ABSTRACT
Doublesex is highly conserved and sex-specifically spliced in insect sex-determination pathways, and its alternative splicing (AS) is regulated by Transformer, an exonic splicing activator, in the model system of Drosophila melanogaster. However, due to the lack of a transformer gene, AS regulation of doublesex remains unclear in Lepidoptera, which contain the economically important silkworm Bombyx mori and thousands of agricultural pests. Here, we use yeast three-hybrid system to screen for RNA-binding proteins that recognize sex-specific exons 3 and 4 of silkworm doublesex (Bm-dsx); this approach identified BxRBP1/Lark binding to the exon 3, and BxRBP2/TBPH and BxRBP3/Aret binding to the exon 4. Investigation of tissues shows that BxRBP1 and BxRBP2 have no sex specificity, but BxRBP3 has – three of its four isoforms are expressed with a sex-bias. Using novel sex-specific silkworm cell lines, we find that BxRBP1 and BxRBP3 directly interact with each other, and cooperatively function as splicing repressors. Over-expression of BxRBP1 and BxRBP3 isoforms efficiently inhibits splicing of the exons 3 and 4 in the female-specific cells and generates the male-specific isoform of Bm-dsx. We also demonstrate that the sex-determination upstream gene Masc regulates alternatively transcribed BxRBP3 isoforms. Thus, we identify a new regulatory mechanism of doublesex AS in the silkworm, revealing an evolutionary divergence in insect sex-determination.
Introduction
Sex determination in animals is determined by both the composition of sex-chromosomes and the environment [Citation1]. Pathways of sex determination in insects, the most abundant and diverse animals on the earth, can be divided into three stages: the primary signal on sex-chromosomes, a cascade of alternatively spliced genes, and the expression of sex-specific developmental genes [Citation2,Citation3].
Many types of sex-chromosome composition have been identified in insects, such as the XY type in the fruit fly, mosquito, and housefly, the XO type in the grasshopper, and the ZW type in silkworm, and no sex chromosome in the honeybee [Citation4,Citation5]. Primary sex-determining signals are different, even in insects with the same type of sex-chromosome composition. For example, the initial signal is believed to be the dosage of XSE (X chromosome-encoded signal element) in Drosophila melanogaster [Citation6,Citation7], Nix (a dominant male-determining factor) in mosquito Aedes aegypti on Y chromosome [Citation8], Mdmd/CWC22 (Musca domestica male determiner) in housefly [Citation9], CSD (complementary sex determiner) in the honeybee Apis mellifera [Citation10], and piRNA in the silkworm Bombyx mori on W chromosome [Citation11].
Alternative splicing (AS) cascades have been well characterized in Drosophila melanogaster, in which XSE triggers a cascade of genes that are sex-specifically spliced. These include sex-lethal (Sxl) [Citation12], transformer (tra) [Citation13], and doublesex (dsx) [Citation14]. Most insects use tra as the first regulator in the alternatively spliced cascade instead of Sxl, such as medfly (Ceratitis capitata) and red flour beetle (Tribolium castaneum), although the protein sequence of Tra homologues is not well conserved among insects [Citation15,Citation16]. In honeybee, the AS cascade contains a tra homologous gene feminizer (fem) and dsx, while the initial signal CSD encodes a tra-like protein and is a potential splicing regulator for fem [Citation17]. Tra/tra-2 complex binds to the exon 4 of dsx and acts as a splicing activator in Drosophila [Citation14], while the detail regulation mechanism still unknown in other insects. So far, the only conserved gene in the insect sex-determination pathway is the downstream gene dsx, which is sex-specifically spliced and encodes transcription factors in both female and male insects for regulation of downstream sexual development genes [Citation2,Citation4,Citation18].
