ABSTRACT
Ribosomal RNA precursors undergo a series of structural and chemical modifications to generate matured RNA molecules that will comprise ribosomes. This maturation process involves a large set of accessory proteins as well as ribonucleases, responsible for removal of the external and internal transcribed spacers from the pre-rRNA. Early-diverging eukaryotes belonging to the Kinetoplastida class display several unique characteristics, in particular in terms of RNA synthesis and maturation. These peculiarities include the rRNA biogenesis and the extensive fragmentation of the large ribosomal subunit (LSU) rRNA. The role of specific endo- and exonucleases in the maturation of the unusual rRNA precursor of trypanosomatids remains largely unknown. One of the nucleases involved in rRNA processing is Rrp44, an exosome associated ribonuclease in yeast, which is involved in several metabolic RNA pathways. Here, we investigated the function of Trypanosoma brucei RRP44 orthologue (TbRRP44) in rRNA processing. Our results revealed that TbRRP44 depletion causes unusual polysome profile and accumulation of the complete LSU rRNA precursor, in addition to 5.8S maturation impairment. We also determined the crystal structure of TbRRP44 endonucleolytic domain. Structural comparison with Saccharomyces cerevisiae Rrp44 revealed differences in the catalytic site and substitutions of surface residues, which could provide molecular bases for the lack of interaction of RRP44 with the exosome complex in T. brucei.
Introduction
Ribosome synthesis is one of the most energy consuming processes of the cell. It involves a multitude of factors to ensure rRNA folding, nucleotide modifications, maturation of rRNA precursors, assembly with ribosomal proteins and export to the cytoplasm. In Saccharomyces cerevisiae more than 200 proteins were identified as necessary for efficient assembly of the 40S and 60S ribosomal subunits. Over the past years, major advances in the field of ribosome biogenesis have been achieved (reviewed in [Citation1–Citation4]), including the three-dimensional structures of pre-ribosome complexes obtained by cryo-electron microscopy, which revealed molecular details of the ribosome assembly pathway [Citation5–Citation8].
Processing of rRNA precursors (pre-rRNA) into mature molecules involves a wide spectrum of endo- and exoribunucleases, which function in a concerted and cooperative manner (reviewed in [Citation9]). In the great majority of eukaryotes, the primary rRNA precursor contains external transcribed spacers (5ʹ- and 3ʹ-ETS) at both ends and internal transcribed spacers (ITS1 and ITS2) that must be excised during the rRNA maturation process.
Many aspects of the rRNA biogenesis in Kinetoplastida, a class of unicellular flagellate protozoans that includes Trypanosoma and Leishmania species, differ from other eukaryotes. One of these peculiarities is fragmentation of the large ribosomal subunit (LSU) rRNA into six matured molecules in Trypanosoma and seven in Leishmania. Consequently, trypanosomatids pre-rRNA contains seven/eight ITSs rather than two, usually present in eukaryotes [Citation10,Citation11]. Moreover, Trypanosoma brucei 18S rRNA is one of the largest mature 18S described and the cleavage that separates SSU (small ribosomal subunit) and LSU precursors precedes the 5´ETS cleavage [Citation12].
Additional unique features of the T. brucei ribosome were revealed by its high-resolution cryo-electron microscopy structure [Citation13]. Large rRNA expansion segments and ribosomal-protein extensions form four additional inter-subunit bridges. Moreover, rRNA insertions were found, including one large rRNA domain not observed in other eukaryotes. In terms of the unusual processing of the large subunit pre-rRNA into six pieces, the structure of the T. brucei ribosome revealed that this extensive cleavage is, at least in part, a structural necessity. The existence of several expansion segments in the conformation observed is possible only because several pieces of rRNA are cleaved, otherwise they would collide [Citation13].
Along with the differences in rRNA species, differences in trypanosomes pre-rRNA processing mechanism are expected. Notably, the Euglenozoa, which comprises Leishmania and Trypanosoma, is the only phylogenetic group where neither RNase P or MRP protein subunits nor the MRP RNA subunit were identified [Citation14,Citation15]. Therefore, such ribonucleoprotein complex, which performs a central function in yeast and human rRNA biogenesis [Citation16–Citation18] is apparently missing in trypanosomatids.
The exosome complex is a major eukaryotic 3ʹ–5ʹ exoribonuclease involved in several metabolic RNA pathways. The eukaryotic exosome is composed by a nine-protein catalytically inert ‘core’ (Exo9) arranged in a two-layers ring architecture. Association of the ribonucleolytic subunits Rrp44 and Rrp6 to the Exo9 core provides the exosome activity towards diverse RNA substrates (reviewed in [Citation19,Citation20]). The exosome participates in different steps of ribosomal RNA processing such as degradation of pre-rRNA 5ʹ-ETS, maturation of 5.8S rRNA [Citation21–Citation25] and, in human cells, the trimming of the 3ʹ-extended pre-18S [Citation26].
Estevez et al. [Citation27,Citation28] have characterized the exosome complex in T. brucei and have shown its role in the 3ʹ trimming of the 7S pre-rRNA intermediate to generate mature 5.8S rRNA [Citation27]. Their findings corroborated the function initially characterized in yeast. However, a striking difference is that there is no evidence for an association between RRP44 and the exosome complex in trypanosomatids so far [Citation27–Citation29].
In contrast to the large amount of data available on yeast ribosome synthesis factors and, to a lesser extent, on other higher eukaryotes, the role of specific endo- and exonucleases in the maturation of the unusual rRNA precursor of trypanosomatids remains largely unknown. In this work, we have investigated the function of T. brucei RRP44 in ribosome biogenesis. Analysis of the effects of the nuclease depletion in T. brucei cells indicate the requirement of TbRRP44 for an initial event in the maturation of the LSU rRNA. Moreover, the crystal structure of TbRRP44 endonucleolytic domain revealed molecular details that suggest a basis for the lack of interaction of RRP44 with the exosome complex, previously reported in Trypanosoma and Leishmania.
