ABSTRACT
Pax3 plays an essential role in myogenesis. Previously, we found a tumor-signature chimeric fusion RNA, PAX3-FOXO1 also present during muscle differentiation, raising the possibility of its physiological role. Here we demonstrated that the fusion is needed transiently for muscle lineage commitment. Interestingly, the fusion ortholog was not found in seven mouse muscle differentiation/regeneration systems, nor in other stem cell differentiation systems of another three mammal species. We noticed that Pax3 is expressed at a much lower level in human stem cells, and during muscle differentiation than in other mammals. Given the fact that the fusion and the parental Pax3 share common downstream targets, we reasoned that forming the fusion may be a mechanism for human cells to escape certain microRNA regulation on Pax3. By sequence comparison, we identified 16 candidate microRNAs that may specifically target the human PAX3 3ʹUTR. We used a luciferase reporter assay, examined the microRNAs expression, and conducted mutagenesis on the reporters, as well as a CRISPR/Cas9 mediated editing on the endogenous allele. Finally, we identified miR-495 as a microRNA that specifically targets human PAX3. Examining several other fusion RNAs revealed that the human-specificity is not limited to PAX3-FOXO1. Based on these observations, we conclude that PAX3-FOXO1 fusion RNA is absent in mouse, or other mammals we tested, the fusion RNA is a mechanism to escape microRNA, miR-495 regulation in humans, and that it is not the only human-specific fusion RNA.
Introduction
Transcription factor, Pax3 plays an essential role in myogenesis. In a subset of alveolar rhabdomyosarcoma (ARMS), a recurrent t(2;13) chromosomal translocation results in the formation of a gene fusion, PAX3-FOXO1. The fusion consists of the first seven exons of PAX3 and the last two exons of FOXO1. The common belief that the chimeric RNA is unique to the tumor, provides the theoretical foundation for current ARMS diagnosis, as well as some ongoing therapeutics attempts. However, we recently found that the fusion RNA and protein products are also transiently expressed during muscle differentiation in vitro and in fetal muscle tissue samples [Citation1], raising the possibility that the fusion may play some normal physiological role during myogenesis.
It has been demonstrated that Pax3 is necessary and sufficient to induce myogenesis in mouse pluripotent stem cells [Citation2]. Even though PAX3-FOXO1 has been shown to have greater transcriptional activity than wild-type PAX3 [Citation3,Citation4], a genome-wide study of the PAX3-FOXO1 binding identified the same sites as parental PAX3 at both MYF5 and MYOD enhancers. Additionally, the PAX3 motif was found to be highly enriched [Citation5], suggesting that the fusion essentially targets the same downstream genes as the parental PAX3. Studies in fibroblast and rhabdomyosarcoma cell lines have demonstrated that PAX3-FOXO1 simultaneously induces myogenesis while blocking differentiation into mature muscle [Citation6,Citation7]. When we forced the constant expression of the fusion in mesenchymal stem cells, even under the differentiation condition, myogenic factors, MYOD and MYOG were continuously expressed throughout the process, in contrast to their normally transient expression pattern [Citation1]. No mature muscle marker MYH expression was observed at any time point, consistent with the fusion’s role in suppressing terminal differentiation [Citation1]. Interestingly, if we silenced the fusion RNA during the muscle differentiation, we observed much reduced MYOD and no MYOG or MYH expression. We thus speculate that PAX3-FOXO1 can commit precursor cells to the myogenic lineage by transactivating the expression of essential myogenesis factors MYOD. On the other hand, the prolonged expression of PAX3-FOXO1 leads to the constant expression of MYOD and MYOG, arresting cells at a premature stage, thus blocking terminal differentiation. These activities of the fusion are consistent with its transient expression pattern during normal myogenesis and its oncogenic role in RMS.
Surprisingly, Pax3-Foxo1 was not detected in seven mouse differentiation systems and tissues, including myoblast, mesenchymal stem cell (MSC), adipose-derived stromal/stem cells (ASC), embryonic carcinoma cells, embryonic stem cells, and fetal muscles, as well as in a toxin-induced muscle regeneration system. In addition, we also failed to detect the fusion in pig, sheep, and horse muscle differentiation systems. Interestingly, the expression of PAX3 in human stem cells, and during muscle differentiation is much lower than its orthologs in other mammals. We then reasoned that in humans, where PAX3-FOXO1 is expressed, the fusion might substitute for the role Pax3 plays in other species.
Given the fact that the PAX3-FOXO1 fusion swaps the 3ʹUTR of PAX3 to that of FOXO1, we wondered whether forming the fusion is a mechanism for human cells to escape certain microRNA regulation. We reasoned that this microRNA regulation must be unique to humans, and not present in the mice. We started with 16 microRNAs that are predicted to target only the human PAX3 3ʹUTR, but not that of the mouse. With luciferase reporter assays, we found six candidates that only affect the human construct. Three were eliminated due to the absence of expression during the human muscle differentiation process. By swapping the seed sequence between human and mouse orthologs, we narrowed down the list to miR-382 and miR-495. We then used CRISPR/Cas9 mediated genome editing to change the human PAX3 miR-495 seed sequence to the corresponding mouse sequence, and observed the upregulation of the PAX3 gene.