Lepidoptera is the second largest order in insects, comprised of hundreds of thousands of species, including the economically important silkworm Bombyx mori and thousands of agricultural pests. For silk production, the male produces better quality silk and is more efficient due to less consumption of mulberry leaves. A female-specific lethal system was previously used to control insect pests based on the sex-specific alternative splicing of genes from insect sex-determination pathway [Citation19,Citation20]. Thus, sex-determination genes could be applied as targets for optimized silk production as well as targets for pest control. The female silkworm contains a Z and a W sex-chromosome, whereas the male silkworm has two Z chromosomes. Despite a long history of silkworm domestication, the sex-determination pathway has not been well understood except for a predicted Feminizer (Fem) gene on the W chromosome [Citation21,Citation22] and the downstream Bm-dsx gene [Citation23,Citation24]. A few years ago, a PSI was identified to facilitate the male-specific AS of dsx, but it is not sex specific [Citation25,Citation26]. Recently, the Fem gene on the W chromosome was revealed to be a piRNA [Citation11,Citation27], which inhibits a downstream gene Masculinizer (Masc) on the Z chromosome in the female silkworm, whereas in the males, Masc expression results in the male-specific AS of dsx [Citation28–Citation30].
The Bm-dsx gene consists of 6 exons, of which exons 3 and 4 are sex specific through alternative splicing, retained in females but skipped in males [Citation23,Citation31]. This is different from Drosophila, in which only exon 4 is sex specific [Citation32]. It remains unclear whether Masc directly regulates AS of Bm-dsx; if not, the additional factors are unknown. Three regulatory cis-acting RNA elements have been found in Bm-dsx exon 4 [Citation25], and our previous studies found that the exon 3 in the silkworm dsx also contains conserved elements required for its splicing regulation [Citation33]. Based on the presence of these cis-acting RNA elements, we performed yeast three-hybrid screens using sex-specific silkworm cDNA libraries to search for proteins that bind to exon 3 or exon 4 of dsx, investigated their regulatory functions in the alternative splicing of dsx, and demonstrated that expression of one of the RNA-binding proteins is controlled by Masc. Thus, here we reveal a novel AS regulatory mechanism in the sex-determination pathway of Lepidoptera.
Results
Screen for RNA-binding proteins that bind to sex-specific exons of Bm-dsx
We previously found that the two sex-specific exons (3 and 4, ) in Lepidoptera dsx mRNAs are more conserved than the four common exons and have three cis-RNA elements, mutations in which affect AS of Bm-dsx [Citation33]. Thus, we used exons 3 and 4 in a yeast three-hybrid system [Citation34] to screen for binding proteins that would regulate AS of Bm-dsx. We constructed two RNA bait plasmids, in which an RNA-binding sequence of MS2 was linked with either Bm-dsx exon 3 or 4; and we prepared three sex-specific libraries of cDNA linked to an activation domain (), including two libraries from male and female embryos of a sex-limited B. mori strain R01 [Citation35] and one from testis of the WT silkworm p50 strain. After selection for HIS3+/β-gal+ colonies, there were 113 positive clones eventually obtained in the 6 screens (2 plasmids x 3 libraries). All clones contained full or partial coding sequences (CDS) from three silkworm genes. We designated these as BxRBP1, BxRBP2, and BxRBP3 (Bombyx mori dsx RNA-binding proteins). BxRBP2 was identified using exon 4 as RNA bait and was frequently obtained from all three screened cDNA libraries, whereas BxRBP1 and BxRBP3 were obtained only from the testis cDNA library and were identified from exon 3 and exon 4 RNA baits, respectively ( and Table S2). These RNA–protein interactions were further confirmed by additional yeast three-hybrids tests using the isolated plasmids ().
Figure 1. Identification of RNA-binding proteins of Bm-dsx sex-specific exons by yeast three-hybrid screens.
(A) Schematic diagram of alternative splicing of Bm-dsx in the female and male silkworm. Introns are in lines and exons are in boxes (orange: female specific, white: common). Lengths of each intron are indicated. (B) Strategy to identify RNA-binding proteins of Bm-dsx by yeast three-hybrid screening. Sex-specific exons 3 and 4 of Bm-dsx were fused with the MS2 RNA stem-loop to form an RNA bridge between factors that bind to the LexA operator and the HIS3/LacZ site. MS2 coat protein (MCP)-LexA fusion protein was used to recruit RNA to the LexA operator, and the cDNA library would express Bm-dsx RNA-binding proteins that fused to a transcription activation domain (AD) to induce expression of β-galactosidase (LacZ) and HIS3. (C) Summary of the six yeast three-hybrid screens. Final positive clones were sequenced and are listed in Table S2. (D) LacZ assays of representative clones that interact with exons 3 and 4 of Bm-dsx. Plasmids from the positive colonies were isolated and re-tested in a clean yeast three-hybrid strain. Clone numbers and CDS coverage are indicated. Ctrl: MS2-binding sites only.
