ABSTRACT
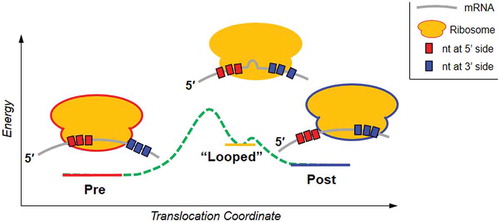
The precise 3-nucleotide movement of mRNA is critical for translation fidelity. One mRNA translocation error propagates to all of the following codons, which is detrimental to the cell. However, none of the current methods can reveal the mRNA dynamics near the ribosome entry site, which limits the understanding of this important issue. We have developed an assay of dual DNA rulers that provides such capability. By uniquely probing both the 3ʹ- and 5ʹ-ends of mRNA, we observed an antibiotic-trapped intermediate state that is consistent with a ribosomal conformation containing mRNA asymmetric partial displacements at its entry and exit sites. Based on the available ribosome structures and computational simulations, we proposed a ‘looped’ mRNA conformation, which suggested a stepwise ‘inchworm’ mechanism for ribosomal translocation. The same ‘looped’ intermediate state identified with the dual rulers persists with a ‘-1’ frameshifting motif, indicating that the branching point of normal and frameshifting translocations occurs at a later stage of translocation.
Introduction
One essential step of protein biosynthesis is the ribosomal translocation on the mRNA by exactly three nucleotides to decode the correct amino acid. There is no gap between codons. Therefore, one translocation error impacts all of the downstream codons, which is lethal and worse than single amino acid misincorporation errors [Citation1]. However, the mechanism of accurate mRNA movement remains elusive, even after multiple seminal structural studies revealed an extensive rRNA to tRNA guiding network [Citation2–Citation5] because mRNAs lack well-defined conformations that can be aligned among different structures. Conversely, a method for objectively probing both the entrance and exit sites of the mRNA with single-nt resolution is necessary and has not been reported in the literature. For example, the toe-printing assay uses a reserve transcriptase primed at the 3ʹ-distal end to transcribe the mRNA toward the ribosome [Citation6]. The residue at the 3ʹ end of the mRNA exiting the ribosome is deduced by the cDNA length that was limited by the clash between the reserve transcriptase and the ribosome. The ribosome profiling method maps ribosome-covered mRNA with high throughput sequencing, which reveals the global ribosome distribution but lacks single codon precision [Citation7]. Among indirect translocation assays, the tandem LC/MS/MS analysis of oligopeptide composition deduces the ribosome reading frame based on the synthesized peptide [Citation8]. Fluorescently labeled tRNA binding/moving events reveal the translating codons [Citation9], and the puromycin reactivity assay is the conventional method for confirming the A-site vacancy resulting from translocation [Citation10]. None of these methods can reveal the movement at the 5ʹ end of the mRNA that is entering the ribosome.
Recently, we developed a force-induced remnant magnetization spectroscopy (FIRMS) technique that used DNA rulers to precisely determine the positions of the 3ʹ end of mRNA during ribosome translocation and frameshifting [Citation11,Citation12]. Briefly, the DNA rulers, which were labeled with magnetic beads, formed duplexes with a portion of the mRNA uncovered by the ribosome, and the length of the duplexes was then obtained from the dissociation force of the duplexes, which was measured by a sensitive atomic magnetometer through the decrease in magnetic signals resulting from the DNA ruler’s removal from the surface under centrifugal forces. Single-nt resolution for the duplex length has been routinely achieved, resolving all the three reading frames of the ribosome on the mRNA. However, we have not shown effective probing at the 5ʹ end of mRNA and thus have not been able to provide the whole picture of mRNA movement.
In this report, we have developed a dual ruler assay that probes the ribosome-uncovered mRNA from both sides with single-nt resolution. For the first time, our results indicate a novel intermediate state that implied a ribosomal conformation in which the mRNA had moved 2 nt at the exit site (3ʹ-end) but only 1 nt at the entrance site (5ʹ-end). Based on structural and computational studies, we have suggested that the ribosome may translocate in a stepwise manner via an inchworm-like mRNA ‘looping’ mechanism.