Results
PAX3-FOXO1 in human muscle differentiation
Previously, we observed the transient expression of PAX3-FOXO1 during MSC and ASC muscle differentiation [Citation1] (example in Figure S1A). When we forced the fusion to be constantly expressed, myoblast marker MYOD and myoblast initial differentiation marker MYOG were also constantly overexpressed instead of being transiently expressed, and no mature muscle marker MYH was detected [Citation1]. When we used a RNAi that has been previously reported to specifically silence the fusion RNA and protein [Citation8] ( and Figure S2A), reduced MYOD was observed, and more significantly, MYOG and MYH1 were delayed or not detected () and Figure S2B). The MSC cells that constantly express PAX3-FOXO1 grew faster in regular growth media ()), and continued to proliferate in differentiation media ()). In addition, they had less apoptosis and survived more under serum-deprived condition () and Figure S2C), consistent with the previously reported activity of pro-growth, and pro-survival of the fusion [Citation9,Citation10]. These findings support the idea that PAX3-FOXO1 is transiently expressed to commit pluripotent cells to the muscle lineage, and its constant expression will lead to arresting at myoblast/myogenic precursor stages.
Figure 1. PAX3-FOXO1 during human MSC muscle differentiation. (a) shRNA targeting PAX3-FOXO1 (shPF) specifically silenced the fusion RNA (PF), but not the parental genes. The target sequence has been reported before [Citation8]. The levels of the transcripts were measured by quantitative RT-PCR, and normalized to that of an internal control, GAPDH. (b) MSC cells stably infected with the virus expressing shPF resulted in reduced expression of MYOD, and no expression of MYOG and MYH1. (C-E) Overexpressing PAX3-FOXO1 promotes human MSC cells proliferation and survival. Cell proliferation and viability was measured by MTT. Compared with empty vector transfected cells (hMSCs/CT), human MSC cells stably expressing PAX3-FOXO1 (hMSCs/PF) grew faster in growth media (GM) (c), continued to proliferate in differentiation media (DM) (d), and survived more in serum-free media (SF) (e). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
![Figure 1. PAX3-FOXO1 during human MSC muscle differentiation. (a) shRNA targeting PAX3-FOXO1 (shPF) specifically silenced the fusion RNA (PF), but not the parental genes. The target sequence has been reported before [Citation8]. The levels of the transcripts were measured by quantitative RT-PCR, and normalized to that of an internal control, GAPDH. (b) MSC cells stably infected with the virus expressing shPF resulted in reduced expression of MYOD, and no expression of MYOG and MYH1. (C-E) Overexpressing PAX3-FOXO1 promotes human MSC cells proliferation and survival. Cell proliferation and viability was measured by MTT. Compared with empty vector transfected cells (hMSCs/CT), human MSC cells stably expressing PAX3-FOXO1 (hMSCs/PF) grew faster in growth media (GM) (c), continued to proliferate in differentiation media (DM) (d), and survived more in serum-free media (SF) (e). *, p < 0.05; **, p < 0.01; ***, p < 0.001.](/cms/asset/800ca72a-1567-499b-9c4d-20cd9c1938d3/krnb_a_1564464_f0001_oc.jpg)
PAX3-FOXO1 is absent in mouse
Overall human PAX3 mRNA (NM_001127366.2) is 90.7% similar to mouse Pax3 (NM_001159520.1), with 92.5% in the coding region and 87.8% similarity in the 3ʹ untranslated region (3ʹUTR). PAX3-FOXO1 in humans involves the first seven exons of PAX3 joining to the last two exons of FOXO1. The flanking exons are very similar in sequence between human and mouse as well as other mammals (Figure S3). We designed primers according to the putative fusion sequence to detect the fusion RNA in the mice.
We first attempted to detect Pax3-Foxo1 in a common skeletal muscle differentiation model, using mouse myoblast cells C2C12. C2C12 can be induced to differentiate into myocytes and forming myotubes with partial serum withdraw. However, no Pax3-Foxo1 was detected during the differentiation ().
In our previous experiments, where we started the differentiation from bone marrow or embryonic stem cell-derived MSCs and ASCs, the time points PAX3-FOXO1 were detected presided myoblast marker, MYOD. We reasoned that C2C12 being a myoblast cell line may be past the stage in which the fusion is expressed. We then differentiated mouse bone marrow-derived mesenchymal stem cells, D1-MSCs, and a mouse ASC cell line along muscle lineage up to 22 days. In neither situation did we observe the fusion RNA () and Figure S4).