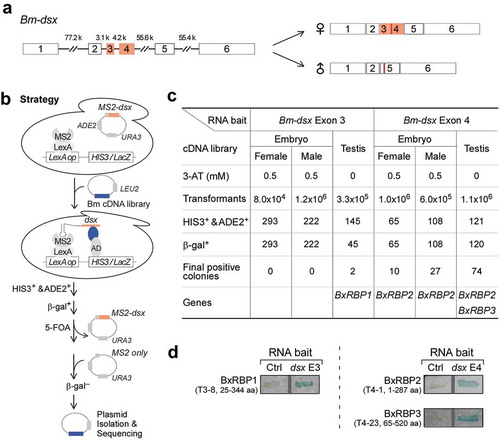
In B. mori, BxRBP1 is located on chromosome 3 and is a homologue of Drosophila lark and mammalian RBM4 (Fig. S1). BxRBP2 is located on chromosome 11 and is a homologue of Drosophila TBPH and mammalian TDP-43 (Fig. S2). BxRBP3 is located on chromosome 12 is and a homologue of Drosophila aret/bruno and mammalian ETR-3/CELF-2 (Fig. S3). All three proteins contain multiple RNA recognition motifs (RRMs), consistent with a role in RNA binding.
Identification of mRNA isoforms and sex specificity of BxRBP genes
To obtain full-length transcripts and all isoforms of the three BxRBP genes, we performed 5ʹ- and 3ʹ-RACE using tissues from both male and female silkworms. We found three alternatively spliced isoforms of BxRBP1, of which two were previously reported [Citation36]. The two long BxRBP1 isoforms are slightly different in their 5ʹ-UTR regions and encode the same protein with two RRMs and one Zinc-Finger (ZnF) domain, while the short isoform would encode a small protein without any potential RNA-binding domain (). As previously described [Citation37], only one transcript was obtained by RACE for BxRBP2, which is an intron-less gene and encodes a protein with two RRMs (). We identified four isoforms of BxRBP3, designated as BxRBP3-A, -B, -C and – D; none of these have been annotated or reported. They share most downstream exons but vary in the upstream transcription start sites, which are scattered across a 406-kb locus according to the genome sequence in SilkDB [Citation38]. Isoforms A, B, and C of BxRBP3 have the same three RRMs with 15~45 aa variations at their N-termini, while the D isoform, the shortest one, lacks the first RRM ().
Figure 2. Gene structures and transcripts of the three identified RNA-binding proteins in Bombyx mori.
Exons and introns of the BxRBP1 (A), BxRBP2 (B) and BxRBP3 (C) transcripts are defined according to results from 5ʹ- and 3ʹ-RACE. Functional protein domains of each transcript are generated according to NCBI information based on their coding sequences (black boxes), in which RRMs are highlighted in orange. Translation start sites (black arrows) and alternative splicing isoforms (V-shape lines) are also indicated. (D) Interaction between full-length of the three BxRBPs with Bm-dsx sex-specific exons by Lac-Z assay. All the four isoforms of BxRBP3 are tested. (E) Quantitation of interactions by β-galactosidase assay. The luminescent signal output from LacZ gene was normalized to cell number. Mean values ± s.d. of three technical replicates from one of three independent experiments are shown.
