ABSTRACT
Porcine OTX2 was found to be highly activated in porcine iPS cells (piPSCs) that were reported by different laboratories worldwide. To reveal the regulatory function of OTX2 in porcine reprogrammed cells, we screened porcine miRNA-seq databases and found two miRNAs, miR-1343 and miR-545, that could specifically bind to 3ʹUTR of OTX2 and suppress endogenous OTX2 expression in piPSCs. Knockdown of OTX2 by miR-1343 and miR-545 could significantly increase the expression of SOX2 and ESRRB, but did not alter the expressions of OCT4 and KLF4, and improve the pluripotency of piPSCs. The promoter-based assays showed that OTX2 potentially bound to the promoter region of SOX2 and ESRRB and suppressed their expression. On the other hand, SOX2 could interact with OTX2 promoter. Ectopic expression of SOX2 could significantly decrease OTX2 promoter activity, showing that there is a negative feedback loop between SOX2 and OTX2. Additionally, SOX2 and ESRRB significantly stimulated miR-1343 expression in piPSCs, but OTX2 down regulated the expression of miR-1343 in either direct or indirect manners. In summary, this study demonstrates that there is a regulatory network mediated by miR-1343, in which downregulation of OTX2 by miR-1343 can elevate the expression of pluripotent genes that were then sustain the pluripotency of piPSCs.
KEYWORDS:
Introduction
Two pluripotent states, naïve state and primed state, of pluripotent stem cells (PSCs) have been reported [Citation1], in which the naïve PSCs, for instance mouse ESCs derived from the inner cell mass and early pre-implantation blastocysts, have the potential in generating germline transmitting chimeras [Citation1,Citation2]. However, the primed PSCs, for instance human ESCs and mEpiSC derived from the post-implantation epiblasts, have the limited capacity to contribute to chimeric offspring [Citation1,Citation3,Citation4]. Different pluripotent state factors may play diverse roles in the pluripotency regulating of PSCs. Some naïve state factors, such as Esrrb, Nanog, and Klf2, were found to maintain PSCs pluripotency with distinct regulatory circuitries [Citation5–Citation7]. Esrrb is a principal target of Gsk3/Tcf3 axis in the pluripotency regulation network, and potently suppresses ESCs differentiation and maintains its self-renewal [Citation6]. In addition, Nanog can bind directly to Esrrb and stimulate Esrrb transcription, thus functional replacement between Nanog and Esrrb plays an important role in maintaining ESCs self-renewal and pluripotency [Citation5]. In mESCs, Klf2 is repressed by Mek/Erk pathway. Klf2 overexpression can replace Mek inhibition and sustain the ground state pluripotency of mESCs [Citation7]. On the other hand, the primed state factors, such as Otx2, Tcf15, and Zfp281, are barriers for maintaining the pluripotency of PSCs [Citation8–Citation10]. Tcf15 is an important primed factor that is dependent on FGF signaling and can repress Nanog expression, resulting in primed PSCs out of pluripotent state [Citation10]. Zfp281, a transcriptional repressor, can restrict the expression of pluripotency genes and promote the differentiation of ESCs [Citation8]. Otx2 directly interacts with many key transfection factors, such as Oct4 and Hmga2, and lead to the exit of ESCs from the pluripotent ground state [Citation9]. A very recent report showed that OTX2 acting as a roadblock functions repressively upstream of PGC transcription factors and controls the entry of epiblast cells to the germline. Downregulation of Otx2 precedes the initiation of the PGC programme both in vitro and in vivo [Citation11].
MicroRNAs (miRNAs) are endogenous small noncoding RNA molecules that have important function to regulate the expression of pluripotent genes. Recent reports showed that miRNAs play an important role in the regulatory network of pluripotency [Citation12–Citation14]. For instance, the miR-145 knocks down the expression of Oct4, Sox2, and Klf4 and represses the pluripotency of human ESCs. Furthermore, miR-145 also shows the ability to inhibit the proliferation of stem cells in vitro via targeting pluripotent genes [Citation15,Citation16]. The miR-302/367 cluster, a stemness regulator in ESCs, promotes the reprogramming efficiency of induced pluripotent stem cells (iPSCs) by targeting multiple genes [Citation17]. Moreover, miR-34a also expands the cell fate potential of PSCs by strongly inducing retroelement MuERV-L/MERVL expression [Citation14].
Currently, multiple attempts have been reported to generate bona fide piPSCs that hold the potential in generating germline transmitting chimeras in vivo [Citation18–Citation23]. However, it is still a challenge for the unclear pluripotency regulatory circuitries of piPSCs. This issue enabled us to pay close attention to the pluripotent state factors and their regulatory network in piPSCs. To address this fundamental question, we made a transcriptome analysis of piPS cell lines, which were reported by the three laboratories worldwide, and found that OTX2 was highly expressed in all tested porcine iPSC lines. Since the previous study indicated that Otx2 was a negative switch in the regulation of pluripotent gene expression [Citation24–Citation27], porcine OTX2 might be a key factor in the pluripotency regulatory network of piPSCs. Thus, we screened the porcine miRNA-seq databases and found that miR-1343, as well as miR-545, could significantly repress OTX2 expression. Downregulation of OTX2 by miR-1343 finally increased the expression of SOX2 and ESRRB, which then elevated the pluripotency of piPSCs.
Results
Conjoint analysis of piPSC RNA and mirna transcriptomes
To uncover the key transcription factors that play the crucial role in porcine cell reprogramming, we analyzed RNA transcriptomes of the three porcine iPSC lines derived from different laboratories, which included bFGF-dependent porcine iPSC line [Citation18], LIF-dependent porcine iPSC line [Citation28], and a porcine iPSC line required both bFGF and LIF [Citation23]. Results showed that transcriptome profiles of piPSCs were significantly different with pig embryonic fibroblast cells (PEFs), but no considerable difference among distinct porcine iPSC lines (). The Venn analysis showed that eight genes, including OTX2, ZIC3, ZSCAN10, TCF15, OCT6, SOX2, NANOG, and ESRRB, were commonly upregulated in piPSCs. However, two genes, RARG and REX1, were downregulated compared to PEFs (, S1A). The normalized estimation of gene expression based on FPKM presented that the number of transcripts of the primed state markers OTX2 and ZIC3 were roughly five to ten-fold higher than that of the naïve state markers NANOG and ESRRB (Fig. S1B). The quantitative RT-PCR assay further confirmed that the proportion of OTX2 expression was nearly ten-fold higher than that of NANOG and ESRRB in the piPSC-dox line that was generated in our lab and reported previously [Citation29] (). These results indicated that the tested piPSCs were in the primed pluripotent state due to the high expression level of OTX2 that might impede the pluripotency of piPSCs through downregulating the expression of naïve state markers [Citation26].