Figure 2. Pax3-Foxo1 fusion RNA is not detected in mouse muscle differentiation/regeneration systems. (a) Mouse myoblast cell line C2C12 were induced to differentiate along a skeletal muscle lineage. RNA was extracted from samples harvested from 4 to 96 hours. (b) Bone marrow-derived mesenchymal stem cells, D1-MSCs, were induced to differentiate along a skeletal muscle lineage. RNA was extracted from samples harvested every other day from day 2 to day 22. (c) Mouse embryonic stem cell line, E14, was induced to differentiate along a skeletal muscle lineage. RNA was extracted from samples harvested from day 1 to day 8. (d) Pax3–Foxo1 RNA is absent in mouse fetal muscle samples. RNAs were extracted from fetal (muscle) samples harvested every other day from embryonic day 8 to day 20. (e) A time course of muscle regeneration was induced in the tibialis anterior (TA) muscles by cardiotoxin (CTX) injection. RNA was extracted from the muscle samples collected from day 1 to day 7. In all above, RNAs from the rhabdomyosarcoma cell line RH30 or mouse myoblast cell line C2C12 were used as controls. Pax3–Foxo1, MyoD, MyoG, Myh1, and Gapdh RNAs were assessed by RT-PCR. Lower panels are immuostaining of EDU on CTX or PBS treated muscle slices.
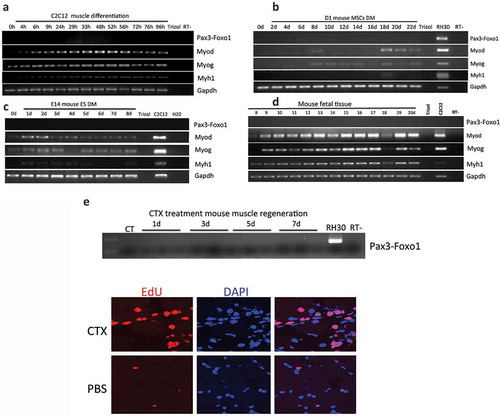
We then conducted muscle differentiation using a mouse embryonic carcinoma cell line, P19EC, and a mouse embryonic stem cell line, E14. Again, even though myoblast marker MyoD, myoblast initial differentiation marker MyoG, and myocyte maker Myh1 were all detected during differentiation, no Pax3-Foxo1 was detected during the time course of 7 or 8 days () and Figure S5). Suspecting that a different isoform of the fusion might be expressed, we used a Foxo1 antibody that recognizes both human and mouse Foxo1 protein. This antibody was used previously to detect the human PAX3-FOXO1 protein [Citation1]. Even though it picked up the signal for parental Foxo1, no Pax3-Foxo1 signal was detected in either mouse differentiation system (Figure S6).
Previously, we also found PAX3-FOXO1 in fetal muscle samples [Citation1]. We then collected mouse embryonic skeletal muscle samples starting from embryonic day 8 and ending on day 20 (the whole embryo was used in the early time points). No Pax3-Foxo1 was found ()). In another muscle regeneration model, where cardiotoxin (CTX) was injected into the tibialis anterior (TA) muscles, EdU staining revealed a high number of proliferating cells, but we failed to detect the fusion RNA ()).
In conclusion, Pax3-Foxo1 was not detected in any of the seven different muscle differentiation/regeneration models.
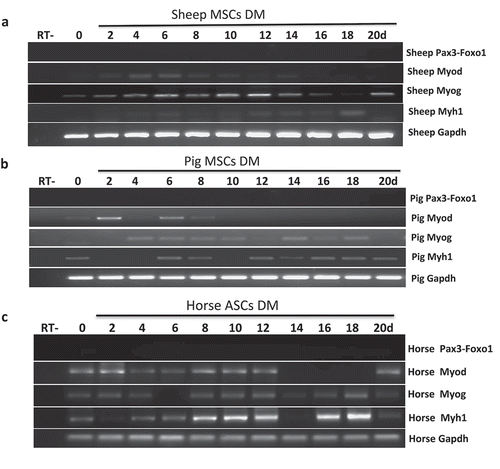
PAX3-FOXO1 is also not found in sheep, pig, or horse muscle differentiation systems
Given the pro-proliferate activity of PAX3-FOXO1, we then reasoned that larger mammals with higher skeletal muscle mass may have developed a need for the fusion. We have established bone marrow-derived MSC cells from pig and sheep, and ASC cells from horse [Citation11,Citation12]. We then induced them into skeletal muscle differentiation for up to 20 days. None of the systems had Pax3-Foxo1 expression at any time point ().
Figure 3. Pax3-Foxo1 fusion RNA is not detected in sheep, pig, or horse muscle differentiation systems. (a-c) Absence of Pax3–Foxo1 RNA during muscle differentiation in sheep bone marrow MSCs (a), pig bone marrow MSCs (b), and horse ASC cells (c). These cells were induced to differentiate along a skeletal muscle lineage. RNA was extracted from samples harvested every other day from day 2 to day 20. Pax3–Foxo1, MyoD, MyoG, Myh1, and Gapdh RNAs were assessed by RT-PCR.