Results
The dual ruler assay is schematically shown in ). Two DNA rulers were designed to form duplexes with the uncovered mRNA: Ruler-In for the 5ʹ-end and Ruler-Out for the 3ʹ-end. The lengths of the duplexes are limited by the ribosome position on the mRNA. Consequently, the number of basepairs (bp) of the DNA-mRNA duplexes will reveal the mRNA movement during translocation. The DNAs are labeled with magnetic beads, so the existence of the DNA-mRNA duplexes is indicated by a magnetic signal (Supplementary Figure S1). The number of bp of each duplex can be determined by FIRMS [Citation11,Citation12]. The challenge is for the DNA rulers to reach the mRNA between the ribosome and the surface, as shown at the 5ʹ-mRNA side. To solve this problem, extra polyT linkers of 0, 10, 30, 50, and 70 T were introduced for Ruler-In. As shown in ), only linkers of at least 50 T led to successful DNA-mRNA hybridization, as indicated by a substantial magnetic signal. The length of 50 T is approximately 17 nm. Considering the approximately 20-nm diameter of ribosomes, 17 nm is approximately a quarter of the circumference of the cross section circle (Supplementary Figure S2). All Ruler-Ins in this report contained 50 T linkers. The linker does not affect the measured force, which is determined only by the binding strength of the mRNA-DNA duplex. At the dissociation force, the DNA probe (to which the magnetic beads are bound) is completely sheared from the surface in one step, in contrast to optical trap experiments in which the displacement of the bead correlates with the force via Hooke’s law. In those experiments, an extra linker may interfere with the bead displacement through stretching of the additional linker. Conversely, a probe DNA with a 70 T linker was tested, and the mRNA-DNA duplex exhibited the same dissociation force as the corresponding probe with a 50 T linker, demonstrating the linker’s trivial effect on the measurement (Figure S3).
Figure 1. Dual ruler assay with single-nt resolution for studying ribosome translocation. a. Schematic of the dual ruler assay, in which two DNA rulers are designed to respectively probe the 5ʹ- and 3ʹ-termini. The red line indicates the polyT linker. b. Optimization of the linker length for Ruler-In. c. FIRMS results to determine the dissociation forces of the duplexes between Ruler-Ins and mRNA. d. Dissociation forces of the duplexes between Ruler-Outs and mRNA.
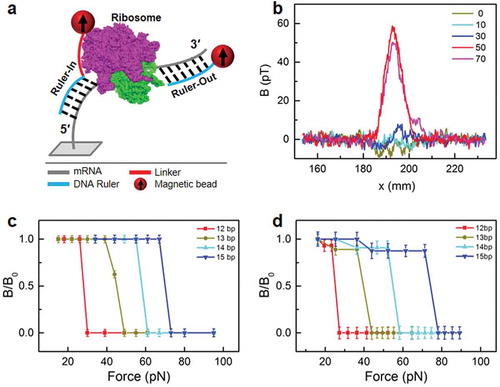
) also shows the typical signal amplitude of 50 pT, which corresponds to 1.1 × 105 magnetic DNA-mRNA duplexes. Detection from the 3ʹ-end with Ruler-Outs is similar to a previous report [Citation11]. To demonstrate single-nt resolution for both rulers, we calibrated the force measurements for the DNA-mRNA duplexes in the absence of ribosomes. ) shows the FIRMS results for the 12-, 13-, 14- and 15-bp duplexes for Ruler-Ins, which were 28, 44, 57, and 70 pN, respectively. The force uncertainty was 3 pN. Because the force difference between duplexes with a 1-bp length difference was usually 12–18 pN, which is much greater than the force uncertainty, single-nt resolution was achieved for the 5ʹ end of the mRNA. Similarly, single-nt resolution was obtained for the 3ʹ end of the mRNA, which yielded 25, 40, 55, and 74 pN for the 12-, 13-, 14- and 15-bp duplexes, respectively, formed between the Ruler-Outs and the mRNA ()). The force values are plotted against the length (bp) of the duplexes in Supplementary Figure S3.
