ABSTRACT
Antisense transcription emerges as a key regulator of important biological processes in the human malaria parasite Plasmodium falciparum. RNA-processing factors, however, remain poorly characterized in this pathogen. Here, we purified the multiprotein RNA exosome complex of malaria parasites by affinity chromatography, using HA-tagged PfRrp4 and PfDis3 as the ligands. Seven distinct core exosome subunits (PfRrp41, PfMtr3, PfRrp42, PfRrp45, PfRrp4, PfRrp40, PfCsl4) and two exoribonuclease proteins PfRrp6 and PfDis3 are identified by mass spectrometry. Western blot analysis detects Dis3 and Rrp4 predominantly in the cytoplasmic fraction during asexual blood stage development. An inducible gene knock out of the PfDis3 subunit reveals the upregulation of structural and coding RNA, but the vast majority belongs to antisense RNA. Furthermore, we detect numerous types of cryptic unstable transcripts (CUTs) linked to virulence gene families including antisense RNA in the rif gene family. Our work highlights the limitations of steady-state RNA analysis to predict transcriptional activity and link the RNA surveillance machinery directly with post-transcriptional control and gene expression in malaria parasites.
Introduction
Malaria remains one of the most devastating infectious diseases in the world with an estimated 400.000 malaria deaths each year globally [Citation1]. The causative agent is a unicellular protozoan pathogen that belongs to the genus Plasmodium (phylum Apicomplexa). In humans, the most virulent form of malaria is caused by P. falciparum. Complex regulatory pathways control parasite development, pathogenesis and transmission during the life cycle stages in the human and mosquito host. In addition to transcription factor networks, previous studies revealed that epigenetic factors control transcription initiation of P. falciparum virulence genes [Citation2]. In the past years, however, post-transcriptional regulation has emerged as another important pathway downstream of the transcriptional control level [Citation3,Citation4]. This includes translational repression in sexual stages [Citation5] and degradation of nascent mRNA by a 3ʹexoribonuclease called PfRNase II [Citation6]. Antisense transcript production was reported for about 25% of genes [Citation7] and recently, it was demonstrated that the developmentally controlled elimination of antisense RNA for a gametocyte development protein 1 gene (GDV1) initiated expression and commitment to sexual stages [Citation8]. The key regulator of RNA processing and turnover in eukaryotic cells, the RNA exosome complex [Citation9] has not been well-studied in malaria parasites and the role of the two exosome associated RNases in post-transcriptional regulation remains elusive.
The RNA exosome is a ubiquitous multi-subunit ribonuclease complex controlling RNA metabolism including RNA maturation and RNA quality surveillance [Citation10]. Moreover, it is emerging as a regulator of expression levels of specific mRNAs and maintenance of genome stability [Citation11]. The architecture and composition of RNA exosomes is highly conserved from archaea to human. In archaea, two subunits with bacterial phosphorolytic exoribonuclease RNase PH-like domain form a hexameric ring-like structure by combination of three Rrp41-Rrp42 heterodimers, whereas three copies of one or both Rrp4 and Csl4, containing S1/KH domains, form a ‘cap’ locating on top of the ring for structure stabilization and extension of the exosome entry tunnel [Citation12,Citation13]. Though the eukaryotic exosome core shares structure similarity with archaea, the subunit composition varies among them. In the well-studied S. cerevisiae and human exosome cores, there are six different RNase PH domain-containing subunits in the basal ring structure known as Rrp41-Ski6, Rrp42, Rrp43-OIP2, Rrp45-PM/Scl75, Rrp46, and Mtr3. These RNase PH subunits are assembled into a toroidal hexamer with three RNA-binding subunits on the top side, Rrp4, Rrp40, and Csl4 to accommodate target RNAs by a RNA entry channel inside the exosome core [Citation9–Citation12]. The eukaryotic RNA exosome is observed in both nuclear and cytoplasmic subcellular compartments, but the two forms of RNA exosome execute different biological functions by interaction with specific co-factors such as the nuclear TRAMP (Trf4–Air2–Mtr4 polyadenylation complex) or the cytoplasmic Ski (superkiller) complex [Citation9,Citation14,Citation15]. Usually, the nuclear RNA exosome processes immature precursors of structured RNAs such as rRNA and controls turnover of non-coding RNAs, cryptic unstable transcripts (CUTs), and rarely nuclear mRNA, whereas the cytoplasmic RNA exosome is mainly implicated in mRNA surveillance, e.g. Nonsense-mediated decay (NMD) pathway that degrades mRNAs containing premature termination codons [Citation16] and general mRNA turnover. Despite the presence of six RNase PH domain-contained proteins, it is now widely accepted that the yeast and human exosome cores are catalytically inactive due to the loss of key catalytic residues of their bacterial homologue. The function of RNA processing and degradation are executed by the two RNA exosome-associated catalytically active subunits, Dis3/Rrp44 and Rrp6. The association of these two exoribonucleases with the RNA exosome core and the targeted substrates vary in different organisms, though evolutionary conserved functions such as rRNA maturation are maintained [Citation17]. For example, Rrp6 is only incorporated into the nuclear exosome in S. cerevisiae but is an integral part of the nuclear and cytoplasmic forms in African trypanosomes [Citation18]. Human hDIS3 is mainly a nuclear protein contributing to shaping the RNA polymerase II transcriptome by degrading unwanted transcripts [Citation19], while its homolog hDIS3L is restricted to cytoplasmic exosome complexes.