Figure 1. Screening miRNAs based on porcine piPSC RNA-seq database. A. Heatmap of pluripotency associated genes in porcine iPSC lines and PEFs. B. The upregulated and downregulated genes in all three porcine iPSC lines (up panel) and the mRNA expression of upregulated pluripotent genes in PEFs and piPSC-dox cells (down panel). C. Conjoint analysis of miRNAs that potentially bind to 3ʹUTR of OTX2 by the three online programs. D. Putative binding sites in 3ʹUTR of OTX2 and the seed sequences (in red) of miRNAs. Data indicate mean ± SD. ** P < 0.01, n = 3.
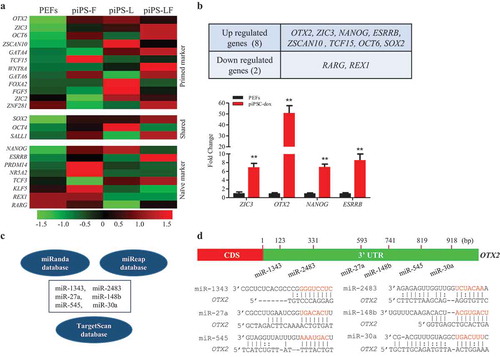
Thereafter, we focused on the regulation of porcine OTX2 expression and applied the 3ʹ untranslated region (3ʹUTR) of OTX2 to align with miRNA-seq database and predict miRNAs that putatively bind to 3ʹUTR of OTX2. The three common online program tools were used to perform the conjoint analysis. The overlapping results showed that six miRNAs, including miR-1343, miR-2483, miR-27a, miR-148b, miR-545, and miR-30a, were potentially bound to the OTX2 (). The putative binding sites and the seed sequences of miRNAs were aligned in the OTX2 3ʹUTR and marked in red ().
miR-1343 and miR-545 bind to 3ʹUTR and repress OTX2 expression
To confirm miRNA-target interactions, a luciferase reporter vector that contains 1.9 kb 3ʹUTR of OTX2 sequence and a lentiviral vector that is able to ectopically express miRNA was constructed. The reporter vector and miRNA lentiviral vector were cotransfected into HEK-293T cells. Luciferase assays showed that two out of six miRNAs, miR-1343 and miR-545, significantly knocked down OTX2 expression (, left). The miRNA mimics could also knock down luciferase activity (, right), indicating that miR-1343 and miR-545 could bind to 3ʹUTR and repress OTX2 expression. Because miR-1343 and miR-545 bound to the different zone of the OTX2 3ʹUTR, application of miR-1343 and miR-545 together could further knocked down OTX2 expression. This synergistic action of miR-1343 and miR-545 on OTX2 knockdown was confirmed by application of miRNA mimics (). We then made the mutant constructs that the base pairs for miR-1343 and miR-545 binding sites were changed, and did luciferase assays. Both miR-1343 and miR-545 did not influence luciferase activity of mutant constructs comparing to the wild type control, suggesting that these two miRNAs could specifically bind to OTX2 3ʹUTR and regulate OTX2 expression ().
Figure 2. Validation of interactions between miRNAs and target OTX2. A. Luciferase assays of OTX2 in HEK-293T cells treated by each individual miRNA (left panel) and miRNA mimic (right panel). B. Luciferase assays of OTX2 in HEK-293T cells treated by cotransfection of miR-1343 and miR-545 (left panel) and miRNA mimics (right panel). C-D. Luciferase assays of OTX2 in HEK-293T cells with the treatments of miR-1343 mimic (C) and miR-545 mimic (D) together with their mutant (MUT) and wild type (WT) constructs. E-F. The mRNA and protein expression of OTX2 in piPSC-dox cells (E) and PS23 cells (F) treated by miRNAs. Ctrl, cells were transfected with the pCD513B-1. β-Actin was as internal control. NC-M, the negative control mimic. Data indicate mean ± SD. ** P < 0.01, n = 3.
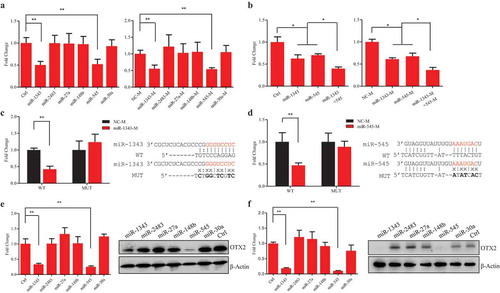
Next, we asked if the miRNAs could repress endogenous OTX2 expression in piPSCs. Two porcine iPSC lines (piPSC-dox and PS23 cells), which were reported in our laboratory previously [Citation22,Citation30], were chosen for the further study. RT-PCR assays indicated that the expression level of miR-1343 and miR-545 in the two piPSC lines was lower than that in control PEFs (Fig. S2). The six miRNA constructs were transfected into piPSCs for 72 h (Fig. S3), and then the expression of OTX2 were detected. The both mRNA and protein levels of OTX2 in piPSC-dox and PS23 cells was significantly downregulated by miR-1343 and miR-545, but was unaffected by other four miRNA treatments (). These results indicated that miR-1343 and miR-545 could directly bind to 3ʹUTR and repress OTX2 expression in vivo.
miR-1343 regulates porcine iPSC pluripotency via repression of OTX2
Since endogenous OTX2 expression was able to be repressed by miRNAs, overexpression of miR-1343 and miR-545 visibly increased the alkaline phosphatase (AP) activity in piPSC-dox cells, however, this effect was completely abolished by overexpression of OTX2 (). The similar phenomenon was also observed in PS23 cells, in which overexpression of miR-1343 and miR-545 could significantly increase AP activity and the number of AP positive clones due to the repression of OTX2 (). The reduction of protein expression was further demonstrated that both miR-1343 and miR-545 negatively regulated OTX2 expression in piPSC-dox and PS23 cells (). Additionally, overexpression of miR-1343 and miR-545 could facilitate the proliferation of piPSC-dox and PS23 cells (Fig. S4). However, this upshot was ended by the ectopic expression of OTX2.
Figure 3. MiR-1343 regulates porcine iPSC pluripotency via OTX2 knockdown. The piPSC-dox and PS23 cells were treated with miRNAs or miRNA plus OTX2 vector for 72 h, and then cells were used for the following experiments. A-B. Alkaline phosphatase (AP) staining of piPSC-dox cells (A) and PS23 cells (B). Scale bar, 200 μm. C-D. The protein expression of OTX2 in piPSC-dox (C) and PS23 (D) cells. E-F. The mRNA expression of pluripotent genes in piPSC-dox (E) and PS23 (F) cells. Ctrl, cells transfected with pCD513B-1. β-Actin was as internal control. Data indicate mean ± SD. * p < 0.05, ** P < 0.01, n = 3.