We first applied the dual ruler assay to investigate normal translocation with and without various antibiotics ()). The pretranslocation and posttranslocation ribosome complexes are denoted Pre and Post, respectively. In these experiments, the MF-Pre complex carries vacant tRNAfMet and MF-tRNAPhe at the P- and A-sites, respectively. MF-Post carries tRNAfMet and MF-tRNAPhe at the E- and P-sites, respectively, with a vacant A-site (, inset).
Figure 2. Probing translocation under the influence of various antibiotics. a. Probing scheme for the MF-Pre and MF-Post complexes. Inset indicates the schematic ribosomes in Pre and Post, which correspond to the solid and dash-lined ovals, respectively. b, c. FIRMS results with no antibiotics. d, e. Results with both fusidic acid and neomycin. f, g. Results with fusidic acid only. h, i. Results with viomycin only. Left panels: Ruler-In from the 5' side; right panels: Ruler-Out from the 3' side.
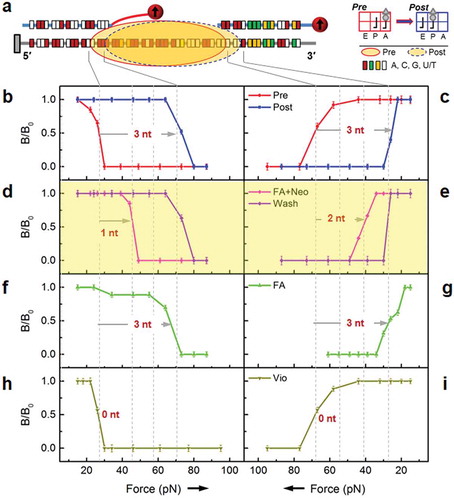
In the absence of antibiotics, the ribosome moved 3 nt toward the 3ʹ-end; therefore, the mRNA-DNA duplexes at the 5ʹ side exhibited 12 bp and 15 bp binding forces in Pre and Post, respectively ()). Conversely, the duplexes at the 3ʹ side exhibited a comparable but reversed change from 15 to 12 bp ()). For clarity, the force axes were oriented in opposite directions for the 3ʹ and 5ʹ sides so that the FIRMS traces for the Pre to Post transition were consistent with the ribosome movement direction. Together, showed that the ribosome covered 27 nt of the mRNA, in excellent agreement with the literature [Citation13]. However, in the presence of both fusidic acid and neomycin (, highlighted), our force spectra suggested that the ribosome moved only 1 nt at the 5ʹ side but 2 nt at the 3ʹ side. This result implied that the ribosome translocated via a stepwise mechanism and that the mRNA formed a loop with an extra nucleotide inside the ribosome. Alternatively, it is possible that the ribosome stretches to cover 28 nt of mRNA. Interestingly, normal translocation was completed after washing away of the antibiotics, as evidenced by 3 nt movements from both the 5ʹ and 3ʹ sides (purple trace in ). For comparison, with only fusidic acid, the force spectra was consistent with the ribosome movement of 3 nt at both sides, similar to the situation for normal translocation (). Similar results were obtained when only neomycin was used (Supplementary Figure S4). Two other antibiotics known to inhibit translocation, viomycin (,i)) and hygromycin B (Supplementary Figure S4), were also studied to confirm the detection method [Citation14]. Both antibiotics inhibited translocation completely, as shown by 0 nt movement on both sides.