Results
Sucrose density gradient reveals an unusual polysome profile upon T. brucei RRP44 depletion
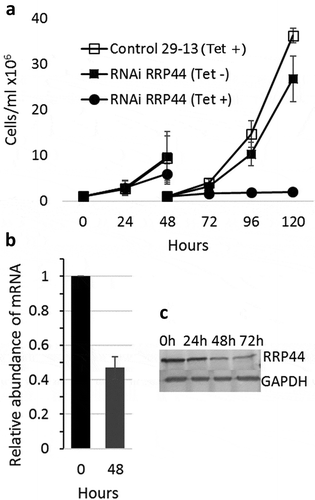
We have generated a procyclic T. brucei cell line containing a p2T7-derived vector (p2T7-177–RRP44), which drives the transcription of a TbRRP44 dsRNA for RNA interference in the presence of tetracycline. Growth curves in the absence or presence of tetracycline show that cell growth is severely reduced 48 hours after RNAi induction and completely halted at 72 hours post induction. Quantitative RT-PCR shows that the levels of TbRRP44 mRNA were decreased to approximately 45% on day 2 after induction, and protein reduction was observed by western blot (). These results are consistent with previous findings of Estevez et al. [Citation27] showing that RRP44 is an essential protein in T. brucei and validate the knockdown.
Figure 1. Depletion of T. brucei RRP44 by inducible RNA interference. (a) Effects on cell growth. T. brucei cells transfected with vector p2T7-177–RRP44 were growth in the presence (circles) or absence (filled squares) of tetracycline. Non-transfected cells in the presence of tetracycline (open squares) were also used as control. Cultures were diluted to 106 cells after 48 hours for the next time points. (b) Analysis of the level of TbRRP44 mRNA by quantitative RT-PCR normalized to GAPDH. Error bars indicate standard deviation of triplicate experiments. (c) Decreasing of RRP44 protein detected on a western blot using polyclonal antiserum at different time-points after RNAi induction.
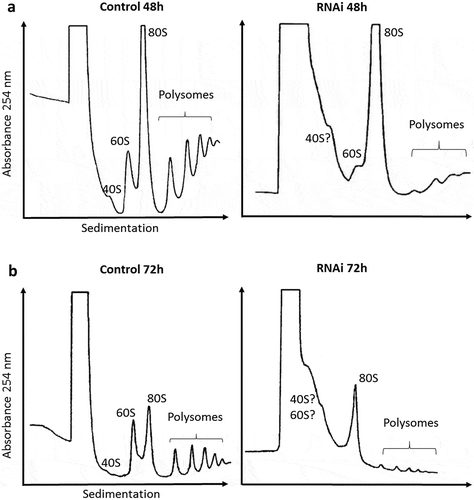
The effects of TbRRP44 depletion on ribosomal assembly were investigated by sucrose gradient polysome fractionation using cycloheximide to block translation, prior to cell extracts preparation. Polysomal profiles revealed unexpected defects in RNAi-induced TbRRP44 cells. Besides an increase in 80S and significant decrease in the polysomal peaks, we observe an enlargement of the peak corresponding to low-molecular weight species sedimentation range, making the 40S and 60S peaks unidentifiable at 72 hours post induction (). Depletion of ribosome biogenesis factors are usually associated with a relative change in 40S, 60S and polysome abundance. However, our analysis shows an intriguing result, which to the best of our knowledge has not been described yet. We hypothesize whether the observed enrichment of lower-molecular weight species and the impairment in 40S and 60S subunits formation upon depletion of TbRRP44 were a consequence of a broader defect in RNA metabolism pathways that goes beyond the pre-rRNA maturation.
Figure 2. Polysomal profile analysis. Cell extracts were submitted to velocity sedimentation on 15 to 55% sucrose gradients. (a) Control (left) and induced TbRRP44 RNAi cells (right) at 48 hours post induction. (b) Control (left) and induced TbRRP44 RNAi cells (right) at 72 hours post induction. 5.8 × 108 and 2.5 × 108 cells were used for the polysomes analysis at 48 and 72 hours post induction, respectively. The experiments were performed at least twice with consistent results.
Depletion of T. brucei RRP44 causes an early blockage in the large ribosomal subunit RNA maturation
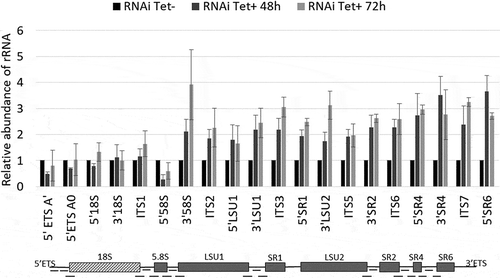
T. brucei exosome component homologs, including RRP44, were shown to be involved in the 3ʹ processing of 7S rRNA to generate the mature 5.8S rRNA [Citation27]. To investigate the role of RRP44 in the cleavage of T. brucei pre-rRNA internal transcribed spacers, we evaluated the efficiency of pre-rRNA processing upon TbRRP44 depletion by quantitative RT-PCR. We used 20 pairs of primers, covering most of the segments that are excised from the rRNA precursor during the maturation process (, Supplementary table 1). The relative amplification of all examined sites was measured at 48 and 72 hours after RRP44 RNAi induction in parallel with control cells. Interestingly, besides the increase of the RNA ratio detected in the 3ʹ region of the 5.8S rRNA, depletion of TbRRP44 resulted in a significant increase of the level of all the amplicons corresponding to the rRNA precursor of the large ribosomal subunit (LSU). We have performed the experiments using two different normalizers (GAPDH and α-actin) and obtained consistent results ( and Supplementary Figure 1).
Figure 3. Depletion of T. brucei RRP44 impairs cleavage steps related to the LSU rRNA maturation. The relative change in the target amplicons was determined by quantitative RT-PCR analysis of total RNA and normalized to the GAPDH mRNA. Control cells (black bars, normalized to 1), cells 48 h post RNAi induction (dark grey bars) and cells 72 h post RNAi induction (light grey bars). Error bars indicate standard deviation of triplicate experiments. At the bottom, a schematic representation of the rRNA precursor, indicating the amplicons analyzed in this work (black dashes).