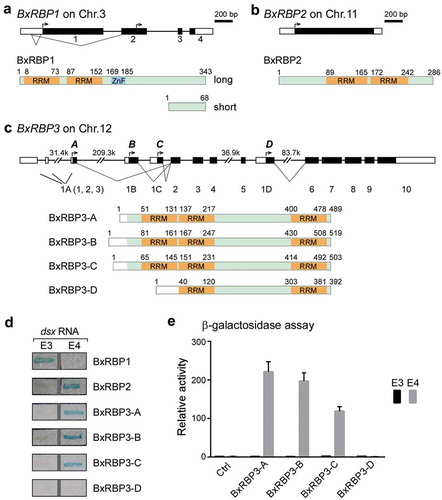
We then tested the RNA-binding abilities of these identified isoforms to Bm-dsx exons. Consistent with the partial sequences found in the three-hybrid screens, the full-length BxRBP1 bound efficiently to exon 3, but not 4; while the full-length BxRBP2 bound efficiently to exon 4 but not 3 (). The full-length A, B, and C isoforms of BxRBP3 bound to exon 4 but not exon 3, while the D isoform could not bind to exon 4, indicating that the first RRM of BxRBP3 is required for binding. We further quantified binding activities of these four isoforms by β-galactosidase assay [Citation34] and found that BxRBP3-A and -B had similar binding activities to the exon 4, whereas the BxRBP3-C’s binding was ~40% lower and BxRBP3-D’s was totally abolished ().
We then investigated sex specificities of the BxRBP1-3 transcripts in different developmental stages and tissues from female and male silkworms. Expression of BxRBP1 isoforms and BxRBP2 were at similar levels in female and male samples ( upper). However, three isoforms of BxRBP3 (-A, -B, and -C) were expressed in sex-biased patterns in gonad-containing samples ( middle). Isoform A was significantly higher in males (testis, pupa, and adult) than in females, whereas the isoform C exhibited an opposite pattern, higher in females than males. Isoform B was slightly higher in male samples and isoform D showed no observable sex-bias.
Figure 3. Expression profiles of all the BxRBP transcripts in stages and tissues of the female and male Bombyx mori.
(A) RT-PCR of BxRBP transcripts in the silkworm stages and tissues. Tissues were dissected from day 3 of the 5th larva stage. Transcripts of Bm-dsx are used as sex-specific marker, and actin is used as loading control. (B) Expression profiles of the BxRBPs in the new generated sex-specific cell lines.
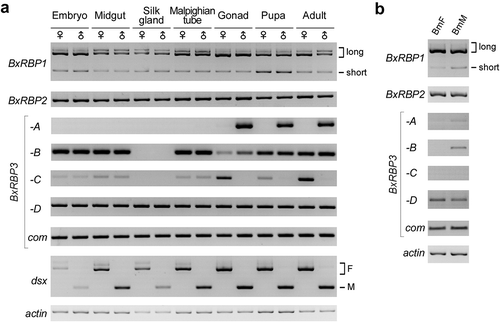
To reveal the function and regulatory roles of the three RBPs on AS of Bm-dsx, we established two sex-specific cell lines from embryos of the above-described R01 silkworm, BmF cells from the female and BmM cells from the male (Fig. S4A). Sex specificities of these two cell lines were confirmed by detection of three random amplified polymorphic DNA (RAPD) markers (Musashi, Sasuke, Bonsai) on the female-specific W chromosome locus [Citation11,Citation21] and by AS patterns of the Bm-dsx (Fig. S4B). Expression patterns of BxRBPs in the two sex-specific cell lines were similar to gonadal tissues, except for a lower level of BxRBP3-C isoform ().
BxRBP1 and BxRBP3 together efficiently inhibit female AS of Bm-dsx
We then tested the effect of expression of the three RBPs on AS of Bm-dsx in the BmF and BmM cell lines. In comparison to over-expression (OE) of the upstream gene Masc, which significantly inhibited splicing of exons 3 and 4 in BmF cells ( lane 9), individual OE of the four BxRBP3 isoforms only slightly inhibited splicing of exons 3 and 4, generating the male-specific AS product of Bm-dsx in BmF cells (, cf. lanes 5–8 to 9). OE of BxRBP1 or BxRBP2 had no detectable effects in BmF cells; as well, none of the OE of RBPs had detectable effects on the AS of Bm-dsx in BmM cells ( lanes 10–18).
Figure 4. BxRBP1 and BxRBP3-B together efficiently inhibit splicing of the sex-specific exons of Bm-dsx.