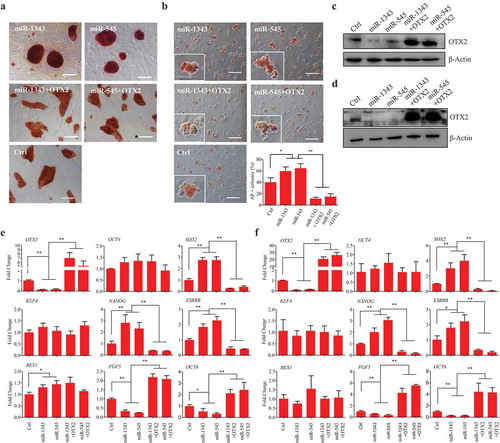
To further investigate the regulatory effect of miRNAs, we examined the expression of pluripotent genes in piPSCs treated by miRNAs. Results showed that miR-1343 and miR-545 could significantly activate the expression of SOX2, NANOG, ESRRB, and REX1 in piPSC-dox cells, however, the expression of OCT4 and KLF4 were not stimulated by these miRNAs. Conversely, the expression of FGF5 and OCT6, which are the downstream target of OTX2, was significantly reduced by miRNAs. On the other hand, ectopic expression of OTX2 could significantly decrease the effect triggered by miR-1343 and miR-545, and enhance the expression of FGF5 and OCT6 (). In PS23 cells, the expression level of SOX2, NANOG, and ESRRB was also significantly increased by the treatment of miR-1343 and miR-545, meanwhile, FGF5 and OCT6 were significantly downregulated (). These results indicated that miR-1343, as well as miR-545, is a crucial regulator to increase the expression of pluripotent factors of piPSCs via knocking down the endogenous OTX2 expression.
To investigate the function of miR-1343 and miR-545 during porcine iPSC differentiation, the miRNA-treated piPSC-dox cells were used to generate the embryoid bodies (EBs) in vitro. The immunofluorescence of germ layer specific markers showed that overexpression of miR-1343 and miR-545 increased the expression of ectodermal marker TUBB3 (tubulin beta 3 class III), and reduced the expression of mesoderm marker DESMIN and endoderm marker AFP when the miRNA-treated piPSC-dox cells were spontaneously differentiated in vitro. However, overexpression of OTX2 could significantly reduce TUBB3 expression and enhance the expression of DESMIN and AFP (Fig. S5A). The mRNA and protein expression of TUBB3, DESMIN, and AFP further confirmed that both miR-1343 and miR-545 promoted the ectodermal gene expression and suppressed the mesodermal and endodermal gene expression due to the knockdown OTX2 (Fig. S5B-C).
OTX2 represses the expression of SOX2 and ESRRB
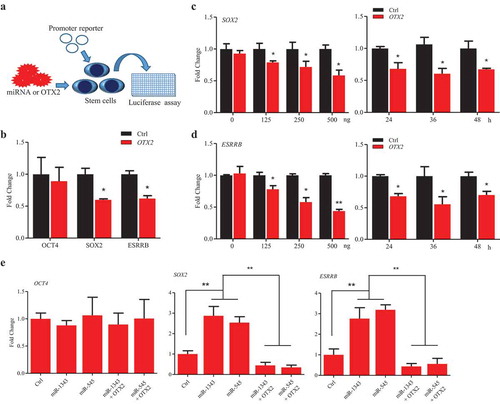
To further investigate that OTX2 regulates porcine pluripotent genes, a screening assay was set up, in which the promoter reporter of pluripotent genes and OTX2 vector were cotransfected into stem cells with or without treatments of miR-1343 and miR-545 (). The luciferase assays showed that OTX2 significantly suppressed the promoter activity of SOX2 and ESRRB, but did not suppress OCT4 promoter activation though it may retain the potential OTX2 binding site (,S6A). The time-course and dose-dependent assays were further confirmed that OTX2 negatively regulated the expression of SOX2 and ESRRB (). Based on our previous study [Citation26] and bioinformatics analysis, the promoter region of SOX2, OCT4, and ESRRB contains the putative OTX2 binding sites (Fig. S6A). The truncated constructs of SOX2 promoter with the predicted OTX2 binding sites were then used to perform the luciferase assay to examine the regulatory role of OTX2 in P19 cells (Fig. S6B). The result showed that OTX2 specifically suppress the promoter activity of SOX2, but no any effect on the truncated construct that is without OTX2 binding site (Fig. S6C). Because of the regulatory interaction between OTX2 and SOX2 and ESRRB promoters, knocking down OTX2 by miR-1343 and miR-545 could significantly increase the promoter activity of SOX2 and ESRRB in piPSC-dox cells (). This result indicates that miR-1343 and miR-545 indirectly elevate the expression of SOX2 and ESRRB and promote the pluripotency of piPSCs through knocking down OTX2 expression.
Figure 4. OTX2 represses the expression of SOX2 and ESRRB. A. Schematic diagram of the screening assay. Stem cells were transfected by miRNAs or miRNA plus OTX2 vector for 36 h. The promoter activation was then determined by luciferase assay. B. Promoter activity of pluripotent genes in P19 cells that were co-transfected with pcDNA3.1 OTX2 and reporter vector of OCT4, SOX2, and ESRRB, respectively. C-D. Dose-dependent (left panel) and time-course (right panel) assays for SOX2 (C) and ESRRB (D) promoter activity in P19 cells. Ctrl, cells were transfected by pcDNA3.1. E. Promoter activity of pluripotent genes in piPSC-dox cells. Ctrl, cells were transfected by pCD513B-1. Data indicate mean ± SD. * p < 0.05, ** P < 0.01, n = 3.
SOX2 inhibits OTX2 promoter activation and increases miR-1343 expression
Our previous study [Citation26] and bioinformatics analysis showed that the promoter region of OTX2 contains the putative binding sites for pluripotent factors. The luciferase assays showed that overexpression of SOX2 and SOX2/OTC4 could significantly reduce the promoter activity of OTX2 in P19 cells, however, overexpression of OCT4 only and ESRRB did not lower the promoter activity of OTX2 (). To further confirm the negative feedback loop between SOX2 and OTX2, several vectors with the truncated OTX2 promoter sequence were constructed, in which there are multiple putative SOX2 binding sites in the promoter region of OTX2 (). Overexpression of SOX2 could significantly decrease OTX2 promoter activity, and this action presented dose-dependent manners, but showed no significant difference over the time-course ().