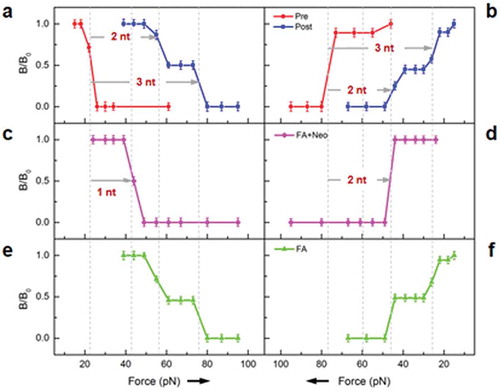
We then studied whether this partially translocated ribosome could form on a ‘-1’ frameshifting motif. The MFNF-Pre and MFNF-Post ribosome complexes carried MFNF oligopeptide at the A- and P-sites, respectively. The translocation took place on the ‘U6A’ motif, which is part of the ‘-1’ frameshifting motif in HIV [Citation15]. As shown in , duplexes shortening and lengthening of 2 and 3 nt were observed at 3ʹ- and 5ʹ-ends, respectively, suggesting both normal and ‘-1’ frameshifting translocations, with nearly 50–50 partitions, as indicated by the corresponding magnetic signals. In the presence of both neomycin and fusidic acid, similar forces consistent with the inferred intermediate state were also detected, which suggested the ribosome moved 2 and 1 nt at the 3ʹ- and the 5ʹ-ends, respectively (). However, in the presence of only fusidic acid (), the result was nearly the same as for MFNF-Post in Panels a and b (blue traces). These results implied that the partially translocated ribosome conformation that is inferred from our assay had formed regardless of the mRNA motif. Therefore, the different outcomes of normal and frameshifting translocations may diverge at a later stage of the process.
Figure 3. FIRMS results of dual rulers to probe frameshifting. a, b. MFNF-Pre and MFNF-Post without antibiotics. c, d. Results with both fusidic acid and neomycin. e, f. Results with only fusidic acid. Left panels: Ruler-In from the 5ʹ side; right panels: Ruler-Out from the 3ʹ side.
Our results correlate well with two important structural studies. An intermediate trapped with fusidic acid and neomycin exhibited a 21° head swivel and 2.7° body rotation (Int1), while another intermediate trapped with fusidic acid alone exhibited an 18° head swivel and 5° body rotation (Int2) [Citation2,Citation3]. In Int2, the A- and P-site tRNAs are in ap/P and pe/E configurations, respectively. The letters separated by the ‘/’ indicate the tRNA binding sites in the 30S and 50S ribosomes, respectively. The two letters (ap) for the 30S binding site indicate the intra-30S hybrid state due to the 30S head swivel [Citation16]. Conversely, in Int1, the tRNAs are in the ap/ap (the two letters for the 50S binding site indicate the intermediate location of the tRNA between the classic A- and P-sites [Citation17,Citation18]) and pe/E configurations. During translocation, the tRNAs move from the A- and P-sites to the P- and E-sites. Therefore, the ap/ap state must occur before the ap/P state along its moving track, i.e., Int1 occurs before Int2. Accordingly, we assign the ribosome states that exhibited force spectra in and 2(f,g) to Int1 and Int2, respectively, based on the use of the same antibiotic conditions. With both antibiotics, the translocation is partial, while complete translocation is observed with fusidic acid alone, which agrees with the transition from Int1 to Int2.
It is known that fusidic acid binds between EF-G domains I and III to prevent protein release, but fusidic acid does not inhibit one round of translocation [Citation19,Citation20], ,are consistent with this circumstance. Conversely, neomycin binds at multiple sites, including h44 and around H69 and H70 [Citation2]. This drug was necessary to trap Int1 for the abovementioned crystallographic study. It is also essential to trap the unique ribosome conformations here. In the structural study, alignment of mRNAs in Int1 and a pre-translocation ribosome indicated that the mRNA had already translocated one codon at the 3ʹ-side in Int1. Our results in agreed with this conclusion within experimental uncertainty, except that only 2 nt were translocated at the 3ʹ-side.