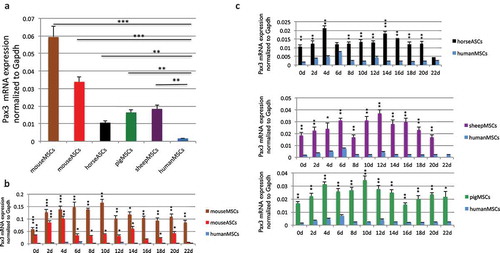
PAX3 is expressed at a much lower level in human cells than its orthologs in other mammals
We then examined the Pax3 expression among the stem cells. We used a primer pair that is conserved in all five species, as well as a conserved Gapdh primer set as the internal control (Figure S7). We found that PAX3 in humans is expressed at a level about 10-fold lower than its orthologs in sheep, pig, or horse, and 20–35 fold lower than Pax3 in mice (). Even during muscle differentiation, PAX3 remained low in humans than in other four mammals (). PAX3-FOXO1 has been reported to bind to the similar sites as wild-type PAX3, suggesting that the fusion targets similar downstream genes as parental PAX3 [Citation5]. Given the critical role of Pax3 in the myogenesis of mice and other mammals [Citation2,Citation13], we then conjectured the possibility that the fusion in humans may substitute for the role Pax3 plays during myogenesis in mice.
Figure 4. Comparison of wild type Pax3 RNA level. (a) Comparison of wild-type Pax3 RNA levels in MSCs and ASCs of different species. RNA was extracted from human MSCs, mouse MSCs, mouse ASCs, horse ASCs, pig MSCs, and sheep MSCs. Pax3 was measured by qRT-PCR, and its level was normalized against the internal control, Gapdh. Pax3 and Gapdh primers annealing to the conserved sequences among these species were used. (b) Comparison of Pax3 RNA expression during muscle differentiation in mouse bone marrow-derived MSCs and mouse ASCs, with human MSCs. (c) Comparison of Pax3 RNA expression during muscle differentiation in sheep MSCs, pig MSCs, and horse ASCs, with human MSCs. RNA was extracted from samples harvested every other day from day 2 to day 22. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Identification of micrornas targeting PAX3 in a human-specific manner
We then speculated that by forming the PAX3-FOXO1 fusion, human cells could escape the microRNA regulation of the parental PAX3. If this is the case, then the regulation must be targeted to the human PAX3, but not the mouse Pax3. We aimed to identify the microRNA(s) (). We first used TargetScan to identify 16 candidate microRNAs that are predicted to have seed sequences only in the human PAX3 3ʹUTR, but not in the corresponding region of mice (examples in Figure S8). Since the Pax3 expression levels in sheep, pig, and horse fall in between that of human and mouse, we did not use the presence or absence of the microRNA seed sequence in their 3ʹUTR sequences as a filtering criterion.
With the 16 candidates, we performed a luciferase reporter assay. The human or mouse Pax3 3ʹUTR was cloned into the 3ʹUTR region of a renilla luciferase gene in the miR-Sens plasmid, which also contains a separate firefly luciferase gene as the internal control [Citation14]. We transfected such a reporter into HEK293T cells, and then transfected in one of the 16 microRNAs mimics. The reduced ratio of renilla to firefly luciferase reflected the downregulation caused by a particular microRNA. We found six microRNAs, which significantly downregulated the human reporter, but not on the mouse reporter ()). These six became the candidates for the next step in validation.
Figure 5. PAX3 is subject to human–specific regulation of microRNA(s) during muscle differentiation. (a) Flowchart for narrowing down the microRNAs that specifically regulate human PAX3, but not mouse Pax3. (b) 16 candidates predicted by TargetScan were examined by luciferase reporter assays. Human PAX3, or mouse Pax3 3ʹUTR was cloned into the miR-Sens plasmid. These plasmids were transfected into HEK293T cells, and subsequently, microRNA mimics were transfected. Six microRNAs that showed suppression on the human construct, but not on the mouse construct are marked with stars. (c) Three microRNAs were further eliminated due to the absence of expression during the human MSC muscle differentiation. qRT-PCR was performed to quantify the level of microRNAs throughout the differentiation process. (d) Mutagenesis at the microRNAs target site. Both miR-495 and miR-382 downregulated the mouse Pax3 reporter when the sequence was changed into the seed sequence of humans. Both also caused upregulation of the human PAX3 reporter when the sequence was mutated to that of mice. miR-186 did not cause downregulation of mutated mouse reporter, even though it did not suppress the mutated human reporter. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
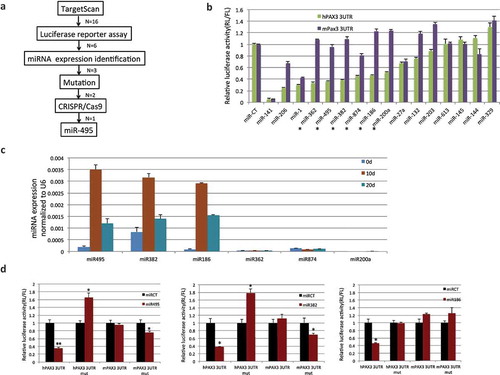
We then examined the expression of the microRNAs throughout human muscle differentiation. Three microRNAs were discarded, as they were hardly expressed. ) showed examples of microRNAs expression at three time points. Figure S9 showed the expression of PAX3-FOXO1 and miR-495 throughout the whole muscle differentiation process.