Figure 5. SOX2 represses OTX2 and stimulates miR-1343 expression. A. Luciferase assay of OTX2 promoter activity in P19 cells that were transfected by pluripotent gene constructs. B. Truncated constructs of OTX2 promoter with the predicted SOX2 binding sites. C. Luciferase assay of OTX2 promoter activity in P19 cells treated by overexpression of SOX2. D. Dose-dependent (left panel) and time-course (right panel) assays of OTX2 promoter activity in P19 cells treated by overexpression of SOX2. E-F. Pluripotent factors regulate the expression of miR-1343 and miR-545 in piPSC-dox (E) and PS23 (F) cells. Ctrl, cells were treated with pcDNA3.1 basic. Data indicate mean ± SD. * p < 0.05, ** P < 0.01 n = 3.
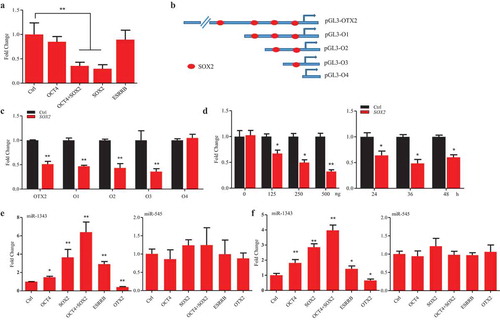
Then, we ask if the pluripotent genes could influence the expression of miR-1343 and miR-545. In piPSC-dox and PS23 cells, overexpression of OCT4, SOX2, and ESRRB could significantly increase miR-1343 expression, but no effect on miR-545. Interestingly, OTX2, as a negative feedback loop, could significantly lower the miR-1343 expression, but it did not significant change the miR-545 expression (). These observations indicate that there is a miRNA/OTX2 regulatory network in piPSCs. This regulation network is mainly operated by miR-1343, but not by miR-545 although miR-545 also targeting on OTX2 and repressing OTX2 expression, via knocking down OTX2 expression and elevating the expression of pluripotent genes. In addition, the pluripotent genes SOX2 and ESRRB promote miR-1343 expression, and OTX2 inhibits miR-1343 expression by either direct or indirect manners (). This study confirms that miR-1343 is a crucial post-transcriptional regulator to maintain the pluripotency of porcine iPS cells by suppressing the expression of OTX2.
Discussion
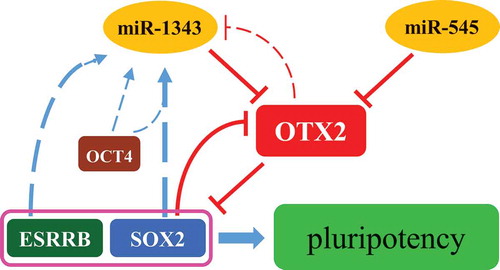
In this study, we find that porcine OTX2 is a key primed pluripotent factor and is able to be knocked down by miR-1343 in piPSCs. The expression of miR-1343 was negatively regulated by OTX2 and positively regulated by SOX2/OCT4 and ESRRB. Thus, we demonstrated that miR-1343, OTX2, and pluripotent factors SOX2 and ESRRB formed a regulatory network to maintain the pluripotency of piPSCs ().
Figure 6. Regulation network of miR-1343-OTX2-SOX2 in piPSCs. The miR-1343 and miR-545 directly bind to OTX2 3ʹUTR and knock down OTX2 expression. OTX2 inhibits the expression of SOX2 and ESRRB. SOX2 is able to interacted with OTX2 promoter and represses OTX2 promoter activity. Both SOX2 and ESRRB upregulate miR-1343 expression, but OTX2 downregulates miR-1343 expression. MiR-1343-OTX2-SOX2 forms a regulatory network to maintain the pluripotency of piPSCs.
According to the transcriptome analysis, we found that the high-level expression of OTX2 might be one of key barriers to impede the maintenance of pluripotency in piPSCs [Citation26] . Additionally, the primed factors ZIC3 and TCF15 were also significantly upregulated in all three reported piPS cell lines. Zic3 is a member of the Zic family of C2H2-type zinc finger protein and represses the oxidative phosphorylation, which is important for the pluripotent state maintenance [Citation31]. Tcf15, the basic helix-loop-helix transcription factor, is driven by FGF signaling and acts as a switch for the pluripotent cell differentiation. Ectopic expression of Tcf15 can repress Nanog expression and oppositely upregulate the epiblast determinant Otx2 [Citation10]. Thus, the high level of ZiC3 and TCF15 in piPSCs seems to prime porcine stem cells out of the pluripotent state. As the key pluripotent factor, Rex1 was directly transactivated by Nanog and Sox2 [Citation32]. We noticed that REX1 was not activated in piPSCs though the naïve markers NANOG and ESRRB were significantly upregulated in piPSCs. Therefore, the inactivation of naïve markers is likely to be another obstacle to decline the pluripotency of piPSCs. Overall, current porcine iPSC lines reported by the different laboratories are in the primed pluripotent state, and studying the pluripotency associated regulators may help us to promote the pluripotency of piPSCs.
The crucial role of Otx2 in the pluripotent regulation has recently been recognized [Citation9]. Previous studies showed that OTX2 could significantly repress the promoter activation of NANOG [Citation26], and the expression level of Nanog and Esrrb was rapidly decreased when Otx2 upregulated during the transition to EpiSC [Citation33]. Oct4 and Sox2 have the similar binding sequence, and the Oct4/Sox2 complex is important for maintaining the pluripotency of ESCs [Citation34]. In our previous report [Citation26] and current study, we found that OTX2 could significantly reduce the expression of SOX2 and ESRRB by directly repressing the promoter activity, but did not inhibit OCT4 expression. On the other hand, SOX2 could negatively regulate OTX2 expression by repressing OTX2 promoter activity, but ESRRB did not regulate OTX2 expression. Thus, the OTX2 and SOX2 form a negative feedback loop to regulate self-renewal and pluripotency of piPSCs.