The RT-qPCR experiments were complemented by Northern blots with probes annealing to different regions of the T. brucei pre-rRNA (Supplementary table 2) to evaluate the steady state of the LSU RNA precursors. Two of the probes, hybridizing in the 5.8S rRNA and in the 5.8S rRNA-ITS2 junction, were selected based on the previous studies by Estevez et al. [Citation27]. As expected, these probes confirmed accumulation of the 7S pre-rRNA, comprising the 5.8S rRNA with part of ITS2 ()). Interestingly, these probes also revealed accumulation of the 5.8 kb precursor, which contains the seven rRNA segments that form the mature large subunit. Thus, a third probe that hybridizes in ITS3 was used, confirming the accumulation of the 5.8 kb LSU precursor and revealing the accumulation of two additional intermediates, of 5.0 kb and 3.9 kb, which are also precursors of the large subunit rRNAs. An additional band migrating in the range of the mature LSU rRNAs was detected by the probe that hybridizes in ITS3 ()). This was an unexpected result not observed in previous studies using probes hybridizing in the same region [Citation30]. However, this probe shows complementarity to 11 nucleotides of the LSU1 rRNA and the band might correspond to LSU1, which might have been detected due to low stringency of the hybridization conditions. The 9.2 kb transcript was not detected using these probes and with the hybridization conditions used in this work.
Figure 4. Accumulation of LSU rRNA precursors in TbRRP44 knockdown cells as revealed by Northern blot analysis. (a) Total RNA was extracted at 0, 24, 48, 72 and 96 hours after induction of RNAi for TbRRP44 depletion and analyzed by Northern hybridization with fluorescently labelled oligonucleotides complimentary to the regions of the T. brucei pre-rRNA corresponding to the 5.8S rRNA (left panel), to the 5.8S rRNA-ITS2 junction (central panel) and to the ITS3 (right panel). The positions of the precursors as wells as of the 5.8S rRNA are indicated on the right-hand side of each panel. (b) Schematic organization of the 9.2 kb precursor of T. brucei ribosomal RNA with the intermediates identified by Northern blotting. Dashed box,18S/SSU rRNA; grey boxes LSU rRNA segments; ETS, external transcribed spacers; ITS internal transcribed spacers; SR, small RNA; * indicates an additional band that might correspond to LSU1 due to the relatively low stringency of the hybridization conditions as probe ITS3 contains 11 nucleotides complementary to the region of LSU1. The three probes reveal accumulation of the 5.8 kb precursor comprising the regions of the seven rRNA segments that form the mature large subunit. In addition, the probes 5.8S rRNA and 5.8S rRNA-ITS2 junction reveal accumulation of the 0.61 kb segment, which corresponds to the 7S pre-rRNA comprising the 5.8S with part of ITS2. The probe that hybridizes in ITS3 reveals also accumulation of the 5.0 kb and 3.9 kb precursors of the large subunit rRNAs.
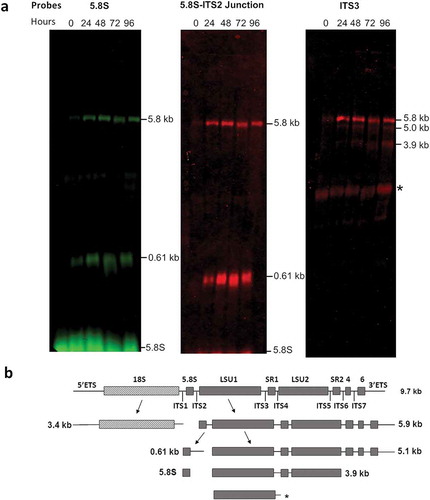
According to the current model of pre-rRNA processing in T. brucei, early events result in the separation of the 18S rRNA precursor (~3.4 kb SSU intermediate) from the 5.8S/LSU rRNA precursor (~5.8 kB intermediate). The latter is subsequently cleaved at the ITS2 generating two intermediates of 0.61 kb (5.8S precursor) and 5.0 kb (LSU/SR precursor) [Citation12,Citation31]. Accumulation of 5.8 kb, 5.0 kb and 3.4 kb species has been previously observed in T. brucei upon depletion of the large ribosomal subunit protein L5 [Citation32]. Our results are consistent with this model ()) and indicate that TbRRP44 is also required for processing of the 5.8 kb precursor, being involved in an earlier event in the LSU rRNA maturation.
The crystal structure of T. brucei RRP44 endonuclease domain reveals differences compared with S. cerevisiae Rrp44 that could account for the lack of interaction with the exosome
Full-length TbRRP44 and a truncated construct (TbRRP44-NPIN), including the N-terminal and the endonuclease domain (PIN domain), were produced and submitted to crystallization. However, suitable crystals were obtained only for the TbRRP44-NPIN construct (residues 1 to 219). TbRRP44-NPIN crystals diffracted to 2.3 Å resolution and the structure was determined by single anomalous diffraction, using the zinc atom endogenously bound to the protein as anomalous scatterer. The six monomers present in the asymmetric unit were refined to final Rwork/Rfree of 20%/23%, respectively (). Lack of electron density did not allow modeling of flexible regions of the polypeptide chains corresponding to N-terminal residues 1 to 27, residues 40 to 50, residues 127 to 136 and residues 180–190. Superposition of the asymmetric unit monomers resulted in RMSD values varying from 0.25 to 0.55 Å.
Table 1. Crystallographic data and refinement statistics. Values in parentheses are for the highest resolution shell.
