ABSTRACT
Deprotection of the 5′ end appears to be a universal mechanism for triggering the degradation of mRNA in bacteria and eukaryotes. In Escherichia coli, for example, converting the 5′ triphosphate of primary transcripts to a monophosphate accelerates cleavage at internal sites by the endonuclease RNase E. Previous studies have shown that the RNA pyrophosphohydrolase RppH catalyzes this transformation in vitro and generates monophosphorylated decay intermediates in vivo. Recently, we reported that purified E. coli RppH unexpectedly reacts faster with diphosphorylated than with triphosphorylated substrates. By using a novel assay, it was also determined that diphosphorylated mRNA decay intermediates are abundant in wild-type E. coli and that their fractional level increases to almost 100% for representative mRNAs in mutant cells lacking RppH. These findings indicate that the conversion of triphosphorylated to monophosphorylated RNA in E. coli is a stepwise process involving sequential phosphate removal and the transient formation of a diphosphorylated intermediate. The latter RNA phosphorylation state, which was previously unknown in bacteria, now appears to define the preferred biological substrates of E. coli RppH. The enzyme responsible for generating it remains to be identified.
Messenger RNA degradation is among the principal mechanisms by which protein synthesis is regulated in all living organisms. For example, in bacterial cells, mRNA half-lives range from seconds to as much as an hour, with corresponding effects on gene expression [Citation1].
Over the past decade, it has become increasingly clear that the diverse lifetimes of bacterial transcripts are often governed by modifications at the 5′ terminus. In both Gram-negative and Gram-positive species, mRNA degradation can proceed via either of two pathways, one of which begins with endonucleolytic cleavage at internal sites and the other of which is triggered by conversion of the 5′-terminal triphosphate to a monophosphate [Citation2−Citation6]. The consequences of the latter modification in any particular bacterial species depend on the ribonucleases present in that organism. In Escherichia coli, a monophosphorylated 5′ end increases the susceptibility of transcripts to internal cleavage by the endonuclease RNase E [Citation7], while in Bacillus subtilis it facilitates 5′-to-3′ exonucleolytic degradation by RNase J [Citation4,Citation8].
The RNA pyrophosphohydrolase RppH is thought to be the enzyme principally responsible for transforming the phosphorylation state of bacterial transcripts. In vitro, RppH reacts with triphosphorylated RNA to yield a monophosphorylated product [Citation3, Citation4]. Consistent with the biochemical activity of the purified enzyme, full-length mRNAs bearing a 5′ monophosphate are common in wild-type E. coli cells but scarce in mutant cells lacking RppH, where the lifetimes of transcripts that are ordinarily targeted by this enzyme are often prolonged as a consequence of its absence [Citation3].
Purified E. coli RppH is able to generate monophosphorylated RNA not only from triphosphorylated in vitro transcripts but also from their diphosphorylated counterparts by cutting between the α and β phosphates to release pyrophosphate or orthophosphate, respectively [Citation3,Citation9]. In a recent article, we reported the unexpected finding that diphosphorylated RNA is significantly more reactive than triphosphorylated RNA as a substrate for E. coli RppH [Citation10]. This surprising observation raised the possibility that diphosphorylated, not triphosphorylated, RNA is the natural biological substrate of RppH in E. coli cells. If so, this inference would imply that the conversion of 5′ triphosphates to monophosphates usually occurs in vivo by a stepwise process involving a diphosphorylated intermediate.
To test this hypothesis, we set out to determine whether E. coli cells contain RNA molecules bearing two phosphates at the 5′ end, an RNA phosphorylation state never before observed in bacteria. A combination of two quantitative assays, one previously developed and the other new, made it possible to precisely determine the phosphorylation state of E. coli transcripts of interest. The established assay, PABLO (Phosphorylation Assay By Ligation of Oligonucleotides; ), detects monophosphorylated RNA 5′ ends on the basis of their ability to undergo splinted ligation to a DNA oligonucleotide when the two are brought into close proximity by annealing to adjacent sites on a second, complementary oligonucleotide [Citation2,Citation11]. By contrast, RNAs with triphosphorylated or diphosphorylated 5′ ends undergo little if any ligation; therefore, they can be separated from the ligation product of their monophosphorylated counterparts by gel electrophoresis and blotting. The percentage of monophosphorylated 5′ ends can then be quantified by comparing band intensities.