(A) Individual over-expression of BxRBP3 isoforms in BmF cells slightly inhibits splicing of Bm-dsx exons 3 and 4. (B) Co-OE of BxRBP1 with BxRBP3 isoforms significantly inhibits splicing of Bm-dsx exons 3 and 4, resulted in male-specific isoform of Bm-dsx in the BmF cells.Empty vector and Masc-OE are tested as negative and positive controls, respectively. Semi-quantitative data shown are mean ± s.d. of three independent experiments.(C) Fluorescence assay of BxRBP cellular localizations. BxRBPs are fused with EGFP and expressed in BmF cells. Nuclei are visualized by DAPI.
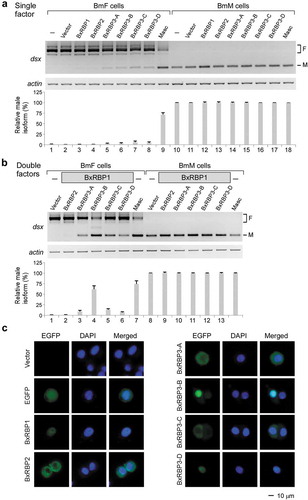
Since the inhibition of female-specific splicing of Bm-dsx by single-RBP-OE was subtle, we then tested all possible combinations of double-RBP-OEs ( and S5A). We found that BxRBP1 strongly enhanced the splicing inhibition by BxRBP3 isoforms and resulted in more male-specific products in BmF cells ( lanes 3–6). The most significant combination was BxRBP1 with BxRBP3-B (lane 4), which shifted ~65% of the female-specific Bm-dsx isoforms to the male-specific isoform, similar to the effect of OE of Masc. This BxRBP1 enhancement was also observed in BmN cells (Fig. S5B), a frequently used female silkworm cell line [Citation39]. In contrast, OE of BxRBP2 did not show any enhancement of exon-skipping of Bm-dsx in combination with BxRBP3 isoforms, nor did any combination of two BxRBP3 isoforms (Fig. S5A).
BxRBP2 lacks a nuclear localization signal
To further understand the function of the three RBPs in regulation of Bm-dsx alternative splicing, we analysed their phylogenetic conservation. Their homologues in other species have been reported as splicing regulators. For example, RBM4 regulates alternative splicing of alpha-tropomyosin, PTB, pyruvate kinase M and Disabled-1 genes in human [Citation40–Citation43]; skipping of exon 9 of the CFTR gene was regulated by both TDP-43 and ETR-3/CELF-2 [Citation44–Citation48]; and Aret regulates alternative splicing of various genes including Stretchin and wupA in Drosophila flight muscles [Citation49,Citation50].
In general, these three RBPs are highly conserved from insects to mammals; however, one notable feature is that a nuclear localization signal (NLS), present in the N-terminus of fly and mammalian homologues, is absent in BxRBP2 (Fig. S2B). Therefore, we tested the cellular location of all three RBPs in silkworm cell lines using EGFP-fusion proteins. As indicated by EGFP fluorescence, BxRBP1, BxRBP3-B, and BxRBP3-D are mostly located in the nucleus of BmF cells, whereas BxRBP3-A and -C preferentially locate in the cytoplasm. In contrast, BxRBP2 is exclusively located in the cytoplasm (). These results are consistent with the above findings that BxRBP1 and BxRBP3 isoforms facilitate the inhibition of splicing of Bm-dsx exons 3 and 4 in BmF cells, whereas BxRBP2 has no function on splicing of Bm-dsx.
We also knocked down the three BxRBPs in the silkworm sex-specific cells using various sets of siRNAs. In comparison to the upstream gene Masc knockdown, knockdown of these BxRBPs did not change AS of the Bm-dsx in BmM cells, in contrast to our expectation (). Two possibilities could explain these results. First, the remaining of BxRBPs after RNAi knockdown (about 20–30%) could still be sufficient to inhibit splicing of exons 3 and 4 of Bm-dsx. Second, the role of BxRBPs in the AS regulation of Bm-dsx could be taken over by other RBPs, which we don’t know.