Several reports have showed that Otx2 is able to be regulated by miRNAs. In mouse neural cells, overexpression of miR-206 regulated cell proliferation and apoptosis by knocking down Otx2 expression [Citation35]. The miR-410 could directly knock down OTX2 and impede the retinal pigment epithelium differentiation from human amniotic epithelial stem cells [Citation36]. In this study, we prove that miR-1343 and miR-545 are able to bind to porcine OTX2 and repress its expression. MiR-1343 was reported to repress the expression of TGF-β receptor genes and block TGF-β induced epithelial to mesenchymal transition [Citation37]. TGF-β signaling plays an important role in the pluripotency regulation [Citation38]. It may be worthy to investigate the regulatory effect of miR-1343 on the TGF-β signaling in piPSCs. MiR-545 has been reported in several carcinomas and cancer cells. Overexpression of miR-545 could suppress colorectal cancer cells proliferation through regulating epidermal growth factor receptor expression [Citation39]. Additionally, miR-545 could promote hepatocellular carcinoma proliferation by activating PI3K/Akt signaling through downregulation of RIG-1 [Citation40]. We found that miR-1343 and miR-545 could promote porcine iPSC proliferation by downregulation of OTX2. Besides, we discovered that the primed markers FGF5 and OCT6 were also significantly downregulated by miR-1343 and miR-545. These observation proved the concept similar to the recent reports that prolonged overexpression of Otx2 leads to cell death when provide with differentiation cues [Citation33,Citation41]. We also found that knockdown of OTX2 by miR-1343 and miR-545 increased the expression of SOX2, NANOG, and ESRRB, but did not significantly change the OCT4 expression. Alternatively, SOX2 and ESRRB, as well as OCT4, were able to activate miR-1343 expression, on the contrary, OTX2 inhibited miR-1343 expression in piPSCs. However, the expression of miR-454 was not regulated by pluripotent factors, suggesting that it does not directly involved in cell reprogramming. Consequently, the feedback loops of OTX2/SOX2, miR-1343/OTX2, and SOX2/miR-1343 form a regulatory network to regulate the pluripotency of piPSCs.
Interestingly, we found that when piPSCs spontaneously differentiated via the formation of embryoid bodies, miR-1343 and miR-545 could increase the expression of ectoderm maker TUBB3 and inhibit the expression of mesodermal and endodermal genes due to the knockdown of OTX2. Our observation proved the concept similar to the previous reported result that Otx2 prevented the accumulation of neural progenitors [Citation41]. Although Sox2 is the key pluripotent transcription factor to stabilize ESCs in a pluripotent state by maintaining the expression level of Oct4 [Citation42,Citation43], it also is an important factor regulating neurogenesis and is essential for neuronal progenitor formation and maintenance [Citation44]. In human embryonic neural precursor cells, Sox2 is reported to repress Otx2 expression, whereas the ChIP-qPCR shows that Otx2 is able to target the promoter region of Sox2 and reduce Sox2 expression [Citation27]. Downregulation of Sox2 in human ESCs results in lowing expression of key stem cell factors and initiating the trophectoderm differentiation [Citation45]. Moreover, the study of lineage segregation of trophectoderm and pluripotent inner cell mass showed that SOX2 is the faithful maker for pluripotency in pig [Citation46]. Therefore, we assume that miR-1343 and SOX2 may act as the important repressors to regulate OTX2 and play an important role in balancing the pluripotency and ectoderm differentiation in piPSCs.
The expression of miRNAs is controlled by its own promoter sequence that can be bound and regulated by transcription factors. For instance, the transcription factor LXRα was proved to activate miR-378 expression by bind to its promoter region [Citation47] and MYC could recruited DNMT3A to the promoter region of miR-200b and repress its expression [Citation48]. In this study, we found that pluripotent factors SOX2 and ESRRB could promote miR-1343 activation, but did not affect the activity of miR-545. We speculate that these transcription factors may directly or indirectly interact with miR-1343 promoter, but not with miR-545 promoter, and enhance its expression. However, the more precise experiments, such as promoter binding assay, may be applied to confirm this speculation in the future. OTX2 downregulation of miR-1343 may also be acted through the repression of miR-1343 promoter activity directly or indirectly since previous studies have showed that Otx2 could bind to the promoter region of target genes and repress their expressions [Citation27,Citation49]. Additionally, OTX2 may also be through down regulation of SOX2 expression, which is confirmed in this study, to reduce miR-1343 expression indirectly.
In summary, we found the miR-1343-OTX2-SOX2 regulation network that enhances pluripotency of piPSCs via knocking down OTX2 by miR-1343 and miR-545. This finding indicates the potential new strategy to maintain porcine iPSC pluripotency through downregulation of OTX2 by miRNAs, which will provide insights into the molecular mechanism of pluripotency regulation in piPSCs.
Materials and methods
Gene cloning and vector construction
Genomic DNA (gDNA) was isolated from PEFs. DNA concentration and the purity were examined using Nanodrop (Thermo Scientific). The 1.9 kb 3ʹUTR of the OTX2 was amplified from pig gDNA by PCR and cloned into pGEM-T Easy Vector (Promega, USA). The 1.9 kb sequence was confirmed by DNA sequencing, and then subcloned into pSICHECK2 vector (Promega, USA). The 293 bp 3ʹUTR fragment with the seed sequence of miR-1343 and the 351 bp 3ʹUTR fragment with the seed sequence of miR-545 were subcloned, and their mutations were generated by the overlap PCR. The promoter sequences of porcine OCT4, ESRRB, and OTX2 were constructed into pGL3-Basic vector (Promega, USA), which were reported previously [Citation50]. A 4.2 kb porcine SOX2 promoter sequence was amplified by PCR and subcloned into the pGL3-Basic. To dissect the core regulatory region of SOX2 promoter, two truncated SOX2 promoter fragments were amplified based on 4.2 kb SOX2 promoter sequence and constructed two reporter vectors. A series of deletion constructs of OTX2 promoter were also constructed based on the 2.1 kb OTX2 promoter sequence. The pri-miRNAs were amplified from pig gDNA, and then cloned into pGEM-T Easy vector, confirmed by DNA sequencing, and subcloned into pCD513B-1 vector (System Biosciences, USA). To construct OTX2 expression vector, the OTX2 CDS was cloned into pGEM-T Easy vector for DNA sequencing, and then subcloned into pCD513B-1 vector. The CDS of porcine OCT4, SOX2, ESRRB, and OTX2 were constructed into pcDNA3.1 vector as described previously [Citation50]. All constructed vectors are listed in Table S1.