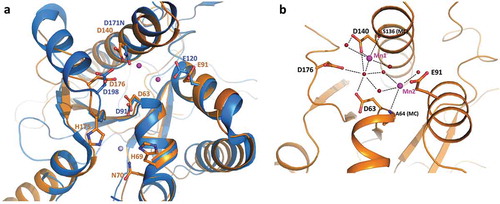
The 3D structure of full length Rrp44 in complex with exosome subunits was previously described for the yeast counterpart (ScRrp44) [Citation33–Citation36]. As expected, TbRRP44-NPIN conserves the overall architecture of the ScRrp44 PIN domain, composed by a five-stranded β-sheet flanked by α-helices (Supplementary Figure 2). The catalytic site is formed by four acidic residues, D63, E91, D140 and D176 and the zinc binding motif (CCCH) present in the N-terminal end of ScRrp44 is also conserved in TbRRP44. Interestingly, in TbRRP44-NPIN structure, the anomalous difference map clearly showed a second zinc atom bound in the neighborhood of the endonucleolytic site (), Supplementary Figure 3A). This second zinc interaction site is not conserved in ScRrp44, where the TbRRP44 histidine residues H60 and H175 are replaced by Q97 and N197, respectively. No zinc salts were added to the solutions used in protein purification or crystallization, thus the Zn atoms bound to the protein are likely to come from the bacterium culture medium, indicating a strong interaction of the metals at both sites. Zinc is a common cofactor of nucleic acids interacting proteins and we speculate whether the second Zn site could be related to a TbRRP44 substrate binding regulation mechanism. Additionally, during the structure refinement the electron density maps indicated the presence of two metal ions bound to the protein catalytic site. We have initially modeled magnesium atoms at both sites, since magnesium chloride is present in the crystallization solution. However, strong positive peaks in the difference density map indicated the presence of a larger atomic number metal. To investigate the binding of manganese ions, which are also possible cofactors of nucleases, we have collected diffraction data from a second crystal at the Mn-edge energy (Supplementary table 3). The anomalous difference map unambiguously showed the presence of manganese at one of the sites (named site 1) in all six monomers of the asymmetric unit, but no significant anomalous peak was observed for the second site (Supplementary Figure 3B). When we modeled Mg atoms at site 2, positive peaks in the difference density map appeared (Supplementary Figure 3C). Although we cannot exclude a partial occupancy of manganese and magnesium atoms in the crystal, modeling Mn atoms at both sites resulted in better-quality electron density maps, without positive or negative peaks in the difference map (Supplementary Figure 3D). Manganese ions were most possibly retained from the expression host and are stably bound to the protein, coordinated by acidic groups and water molecules at the catalytic site ()).
Figure 5. TbRRP44 endonucleolytic site. (a) Superposition of TbRRP44 (orange) and ScRrp44 (blue, PDB 5K36) structures. TbRRP44 catalytic residues and the corresponding residues in ScRrp44 are shown in sticks and labeled. The residues forming the neighboring Zn interaction site in TbRRP44 (H69, N70 and H175) are also shown. The Mn ions bound to the TbRRP44 active site are represented as purple spheres while the Zn atom is shown in light grey. (b) Detail of the interactions involving the manganese ions. Water molecules are represented as red spheres. TbRRP44 catalytic residues and main chain (MC) atoms involved in the interactions are identified.
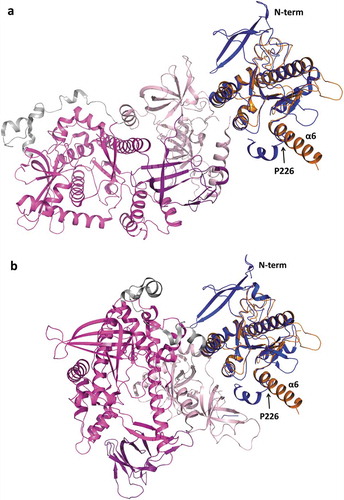
Superposition of TbRRP44-NPIN structure with the ScRrp44 PIN domain (PDB 5K36) results in RMSD of 1.62 Å for 146 C-alpha atoms aligned. Interestingly, in TbRRP44 we observe a long α-helix at the C-terminal of the PIN domain (helix α6), whereas in ScRrp44 the corresponding helix is broken by the presence of a proline residue (P226) (). Downstream to this helix, the region of connection with the next structural domain (CSD1) confers flexibility to ScRrp44 to perform a significant structural rearrangement, which is associated to distinct RNA degradation mechanisms, depending on whether the substrate reaches the Rrp44 catalytic site through the exosome core (channel route) or directly (direct route) [Citation35,Citation37]. Even though the TbRRP44-NPIN construct is truncated at the end of the helix α6, which prevents the full comparison of the structures, the orientation of helix α6 and absence of a ‘helix breaker’ residue in TbRRP44 (ScRrp44 P226 is replaced by a valine), indicate a distinct overall arrangement of the protein domains.
Figure 6. Superposition of TbRRP44-NPIN and ScRrp44 structures in two functional conformations. TbRRP44-NPIN is shown in orange. The ScRrp44 coordinates are from PDB 5K36 and 4IFD, corresponding to the direct route (a) and channel route (b) conformations, respectively. ScRrp44 is colored by domain. NPIN is shown in blue. CSD, RNB and S1 domains are represented in light pink, magenta and dark purple, respectively. TbRRP44 helix α6 and ScRrp44 Pro226 are indicated.
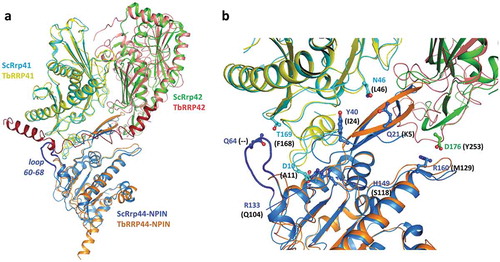
Interaction of S. cerevisiae Rrp44 with the exosome core is mainly mediated by the N-terminal PIN domain, which participates in intermolecular contacts with subunits Rrp41 and to a lesser extent, Rrp42 [Citation34–Citation36]. To investigate whether similar molecular interfaces between T. brucei exosome subunits counterparts would be likely to occur, we have generated structure models of T. brucei RRP41A and RRP42 using the I-TASSER server. We have also generated a model for TbRRP44-NPIN to include the N-terminal region and flexible loops that are missing in the crystal structure. T. brucei models were aligned with the corresponding yeast exosome subunits (PDB code 4IFD). Superposition of TbRRP41A and ScRrp41 resulted in RMSD of 1.03 Å for 236 aligned residues and 24% sequence identity. The TbRRP42 model shows lower similarity to the yeast counterpart. Structural alignment resulted in RMSD of 2.36 Å for 223 aligned residues and 18% sequence identity. Superposition of TbRRP44 and ScRrp44 NPIN domains resulted in RMSD of 1.83 Å for 187 aligned residues and 24% sequence identity. Comparison of ScRrp44-Rrp41 and ScRrp44-Rrp42 interfaces with an equivalent molecular arrangement in T. brucei shows significant global and local differences (, Supplementary Figure 4). ScRrp44 N-terminal region, which encompasses residues in the interface with ScRrp41 is 16 residues longer than TbRRP44 N-terminal. ScRrp44 residues 60–68 which also form an interaction surface with ScRrp41 are replaced by a shorter loop in TbRRP44. Moreover, TbRRP42 presents a large C-terminal extension, which according to the structural prediction would collide with TbRRP44-NPIN () in the context of the subunits’ assembly. Structure based sequence alignment of ScRrp44, ScRrp41 and ScRrp42 with T. brucei counterparts shows a large number of non-conservative substitutions of ScRrp44-Rrp41 and ScRrp44-Rrp42 interface residues (Supplementary Fig. 4). Analysis of evolutionary conservation of amino acid positions using the ConSurf server reveals high degree of variability of TbRRP44 surface residues that would nominally interact with the exosome (Supplementary Figure 5).