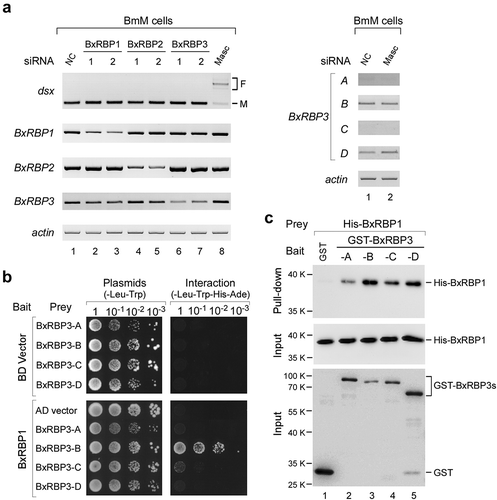
Figure 5. BxRBP1 strongly interacts with BxRBP3-B.
(A) Down-regulation of BxRBPs do not facilitate female splicing of Bm-dsx in the male cells, and down-regulation of Masc does not affect the mRNA level of BxRBP3 isoforms in male cells. Both yeast two-hybrid (B) and in vitro protein interactions (C) show that BxRBP1 directly interacts with BxRBP3 isoforms, in which the BxRBP3-B is the strongest. In the yeast two-hybrid, BxRBP1 is used as bait and BxRBP3 isoforms are used as preys, interactions are visualized by reporter of HIS3 and ADE2. In the in vitro protein interaction assay, GST-BxRBP3s and His-BxRBP1 are incubated together and pulled down by glutathione beads. GST alone protein is used as negative control.
BxRBP1 directly interacts with BxRBP3 isoforms
Above, we demonstrated that BxRBP1 binds to the Bm-dsx exon 3 and BxRBP3 isoforms bind to exon 4, and they coordinately facilitate the skipping of exons 3 and 4 in B. mori female-specific cells, resulting in the male-specific Bm-dsx isoform. Therefore, we asked whether BxRBP1 and BxRBP3 isoforms have direct interaction by two kinds of assays, the yeast two-hybrid and in vitro protein–protein interaction. In the yeast two-hybrid assay, we found that BxRBP1 (bait) with BxRBP3-B (prey) effectively induced expression of reporter genes (His3 and Ade2), whereas the bait with binding domain (BD) alone could not, showing the interaction between BxRBP1 and BxRBP3-B (). Interactions of BxRBP1 with the other three isoforms of BxRBP3 are weak, especially the -A and -D isoforms. Similarly, using purified recombinant proteins, all four GST-tagged BxRBP3 isoforms could pull down His-tagged BxRBP1, in which BxRBP3-B was the most efficient, whereas GST alone could not (). Together, these results demonstrate that BxRBP1 directly interacts with BxRBP3 isoforms, and imply a cooperative mechanism for skipping of exons 3 and 4 in the male-specific splicing of Bm-dsx that is ensured by protein-protein interactions between BxRBP1 and BxRBP3-B.
It was previously identified two proteins, PSI and IMP, that bind with exon 4 of Bm-dsx [Citation25,Citation26]. Therefore, we asked whether these two proteins directly interact with our identified BxRBPs. Using yeast two-hybrid and in vitro pulldown assays, we found that PSI interacts with BxRBP3, but not BxRBP1 or BxRBP2. No interaction was found between IMP and the three BxRBPs (Fig. S6).
Masc regulates transcription of BxRBP3 isoforms
When either Masc or BxRBP1+ BxRBP3-B were over-expressed in the female-specific BmF cells, we observed the same AS changes of Bm-dsx: efficient skipping of exons 3 and 4 generating the male dsx isoform in female-specific silkworm cells. Masc is a zinc-finger motif-containing protein and regulates transcription of a number of genes [Citation11]. To test whether Masc regulates transcription of the three identified BxRBPs, we measured expression levels of the RBPs in both BmF and BmM cells, with or without over-expression of Masc. We found that Masc-OE resulted in significantly increased BxRBP3-A (up to more than 25 folds) and a mild increase of BxRBP3-B, but no significant change in BxRBP3-C and -D isoforms ( & ). Expression of total BxRBP3 was also significantly increased. However, Masc-OE had no detectable effect on the other two BxRBPs, BxRBP1 and BxRBP2 ( & ). These results suggested that Masc regulates alternative transcription of BxRBP3, activating transcription of the -A and -B isoforms of BxRBP3.