The study of protozoan eukaryotes such as Tetrahymena, Trichomonas, Trypanosome, and Giardia have revealed slightly distinct types of organizations [Citation20]. In P. falciparum, only eight putative exosome subunits, PfRrp41, PfRrp42, PfRrp45, PfRrp4, PfRrp40, PfCsl4, PfRrp44/PfDis3, and PfRrp6 were identified in silico with three exosome core subunits, Rrp43, Prp46, and Mtr3 missing in the parasites [Citation6]. In this study we sought to experimentally identify and functionally characterize the plasmodial RNA exosome. We generated HA-tagged transfectant lines for two putative subunits (Rrp4 and Rpr44/Dis3). Affinity purification of the RNA exosome complex followed by mass spectrometry analysis consistently detected eight exosome subunits, but also identifies a putative additional subunit, PfMtr3. We observe a predominant cytoplasmic location of the RNA exosome in asexual blood stage parasites. Conditional knock out of PfDis3 revealed dramatically more RNA transcripts then predicted by steady state RNA analysis. We identify the upregulation of more than 1000 antisense RNAs and detect cryptic transcripts made from virulence gene loci. This finding opens new experimental avenues into our understanding of transcriptionally active genome regions and directly links exosome-mediated RNA decay to gene regulation in this fatal pathogen.
Results
Characterization of P. falciparum exosome subunit Rrp4 and Dis3
Our first aim was to identify the composition of the P. falciparum exosome complex, including core exosome subunits and associated catalytic factors, by Co-immunoprecipitation (Co-IP) followed by Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. As antibodies generated against recombinant proteins or synthetic peptides of P. falciparum exosome subunits displayed non-specific background signal, we decided to generate transgenic parasite lines with terminal epitope-tags (3xHA-3xTy1), both for the putative core exosome component Rrp4 (called here PfRrp4, PF3D7_0410400) and the associated catalytic subunit Dis3/Rrp44 (called here PfDis3, PF3D7_1359300). Utilizing the CRISPR/Cas9 gene editing technique to achieve fast endogenous integration of the tag, plasmids targeting either the N- or C-terminus of the PfRrp4 or PfDis3 gene were constructed (Table S1) and co-transfected with Cas9 expression vector pUF1-cas9 ()). An episomal expression construct (pARL-gfp-HA-Ty1) was used as negative control for the Co-IP assay (Figure S1(a)). Three weeks after transfection, parasites carrying pL6-Ty1-HA-PfRrp4/pUF1-cas9, pL6-PfDis3-HA-Ty1/pUF1-cas9, and pARL-gfp-HA-Ty1 control were obtained by drug selection with both WR and BSD. C-terminal tagging of the PfRrp4 gene and N-terminal tagging of the PfDis3 gene was not successful after three attempts of transfection, suggesting that the C-terminal and N-terminal sequences are important for conformation-dependent functions of PfRrp4 and PfDis3, respectively. However, failure to genetically modify these genes may also be due to technical reasons. Clones of the Ty1-HA-PfRrp4, PfDis3-HA-Ty1 and GFP control transfectant were obtained by limiting dilution, and the integration verified by PCR and Western blot. Epitope-tagged proteins were detected as a single band by western blot assay, confirming the specificity of the antibodies against Ty1 or HA-tagged proteins in P. falciparum parasite asexual blood stage extracts (Figure S1).
Figure 1. Cytoplasmic localization of RNA exosome in P.falciparum parasites.
(a) Schematic representation of constructs for generation of epitope-tagging PfRrp4 (left) and PfDis3 (right) transfectant lines by the CRISPR-Cas9 system. For both strains, the plasmids carrying a single guide RNA (sgRNA) and the Cas9 endonuclease were co-transfected. bsd, blasticidin S deaminase. hdhfr, human dihydrofolate reductase. HR, homology region.(b) Western blot analysis of Ty1-HA-PfRrp4 and PfDis3-HA-Ty1 lines with nuclear and cytoplasmic extracts, respectively. Aldolase and Histone 3 were used as loading control. CE, cytoplasmic extract. NE, nuclear extract. R, ring. T, trophozoite. S, schizont.(c) Immunofluorescence assay (IFA) of Ty1-HA-PfRrp4 and PfDis3-HA-Ty1 lines in Ring, Trophozoite, Schizont, respectively. The nuclei were stained by DAPI. The bar represents 1 μm.
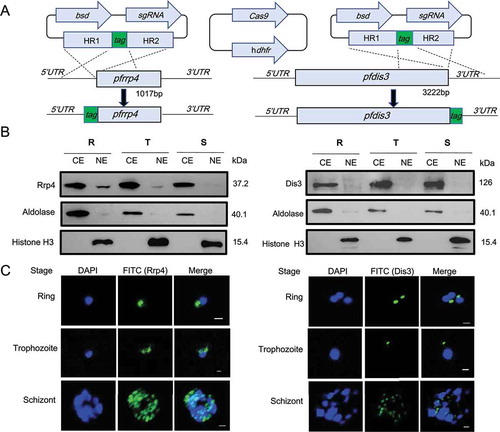
Subsequently, we analyzed the expression pattern of the core exosome subunit PfRrp4 and the exosome associated factor PfDis3 across the asexual blood stage by western blot. We prepared nuclear and cytoplasmic protein extracts from highly synchronized ring, trophozoite, and schizont-stage parasites of epitope-tagged transfectant lines. Western blot analysis shows that PfRrp4 and PfDis3 proteins are expressed in all intraerythrocytic stages (ring, trophozoite and schizont). PfDis3, however, was only detected in the cytoplasmic fraction even after overexposure of the blot ()). PfRrp4 was also mainly in the cytoplasmic fraction but a very minor fraction was detected in the nuclear extract only after overexposure ()). Antibodies against histone H3 and aldolase were used as markers of the two cellular compartments. Our cellular fractionation data suggest a predominant cytoplasmic role of the exosome. This observation was further validated by IFA with synchronized parasites ()). In rings and trophozoites the antibody signal for both tagged proteins is clearly distinct from the nucleus. In the schizont stage the IFA images are more difficult to interpretate due to the high parasite load at this stage.