Figure 7. Superposition of TbRRP44-NPIN, TbRRP41 and TbRRP41 models and the yeast exosome subunits counterparts ScRrp44 (NPIN domain), ScRrp41 and ScRrp42 (PDB 4IFD, channel route conformation). (a) Overall structure of the assembly. The proteins are identified with different colors. The loop formed by residues 60–68 in ScRrp44 is highlighted in dark blue. TbRRP42 C-terminal extension (dark red) collides with TbRRP44-NPIN according to the structural prediction. (b) Detail of the subunits interface. TbRRP42 C-terminal was omitted for clarity. ScRrp44 side chains involved in intermolecular hydrogen bonds and salt bridges which are not conserved in TbRRP44-NPIN are shown in sticks and labeled. Substitutions in TbRRP44 are indicated in parenthesis.
Together with a possible loss of inter-domain flexibility linked to TbRRP44 α6 structure (described above), modifications in TbRRP44-NPIN and the exosome subunits surfaces suggest a molecular basis for the lack of association of RRP44 with the exosome complex previously reported in Trypanosoma and Leishmania [Citation27–Citation29].
Discussion
Several nucleases involved in the processing of ribosomal RNA precursor molecules in eukaryotes have been characterized. Among them, the enzymes responsible for pre-rRNA endoribonycleolytic cleavages such as Rnt1, which belongs to the RNase III family; Rcl1, a member of the RNA 3ʹ-terminal cyclase family; Las1, a metal-independent nuclease belonging to the HEPN superfamily, the ribonucleoprotein complex MRP and the PIN domain-containing nucleases Utp23, Utp24, Nob1 and Dis3/Rrp44 (reviewed in [Citation9]). The function of Trypanosoma brucei Nob1 ortholog in 18S rRNA maturation was recently characterized, showing a conserved role across eukaryotes [Citation38]. Besides Nob1 and Rrp44, potential orthologs of Rcl1, Utp24 and Las1 endonucleases can be identified in Kinetoplastida genomes. However, based on our databases searches, in addition to the MRP complex, Rnt1 and Utp23 are apparently also missing in trypanosomes.
The exosome complex has been extensively studied and its role in rRNA maturation has been well established [Citation22–Citation25]. Although the exosome core is catalytically inert, in vivo assays showed that its central channel is essential for both Rrp44 endo and exonucleolytic activities [Citation39,Citation40]. Moreover, Rrp6 and Rrp44, interacting at opposite sides of the Exo-9 core, act in a concerted manner and conformational changes in both proteins mediate substrate selection and degradation by the exosome [Citation35]. The fact that there is no evidence of association of RRP44 with the exosome complex in trypanosomatids [Citation27–Citation29], raises questions about the enzyme mechanism for substrate binding and degradation. As revealed by ScRrp44 three-dimensional structures [Citation33–Citation36], both endo and exo catalytic sites are exposed to the solvent and accessible to the RNA substrate by a direct route. Thus, it is plausible to think of an exosome-independent activity of RRP44 in trypanosomatids, encompassing multiple functions in RNA degradation.
Our analyses of the pre-rRNA processing indicate that depletion of TbRRP44 blocks most, if not all, steps in the LSU rRNA maturation, suggesting that TbRRP44 is required for endonucleolytic cleavages of the ITSs. Despite of its PIN domain and endonucleolytic activity, Rrp44 is not the endonuclease responsible for direct ITS2 cleavage in yeast. Instead, ITS2 processing is initiated by endonuclease Las1 cleavage of the 27SB pre-rRNA at site C2 [Citation41,Citation42], which is followed by 5ʹ trimming of the 26S intermediate by Rat1-Rai1 and 7S to 5.8S processing by the exosome [Citation25]. The endo and exoribonucleolytic activities of ScRrp44 together with ScRrp6 activity are necessary to the efficient 3ʹ trimming of the 7S pre-rRNA intermediate [Citation25]. Our results indicate a different scenario in T. brucei, where depletion of TbRRP44 causes accumulation of the complete LSU pre-rRNA intermediate, affecting not only 7S but also 5.8 kb processing. These defects are more similar to the ones caused by Las1 depletion in yeast, which leads to accumulation of the 27SA3/27SB and 7S LSU precursors along with the full length 35S pre-rRNA [Citation41]. Genes encoding proteins containing Las1-like domain are found in trypanosomatids (such as T. brucei Tb927.8.5820) although so far, their function has not been characterized. We speculate whether TbRRP44 has a broader function in ITS2 processing, or eventually participates in additional steps required for processing of the multiple internal transcribed spacers present in trypanosomatids LSU rRNA precursor.
The crystal structure of T. brucei RRP44 PIN domain also revealed interesting features of TbRRP44. In yeast Rrp44 the Zn binding motif C3H that is present at the protein N-terminal was shown to be important for interaction with the exosome core and exosome function [Citation43]. In addition to this conserved motif, the second Zn binding site located nearby the catalytic center of TbRRP44 PIN domain may indicate a substrate interaction regulation mechanism mediated by Zn, which is not conserved in yeast. Structural differences observed at the C-terminal helix of TbRRP44 PIN domain and modifications in the corresponding surface that is involved in Rrp44 interaction with the exosome core in yeast provide molecular support for the exosome-independent mechanism of TbRRP44. A cooperative function between TbRRP44 and the RRP6-containing exosome is not excluded. However, considering its dual endo and exoribonucleolytic activities, we speculate whether trypanosomatids RRP44 may target a wider range of RNA substrates by itself.