Figure 6. Masc regulates expression of BxRBP3 isoforms but not Bm-dsx.
(A) OE of Masc in the silkworm cell lines stimulates expression of BxRBP3-A and -B. Empty vector is tested as a negative control. Expression levels of Masc were shown by RT-PCR and western blotting. Actin and Tubulin are used as loading controls. (B) Semi-quantitative detection of RT-PCR products in panel (A), which are mean ± s.d. of three independent experiments. (C) Schematic of Bm-dsx mini-gene. The arrows indicate locations of primers used for amplification. OpIE2, Orgyia pseudotsugata immediate early 2 promoter sequence; OpIE2 pA, Orgyia pseudotsugata immediate early 2 polyadenylation sequence. (D) Alternative splicing of Bm-dsx mini-gene in BmF cells with OE of BxRBPs or Masc. (E) Alternative splicing of Bm-dsx mini-gene in human HEK293T cells with OE of BxRBPs or Masc.Actin and GAPDH are used as loading controls of total RNA in BmF and HEK293T, respectively, and Tubulin is used as loading control of total protein for western blotting. Isoforms of Bm-dsx mini-gene are schematically indicated. Asterisk: non-specific signal.
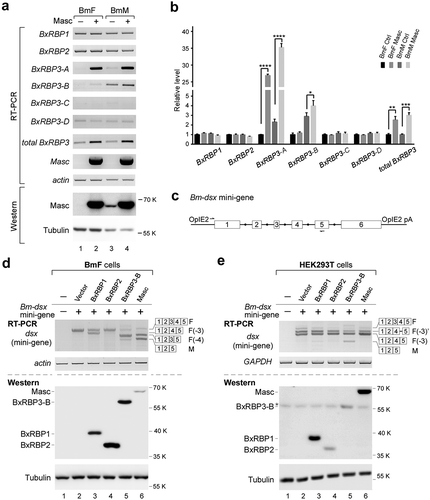
To further distinguish whether Masc and BxRBPs directly regulate AS of Bm-dsx, we constructed a plasmid-borne Bm-dsx mini-gene that contains all exon sequences but shorter introns sequences (), and tested its AS in both human HEK293T cells and silkworm BmF cells with expression of Masc or BxRBPs. In silkworm BmF cells, the Bm-dsx mini-gene was spliced in a female-specific pattern, recapitulating the effects on the endogenous Bm-dsx gene previously observed ( lane 2, compare to lane 1). Expression of Masc efficiently inhibited splicing of exons 3 and 4 and resulted in male-specific splicing, whereas expression of BxRBP2 had no effect. However, unlike the effects on the endogenous Bm-dsx gene, expression of BxRBP1 alone or BxRBP3-B alone was sufficient to inhibit splicing of exon 3 or 4 of Bm-dsx mini-gene, resulting in F(−3) isoform (female isoform without exon 3) or F(−4) isoform (female isoform without exon 4), respectively (, lanes 3 and 5). In human HEK293T cells, the Bm-dsx mini-gene was spliced into two groups of female-isoform-like products: one was the silkworm female-specific product (F isoform), the other was products of F(−3) isoform and F(−3)’ isoform which is F(−3) isoform with an extra 59-nt exon from intron 4 ( lane 2). In contrast to the silkworm BmF cells, expression of Masc in human HEK293T cells did not inhibit splicing of exon 4 in the Bm-dsx mini-gene, neither BxBRP1 nor BxRBP2, implying that Masc is a direct factor to regulate AS of Bm-dsx. However, expression of BxRBP3-B alone was sufficient to inhibit splicing of exon 4 in the human cells ( lane 5). Taken together, these results demonstrate that in the sex-determination pathway of silkworm, Masc is an upstream gene, which does not directly regulate AS of Bm-dsx, whereas BxRBP3-B is a direct regulator of AS.
