858–66. doi:10.1074/jbc.R112.397760
- Chiu TK, Davies DR. Structure and function of HIV-1 integrase. Curr Top Med Chem. 2004;4:965–77. doi:10.2174/1568026043388547
- Ojwang JO, Buckheit RW, Pommier Y, et al. T30177, an oligonucleotide stabilized by an intramolecular guanosine octet, is a potent inhibitor of laboratory strains and clinical isolates of human immunodeficiency virus type 1. Antimicrob Agents Chemother. 1995;39:2426–35. doi:10.1128/AAC.39.11.2426
- Métifiot M, Leon O, Tarrago-Litvak L, et al. Targeting HIV-1 integrase with aptamers selected against the purified RNase H domain of HIV-1 RT. Biochimie. 2005;87:911–9. doi:10.1016/j.biochi.2005.03.013
- Jing N. Developing G-quartet oligonucleotides as novel anti-HIV agents: focus on anti-HIV drug design. Expert Opin Investig Drugs. 2000;9:1777–85. doi:10.1517/13543784.9.8.1777
- Dupont DM, Andersen LM, Botkjaer KA, et al. Nucleic acid aptamers against proteases. Curr Med Chem. 2011;18:4139–51. doi:10.2174/092986711797189556
- Dupont DM, Bjerregaard N, Verpaalen B, et al. Building a molecular trap for a serine protease from aptamer and peptide modules. Bioconjug Chem. 2016;27:918–26. doi:10.1021/acs.bioconjchem.6b00007
- Woodruff RS, Sullenger BA. Modulation of the Coagulation Cascade Using AptamersSignificance. Arterioscler Thromb Vasc Biol. 2015;35:2083–91. doi:10.1161/ATVBAHA.115.300131
- Duclair S, Gautam A, Ellington A, et al. High-affinity RNA aptamers against the HIV-1 protease inhibit both in vitro protease activity and late events of viral replication. Molecular Therapy-Nucleic Acids. 2015;4:e228. doi:10.1038/mtna.2015.1
- Nair M, Guduru R, Liang P, et al. Externally controlled on-demand release of anti-HIV drug using magneto-electric nanoparticles as carriers. Nat Commun. 2013;4:1707. doi:10.1038/ncomms2717
- Sagar V, Pilakka-Kanthikeel S, Pottathil R, et al. Towards nanomedicines for neuroAIDS. Rev Med Virol. 2014;24:103–24. doi:10.1002/rmv.1778
- Li N, Nguyen HH, Byrom M, et al. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS One. 2011;6:e20299. doi:10.1371/journal.pone.0020299
- Yan X, Gao X, Zhang Z. Isolation and characterization of 2'-amino-modified RNA aptamers for human TNFalpha. Genomics Proteomics Bioinformatics. 2004;2:32–42. doi:10.1016/S1672-0229(04)02005-4
- Perrone R, Butovskaya E, Lago S, et al. The G-quadruplex-forming aptamer AS1411 potently inhibits HIV-1 attachment to the host cell. Int J Antimicrob Agents. 2016;47:311–6. doi:10.1016/j.ijantimicag.2016.01.016
- Sullenger B, Woodruff R, Monroe DM. Potent anticoagulant aptamer directed against factor IXa blocks macromolecular substrate interaction. J Biol Chem. 2012;287:12779–86. doi:10.1074/jbc.M111.300772