The effect of TbRRP44 depletion on ribosomal and polysomal profiles was unexpected. Depletion of ribosome biogenesis factors involved in LSU formation usually causes a decrease in polysomes peaks and an increase in 40S/60S ratio, observed in sucrose gradient fractionation experiments, and T. brucei has not been an exception [Citation30,Citation44,Citation45]. The observed ‘shift’ of 40S/60S to lower-molecular weight sedimentation range in TbRRP44 knockdown cells () indicates maturation impairment of both ribosomal subunits, affecting not only their abundance but also the subunits integrity. Protein content analysis of the gradients fractions by mass spectrometry were inconclusive so far (data not shown). TbRRP44 knockdown cells gradients showed an enrichment of uncharacterized proteins in the fractions corresponding to the ‘shoulder’ observed in the low-molecular weight peak, but not a significant enrichment of ribosomal proteins. The relative increase in 80S population is also intriguing. We hypothesize whether multiple failures in rRNA and mRNA maturation caused by TbRRP44 depletion could be the explanation for 40S and 60S impairment and accumulation of translationally inactive monosomes.
Early-diverging eukaryotes belonging to the kinetoplastidae class display several unique characteristics, in particular in terms of RNA synthesis and maturation. The absence of control of transcription initiation in individual genes displaces the gene expression regulation to the level of RNA metabolism (reviewed in [Citation46]). In this context, ribonucleases such as RRP44 may have a wider function in RNA metabolic pathways and affect RNA maturation in different levels. The lack of association of RRP44 with the exosome complex in trypanosomatids may have an implication for its catalytic mechanism and substrate selection.
Materials and methods
Cell growth, transfection and induction
Procyclic T. brucei strain 29–13 were grown in SDM-79 medium containing 10% fetal bovine serum, 15 µg/mL of G418 and 50 µg/mL of hygromycin. The Tetracycline-inducible RRP44 RNAi vector p2T7-177-RRP44 was constructed by cloning a 472 bp fragment from T. brucei genomic DNA using the primers TbRRP4_xba_RNAiF (CATCTCTAGATGTCACTAGATGTCTGCGGCAGGAC) and TbRRP4_xho_RNAiR (GATCCTCGAGCCGCACTTCAATACCAACCTTCACC). The product was inserted into the p2T7-177 plasmid [Citation47]. NotI linearized p2T7-177-RRP44 was transfected by electroporation into procyclic T. brucei 29–13 cells and positive transformation was selected by addition of phleomycin (2.5 µg/mL) to the growth medium. Transcription of TbRRP44 dsRNA was induced with 2 µg/mL of tetracycline on the first day and 1 µg/mL subsequently.
Sucrose gradient analysis
Control (RNAi Tet-) and TbRRP44 RNAi (RNAi Tet+) cells were harvested by centrifugation at 3,000 × g for 5 minutes. 5.8 × 108 and 2.5 × 108 cells were used for the polysomes analysis at 48 and 72 hours post induction, respectively. The cells were suspended in 5 mL of medium with cycloheximide (100 µg of cycloheximide for 1 × 108 cells), incubated for 10 minutes at 28°C and centrifuged at 3,000 × g for 5 minutes. Then, cells were washed with 8 mL of TKM buffer (10 mM Tris-HCl pH 7.5, 300 mM KCl, 10 mM MgCl2) with cycloheximide (100 μg/mL). After centrifugation, the cells were suspended in 450 μL of buffer TKMI (440 µL of TKM buffer, 5 μL cycloheximide [stock 10 mg/mL], 5 µL PMSF [stock 100 mM]) added by 50 µL of lysis buffer 10x (10 mM Tris-HCl pH 7.5, 300 mM KCl, 10 mM MgCl2, 10% (v/v) NP-40, 2 M sucrose). The samples were vigorously homogenized for about 30 seconds and the lysate was cleared by centrifugation at 16,000 x g for 10 minutes at 4°C. The supernatant was loaded on a 15–55% sucrose gradient (% [w/v] sucrose, 10 mM Tris-HCl pH 7.5, 300 mM KCl, 10 mM MgCl2, 100 µg/mL cycloheximide, 1 mM PMSF) and centrifuged for 2.5 hours at 39,000 rpm using a HITACHI Himac CP80WX centrifuge. A Teledyne ISCO gradient fractionator was used to collect 1 mL fractions with UV (A254) profile recording.
qRT-PCR
Total RNA was isolated from 1.5 × 107 control and RNAi induced cells using Trizol reagent according to the manufacturer’s instructions (Invitrogen). RNA samples were submitted to quality control and quantification using a Bioanalyzer (Agilent). cDNA was prepared from 1 μg RNA using random hexamers and SuperScript IV (Invitrogen) according to the manufacturer’s instructions. qPCR was performed in 96 wells plates in triplicate. The PCR reaction contained 10 µL of SYBR Green Master Mix, 0.8 µL of forward and reverse oligonucleotides (Supplementary table 1), 1 µL of cDNA template and 7.4 µL of water. Relative amount of RNA was calculated using the Pfaffl method [Citation48] and data were normalized to GAPDH and α-actin.
Northern blot
RNA was extracted as described for qRT-PCR. 8 μg of total RNA were fractionated on 1% agarose-formaldehyde gels and transferred overnight to Hybond N+ nylon membranes (Amersham Biosciences) by capillary transfer. The membranes were prehybridized for two hours at 37°C on a rotating shaker in modified Church-Gilbert buffer [Citation49] containing 0.5 M phosphate buffer pH 7.2, 1 mM EDTA, 5% SDS (v/v) and 1% BSA (v/w) using 0.3 mL of solution per cm2. Subsequently the membranes were hybridized overnight at 37°C on a rotating shaker in modified Church-Gilbert buffer containing 200–400 ng/mL of each oligonucleotide probe. The probes (Supplementary table 2) were acquired from IDT (Integrated DNA Technologies) fluorescently labelled with either IRDye-700 or IRDye-800 at the 5ʹ end. After hybridization, the membranes were washed three times for 10 min at 37°C with 2x SSC buffer (1x SSC is 0.15 M NaCl, 0.015 M sodium citrate) containing 0.5% SDS (v/v) and the images acquired on an Odyssey scanner (LI-COR Biosciences).