For the remaining three candidates, miR-186, miR-382, and miR-495, we performed mutagenesis on the luciferase reporters. To the human PAX3 3ʹUTR reporter, we specifically mutated the seed sequences of the microRNAs in human to the corresponding sequences in mouse. Reciprocally, we mutated the mouse Pax3 3ʹUTR reporter construct at the corresponding sites to the human microRNA seed sequence. This way, we expect to see escaping of the mutated human reporter, and suppression of the mutated mouse reporter from a candidate microRNA. Indeed, we observed the loss of suppression in the mutated human reporter from the three microRNAs. Interestingly, the renilla/firefly luciferase ratio was even upregulated in both miR-382 and miR-495 transfected cells with the mutated reporter. For the mutated mouse reporter, both miR-382 and miR-495, but not miR-186 suppressed the ratio of renilla/firefly luciferase ()).
CRISPR/CAS9 mediated genome editing at miR-495 seed sequence
We then investigated whether mutating the endogenous sequence of the seed sequence of the two remaining microRNAs to the corresponding mouse sequence would enable the cells to escape the microRNA regulation. To do so, we used CRISPR/Cas9 mediated genome editing, by introducing two sgRNAs flanking the targeted sequence, and a ssDNA oligo as the template (). The reason we chose to convert the human sequence to the mouse sequence is that the human PAX3 is expressed at an almost undetectable level; we would have a higher chance of detecting the PAX3 upregulation, even with a small percentage of cells successfully converted and escaping the microRNA regulation. In contrast, we would have a harder time detecting a downregulation, if we chose to mutate the mouse sequence to the human sequence. Indeed, we successfully made the conversion for miR-495 seed sequence. With specific primers, we could detect the mutant allele, only when both sgRNAs and the ssDNA were included ()). Quantitatively, we estimated the efficiency of the conversion is about 1/1000 ()). Even with this low efficiency, we observed an upregulation of PAX3, and it was most evident around differentiation day 4 ()). We failed to convert the seed sequence for miR-382, leaving it still a possible microRNA for regulating human PAX3.
Figure 6. CRISPR/Cas9 mediated genome editing at miR-495 seed sequence. (a) The location and sequence of the sgRNAs, and the template ssOligo for CRISPR/Cas9 editing. miRNA-495 seed sequence and primers specific to the wild-type and mutated alleles were also plotted. (b) Allele-specific primers picked up the mutant form only when both sgRNAs and the ssOligo template were included. (c) Relative number of the mutated alleles to the wild-type alleles after CRISPR/Cas9 mediated genome editing. Quantitative PCR revealed that the relative number of mutated alleles to the wild-type alleles is close to 1/1000. Purified PCR products were serial diluted, and used as templates to construct the standard curve. (d) PAX3 RNA was increased during human MSC muscle differentiation when miR-495 seed sequence was changed to the corresponding sequence of mice. (e) miR-495 suppressed endogneosu PAX3, but not the fusion. RH30 cells were transfected with miR-495 mimics or the control miR-CT. Western blot analysis were performed using FOXO1, PAX3, and GAPDH antibodies (left). Quantification of protein signal intensity was conducted using densitometry, and plotted on the right. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
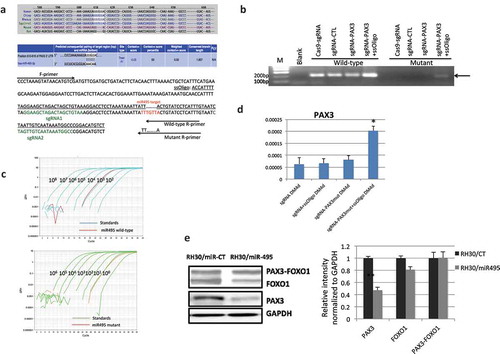
To confirm that miR-495 targets endogenous PAX3, but not PAX3-FOXO1 fusion, we transfected miR-495 mimics into RH30 cells. Indeed, the wild-type PAX3 was reduced when miR-495 was transfected compared with miR-CT transfection, whereas wild-type FOXO1 or the fusion protein, PAX3-FOXO1 had no obvious difference ()).
Several other human chimeric RNAs are also absent in other mammals
To investigate whether the human-specific expression pattern is unique to PAX3-FOXO1, we examined several other chimeric RNAs previously identified during human MSC differentiation. Similar to Pax3-Foxo1, Fbxo11-Map2k5, Eef1d-Naprt1, Megf11-Tipin, Setd5-Nudt9, and Tbcel-Tbcta, were not found in the mouse mesenchymal stem cell differentiation process (Figure S10A). These results are also consistent with our previous observation that human harbors more chimeric RNAs than mice [Citation15]. None of the above chimeric RNAs were detected in pig and sheep MSCs, or in ASCs from horse (Figure S10B-S10D), indicating that there are more human-specific chimeric RNAs.