Identification of P. falciparum RNA exosome composition
Although the exosome structure is highly conserved from Archaea to eukaryotic organisms, its subunit composition varies, as Archaea only possesses two subunits in the hexameric ring structure, Rrp41 and Rrp42, whereas studied model organisms such as yeast, human or drosophila use six distinct RNase PH-domain containing proteins. Putative orthologues of two exosome-associated catalytic subunits (Rrp6 and Dis3) and three structure stabilization subunits of the core exosome ‘cap’ (Rrp4, Rrp40, Csl4) were annotated in the P. falciparum genome database (PlasmoDB.org) by bioinformatic analysis according to their conserved functional domains such as RNase II, RNase D, S1, and KH. However, only three candidate subunits of the hexameric RNase-PH ring structure were predicted, i.e. Rrp41, Rrp42, and Rrp45. To experimentally establish the composition of the P. falciparum exosome complex, we performed co-immunoprecipitation (Co-IP) coupled to LC-MS/MS analysis utilizing total extracts from the Ty1-HA-tagged transfectants of PfRrp4 and PfDis3 with GFP-HA-Ty1 as negative control ( and S2). The mass spectrometry results included peptides for virtually the entire predicted exosome complex. We also observed peptides of abundant proteins such as histones, heat-shock proteins (HSP) and ribosomal proteins, however these proteins were also present in both replicates of our GFP unspecific control and excluded by further statistical analysis (p < 0.05) (see Dataset S1 and Materials and Methods section). Importantly, no exosome-related peptides were detected in the GFP-HA-Ty1 pull down.
Figure 2. Composition identification of P.falciparum RNA exosome subunits.
(a) Schematic representation of exosome isolation by Co-IP with antibody against HA fused to PfRrp4 and PfDis3 respectively.(b) Comparative summary of the RNA exosome composition in Archaea, S.cerevisiae, human, and P.falciparum identified by Co-IP and LC-MS/MS in this study.(c) Putative model of RNA exosome complex of P.falciparum parasites compared with that of Archaea, S.cerevisiae, and human.
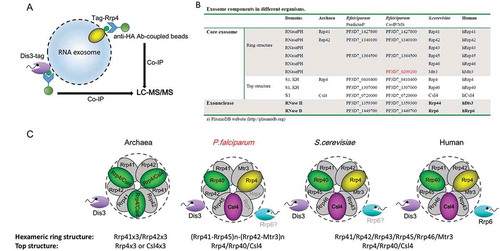
As summarized in ), in the two independent biological replicates we detect three annotated exosome RNase PH-ring structure subunits (Rrp41, Rrp42, Rrp45) and all predicted cap structure subunits (Rrp4, Rrp40, Csl4) of the core exosome, as well as two exosome-associated catalytic enzymes (Dis3 and Rrp6). Interestingly, a protein annotated as putative 3ʹ exoribonuclease (PF3D7_0209200), was also isolated in all replicates, but absent in the GFP control (p < 0.05). It possesses a RNase PH domain (InterPro domain IPR001247) and had previously been informatically predicted as putative exosome subunit [Citation21]. Sequence homology indicates that this protein belongs to the Rrp41-subtype of the RNase PH protein family (comprising eukaryotic Rrp41, Mtr3 and Rrp46 proteins) and is a putative orthologue of Mtr3 (see Figure S3), suggesting two Rrp41-like proteins, called here Rrp41and Mtr3, exist in the genome of P. falciparum. Surprisingly, the bioinformatically predicted Rrp42 subunit was only detected by mass spectrometry analysis in one assay with Dis3-HA-Ty1 line. In all replicates, the three cap structure subunits and PfDis3 were detected whereas PfRrp6 was absent in one assay with the PfRrp4-HA-Ty1 line.
However, since Rrp6 copurified with Dis3 in both replicates, and this interaction is thought to be mediated via the core in other organisms [Citation11], this possibility has to be further investigated. Since we only identified four core members of the hexameric ring structure, this finding suggests an intermediate form of this complex in malaria parasites compared to that of Archaea, S.cerevisiae and human ()).