Expression and purification of TbRRP44-NPIN
The gene encoding a truncated construct of TbRRP44 was synthesized by GeneCust and cloned into the pET28a vector (Novagen) to express TbRRP44-NPIN (residues 1 to 219) in fusion with a C-terminal His-tag. Escherichia coli BL21 Star (DE3) were transformed with the expression vector and cells were grown at 37°C in LB medium containing kanamycin 25 µg/mL. When the culture reached the OD600 ~ 0.6, the temperature was reduced to 18°C and protein expression was induced with 0.2 mM of isopropyl-β-D-thiogalactopyranoside (IPTG), overnight. Cells from 1 L culture were harvested by centrifugation at 6,000 x g for 15 minutes, suspended and lysed in 10 mL of buffer A (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 20 mM imidazole) using a microfluidizer processor (Microfluidics™). The soluble fraction was isolated by centrifugation at 40,000 × g for 30 minutes at 4°C. The extract was loaded onto a 5 mL His-Trap HP column (GE Healthcare Life Sciences) equilibrated with buffer A. TbRRP44-NPIN was eluted with a two steps gradient, a 10 column volumes (CV) linear gradient from 0 to 10% buffer B (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 800 mM imidazole) followed by a 15 CV linear gradient from 10 to 100% buffer B. Fractions containing the target protein were pooled, dialyzed against 50 mM Tris-HCl pH 6.7 and loaded onto a 5mL Hi-Trap Q HP column (GE Healthcare Life Sciences). TbRRP44-NPIN was eluted with a 20 CV linear gradient from 0 to 1 M NaCl. The sample was concentrated to a final volume of 1 mL and loaded onto a Superdex 75 16/60 (GE Healthcare Life Sciences) equilibrated with 30 mM Tris-HCl pH 6.7, 150 mM NaCl. The eluted protein was concentrated to approximately 5 mg/mL for crystallization assays. Purified TbRRP44-NPIN was also used to produce polyclonal antisera in mice.
Crystallization, data collection and processing
Crystallization trials were performed by sitting drop vapor-diffusion method using commercial screens. Optimization of the initial crystallization conditions were performed by varying precipitant and protein concentrations. The best crystals were obtained by hanging drop vapor diffusion at 18°C by mixing the TbRRP44-NPIN protein at 5 mg/mL in 30 mM Tris-HCl pH 6.7, 150 mM NaCl with crystallization buffer containing 0.2 M MgCl2, 0.1 M MES pH 6.0 and 17% (v/v) PEG 6000 or PEG 3350. Before data collection, the crystals were cryo-protected with 20% (v/v glycerol added to the mother liquor prior to flash cooling in liquid nitrogen. X-ray diffraction data were collected at the PROXIMA 1 beam line of the Synchrotron SOLEIL using a PILATUS 6M detector (Dectris) at the Zn-absorption wedge wavelength (1.2824 Å). A second crystal was used for data collection at the Mn-absorption wedge wavelength (1.8920 Å). Diffraction data were processed with XDS package [Citation50].
Structure determination and refinement
The structure of TbRRP44-NPIN was determined by single-wavelength anomalous diffraction using the zinc atoms bound to the protein as scatterers. The heavy atoms positions were determined using the program SHELXD [Citation51] and the initial phases were calculated using PHASER [Citation52]. Density modification was performed using PARROT [Citation53] and the experimental electron density map allowed the automatic construction of approximately 80% of the model by the program Buccaneer [Citation54]. Refinement of the structure was performed alternating cycles of BUSTER [Citation55] with visual inspection and manual rebuilding using COOT [Citation56]. The atomic coordinates and structure factors have been deposited in the Protein Data Bank (code 6MD3), Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Structure predictions and analysis
3D models of T. brucei RRP41A and RRP42 were generated using the I-TASSER server [Citation57]. We have also generated a model for TbRRP44-NPIN to include the N-terminal region and flexible loops that were missing in the crystal structure. Structure based sequence alignments were performed using PROMALS3D [Citation58]. Molecular interface analysis was performed using the PDBePISA server [Citation59,Citation60]. Analysis of the evolutionary conservation of amino acids positions was performed using the ConSurf server [Citation61–Citation63].
Supplemental Material
Download MS Word (2.6 MB)Acknowledgments
The authors acknowledge the Platform for Protein Purification and Characterization of the FIOCRUZ Technical Platform program. The authors are grateful to Synchrotron SOLEIL for beam time awarded at PROXIMA1 beamline (Proposal 20160778) and all the support they benefited as SOLEIL’s users.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Henras AK, Soudet J, Gérus M, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65(15):2334–2359.
- Henras AK, Plisson-Chastang C, O’Donohue MF, et al. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6(2):225–242.
- Woolford JL Jr, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195(3):643–681.
- Turowski TW, Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA. 2015;6(1):129–139.
- Kornprobst M, Turk M, Kellner N, et al. Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016;166(2):380–393.
- Cheng J, Kellner N, Berninghausen O, et al. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat Struct Mol Biol. 2017;24(11):954–964.
- Kater L, Thoms M, Barrio-Garcia C, et al. Visualizing the assembly pathway of nucleolar pre-60s ribosomes. Cell. 2017;171(7):1599–1610.
- Schuller JM, Falk S, Fromm L, et al. Structure of the nuclear exosome captured on a maturing preribosome. Science. 2018;360(6385):219–222.
- Tomecki R, Sikorski PJ, Zakrzewska-Placzek M. Comparison of preribosomal RNA processing pathways in yeast, plant and human cells - focus on coordinated action of endo- and exoribonucleases. FEBS Lett. 2017;591(13):1801–1850.
- White TC, Rudenko G, Borst P. Three small RNAs within the 10 kb trypanosome rRNA transcription unit are analogous to domain VII of other eukaryotic 28S rRNAs. Nucleic Acids Res. 1986;14(23):9471–9489.