Cell culture
The piPSC-dox cells generated in this laboratory were cultured in LF2i medium [Citation29], which included DMEM (Hyclone, USA) supplemented with 15% FBS (SeraPro, Germany), 0.1 mM nonessential amino acids (NEAA, Gibico, USA), 1 mM L-glutaMAX (Gibico, USA), 10 ng/mL LIF (Merck Millipore, USA), 10 ng/mL bFGF (PeproTech, USA), 0.1 mM β-mercaptoethanol (β-met, Sigma Aldrich, USA), 3 µM CHIR99021 (StemRD, USA), 2 µM SB431542 (StemRD. USA), 1 μg/mL doxycycline (Dox, Sigma Aldrich, USA). Porcine induced pluripotent cell line PS23, which was generated in our laboratory [Citation22], was cultured in knock-out DMEM (Invitrogen, USA) supplemented with 20% FBS, 0.1 mM NEAA, 1 mM L-glutaMAX, 10 ng/mL LIF, 10 ng/mL bFGF, 0.1 mM β-met. The porcine embryonic fibroblasts (PEFs) were derived from 35-day-old porcine fetuses and cultured with medium consisting of DMEM supplemented with 15% FBS, 0.1 mM NEAA, 1 mM L-glutaMAX, and 0.1 mM β-met. The mouse carcinoma cell line P19 was grown in alpha-MEM (HyClone, USA) with 2.5% FBS and 7.5% NBCS (HyClone, USA). The human embryonic kidney cell line HEK-293T was cultured in DMEM with 10% FBS. All cells were cultured at 37°C in a 5% CO2 humidified atmosphere. To determine cell growth curve, the 4 × 104 cells/well of piPSC-dox and PS23 cells were seeded in 12-well plate and harvested every 24 h. Cells in triplicate wells were then counted and averaged for each treatment.
Cell transfection
To make lentivirus particles, 2 × 106 HEK-293T cells were seeded on a 60 mm culture dish. When cells reach to 80% confluence, lentiviral vectors (4 μg) carrying with miRNAs and porcine OTX2 were transfected into HEK-293T cells together with 3 μg pSPAX2 and 2 μg pVSVG, respectively. After 48 h post-transduction, medium with viral particles was collected and filtered through 0.45 μm filter (Millipore, USA). The viral supernatants mixed with 8 μg/mL Polybrene were infected into piPSCs for 12 h. The infected piPSCs could be treated with 4 μg/mL puromycin (Sigma Aldrich, USA) for 2 days to enhance the proportion of infected cells. To investigate the regulation network of miR-1343 in piPSCs, cells were plated on a 6-well plate before transfection. After reaching 60% confluence, 4.0 μg vectors carrying with OCT4, SOX2, ESRRB, and OTX2 were transfected with Lipofectamine 3000 Reagent (Invitrogen, USA), respectively. Samples were analyzed at 36 h after transfection.
Luciferase assay
To investigate promoter activity of pluripotent genes, piPSCs transfected with miRNAs vector or miRNAs/OTX2 vector were seeded in a 48-well plate 36 h prior to transfection. When reached 60% confluence, cells were transfected by 250 ng reporter vectors (pGL3-OCT4, pGL3-SOX2, and pGL3-ESRRB), and 0.1 μg pRT-TK control vector with Lipofectamine 3000 (Invitrogen, USA). To investigate promoter activity of pluripotent genes in P19, cells were seeded in a 48-well plate 24 h prior to transfection. When reached 80% confluence, cells were transfected by vectors carrying OCT4, SOX2, ESRRB, and OTX2, 250 ng reporter vector (pGL3-OCT4, pGL3-SOX2, pGL3-ESRRB, and pGL3-OTX2), and 0.1 μg pRT-TK control vector with Lipofectamine 2000 Reagent (Invitrogen, USA), respectively. For 3ʹUTR luciferase assay, the miRNA mimics and mimic negative control were synthesized by GenePharma. HEK-293T cells were seeded in a 48-well plate 24 h prior to transfection. When 80% confluence reached, cells were cotransfected by miRNA lentiviral vector and miRNA mimics with pSICHECK2-3ʹUTR vector by using Lipofectamine 3000. Cells were harvested 36 h after transfection and lysed for 15 min at room temperature. The firefly and renilla luciferase activities were analyzed using the commercial Dual-luciferase reporter assay system (Promega, USA,) on a microplate luminometer, and the firefly luciferase activity was normalized to the renilla luciferase activity.
Reverse transcription polymerase chain reaction
Total RNAs from piPSC-dox, PS23, and PEF cells were extracted by TRIzol Reagent (Invitrogen, USA) according to the manufacturer’s procedure. RNA samples were examined by measuring the 260/280 ratio, samples with an optical density ratio of 2.0 were used for reverse transcription. One microgram RNA was reverse transcribed to cDNAs using Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, USA) and Mir-X™ miRNA First-Strand Synthesis and SYBR qRT-PCR (Clontech, USA) according to the manufacturer’s procedure. The PCR was performed for 33 cycles at 94°C 10 s, 58°C 30 s, and 72°C 60 s. The PCR products were analyzed on 1.0% agarose gel. For qRT-PCR, reactions were performed for 95°C 30 s at the first cycle, following another 40 cycles of 95°C 5 s and 60°C 30 s in triplicate using SYBR Green PCR Master Mix (TransGen Biotech, Peking, China), and detected by the CFX96 real-time PCR system (Bio-Rad, Hercules, USA). Data were normalized to either β-Actin (for mRNA) or U6 (for miRNAs). The relative expression levels were calculated using 2−ΔΔCt. Primer sequences are listed in Table S2.
Western blotting
To determine the expression of OTX2, TUBB3, DESMIN, and AFP, cells were lysed by RIPA buffer (Thermo Scientific, USA) supplemented with 1 mM PMSF (Sigma-Aldrich, St Louis, USA) at 0°C; for 10 min. Protein samples were mixed with 5 × loading buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 2% β-met and 0.05% bromophenol blue) and heated at 100°C for 5 min. The protein samples were then loaded into 12% SDS-PAGE. After electrophoresis, proteins were transferred to PVDF membrane (Millipore, Bedford, MA) by semidry electrophoretic transfer for 45 min at 15 V. The membrane was blocked with blocking buffer (5% skim milk, 20 mM Tris-HCl, pH 7.6, 137 mM NaCl, 0.1% Tween 20) at 37°C for 1 h, and then incubated with the primary anti-OTX2 antibody (13497–1-AP, Proteintech, China), anti-TUBB3 antibody (MMS-435P, Covance, USA), anti-DESMIN antibody (16520–1-AP, Proteintech, China), anti-AFP antibody (MAB1368, R&D System, USA), and anti-β-ACTIN antibody (KM9001, Sungene Biotech, China) at 4°C overnight, respectively. After washing three times with TBS-T buffer (20 mM Tris-HCl, pH 7.6, 137 mM NaCl, 0.1% Tween 20), the membrane was incubated with the HRP-conjugated secondary antibody for 1 h at 37°C. After washing three times in TBS-T for 5 min at room temperature, the membrane was incubated in the enhanced chemiluminescent substrate (Pierce, USA) for 1 min and detected with a Chemiluminescent Imaging System (Tanon, China).