Discussion
Using the FIRMS technique to probe both the 3ʹ and 5ʹ sides of the ribosome-uncovered mRNA, we have obtained evidence for a unique ribosome conformation that is consistent with the mRNA movement of 2 and 1 nt at the 3ʹ- and 5ʹ-ends, respectively. These findings imply that the translocation is asymmetric and stepwise. In this conformation, the ribosome covered 28 nt of the mRNA, which is 1 nt longer than the normal 27 nt coverage in both the Pre and Post complexes. This conformation could be a consequence of either a ‘looped’ mRNA or ribosome expansion. A computational simulation indicated that the mRNA was clamped at 3ʹ- and 5ʹ-ends by two groups of proteins, (S3-5) and (S7, 8, 11), respectively. The clamps opened and closed alternately to keep the mRNA reading frame [Citation21]. More recently, Nguyen and Whitford identified a 30S head tilting motion that is essential to release the steric hindrance along the mRNA pathway [Citation22]. Both simulations indicated that the mRNA motion and the correct codon reading frame were tightly controlled by the ribosome internal structures. In addition, the Int1 state that was identified by X-ray crystallography has exhibited multiple expected intermediate rRNA-tRNA interactions that can guide tRNA movements, such as the approach of the 16S 966 loop to the A-tRNA and the simultaneous interaction of the A and P loops with the A-tRNA. Based on this information, it seems reasonable to assume that the inferred asymmetrically and partially translocated ribosome conformation in our study corresponds to an intermediate state in which the 5ʹ-clamp is closed, the head tilting has not finished, the 3ʹ-clamp is opened, and the head has swiveled through a large angle. In Int1, the codon-anticodon interaction remained intact while the tRNAs adopted a compact conformation [Citation2]. Therefore, it is possible that when the tRNAs become closer to each other, the attached mRNA is forced to form a loop.
Because this unique ribosome conformation is only observed in the presence of both fusidic acid and neomycin, it is unclear whether this conformation is kinetically sampled during normal translocation. However, two available observations support its biological significance. First, the abovementioned Int1 ribosome complex was prepared with the same two antibiotics with which many ribosomal components changed in the correct direction for the proper tRNA transition. Second, after antibiotics were washed away in our experiments, full translocation was achieved, which implies that the trapped intermediate state was along the true translocation pathway. Time-resolved measurements should be able to clarify this issue in the future.
Our result and its consistency with the literature enable us to propose a novel translocation mechanism (). The ribosome conformation that exhibited the specific force spectra in and was probably the abovementioned Int1, in which the mRNA is transiently packed to form a loop due to the nonsynchronous dynamics at the two ribosome edges, as well as complex internal rearrangements. Washing away the antibiotics led this intermediate state to switch to Post, implying that the translocation steps after this intermediate state did not require energy. Therefore, the energy barrier before this intermediate state should be higher and probably needs GTP hydrolysis energy from EF-G. We have measured an 89 pN force generated by EF-G. Given the approximately 75% usage of the hydrolysis energy of one GTP, this force should correspond to a distance of 1 but not 3 nt (0.5 nm for 1 nt movement) [Citation23] . Interestingly, this predicated working distance of 0.5 nm agreed well with the conformational change in Int1, which underwent a 0.6 nm A-site tRNA displacement concurrent with the movement of EF-G domain IV [Citation2]. More simulations and experiments are desired to test this model.
Methods
All of the mRNAs and DNA probes are purchased from IDTDNA (Integrated DNA Technologies). The sequence of the mRNA was 5′-Bio- CAA CUG UUA AUU AAA UUA AAU UAA AAA GGA AAU AAAA AUG UUU AAU UUU UUA GGG CGC AAU CUA CUG CUG AAC UC-3′ (Bio: biotin functionalized). The underline indicates the slippery U6A motif that causes frameshifting.
The probes are tabulated below:
The magnetic beads were purchased from Invitrogen, under commercial name M280 that represents the particle diameter of 2.8 μm. All antibiotics were purchased from Sigma-Aldrich and used directly.
Preparation of ribosome complexes
All the mixtures were in TAM10 buffer, which consisted of 20 mM tris-HCl (pH 7.5), 10 mM Mg (OAc)2, 30 mM NH4Cl, 70 mM KCl, 5 mM EDTA, and 7 mM BME (2-mercaptoethanol), and 0.05% Tween20. Five mixtures were prepared: the ribosome mix, TuWG mix, Tu0G mix, Phe mix and Asnmix. The ribosome mix contains 1 μM ribosome, 1.5 μM each of IF1, IF2, IF3, 2 μM of mRNA, 4 μM of charged fMet-tRNAfMet, and 4 mM of GTP. The TuWG mix contained 4 μM EF-Tu, 0.4 μM EF-Ts, 2 μM EF-G, 4 mM GTP, 4 mM PEP, and 0.02 mg/mL Pyruvate Kinase. The Tu0G mix contained no EF-G but all the other components in the TuWG mix. The Phe mix contained 100 mM Tris (pH 7.8), 20 mM MgAc2, 1 mM EDTA, 4 mM ATP, 7 mM BME, 0.1 mg/mL total synthetase, 50 A260/ml total tRNA, and 0.25 mM phenylalanine. The Asn mix contained the same components except that the phenylalanine was replaced by Asparagine.