Inducible gene knock out shows that PfDis3 is essential
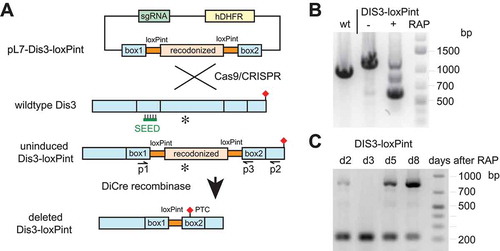
To assess the physiological role of the RNA exosome in P. falciparum, we wanted to delete its catalytic subunit PfDis3. We failed to obtain mutant parasites with a traditional knock-out approach and mutagenesis of the putative critical aspartate residues, D607A, D613A, D615A, and D616A, accounting for the catalytic activity of PfDis3 [Citation6,Citation22] (data not shown), indicating an essential function of this gene. For this reason, a conditional knockout strategy that allows targeting of essential genes was utilized [Citation23,Citation24]. Using parasites expressing inducible DiCre recombinase (3D7A DC) we introduced a cassette containing artificial introns which contain loxP sites (loxPint) flanking the exonuclease domain of PfDis3 CDS ()) [Citation23]. This modification should be phenotypically silent, as after splicing of the introns the correct coding sequence is reconstituted. The integration of the cassette was verified by PCR ()) and two clones were isolated using the limited dilution technique. We used synchronized ring stage parasites to induce Pfdis3 gene deletion by addition of 100 nM rapamycin for 4 hours. In the next cycle the genotype of the induced KO culture was compared to the wildtype and uninduced KO by PCR ()). We observed the slightly higher band in the uninduced KO parasites compared to the wild type, corresponding to the integration of two loxP introns. Upon induction by rapamycin, a lower band corresponding to the expected excision product is detected. Sanger sequencing of the PCR amplified fragment confirmed the correct recombination event. Since the Pfdis3 deletion was not complete in the culture after 4 hours, we increased the rapamycin concentration and the incubation time. These changes did not increase the deletion efficiency. Even repeating the induction in the next cycle after the initial rapamycin treatment did fail to give complete excision, as the undeleted genotype was reappearing in the following cycles ()). Furthermore, our attempts to obtain a Pfdis3 knock out clone by limiting dilution technique directly after the initial rapamycin induction, failed. We performed the growth rate analysis with and without rapamycin induction as well as cell cycle progression analysis. No major difference was observed for the first two cycles (see Figure S4(a,b)). Taken together, these data further indicate that the Pfdis3 KO has an important growth defect that is overgrown by parasites with intact Pfdis3 after several blood stage cycles.
Figure 3. Generation of DiCre recombinase-mediated conditional PfDis3 KO line.
(a) Schematic representation of the PfDis3-loxPint conditional KO strategy. Co-transfection of the plasmid pL7-dis3-loxPint with pUF1-Cas9 leads to integration of the loxPint containing cassette in the endogenous locus. Activation of DiCre recombinase by rapamycin induces the excision of the loxP flanked CDS region that contains the exonuclease domain (asterisk) and by frameshift mutation to a premature termination codon (PTC, red diamond). The sgRNA target sequence is marked with SEED.(b) PCR verification of the conditional KO using oligonucleotides in homology box1 and the 3ʹCDS (p1-p2). Bands of the expected size were observed (wildtype-1020bp, undeleted Pfdis3-loxPint 1250bp, deleted Pfdis3-loxPint 610bp; gDNA harvested day 2 after induction).(c) Repeated induction and outgrowth of KO cultures. On day 2 after the first induction, rapamycin treatment was repeated and gDNA from the following days analyzed by PCR (p1-p3, expected size undeleted: 855bp, deleted: 217bp).
PfDis3 degrades mRNA but targets predominantly antisense RNA
To evaluate the effect of exosome disruption by depletion of Pfdis3, the global RNA transcriptome was analyzed by RNA-seq. We examined poly (A) enriched RNA from two clones (E2/F10) of the conditional Dis3-loxPint KO. RNA from uninduced (DMSO treated) and induced (rapamycin treated) KO cultures was isolated in the cycle after induction (day 2, late ring stage parasites, 20 ± 5 hours post-infection, see Figure S4(c)). Differential gene expression was evaluated by combining the results from the two clones in replicate experiments (2x E2+/- and 2x F10+/-). Analysis of expression in the induced KO vs uninduced culture revealed a large number of transcripts with increased expression and only very few transcripts displayed a significantly decreased abundance ()). In Dataset S2 all transcripts with at least 2-fold differential expression between the two conditions are listed and the raw data is accessible on NCBI BioProject (accession: PRJNA453694).
Figure 4. Differential gene expression and accumulation of virulence gene-associated cryptic unstable transcripts (CUTs) in Dis3-loxPint KO line.
(a) Scatter plot of differential gene expression in the Pfdis3-loxPint KO. Log2 RPKM values of the uninduced Pfdis3-KO are plotted against the log2 RPKM values of the induced KO (combined results from two clones in replicate). The yellow and dark green dots mark differentially expressed (≥ 1.75 fold) antisense and sense transcripts, respectively. The grey dots mark genes that are not significantly changed (combined sense and antisense).(b) Expression profile for the Pfdis3 transcript, with the black box highlighting the KO region (bedgraph coverage illustration of relative expression from IGV). The uninduced expression profile is marked in green and the Pfdis3-KO in red; upper part of the panel show sense (s) and the lower panel antisense (as) expression for both conditions, respectively.(c) Other representative examples of differentially expressed genes: increased sense transcript was observed for Sir2a (PF3D7_1328800), PfEMP1 pseudogene (PF3D7_1480100), snoRNA07 (PF3D7_1109000), while for alpha tubulin 2 (PF3D7_0422300) antisense RNA is increased.(d) Representative expression profiles of two silent var genes and their neighboring rif (PF3D7_1200500 and PF3D7_1300200). As these genes are coded on different strands, sense direction for the var gene and antisense for the rif are in the top part of the panel; antisense for the var gene and sense for the rif in the lower part.