Alkaline phosphatase staining
The alkaline phosphatase (AP) activity of piPSC-dox and PS23 cells was determined by AST Fast Red TR (Sigma-Aldrich, USA) and a-Naphthol AS-MX Phosphate (Sigma-Aldrich, USA) according to the manufacturer’s instruction. Briefly, cells were washed twice with ice-cold PBS, fixed with 4% paraformaldehyde (pH 7.4) at room temperature for 15 min, and followed by washing three times with ice-cold PBS. Then, cells were incubated with dye solution containing AST Fast Red TR (1.0 mg/mL) and a-Naphthol AS-MX (0.4 mg/mL) in 0.1 M Tris Buffer at room temperature. After 5–10 min incubation, cells were washed three times with PBS and images were documented with a Nikon microscope.
Embryoid body formation and spontaneous differentiation
The piPSCs were dispersed to single cells with 0.25% Trypsin (Invitrogen) and cultured in 60 mm Petri dish for the suspension culture (2 × 105 cell/mL) with the MEM plus 15% FBS without growth factors and small molecules. The culture medium was replaced every 2 days. After 5 days in suspension culture, the embryoid bodies (EBs) were transferred to a gelatin coated culture dish allowing the spontaneous differentiation for another 7 days.
Immunocytochemistry
To perform immunocytochemistry, cells were fixed with 4% PFA for 15 min at room temperature, then washed three times with ice-cold PBS and permeabilized with PBS containing 0.1% Triton X-100 for 15 min at room temperature. After three washes with PBS, cells were blocked with 1% BSA for 60 min at room temperature, and then incubated with antibodies of anti-TUBB3 (MMS-435P, Covance, USA), anti-DESMIN (16,520–1-AP, Proteintech, China), and anti-AFP (MAB1368, R&D System, USA) at 4°C overnight. Next day, cells were washed three times with PBS and incubated with FITC conjugated secondary antibody for 60 min at room temperature. Nuclei were stained by DAPI for 5 min. Images were documented by an EVOS fluorescence microscope.
Data sources and bioinformatics analysis
For the transcriptome analysis, RNA-seq databases of piPS-L, piPS-LF, and PEFs were generated in this laboratory and were deposited in the European Bioinformatics Institute (www.ebi.ac.uk/) under accession number E-MTAB-2634 [Citation23]. RNA-seq database of piPS-F was retrieved from National Center for Biotechnology Information [Citation51]. The mRNA profiles were clustered by using R package edgeR. The FDR < 0.05 was used as threshold to define differences of gene expression as the significance. The miRNA-seq of piPS-L, piPS-F, piPS-LF, and PEFs were generated in this laboratory [Citation52], which were deposited in EMBL-EBI database under accession number E-MTAB-2631. The significantly downregulated miRNAs were predicted by bioinformatics tools, including miRanda, miReap, and TargetScan from miRNA-seq database [Citation53,Citation54].
Statistical analysis
The calculated data are presented as mean ± S.D. The differences between two groups were determined by the Student t-test. Differences among grouped data were determined by two-way ANOVA. All statistical analyses were done with GraphPad Prism 5.0 (GraphPad, USA). Statistical significance was accepted at P < 0.05.
Author Contributions
Conceived and designed the experiments: HW. Performed the experiments: YX, HC, ZZ. Contributed reagents/materials/analysis tools: YX, HC. Analyzed the data and wrote the paper: YX, SZ, HW.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download MS Word (1.3 MB)Acknowledgments
We think Qiang Zhao for her help on constructing lentiviral vectors. We also think Ziyu Ma, Yuanxing Cai for their help on the constructing promoter vectors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. PMID:19497275.
- Niwa H, Ogawa K, Shimosato D, et al. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. PMID:19571885.
- Xiao J, Mai DH, Xie L. Resetting human naive pluripotency. Genet Epigenet. 2016;8:37–41. PMID:27512340.
- Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. PMID:20876656.
- Festuccia N, Osorno R, Halbritter F, et al. Esrrb is a direct nanog target gene that can substitute for nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. PMID:WOS:000309896500009.
- Martello G, Sugimoto T, Diamanti E, et al. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. PMID:23040478.
- Yeo JC, Jiang J, Tan ZY, et al. Klf2 is an essential factor that sustains ground state pluripotency. Cell Stem Cell. 2014;14:864–872. PMID:24905170.
- Fidalgo M, Shekar PC, Ang YS, et al. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells. 2011;29:1705–1716. PMID:21915945.
- Navarra A, Musto A, Gargiulo A, et al. Hmga2 is necessary for Otx2-dependent exit of embryonic stem cells from the pluripotent ground state. BMC Biol. 2016;14:24. PMID:27036552.
- Davies OR, Lin CY, Radzisheuskaya A, et al. Tcf15 primes pluripotent cells for differentiation. Cell Rep. 2013;3:472–484. PMID:23395635.
- Zhang J, Zhang M, Acampora D, et al. OTX2 restricts entry to the mouse germline. Nature. 2018;562:595–599. PMID:30283136.
- Lee YJ, Ramakrishna S, Chauhan H, et al. Dissecting microRNA-mediated regulation of stemness, reprogramming, and pluripotency. Cell Regener. 2016;5:2. PMID:27006752.
- Judson RL, Greve TS, Parchem RJ, et al. MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells. Nat Struct Mol Biol. 2013;20:1227–1235. PMID:24037508.
- Choi YJ, Lin CP, Risso D, et al. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science. 2017;355. PMID: 28082412. DOI:10.1126/science.aag1927
- Xu N, Papagiannakopoulos T, Pan G, et al. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. PMID:19409607.
- Adammek M, Greve B, Kassens N, et al. MicroRNA miR-145 inhibits proliferation, invasiveness, and stem cell phenotype of an in vitro endometriosis model by targeting multiple cytoskeletal elements and pluripotency factors. Fertil Steril. 2013;99:1346–1355e5. PMID: 23312222.
- Subramanyam D, Lamouille S, Judson RL, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. PMID:21490602.
- West FD, Terlouw SL, Kwon DJ, et al. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;19:1211–1220. PMID:20380514.
- Wu Z, Chen J, Ren J, et al. Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol. 2009;1:46–54. PMID:19502222.
- Esteban MA, Xu J, Yang J, et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2009;284:17634–17640. PMID:19376775.
- Ezashi T, Telugu BP, Alexenko AP, et al. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci U S A. 2009;106:10993–10998. PMID:19541600.
- Cheng D, Guo Y, Li Z, et al. Porcine induced pluripotent stem cells require LIF and maintain their developmental potential in early stage of embryos. PloS one. 2012;7:e51778. PMID:23251622.