Figure 1. Assays for examining the 5′-terminal phosphorylation state of RNA. (Left) Analysis by PABLO to quantify the percentage of an RNA of interest that is monophosphorylated. (Right) Analysis by PACO to quantify the percentage of an RNA that is diphosphorylated. The colored lines represent transcripts that are initially triphosphorylated (blue), diphosphorylated (red), or monophosphorylated (green). The black line represents a DNA oligonucleotide selectively joined to the 5′ end of monophosphorylated RNA by splinted ligation with T4 DNA ligase. In both assays, the ligated and unligated RNA products are separated by gel electrophoresis and detected by Northern blotting. p, phosphate; Gppp, unmethylated cap; HO, hydroxyl.
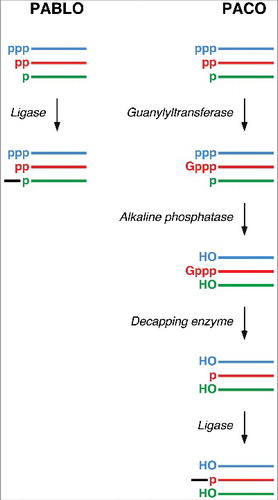
To detect diphosphorylated RNA 5′ ends, a novel assay was devised. This assay, PACO (Phosphorylation Assay by Capping Outcome; ), relies on an RNA guanylyltransferase that catalyzes the reaction of GTP with diphosphorylated RNA 5′ termini to add a Gppp cap, a key step in the capping of eukaryotic mRNAs [Citation12]. Unlike the exposed phosphates at the 5′ ends of their uncapped counterparts, the phosphates of the resulting caps cannot be removed by alkaline phosphatase. These properties make it possible to detect diphosphorylated RNAs by sequential treatment with Pce1 (the RNA guanylyltransferase of Schizosaccharomyces pombe), alkaline phosphatase, and a decapping enzyme to generate a monophosphorylated product that can be quantified by PABLO.
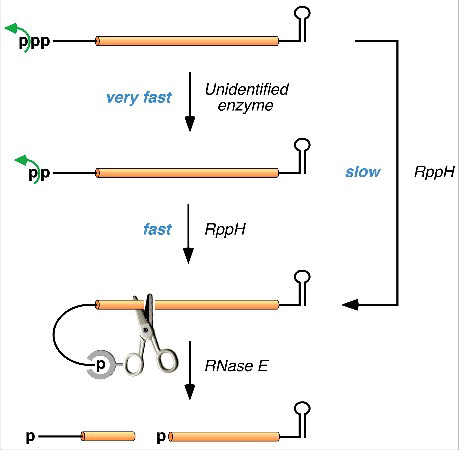
Together, these assays were used to examine the 5′-terminal phosphorylation state of two representative E. coli transcripts that are degraded by a 5′-end-dependent mechanism: yeiP, which encodes a paralog of translation elongation factor EF-P, and metE, which encodes a methyltransferase important for methionine biosynthesis. This investigation revealed that a substantial fraction of each is diphosphorylated at steady state in wild-type cells (34% and 51%, respectively), with most of the remainder being monophosphorylated, and that the diphosphorylated fraction approaches 100% in mutant cells lacking RppH. These findings indicate that, for each of these transcripts, conversion of the 5′ triphosphate to a monophosphate generally occurs in E. coli not in one step as was previously thought but rather by sequential removal of the γ and β phosphates (). They further indicate that RppH is needed to release the second of these phosphates from the diphosphorylated intermediate, which otherwise accumulates to a high level. The finding that two E. coli mRNAs with distinct 5′-terminal sequences undergo sequential removal of these phosphates suggests that this is likely to be the principal mechanism by which the 5′ end of E. coli transcripts is converted from a triphosphate to a monophosphate. Nevertheless, it remains possible that this destabilizing modification may sometimes be achieved in a single step in which RppH converts triphosphorylated RNA directly to its monophosphorylated counterpart while releasing pyrophosphate as a by-product.