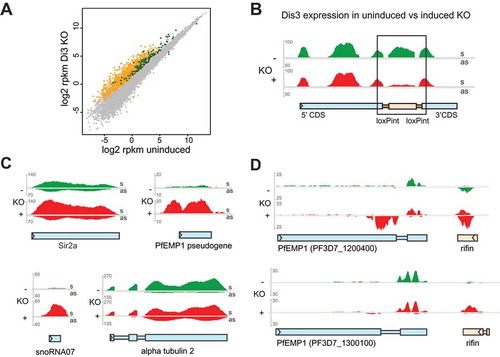
We were also interested to see the expression of the Pfdis3 transcript itself. When focusing on the loxP flanked region of the Pfdis3 gene the decrease of read coverage is evident in the induced KO, but as could be expected from the previous PCR result, the deletion is not complete and a few reads remain for the floxed part ()). Nevertheless, the KO reveals a major effect on the RNA homeostasis of the parasite.
When the reads are separated into sense and antisense alignments, we observe a striking global increase of antisense transcripts (n = 1046) and to a lesser extend sense transcripts (n = 131) (Dataset S2, visualized in )). Many of these are cryptic transcripts not detectable in parasites before disruption of Pfdis3.
We detected increased transcripts in mRNAs but also in different species of pseudogenes and noncoding RNAs (snRNAs, snoRNAs, RNA of unknown function RUF1, RUF2). Some representative examples taken from the integrative genome viewer (IGV) are shown in ) to illustrate the different types of transcriptional changes in Pfdis3 mutant parasites.
When analyzing the antisense dataset, we noticed a remarkable number of highly expressed rif antisense transcripts. Rifins represent a class of surface proteins that are encoded by a multicopy gene family organized in the genome in arrays often associated tail to tail with neighboring var genes, coding for another P. falciparum virulence factor [Citation25]. In many cases the rif antisense RNA is barely detectable before the KO, whereas its expression is strongly increased in the Pfdis3 mutant, often to a higher expression level than the corresponding rif sense transcript ()). Furthermore, we observe bidirectional promoter activity from the var intron that has been reported previously [Citation26] and has been associated to transcribed var genes [Citation27,Citation28]. The intron derived antisense RNA is significantly increased in 22 silent var genes upon Pfdis3 disruption (see Dataset S2 and example in )), indicating that the var antisense steady state level is controlled by PfDis3.
Discussion
Malaria parasites use post-transcriptional regulation to fine tune gene expression and to transition between different developmental stages. The mechanism by which RNA is targeted for turnover is still not understood in this pathogen. Here we present functional studies that reveal canonical and parasite specific contributions of the plasmodial RNA exosome to RNA surveillance and establishment of antisense RNA levels. By bioinformatic prediction, six members of the exosome core and two exoribonuclease members, PfDis3 and PfRrp6, were previously identified in the P. falciparum genome. The 3ʹ-5ʹ exoribonuclease activity of PfDis3 had previously been validated by using a recombinant Dis3 [Citation6]. In this work, we generated tagged exosome members to investigate experimentally the composition of the RNA exosome complex. We detected all predicted plasmodium members of the exosome complex and identified an additional member being part of the complex. We call this protein Mtr3-like core member (PfMtr3).
Eukaryotic exosomes of studied model organisms, from trypanosomes, yeast, fly and human, canonically possess at least six distinct RNase PH domain proteins, three of the Rrp41-subtype (Rrp41, Rrp46 and Mtr3) and three of the Rrp42-subtype (Rrp42, Rrp43, Rrp45). Even including the newly identified exosome protein PfMtr3, P. falciparum is still short two RNase PH domain proteins and no additional proteins could be identified in the published genome. Furthermore, genome database searches reveal the same situation for all other published Plasmodium species (PlasmoDB.org). Interestingly, the apicomplexan phylum seems heterogenous in this regard, as the genomes of some species like Toxoplasma gondii and Cryptosporidium parvum encode six distinct proteins, while others, for example Giardia, Theileria parva or Gregarina niphandrodes, like Plasmodium, encode only four (Table S2). The consequences on the stoichiometry of the different subunits present in the exosome complex or possible specialized subcomplexes will have to be examined in future studies.
P. falciparum shows another unusual feature. We observed an uncommon RNA exosome complex distribution between cytoplasmic and nuclear fraction in asexual blood stage parasites. Only a very minor fraction was detected in the nucleus. This may indicate that the cytoplasmic exosome activity has been amplified in P. falciparum to perform extensive RNA turnover in blood stage parasites. This idea is supported by our finding, that the majority of antisense RNA (> 1000) expression levels are affected by the depletion of PfDis3. The general role of the widespread abundance of antisense in malaria parasites is still obscure. However, for a few genes experimental evidence supports a role in the control of expression. For example, the var gene family, that encodes the major virulence factor of P. falciparum, produces antisense lncRNA from its intron, that have been suggested to play a role in the activation process [Citation27,Citation28]. Another recent study on the GDV1 gene provided solid experimental evidence that the abolition of antisense RNA of the GDV1 gene leads to the expression of the protein with subsequent activation of the sexual stages [Citation8]. Furthermore, several studies have shown extensive dynamics in nascent mRNA transcription, mRNA stabilization and mRNA decay during asexual development [Citation29,Citation30]. In line with that, the RNA exosome might be one effector complex mediating mRNA decay as observed by the increase of mRNA transcripts following PfDis3 KO in addition to its role in antisense transcript degradation. Overall, we observe an important increase of the steady state level of antisense RNA. We hypothesize that the RNA exosome plays a prominent role in P. falciparum gene regulation via its role in the control of antisense RNA degradation. Future studies will explore if the increase of antisense RNA in PfDis3 mutants translates into different levels of protein expression.