- Zhang S, Guo Y, Cui Y, et al. Generation of intermediate porcine iPS cells under culture condition favorable for mesenchymal-to-epithelial transition. Stem Cell Rev. 2015;11:24–38. PMID:25134796.
- Acampora D, Omodei D, Petrosino G, et al. Loss of the Otx2-binding site in the nanog promoter affects the integrity of embryonic stem cell subtypes and specification of inner cell mass-derived epiblast. Cell Rep. 2016;15:2651–2664. PMID:27292645.
- Yang SH, Kalkan T, Morissroe C, et al. Otx2 and Oct4 drive early enhancer activation during embryonic stem cell transition from naive pluripotency. Cell Rep. 2014;7:1968–1981. PMID:24931607.
- Wang N, Wang Y, Xie Y, et al. OTX2 impedes self-renewal of porcine iPS cells through downregulation of NANOG expression. Cell Death Discov. 2016;2:16090. PMID:27924227.
- Kaur R, Aiken C, Morrison LC, et al. OTX2 exhibits cell-context-dependent effects on cellular and molecular properties of human embryonic neural precursors and medulloblastoma cells. Dis Model Mech. 2015;8:1295–1309. PMID:26398939.
- Fujishiro SH, Nakano K, Mizukami Y, et al. Generation of naive-like porcine-induced pluripotent stem cells capable of contributing to embryonic and fetal development. Stem Cells Dev. 2013;22:473–482. PMID:22889279.
- Rosati J, Ferrari D, Altieri F, et al. Establishment of stable iPS-derived human neural stem cell lines suitable for cell therapies. Cell Death Dis. 2018;9:937. PMID:30224709.
- Ma Y, Yu T, Cai Y, et al. Preserving self-renewal of porcine pluripotent stem cells in serum-free 3i culture condition and independent of LIF and b-FGF cytokines. Cell Death Discov. 2018;4:21. PMID:29531818.
- Sone M, Morone N, Nakamura T, et al. Hybrid cellular metabolism coordinated by Zic3 and Esrrb synergistically enhances induction of naive pluripotency. Cell Metab. 2017;25:1103–1117e6. PMID: 28467928.
- Shi W, Wang H, Pan G, et al. Regulation of the pluripotency marker Rex-1 by Nanog and Sox2. J Biol Chem. 2006;281:23319–23325. PMID:16714766.
- Buecker C, Srinivasan R, Wu Z, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. PMID:24905168.
- Pan X, Cang X, Dan S, et al. Site-specific disruption of the Oct4/Sox2 protein interaction reveals coordinated mesendodermal differentiation and the epithelial-mesenchymal transition. J Biol Chem. 2016;291:18353–18369. PMID:27369080.
- Wang R, Hu Y, Song G, et al. MiR-206 regulates neural cells proliferation and apoptosis via Otx2. Cell Physiol Biochem. 2012;29:381–390. PMID:22508046.
- Choi SW, Kim JJ, Seo MS, et al. miR-410 inhibition Induces RPE differentiation of amniotic epithelial stem cells via overexpression of OTX2 and RPE65. Stem Cell Rev. 2015;11:376–386. PMID:25351180.
- Stolzenburg LR, Wachtel S, Dang H, et al. miR-1343 attenuates pathways of fibrosis by targeting the TGF-beta receptors. Biochem J. 2016;473:245–256. PMID:26542979.
- Mullen AC, Wrana JL. TGF-beta family signaling in embryonic and somatic stem-cell renewal and differentiation. Cold Spring Harb Perspect Biol. 2017;9. PMID: 28108485. DOI:10.1101/cshperspect.a022186
- Huang X, Lu S. MicroR-545 mediates colorectal cancer cells proliferation through up-regulating epidermal growth factor receptor expression in HOTAIR long non-coding RNA dependent. Mol Cell Biochem. 2017;431:45–54. PMID:28364379.
- Liu Z, Dou C, Yao B, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350–25365. PMID:26992218.
- Acampora D, Di Giovannantonio LG, Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2013;140:43–55. PMID:23154415.
- Adachi K, Suemori H, Yasuda SY, et al. Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes Cells. 2010;15:455–470. PMID:20384793.
- Masui S, Nakatake Y, Toyooka Y, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. PMID:17515932.
- Panaliappan TK, Wittmann W, Jidigam VK, et al. Sox2 is required for olfactory pit formation and olfactory neurogenesis through BMP restriction and Hes5 upregulation. Development. 2018;145. PMID: 29352015. DOI:10.1242/dev.153791
- Fong H, Hohenstein KA, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells. 2008;26:1931–1938. PMID:18388306.
- Liu S, Bou G, Sun R, et al. Sox2 is the faithful marker for pluripotency in pig: evidence from embryonic studies. Dev Dyn. 2015;244:619–627. PMID:25619399.
- Zhang T, Duan J, Zhang L, et al. LXRalpha promotes hepatosteatosis in part via activation of MicroRNA-378 transcription and inhibition of Ppargc1beta expression. Hepatology. 2018. PMID: 30281809. DOI:10.1002/hep.30301
- Pang Y, Liu J, Li X, et al. MYC and DNMT3A-mediated DNA methylation represses microRNA-200b in triple negative breast cancer. J Cell Mol Med. 2018;22:6262–6274. PMID:30324719.
- Gherzi R, Briata P, Boncinelli E, et al. The human homeodomain protein OTX2 binds to the human tenascin-C promoter and trans-represses its activity in transfected cells. DNA Cell Biol. 1997;16:559–567. PMID:9174161.
- Yu T, Ma Y, Wang H. EpCAM intracellular domain promotes porcine cell reprogramming by upregulation of pluripotent gene expression via beta-catenin signaling. Sci Rep. 2017;7:46315. PMID:28393933.
- Xiao S, Xie D, Cao X, et al. Comparative epigenomic annotation of regulatory DNA. Cell. 2012;149:1381–1392. PMID:22682255.
- Zhang S, Xie Y, Cao H, et al. Common microRNA-mRNA interactions exist among distinct porcine iPSC lines independent of their metastable pluripotent states. Cell Death Dis. 2017;8:e3027. PMID:29048434.
- Zhang X, Liu X, Liu C, et al. Identification and characterization of microRNAs involved in ascidian larval metamorphosis. BMC Genomics. 2018;19:168. PMID:29490613.
- Li Y, Wan L, Bi S, et al. Identification of drought-responsive MicroRNAs from roots and leaves of alfalfa by high-throughput sequencing. Genes (Basel). 2017;8:E119. PMID: 28406444.