Discussion
Chimeric fusion RNAs have been traditionally viewed as hallmarks of cancer cells. Recently, we and others have demonstrated their presence in normal human cell lines, and tissues [Citation15–Citation20]. Some of these chimeras are involved in crucial cellular processes, including: proliferation, apoptosis, regulation of cell cycle, and also pluripotency maintenance [Citation15,Citation21–Citation23]. Even though they were once believed to be solely generated by chromosomal rearrangement, chimeric RNAs may be products of intergenic splicing [Citation24,Citation25]. Previously, we reported that a ‘cancer-signature’ chimeric RNA, PAX3-FOXO1, as being present in normal muscle differentiation [Citation1]. Consistent with its expression in normal cells, no evidence of t(2;13) was seen as in the situation of rhabdomyosarcoma [Citation1], suggesting that it is a product of trans-splicing during muscle differentiation. We found that the expression of the chimeric RNA is transient during myogenesis, and presides other myogenic factors including MYOD, and MYOG. Silencing the chimera resulted in reduced/delayed expression of these factors. On the other hand, forced constant expression of the chimera lead to the constant expression of MYOD and MYOG. These cells proliferate more in growth media, and survive better in serum-deprived media. These findings support the crucial role of the chimera, and its expression pattern during muscle differentiation. Surprisingly, in seven different mouse muscle differentiation, or regeneration systems, we failed to detect the fusion. Considering the importance of Pax3 during myogenesis in mice and other mammals [Citation2,Citation13], and that the fusion shares similar downstream targets with the parental Pax3, we propose that the formation of the fusion may be a mechanism to escape certain microRNA regulation of the wild-type PAX3. Several recent studies also suggest that fusion genes can bypass microRNA-mediated regulation via changes of the 3′-UTR. For example, transcription factor MYB are typically expressed at a low level in adenoid cystic carcinoma (ACC) due to high expression of the MYB repressors miR-15a and miR-16 [Citation26,Citation27]. However, forming the MYB-NFIB fusion gene results in the loss of the MYB 3ʹUTR, leading to elevated levels of the MYB protein [Citation28]. The tumorigenic gene fusion, FGFR3-TACC3, in glioblastoma, is caused by a tandem duplication at 4p16.3. The fusion leads to the loss of the 3ʹ-UTR of FGFR3, blocking gene regulation of miR-99a, which enhances the expression of the fusion gene [Citation29].
The miR-495 gene resides in the microRNA cluster, the GTL2-DIO3 locus. It has been reported that transcription factor MEF2 directly regulates the Gtl2-Dio3 cluster in skeletal muscle [Citation30]. These microRNAs repress secreted Frizzled-related proteins (sFRPs), which in turn regulate the WNT signaling pathway. In addition, miR-410 and miR-495 are known to be regulated by MEF2 in cardiomyocytes, and their overexpression results in increased cardiomyocyte proliferation [Citation31]. Interestingly, PAX3-FOXO1 overexpression leads to an upregulation of microRNAs mir-495 and miR-543 [Citation32], suggesting that there may be additional feedback loops connecting PAX3-FOXO1, miR-495, and other players in myogenesis.
PAX3-FOXO1, together with several other chimeric RNAs were only detected in humans (Figure S10). It is not clear whether these chimeric RNAs are all avoiding microRNA regulation on the parental 5ʹ gene. Direct binding assays are also needed to prove the interaction of microRNAs and the parental genes for these chimeras including PAX3-FOXO1. In a genome-wide study, we also found small overlaps in the chimeric RNAs found in humans and in mice [Citation15]. These observations remind us that the conclusions from multiple studies which revealed that <20% of alternative splicing events are conserved during the ∼80- to 90-million-year interval separating humans and mice [Citation33–Citation36]. It is possible that in humans, chimeric RNAs, similar to alternative spliced RNAs, represent an expanded repertoire of functional genome.
Alu elements are primate specific and inverted Alu elements has been reported to affect the biogenesis of trans-splicing [Citation37]. We wondered whether inverted Alu elements may be responsible for the formation of the six human-specific chimeric RNAs including PAX3-FOXO1. We checked intron 7 of PAX3 and intron1 of FOXO1. Three Alu elements in different directions were found in the intron 7 of PAX3. Due to the large size of intron 1 of FOXO1 (>100kb), we found over 40 Alu elements in both directions. Similarly, we searched the introns of other human-specific chimeras. We found over 60 Alu elements in the large intron1 of FBXO11, and 5 Alu elements in the intron of MAP2K5; 13 Alus in the intron1 of SETD5, and 11 in the intron1 of NUDT9; Over 40 Alus in the intron1 of MEGF11, and 12 Alus in the intron3 of TIPIN; The large number of Alu elements in the relative large introns is not surprising, as the human genome contains over one million Alu elements, and full-length Alu elements are randomly distributed throughout the genome in both forward and reverse orientations. On average, every 3kb fragment contains one Alu. On the other hand, for two chimeras, LINC00545-MEDAG, and TBCEL-TECTA, no Alu was found in LINC00545 or TECTA introns, suggesting that inverted Alu is not the only mechanism for all the chimeric RNAs.