Figure 2. Mechanism of 5′-end-dependent degradation of triphosphorylated RNA in E. coli. In the principal pathway for 5′-end-dependent degradation, the γ and β phosphates of the triphosphorylated primary transcript are sequentially removed by an unidentified enzyme and by RppH, respectively, to generate first a diphosphorylated intermediate and then a monophosphorylated intermediate. The latter intermediate is vulnerable to rapid endonucleolytic cleavage by RNase E (scissors), whose 5′-end binding pocket can selectively accommodate a 5′ monophosphate but not a 5′ triphosphate or diphosphate. The resulting RNA fragments are then swiftly degraded (not shown). Although E. coli RppH prefers RNA substrates that are diphosphorylated, it can also convert triphosphates directly to monophosphates, albeit more slowly, releasing pyrophosphate as a by-product.
Remarkably, the diphosphorylated forms of yeiP and metE mRNA are not only detectable in E. coli but also at least 2–10 fold more abundant than their triphosphorylated precursors in wild-type cells, evidence that the rate constant for γ phosphate removal exceeds that for β phosphate removal by RppH. Two lines of evidence indicate that the enzyme responsible for catalyzing rapid γ phosphate release in E. coli is neither RppH nor DapF, a lysine biosynthesis enzyme (diaminopimelate epimerase) that binds to RppH and stimulates its catalytic activity [Citation13]. First, no diphosphorylated reaction product can be detected when either purified enzyme is added to triphosphorylated RNA in vitro [Citation10,Citation13]. Furthermore, deleting the gene that encodes either of them causes levels of diphosphorylated RNA in E. coli to rise, not fall (D. J. Luciano and J. G. Belasco, unpublished results) [Citation10]. By contrast, B. subtilis RppH, which is only distantly related to the proteobacterial RppH present in E. coli [Citation14], removes the γ and β phosphates of triphosphorylated substrates one at a time in vitro, generating monophosphorylated RNA and orthophosphate (not pyrophosphate) as the only detected reaction products of the purified enzyme [Citation4]. Whether diphosphorylated intermediates can be detected in B. subtilis cells has not yet been investigated.
What, then, is the identity of the enzyme that accelerates the degradation of E. coli mRNAs by removing the γ phosphate from the 5′ end so as to enhance their reactivity with RppH? A genetic screen of an E. coli mutant library lacking each nonessential gene has failed to reveal a promising candidate (M. P. Hui and J. G. Belasco, unpublished results). Therefore, this critical activity appears to derive from either a single gene product that is essential for growth or multiple gene products that are functionally redundant. Although the E. coli genome does not encode an enzyme homologous to the RNA triphosphatases important for RNA capping in eukaryotes, it contains several hundred genes of unknown function, one of which may encode an as yet uncharacterized bacterial RNA triphosphatase. It is also possible that one or more of the known NTPases or kinases in E. coli is able to act not only on the 5′ phosphates of mononucleotides but also on the 5′ phosphates of polynucleotides. Whatever the source of this activity may be, it apparently can remove the γ phosphate of transcripts that begin with either A (yeiP) or G (metE), the two nucleotides most commonly present at the 5′ end of primary transcripts. (Transcripts that begin with a pyrimidine have not yet been tested for a diphosphorylated decay intermediate.)