- Campbell DA, Kubo K, Clark CG, et al. Precise identification of cleavage sites involved in the unusual processing of trypanosome ribosomal RNA. J Mol Biol. 1987;196(1):113–124.
- Hartshorne T, Toyofuku W. Two 5ʹ-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999;27(16):3300–3309.
- Hashem Y, Des Georges A, Fu J, et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature. 2013;494(7437):385–389.
- Rosenblad MA, López MD, Piccinelli P, et al. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34(18):5145–5156.
- Piccinelli P, Rosenblad MA, Samuelsson T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33(14):4485–4495.
- Chu S, Archer RH, Zengel JM, et al. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):659–663.
- Lindahl L, Bommankanti A, Li X, et al. RNase MRP is required for entry of 35S precursor rRNA into the canonical processing pathway. RNA. 2009;15(7):1407–1416.
- Goldfarb KC, Cech TR. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev. 2017;31(1):59–71.
- Mitchell P. Exosome substrate targeting: the long and short of it. Biochem Soc Trans. 2014;42(4):1129–1134.
- Januszyk K, Lima CD. The eukaryotic RNA exosome. Curr Opin Struct Biol. 2014;24:132–140.
- Allmang C, Kufel J, Chanfreau G, et al. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. Embo J. 1999;18(19):5399–5410.
- Allmang C, Mitchell P, Petfalski E, et al. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28(8):1684–1691.
- Lebreton A, Tomecki R, Dziembowski A, et al. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456(7224):993–996.
- Schneider C, Leung E, Brown J, et al. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37(4):1127–1140.
- Fromm L, Falk S, Flemming D, et al. Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat Commun. 2017;8(1):1787.
- Sloan KE, Mattijssen S, Lebaron S, et al. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J Cell Biol. 2013;200(5):577–588.
- Estévez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. Embo J. 2001;20(14):3831–3839.
- Estévez AM, Lehner B, Sanderson CM, et al. The roles of intersubunit interactions in exosome stability. J Biol Chem. 2003;278(37):34943–34951.
- Cristodero M, Böttcher B, Diepholz M, et al. The Leishmania tarentolae exosome: purification and structural analysis by electron microscopy. Mol Biochem Parasitol. 2008;159(1):24–29.
- Jensen BC, Wang Q, Kifer CT, et al. The NOG1 GTP-binding protein is required for biogenesis of the 60 S ribosomal subunit. J Biol Chem. 2003;278(34):32204–32211.
- Hartshorne T, Agabian N. RNA B is the major nucleolar trimethylguanosine-capped small nuclear RNA associated with fibrillarin and pre-rRNAs in Trypanosoma brucei. Mol Cell Biol. 1993;13(1):144–154.
- Umaer K, Ciganda M, Williams N. Ribosome biogenesis in african trypanosomes requires conserved and trypanosome-specific factors. Eukaryot Cell. 2014;13(6):727–737.
- Bonneau F, Basquin J, Ebert J, et al. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139(3):547–559.
- Makino DL, Baumgärtner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495(7439):70–75.
- Makino DL, Schuch B, Stegmann E, et al. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature. 2015;524(7563):54–58.
- Zinder JC, Wasmuth EV, Lima CD. Nuclear RNA exosome at 3.1 Å reveals substrate specificities, RNA paths, and allosteric inhibition of Rrp44/Dis3. Mol Cell. 2016;64(4):734–745.
- Liu JJ, Bratkowski MA, Liu X, et al. Visualization of distinct substrate-recruitment pathways in the yeast exosome by EM. Nat Struct Mol Biol. 2014;21(1):95–102.
- Kala S, Mehta V, Yip CW, et al. The interaction of a Trypanosoma brucei KH-domain protein with a ribonuclease is implicated in ribosome processing. Mol Biochem Parasitol. 2017;211:94–103.
- Wasmuth EV, Lima CD. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol Cell. 2012;48(1):133–144.
- Drazkowska K, Tomecki R, Stodus K, et al. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013;41(6):3845–3858.
- Schillewaert S, Wacheul L, Lhomme F, et al. The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol Cell Biol. 2012 Jan;32(2):430–444.
- Gasse L, Flemming D, Hurt E. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol Cell. 2015;60(5):808–815.
- Schaeffer D, Reis FP, Johnson SJ, et al. The CR3 motif of Rrp44p is important for interaction with the core exosome and exosome function. Nucleic Acids Res. 2012;40(18):9298–9307.
- Sakyiama J, Zimmer SL, Ciganda M, et al. Ribosome biogenesis requires a highly diverged XRN family 5ʹ->3ʹ exoribonuclease for rRNA processing in Trypanosoma brucei. Rna. 2013;19(10):1419–1431.
- Fleming IM, Paris Z, Gaston KW, et al. A tRNA methyltransferase paralog is important for ribosome stability and cell division in Trypanosoma brucei. Sci Rep. 2016;6:21438.
- Clayton CE. Gene expression in kinetoplastids. Curr Opin Microbiol. 2016;32:46–51.
- Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol. 2002;125(1–2):211–216.
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45.
- Church GM, Gilbert W. Genomic sequencing. Proc Nat Acad Sci USA. 1984;81:1991–1995.
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(2):125–132.
- Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(4):479–485.
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(4):658–674.
- Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(4):470–478.
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62(9):1002–1011.
- Bricogne G, Blanc E, Brandl M, et al. BUSTER version 2.10.2. Cambridge (UK): Global Phasing Ltd; 2017.
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt12 Pt 1):2126–2132.
- Yang J, Yan R, Roy A, et al. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8.
- Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36(7):2295–2300.
- Protein interfaces, surfaces and assemblies service PISA [Internet]. European Bioinformatics Institute. Available from: http://www.ebi.ac.uk/pdbe/prot_int/pistart.html
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797.
- Glaser F, Pupko T, Paz I, et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19(1):163–164.
- Landau M, Mayrose I, Rosenberg Y, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302.
- Ashkenazy H, Abadi S, Martz E, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucl Acids Res. 2016;44:W344–W350.