Materials and methods
Cell lines and culture conditions
Human embryonic stem cell–derived mesenchymal stem cells (hESC-MSC) were obtained from Millipore. The rhabdomyosarcoma cell line RH30 was cultured in RPMI-1640 medium with 10% FBS. Mouse bone marrow-derived mesenchymal stem cells D1-MSCs, and adipose-derived stromal/stem cells (ASC) were gifts from Dr. Gary Balian (University of Virginia). Sheep MSC and horse ASC cells were established by us previously [Citation11,Citation12]. Pig MSCs, mouse myoblast cell line C2C12, mouse MSCs, and mouse ASCs were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS (Gibco). Mouse embryonic carcinoma cell line, P19EC was maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 7.5% calf serum, and 2.5% fetal bovine serum (Gibco). E14 mouse ES cells were cultured in ESC medium (DMEM high glucose with 15% FBS, 100 μM nonessential amino acids, 2 mM l‐glutamine, 55 nM β‐mercaptoethanol, and 1500 U/ml LIF). Sheep MSCs and Horse ASCs were cultured in DMEM-F12 supplemented with 10% FBS. Differentiation media is the corresponding base medium with 2% horse serum.
Cloning of 3ʹUTR reporter constructs and luciferase assay
The miR-Sens plasmid was a gift from Dr. Mathijs Voorhoeve [Citation14]. The Pax3 reporters were constructed by ligation of human or mouse Pax3 3ʹUTR fragments at the XhoI and NotI sites on the miR-Sens vector. miRNA seed site mutation was done by site-directed mutagenesis on the wild-type Pax3 3ʹUTR construct, using primers listed in Table S1. Construct identities and orientations were confirmed by sequencing.
The Dual-Luciferase Reporter kit (Promega) was used for the detection of the luciferase activities, according to the manufacturer’s protocol. The experiments were repeated at least three times with similar results, unless otherwise stated.
RT-PCR and sanger sequencing
Total RNA was extracted using Trizol (Invitrogen) and following manufacturer’s instructions, treated with DNaseI (Ambion), and reverse-transcribed (Bioline). Fusion candidates were validated at the RNA level by standard RT-PCR, or qRT-PCR with the primers listed in Table S1. After RT-PCR and gel electrophoresis, all purified bands were sent for Sanger sequencing. Gapdh was used as an internal control. Quantitative PCR (qPCR) was carried out on a StepOne Plus instrument (Applied Biosystems).
Mirna PCR
Total RNA from cultured cells was extracted using TRIzol reagent (Invitrogen). Concentration and quality were determined using the Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.). RNA (1 µg) was reverse transcribed using Ncode™ miRNA First-Strand cDNA Synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. The forward primers are listed in Table S1. The universal reverse primer sequence is 5ʹ-GAGACTGCGGATGTATAGAACTTGA-3. Real-time PCR was performed using the Platinum SYBR Green qPCR SuperMix UDG (Invitrogen) on a StepOne Plus instrument (Applied Biosystems).
Western blot analysis
Cells were lysed in RIPA buffer (25 mM Tris-HCL ph7.4, 1 mM EDTA, 1% NP-40, 5% Glycerol). An equal amounts of total protein were separated by 10% SDS-PAGE, and transferred to a PVDF membrane using the Trans-Blot Turbo Transfer System (Bio-Rad). The following antibodies and dilutions were used for Western blotting: rabbit anti-FoxO1 (1:1000; CST 2880), and mouse anti-GAPDH (1:3000; CST 97166).
Plasmid preparation
shRNA against PAX3-FOXO1 target sequence is: CCTCTCACCTCAGAATTCA. It is cloned into the pSIREN plasmid. The Cas9/sgRNA dual-expression vector and sgRNA vector were gifts from Dr. Mazhar Adli (University of Virginia) [Citation38]. sgRNAs were designed according to the sgRNA prediction tool (http://crispr.mit.edu/).
Statistics
All quantitative data are calculated from at least three independent experiments. Two-tailed t-tests were used for expression and cell number comparisons. p-values < 0.05 were considered statistically significant.
Authors’ contributions
ZX performed most of the experiments, analyzed data, and wrote the manuscript. YT performed some experiments and constructed sgRNA plasmids. XS, JC, and YX generated some MSC and ASC cells. HL conceived the project, designed experiments, performed some analyses, and wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
| ARMS | = | alveolar rhabdomyosarcoma |
| RMS | = | rhabdomyosarcoma |
| MSC | = | mesenchymal stem cell |
| ASC | = | Adipose-derived stromal/stem cells |
Supplemental Material
Download Zip (11.1 MB)Acknowledgments
We thank Loryn Facemire for her assistance in editing the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Yuan H, Qin F, Movassagh M, et al. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013;3:1394–1403.
- Ridgeway AG, Skerjanc IS. Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. J Biol Chem. 2001;276:19033–19039.