Another important observation in PfDis3 mutant parasites, is the detection of cryptic transcripts that are not observed in steady state RNA samples of the corresponding uninduced cultures. For example, the pseudo var gene (PF3D7_1480100) with an incomplete 3ʹ end produces sense transcripts and the members of the rif gene family make antisense transcripts either in the presence or absence of sense transcripts (). Our work demonstrates that the actual reported genome-wide transcription profile for P. falciparum is an underestimation of the actively transcribed regions.
The mutant PfDis3 also reveals changes in levels of RNA molecules that normally are processed in the nucleus in other organisms (eg snoRNAs). This is counter intuitive, given that at best only traces of PfDis3 could be detected in nuclear extracts by Western blot. We hypothesize that a feedback loop from the disrupted exosome in the cytoplasm could affect nuclear processing, for example by affecting correct processing of spliceosomal RNAs (eg snRNA U4) in the cytoplasm that are also required for nuclear processing events [Citation31].
In conclusion, here we experimentally validate the RNA exosome composition and its cellular localization in malaria parasites. An inducible knock out of Dis3 reveals canonical functions reported in other organisms and the control of antisense RNA in malaria parasites. The latter are likely to have important implications because they link the RNA surveillance machinery directly with transcriptional control and gene expression. The next big step in understanding the regulatory function of RNA exosome will be to identify the proteins that target the different types of RNA classes to the exosome either for processing or degradation.
Materials and methods
Plasmid construction for transfection
To generate Rrp4- and Dis3-HA-Ty1 tagged lines, we modified the plasmid pL6-gfp by replacing the eGFP box with a 1-kb homologue sequence flanking the N- or C-terminus of the targeted genes which contained two epitopes (HA-Ty1) each in three copies in tandem, targeting the N- or C-terminal end, and inserting a guide RNA sequence specific to the PfRrp4 gene (PF3D7_0410400) or PfDis3 gene (PF3D7_1359300) by In-Fusion PCR Cloning System, respectively (see Table S1) [Citation32]. The resulting plasmids were pL6-PfRrp4-HA-Ty1, pL6-Ty1-HA-PfRrp4, pL6-PfDis3-HA-Ty1, and pL6-Ty1-HA-PfDis3. The plasmid pUF1-Cas9-infusion carrying Cas9 expression cassette was modified by replacing the original ydhodh gene with hdhfr.
For the Dis3-loxPint conditional knock out construct, a synthetic sequence including the homology boxes, artificial introns containing loxP sites and a recoded exonuclease domain was ordered from Genscript (Table S1) This fragment was cloned into pL6 with the Dis3 gRNA sequence (GTAGATGTATAAGTGTGTTA) to give pL7-Dis3-loxPint.
Parasite culture and transfection
P. falciparum 3D7 strain was maintained in culture in vitro and synchronized as described previously [Citation33]. For transfection, synchronized ring-stage parasites at ~ 5% parasitemia were transfected with ~ 100 μg of plasmid sgRNA and Cas9 by electroporation as described previously. Transfected parasites were selected by blasticidin S deaminase drug and WR99210. After approximately 1 month, the established culture with rrp4- or dis3-tag integration were cloned by limitation cloning, and the integration events were validated by PCR following sequencing.
For the the conditional Dis3 KO, pL7-Dis3-loxPint and pUF1-Cas9 were co-transfected as described above in P. falciparum 3D7A DiCre background [Citation23,Citation34] Transfected parasites were selected with 2.5nM WR99210 and 1µM DSM1. Integration was verified and the culture cloned out by limiting dilution.
Western Blot
Total parasite extracts were prepared by 0.15% saponin treatment and resuspended in 1x SDS-loading buffer (Bio-Rad), then separated on 8 ~ 12% SDS-PAGE gel according to the molecular weight of targeted proteins, and subjected to Western blot analysis. The commercial antibodies used in this study were mouse anti-Ty1 (1:500, Sigma), mouse anti-HA (1:2000, Roche), rabbit anti-PfAldolase (1:1000, Abcam), and rabbit anti-Histone 3 (1:1000, Abcam). ECL western blotting kit (GE healthcare) was used to develop blots.
Immunofluorescence
Immunofluorescence assay was performed on synchronous parasites fixed with 4% paraformaldehyde in 1xPBS as described previously [Citation33]. Antibody dilutions for mouse anti-Ty1 was 1:300, and second antibodies of Alexa-Fluor-488-conjugated anti-rabbit were 1:2000.