The ability of RppH to convert triphosphorylated substrates directly to monophosphorylated products with the concomitant release of pyrophosphate prompts the question as to why this modification evolved in E. coli as a two-step process involving multiple enzymes and a diphosphorylated intermediate rather than a simple one-step process catalyzed solely by RppH. After all, speedier conversion of triphosphates to monophosphates could just as easily have been accomplished by elevating the cellular concentrations of RppH and DapF, which E. coli tolerates quite well [Citation13]. One hypothesis already mentioned is that some NTPases or kinases might lack the specificity needed to distinguish mononucleotides from polynucleotides. If so, E. coli may simply have adapted to the tendency of these rogue enzymes to rapidly convert the 5′ ends of primary transcripts to diphosphates. Alternatively, the two-step conversion of triphosphorylated RNA to monophosphorylated RNA may have evolved to provide bacteria an additional point at which to control rates of RNA processing and degradation by modulating the concentration or activity of the enzyme(s) responsible for γ phosphate removal. A third possibility, suggested by the unexpectedly high cellular concentration of the diphosphorylated forms of yeiP and metE mRNA, is that this RNA phosphorylation state may have an additional, as yet unrecognized biological function distinct from its role in facilitating degradation.
Whatever the evolutionary imperative for this novel phosphorylation state might have been, it joins a diverse and growing list of 5′-terminal modifications on full-length bacterial transcripts that were not previously imagined [Citation2,Citation15,Citation16]. That each of these modifications can substantially impact the cellular lifetime of RNA [Citation3,Citation10,Citation17] demonstrates the importance of the 5′ end as a site for governing gene expression.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Hui MP, Foley PL, Belasco JG. Messenger RNA degradation in bacterial cells. Annu Rev Genet. 2014;48:537–59.
- Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5' pyrophosphate removal. Mol Cell. 2007;27:79–90.
- Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–8.
- Richards J, Liu Q, Pellegrini O, et al. An RNA pyrophosphohydrolase triggers 5'-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol Cell. 2011;43:940–9.
- Durand S, Gilet L, Bessières P, et al. Three essential ribonucleases – RNase Y, J1, and III – control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 2012;8:e1002520.
- Clarke JE, Kime L, Romero AD, et al. Direct entry by RNase E is a major pathway for the degradation and processing of RNA in Escherichia coli. Nucleic Acids Res. 2014;42:11733–51.
- Mackie GA. Ribonuclease E is a 5'-end-dependent endonuclease. Nature. 1998;395:720–3.
- Mathy N, Bénard L, Pellegrini O, et al. 5'-to-3' exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5' stability of mRNA. Cell. 2007;129:681–92.
- Vasilyev N, Serganov A. Structures of RNA complexes with the Escherichia coli RNA pyrophosphohydrolase RppH unveil the basis for specific 5'-end-dependent mRNA decay. J Biol Chem. 2015;290:9487–99.
- Luciano DJ, Vasilyev N, Richards J, et al. A novel RNA phosphorylation state enables 5' end-dependent degradation in Escherichia coli. Mol Cell. 2017;67:44–54.
- Celesnik H, Deana A, Belasco JG. PABLO analysis of RNA: 5'-phosphorylation state and 5'-end mapping. Methods Enzymol. 2008;447:83–98.
- Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40.
- Lee CR, Kim M, Park YH, et al. RppH-dependent pyrophosphohydrolysis of mRNAs is regulated by direct interaction with DapF in Escherichia coli. Nucleic Acids Res. 2014;42:12746–57.
- Foley PL, Hsieh PK, Luciano DJ, et al. Specificity and evolutionary conservation of the Escherichia coli RNA pyrophosphohydrolase RppH. J Biol Chem. 2015;290:9478–86.
- Chen YG, Kowtoniuk WE, Agarwal I, et al. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol. 2009;5:879–81.
- Cahová H, Winz ML, Höfer K, et al. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–7.
- Bird JG, Zhang Y, Tian Y, et al. The mechanism of RNA 5' capping with NAD+, NADH and desphospho-CoA. Nature. 2016;535:444–7.