- Bennicelli JL, Advani S, Schafer BW, et al. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene. 1999;18:4348–4356.
- Fredericks WJ, Galili N, Mukhopadhyay S, et al. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol. 1995;15:1522–1535.
- Cao L, Yu Y, Bilke S, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70:6497–6508.
- Graf Finckenstein F, Shahbazian V, Davicioni E, et al. PAX-FKHR function as pangenes by simultaneously inducing and inhibiting myogenesis. Oncogene. 2008;27:2004–2014.
- Khan J, Bittner ML, Saal LH, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci U S A. 1999;96:13264–13269.
- Kikuchi K, Tsuchiya K, Otabe O, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365:568–574.
- Crose LE, Galindo KA, Kephart JG, et al. Alveolar rhabdomyosarcoma-associated PAX3-FOXO1 promotes tumorigenesis via Hippo pathway suppression. J Clin Invest. 2014;124:285–296.
- Linardic CM. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008;270:10–18.
- Su X, Ling Y, Liu C, et al. Isolation, culture, differentiation, and nuclear reprogramming of Mongolian sheep fetal bone marrow-derived mesenchymal stem cells. Cell Reprogram. 2015;17:288–296.
- Liu -Z-Z, Lin-Gai W, Xiao-Hu S, et al. Differentiation of Mongolia horse adipose tissue-derived mesenchymal stem cells into adipocytes and osteoblasts in Vitro. J South China Agric Univ. 2011;32:1.
- Ling YH, Sui MH, Zheng Q, et al. miR-27b regulates myogenic proliferation and differentiation by targeting Pax3 in goat. Sci Rep. 2018;8:3909.
- Beillard E, Ong SC, Giannakakis A, et al. miR-Sens–a retroviral dual-luciferase reporter to detect microRNA activity in primary cells. Rna. 2012;18:1091–1100.
- Babiceanu M, Qin F, Xie Z, et al. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016;44:2859–2872.
- Magrangeas F, Pitiot G, Dubois S, et al. Cotranscription and intergenic splicing of human galactose-1-phosphate uridylyltransferase and interleukin-11 receptor alpha-chain genes generate a fusion mRNA in normal cells. Implication for the production of multidomain proteins during evolution. J Biol Chem. 1998;273:16005–16010.
- Frenkel-Morgenstern M, Lacroix V, Ezkurdia I, et al. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012;22:1231–1242.
- Carrara M, Beccuti M, Cavallo F, et al. State of art fusion-finder algorithms are suitable to detect transcription-induced chimeras in normal tissues? BMC Bioinformatics. 2013;14(Suppl 7):S2.
- Finta C, Zaphiropoulos PG. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem. 2002;277:5882–5890.
- Chase A, Ernst T, Fiebig A, et al. TFG, a target of chromosome translocations in lymphoma and soft tissue tumors, fuses to GPR128 in healthy individuals. Haematologica. 2010;95:20–26.
- Qin F, Song Z, Babiceanu M, et al. Discovery of CTCF-Sensitive Cis-Spliced fusion RNAs between adjacent genes in human prostate cells. PLoS Genet. 2015;11:e1005001.
- Wu CS, Yu CY, Chuang CY, et al. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res. 2014;24:25–36.
- Li H, Ma X, Wang J, et al. Effects of rearrangement and allelic exclusion of JJAZ1/SUZ12 on cell proliferation and survival. Proc Natl Acad Sci U S A. 2007;104:20001–20006.
- Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–211.
- Chuang TJ, Wu CS, Chen CY, et al. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2016;44:e29.
- Chung EY, Dews M, Cozma D, et al. c-Myb oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol Ther. 2008;7:1758–1764.
- Zhao H, Kalota A, Jin S, et al. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113:505–516.
- Persson M, Andren Y, Mark J, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106:18740–18744.
- Parker BC, Annala MJ, Cogdell DE, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123:855–865.
- Snyder CM, Rice AL, Estrella NL, et al. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development. 2013;140:31–42.
- Clark AL, Naya FJ. MicroRNAs in the myocyte enhancer factor 2 (MEF2)-regulated Gtl2-Dio3 noncoding RNA locus promote cardiomyocyte proliferation by targeting the transcriptional coactivator cited2. J Biol Chem. 2015;290:23162–23172.
- Muir S, Nathanson J, Wilbert M, et al. The role of miRNA in PAX3-FKHR positive rhabdomyosarcoma. Cancer Res. 2014;74(19 Suppl):Abstract nr 3558.
- Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180.
- Nurtdinov RN, Artamonova II, Mironov AA, et al. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum Mol Genet. 2003;12:1313–1320.
- Pan Q, Bakowski MA, Morris Q, et al. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77.
- Yeo GW, Van Nostrand E, Holste D, et al. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci U S A. 2005;102:2850–2855.
- Chuang TJ, Chen YJ, Chen CY, et al. Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res. 2018;46:3671–3691.
- Qin P, Parlak M, Kuscu C, et al. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat Commun. 2017;8:14725.