Co-immunoprecipitation (Co-IP)
The Co-IP was performed as described previously [Citation6]. Briefly, asynchronized asexual-stage parasite cultures were harvested and treated with 0.15% saponin. The released parasites were washed with 1xPBS for three times, then resuspended in six volumes of lysis buffer (25 mM Tris-Cl, pH 7.5, 100 mM KCl, 2 mM EDTA, 0.5 mM PMSF, 0.05% NP-40, 1x protease inhibitor cocktail (Thermo)), and subjected to sonication at highest power for 3 min at 30 sec intervals with a sonicator (Bioruptor). The supernatants of the lysates were isolated and immediately incubated with magnetic beads pre-coupled with HA antibody (Thermo) for 2 h at 4 ℃. After washing the beads twice with IPP500 (500 mM NaCl, 10 mM Tris-Cl, pH 8.0, 0.05% NP-40) and once with 1x PBS, bound proteins were either eluted with sample buffer for SDS-PAGE followed by silver-staining, coomassie-staining, and western blot analysis. After validation of the enrichment of targeting proteins by western blot with anti-Ty1 antibody, the full gel lanes loaded with elution fractions were subject to mass spectrometry (LC-MS/MS) analysis with at least two technical replicates.
LC-MS/MS
After filtration through 0.22 µm membrane the clear solution was subjected to nano LC-MS/MS separation. A volume of 3.0 µL of each sample was desalted by loading on a Thermo C18 PepMap100 precolumn (300 µm × 5 mm) and eluted on a Thermo Acclaim PepMap RSLC analytical column (75 µm × 15 cm). Mobile phase A (0.1% formic acid in H2O) and mobile phase B (0.1% formic acid in acetonitrile) were used to establish the 120 min gradient comprised of 85 min of 4−30% B, 15 min of 30−50% B, and 5 min of 90% B, followed by re-equilibrating at 4% B for 15 min. The flow rate was 0.3 µL/min. Peptides were then analyzed on a Q-Exactive proteomic mass spectrometer (Thermo Scientific) in a data-dependent manner, with automatic switching between MS and MS/MS scans using a top 20 method. MS spectra were acquired at a resolution of 70,000 with a target value of 3 × 106 ions or a maximum integration time of 50 ms. The scan range was limited from 375 to 1400 m/z. Peptide fragmentation was performed via higher-energy collision dissociation (HCD) with the energy set at 32 NCE. The MS/MS spectra were acquired at a resolution of 35,000 with a target value of 1 × 105 ions or a maximum integration time of 100 ms. The fixed first m/z was 100, and the isolation window was 1.2 m/z.
The data were processed with MaxQuant version 1.5.3.30, and the peptides were identified from the MS/MS spectra searched against the protein database (PlasmoDB 38 Released, 19 June 2018) using the Andromeda search engine. The precursor mass tolerance was set to 10 ppm and fragment ion mass tolerance to 0.02 Da. One missed cleavage site of trypsin was allowed. Carbamidomethyl (C) was used as a fixed modification and oxidation (M) was used as variable modifications. All spectra were searched against protein database using a target false discovery rate (FDR) of 1%. Other parameters were used as pre-set in the software. LFQ experiments in MaxQuant were performed using the built-in label-free quantification algorithm (MaxLFQ) [Citation35]. Missing values were imputed to mimic low abundance values (width 0.2, downshift 2.1, total matrix). Statistical analysis was performed with Perseus 1.5.1.6. LFQ ratios were transformed with log2-(x) and then normalized using Z-score, and -log10-(p-value) of all proteins were obtained by a two-sided one sample t-test over two biological replicates. Only proteins identified have average ratios > 2.0 and p-values < 0.05 were considered statistical significant identification.
Conditional knockout induction and sample preparation
For the induction of the Dis3-loxPint KO the cultures were synchronized by Sorbitol/Plasmion treatment and at the ring stage incubated with 100 nM rapamycin (induced) or the same volume of DMSO (uninduced) for 4 hours. The culture was washed and left to the next cycle. On day 2 after induction, gDNA (NucleoSpin® Tissue kit, Macherey-Nagel) and RNA (miRNeasy kit, Qiagen) was harvested from Saponin lysed cells pellets. Oligonucleotides used in PCR to verify integration and recombination were p1, p2, and p3 as shown in Table S1.
For RNAseq two independent Dis3-loxPint clones (E2/F10) were analyzed, from repeated induction rounds (2x E2+/- RAP; 2x F10+/- RAP). Total RNA was poly(A) selected (Dynabeads® mRNA DIRECT™ Kit, Thermo Fischer Scientific) before library preparation with the TruSeq Stranded mRNA kit (Illumina) and paired end sequencing was performed on the Illumina Nextseq 550 machine.
RNA sequencing analysis
Raw Illumina data were converted to fastq files and demultiplexed using Illumina’s bcl2fastq (v2.19.1.403). The paired sequencing reads were mapped to the P. falciparum genome (plasmoDB.org, v3 release 28) [Citation25] using ‘bwa mem’ [Citation36] allowing only unique read mappings to the genome (option -c 1). Optical read mapping duplicates were removed using samtools ‘rmdup’ [Citation37] and only alignments with a mapping quality ≥ 20 were retained using samtools ‘view’.
Read counts for genic regions were obtained using ‘htseq-count’ [Citation38] for both the sense and antisense direction of transcription. The two files (i.e. sense + antisense read counts) were merged for library normalization between replicates and conditions. Differential expression analysis was performed in R (http://www.r-project.org/) using the package edgeR [Citation39] with an FDR value cutoff ≤ 0.05.
For visualization of the RNA-seq data were, bedgraph formatted files were generated from the BAM alignment files using deepTools2 ‘bamCoverage’ [Citation40] with a binsize of 1 nucleotide and RPKM (‘reads pere kilobase per one million mapped reads’) library normalization enabled (option: – normalizeToRPKM) for comarpsion between samples. The data were visualized using the Integrated Genomics Viewer (version 2.3.78) [Citation41].