The five mixes were incubated at 37 °C for 25 min before making the ribosome complexes. The ribosome mix, TuWG mix, and Phe mix were incubated with 1:2:2 volume ratio at 37 °C for 15 min. The resulting MF-Post ribosome complex was added on 1.1 M sucrose cushion and purified by ultra-centrifuge. The ribosome preparation was similar to our previous work [Citation11].
The MF-Pre complex was formed by incubating the ribosome mix, Tu0G mix and A mix in the volume ratio of 1:2:2, at 37 °C for 2 min. The resulting ribosome complex was added on 1.1 M sucrose cushion and purified by ultra-centrifuge.
The MFN-Post was formed by incubating MF-Post (1 μM) with the Asn mix and TuWG mix in the volume ratio of 1:2:2 at 37 °C for 15 min. The resulting ribosome complex was added on 1.1 M sucrose cushion and purified by ultra-centrifuge.
The MFNF-Post was formed by incubating MFN-Post (1 μM) with the Phe mix and TuWG mix, in the volume ratio of 1:2:2, at 37 °C for 15 min. The resulting ribosome complex was added on 1.1 M sucrose cushion and purified by ultra-centrifuge.
The MFNF-Pre was formed by incubating the MFN-Post with Phe mix and Tu0G mix in the volume ratio of 1:2:2, at 37 °C for 2 min. The resulting ribosome complex was added on 1.1 M sucrose cushion and purified by ultra-centrifuge.
To capture the mRNA-looping intermediate state, neomycin was added into the MF-Pre ribosome complexes and incubated for 10 minutes at 37 °C. EF-G solution pre-incubated with fusidic acid for 20 minutes at 37 °C was added to the ribosome-mRNA complex, and incubated at 37 °C for an additional 5 minutes. The final conditions were 0.1 μM ribosome complexes, 2 μM EF-G, 4 mM GTP, 4 mM PEP, 0.02 mg/mL Pyruvate Kinase, 0.2 mM neomycin, and 0.25 mM fusidic acid.
All other antibiotics were prepared similarly. Viomycin, hygromycin B, and fusidic acid pre-incubated with EF-G were incubated with MF-Pre ribosome complexes, respectively. The final concentrations were 0.2 mM viomycin, 0.4 mM hygromycin B, 0.25 mM fusidic acid, respectively.
Sample preparation prior to magnetic and force measurements
A sample well with dimensions of 4 × 3 × 2 mm3 (L × W × D) was glued with a piece of biotin coated glass on the bottom surface. An aqueous solution of 0.25 mg/mL streptavidin was loaded into the sample well and incubated for 40 min. Then the sample well was rinsed twice with TAM10 buffer.
For studies involving no antibiotics, 20 μL of 0.1 μM ribosome complexes were immobilized on the surface via the 5ʹ-end biotin on the mRNA and incubated for 1 hr. After rinsing the surface, 20 μL of 1 μM biotinylated probing DNA strand was added and incubate for overnight. The formed DNA-mRNA duplex was rinsed with TAM10 buffer. Subsequently, the streptavidin-coated magnetic beads were introduced into the sample well and incubated for 2 hr, the non-specific bound magnetic particles were removed from the surface by applying centrifuge with the speed of 1000 rpm for 5 min. The sample was then magnetized for 2 min using a permanent magnet (~ 0.5 T).