Availability of Data
The fastq files supporting the results of this article are available in the NCBI BioProject accession PRJNA453694.
Supplemental Material
Download Zip (46.9 MB)Acknowledgements
We thank Moritz Treeck for the 3D7 DiCre parasites and helpful discussions with the inducible transfection system.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
Additional information
Funding
Notes on contributors
Qingfeng Zhang
Qingfeng Zhang and DD conceived and designed the experiments. G.W. and Y.F. generated HA-Ty1-tagged PfRrp4 and PfDis3 transfectant lines. G.W. and G.G. performed IFA, WB, and Co-IP experiments. Y.Z. and Y.X. performed mass spectrometry experiments. D.D. performed Dis3 KO mutant analysis; S.B. performed RNA-seq data analysis, A.S. Q.Z. and DD wrote the manuscript. All authors read and approved the final manuscript.
References
- World malaria report. Geneva: World Health Organization; November 2017. ISBN: 978 92 4 156552 3. Available from: http://www.who.int/malaria/publications/world-malaria-report-2017/report/en/.2017
- Duraisingh MT, Horn D. Epigenetic regulation of virulence gene expression in parasitic protozoa. Cell Host Microbe. 2016;19:629–640.
- Rai R, Zhu L, Chen H, et al. Genome-wide analysis in Plasmodium falciparum reveals early and late phases of RNA polymerase II occupancy during the infectious cycle. BMC Genomics. 2014;15:959.
- Vembar SS, Droll D, Scherf A. Translational regulation in blood stages of the malaria parasite Plasmodium spp.: systems-wide studies pave the way. Wiley Interdiscip Rev RNA. 2016;7:772–792.
- Mair GR, Braks JA, Garver LS, et al. Regulation of sexual development of Plasmodium by translational repression. Science. 2016;313:667–669.
- Zhang Q, Siegel TN, Martins RM, et al. Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature. 2014;513:431–435.
- Siegel TN, Hon CC, Zhang Q, et al. Strand-specific RNA-Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC Genomics. 2014;15:150.
- Filarsky M, Fraschka SA, Niederwieser I, et al. GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science. 2018;359:1259–1263.
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776.
- Januszyk K, Lima CD. The eukaryotic RNA exosome. Curr Opin Struct Biol. 2014;24:132–140.
- Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol. 2016;17:227–239.
- Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657.
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237.
- Meola N, Domanski M, Karadoulama E, et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol Cell. 2016;64:520–533.
- Halbach F, Reichelt P, Rode M, et al. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell. 2013;154:814–826.
- Schneider C, Tollervey D. Threading the barrel of the RNA exosome. Trends Biochem Sci. 2013;38:485–493.
- Lykke-Andersen S, Tomecki R, Jensen TH, et al. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 2011;8:61–66.
- Estévez AM, Lehner B, Sanderson CM, et al. The roles of intersubunit interactions in exosome stability. J Biol Chem. 2003;278:34943–34951.
- Szczepińska T, Kalisiak K, Tomecki R, et al. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 2015;25:1622–1633.
- Jensen TH. RNA exosome. New York (NY): Landes Bioscience and Springer Science+Business Media, LLC; 2010.
- Williams CW, Elmendorf HG. Identification and analysis of the RNA degrading complexes and machinery of Giardia lamblia using an in silico approach. BMC Genomics. 2011;12:586.
- Zuo Y, Vincent HA, Zhang J, et al. Structural basis for processivity and single-strand specificity of RNase II. Mol Cell. 2006;24:149–156.
- Jones ML, Das S, Belda H, et al. A versatile strategy for rapid conditional genome engineering using loxP sites in a small synthetic intron in Plasmodium falciparum. Sci Rep. 2016;6:21800.
- Knuepfer E, Napiorkowska M, van Ooij C, et al. Generating conditional gene knockouts in Plasmodium - a toolkit to produce stable DiCre recombinase-expressing parasite lines using CRISPR/Cas9. Sci Rep. 2017;7:3881.
- Gardner MJ, Hall N, Fung E, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511.
- Calderwood MS, Gannoun-Zaki L, Wellems TE, et al. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem. 2003;278:34125–34132.
- Amit-Avraham I, Pozner G, Eshar S, et al. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci USA. 2015;112:E982–91.
- Jiang L, Mu J, Zhang Q, et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499:223–227.
- Painter HJ, Chung NC, Sebastian A, et al. Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development. Nat Commun. 2018;9:2656.
- Shock JL, Fischer KF, DeRisi JL. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biol. 2007;8:R134.
- Zhang L, Wan Y, Huang G, et al. The exosome controls alternative splicing by mediating the gene expression and assembly of the spliceosome complex. Sci Rep. 2015;5:13403.
- Ghorbal M, Gorman M, Macpherson CR, et al. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32:819–821.
- Zhang Q, Huang Y, Zhang Y, et al. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 2011;10:451–463.
- Collins CR, Das S, Wong EH, et al. Robust inducible Cre recombinase activity in the human malaria parasite Plasmodium falciparum enables efficient gene deletion within a single asexual erythrocytic growth cycle. Mol Microbiol. 2013;88:687–701.
- Cox J, Hein MY, Luber CA, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760.
- Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079.
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140.
- Ramírez F, Ryan DP, Grüning B, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–65.
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012;14:178–192.