Magnetic and force measurements
Magnetic signal of the samples was measured by an atomic magnetometer as a function of mechanical force, using the force-induced remnant magnetization spectroscopy (FIRMS) technique [11,12]. The atomic magnetometer had a sensitivity of ~ 100 fT/(Hz)1/2. The force was provided by a centrifuge (5427R from Eppendorf), with the speed increasing by 100 rpm (revolution per minute) per step. The dissociation of the DNA-mRNA duplexes was indicated by a decrease in the magnetic signal, which occurred when the centrifugal force reached the dissociation force of the DNA-mRNA duplex. This is because the dissociated magnetic beads were removed from the sample. The typical force range in this work was 90 pN, after which the residual magnetic signal was taken as the background. FIRMS profiles were obtained by normalizing the overall magnetic signal decrease (B0) to be 100% and then plotting the relative magnetic signal decrease (B/B0) vs. the external force. The force values were calculated according to mω2r, in which m is the buoyant mass of M280 magnetic beads (4.6 × 10−15 kg), ω is the centrifugal speed, and r is the distance of the magnetic beads from the rotor axis (7.5 cm for 5427R). The typical force resolution was 3–4 pN in this work. Each profile reported in this work was repeated at least three times to assure reproducibility.
Author contributions
H.Y., S.X., and Y. W. conceived the experimental concepts and designed the experiments. H. Y. and Y.W. performed the experiments. All authors contributed to data analysis and manuscript preparation.
Supplemental Material
Download PDF (219.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
Additional information
Funding
References
- Noller HF, Lancaster L, Mohan S, et al. Ribosome structural dynamics in translocation: yet another functional role for ribosomal RNA. Q Rev Biophys. 2017;50:e12.
- Zhou J, Lancaster L, Donohue JP, et al. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345:1188–1191.
- Ramrath DJ, Lancaster L, Sprink T, et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci USA. 2013;110:20964–20969.
- Schuwirth BS, Borovinskaya MA, Hau CW, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834.
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322.
- Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. Rna. 2007;13:1473–1482.
- Woolstenhulme CJ, Guydosh NR, Green R, et al. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 2015;11:13–21.
- Yan S, Wen JD, Bustamante C, et al. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881.
- Caliskan N, Katunin VI, Belardinelli R, et al. Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014;157:1619–1631.
- Kemkhadze KS, Odintsov VB, Semenkov YP, et al. Quantitative study of the interaction of aminoacyl-tRNA with the a site of Escherichia coli ribosomes: equilibrium and kinetic parameters of binding in the absence of EF-Tu factor and GTP. FEBS Lett. 1981;125:10–14.
- Tsai TW, Yang H, Yin H, et al. High-efficiency “-1” and “-2” ribosomal frameshiftings revealed by force spectroscopy. ACS Chem Biol. 2017;12:1629–1635.
- De Silva L, Yao L, Wang Y, et al. Well-defined and sequence-specific noncovalent binding forces of DNA. J Phys Chem B. 2013;117:7554–7558.
- Qu X, Wen JD, Lancaster L, et al. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121.
- Peske F, Savelsbergh A, Katunin VI, et al. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194.
- Jacks T, Power MD, Masiarz FR, et al. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283.
- Ratje AH, Loerke J, Mikolajka A, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716.
- Brunelle JL, Youngman EM, Sharma D, et al. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. Rna. 2006;12:33–39.
- Samaha RR, Green R, Noller HF. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature. 1995;377:309–314.
- Belardinelli R, Rodnina MV. Effect of fusidic acid on the kinetics of molecular motions during EF-G-induced translocation on the ribosome. Sci Rep. 2017;7:10536.
- Okura A, Kinoshita T, Tanaka N. Formation of fusidic acid-G factor-GDP-ribosome complex and the relationship to the inhibition of GTP hydrolysis. J Antibiot. 1971;24:655–661.
- Kurkcuoglu O, Doruker P, Sen TZ, et al. The ribosome structure controls and directs mRNA entry, translocation and exit dynamics. Phys Biol. 2008;5:046005.
- Nguyen K, Whitford PC. Capturing transition states for tRNA hybrid-state formation in the ribosome. J Phys Chem B. 2016;120:8768–8775.
- Yao L, Li Y, Tsai TW, et al. Noninvasive measurement of the mechanical force generated by motor protein EF-G during ribosome translocation. Angew. Chem. Int. Ed. 2013;52:14041–14044.