Discussion
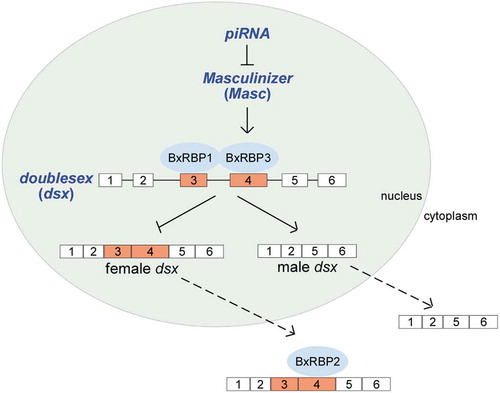
Regulation of alternative splicing is determined by various cis-acting RNA elements in pre-mRNA and many trans-acting protein factors [Citation51]. To elucidate the sex-specific AS regulation of doublesex gene in the silkworm and in important agricultural pests of Lepidoptera, it is necessary to identify Bm-dsx RNA binding proteins that would function as splicing trans-acting factors. In this study, we identified three BxRBPs that bind to exon 3 or exon 4 RNA of the silkworm doublesex and found two of them are important for AS regulation. Based on our data, we propose a cooperative model for AS regulation of Bm-dsx in the silkworm sex-determination pathway (). BxRBP1 and BxRBP3 bind to sex-specific exons 3 and 4 of Bm-dsx, respectively, and cooperatively function as trans-acting repressors to inhibit the spliceosomal recognition of these two exons, resulting in male-specific splicing of Bm-dsx. In addition, in the absence of the primary sex-determination signal piRNA that is located on the female W chromosome, the upstream Masc gene is expressed in the male silkworm and stimulates transcription of BxRBP3-A and B isoforms, resulting in inhibition of splicing of Bm-dsx exons 3 and 4 in males.
Figure 7. A proposed model for sex-determination regulation in B. mori.
In the absence of the primary sex-determination signal piRNA on W chromosome, Masc facilitates the expression of BxRBP3 isoforms, which can bind exon 4 of Bm-dsx. BxRBP3 isoforms function together with BxRBP1 that bind exon 3 of Bm-dsx, efficiently inhibit the female splicing of Bm-dsx, and induce male-specific splicing. BxRBP2, lacking an NLS, binds exon 4 of Bm-dsx in the cytoplasm.
In the well-known sex-determination pathway of Drosophila melanogaster, AS of doublesex (Dm-dsx) is regulated by Tra and Tra-2, which bind to exon 4 of Dm-dsx in the female fruit fly and function as trans-acting splicing activators [Citation52,Citation53]. However, homologues of tra have not been found in silkworm and other Lepidoptera; and it has been shown that tra-2 homolog in silkworm is not involved in the regulation of AS of Bm-dsx [Citation54]. Therefore, AS regulation of doublesex genes in the fruit fly and the silkworm (Lepidoptera) is different [Citation31]. The model supported by our data suggests a distinctly different mechanism of regulation of the sex-specific exon(s) of doublesex genes in the two insects, implying an evolutionary divergence between these two species.
The three RNA-binding proteins of Bm-dsx identified here are highly conserved from insects to mammals. As described above, their homologous proteins have been characterized as splicing trans-acting activators or repressors in many species [Citation40,Citation41,Citation44,Citation45,Citation49,Citation50]. In comparison to their homologues, BxRBP2 is special in having lost its NLS and thus cannot be imported into the nucleus. Therefore, it has no effective function as a splicing regulator. We hypothesize that BxRBP2 might regulate translation or other events of the female Bm-dsx mRNA in the cytoplasm, but not the splicing of exon 4 in the Bm-dsx (). Homologues of BxRBP2 without an NLS can only be found in the Lepidoptera, not other insects or mammals (Fig. S7A upper). In the silkworm, we found two BxRBP2-like proteins, BxRBP2-L1 and BxRBP2-L2, which have NLS sequences (Fig. S7A lower) and can be localized in the nucleus (Fig. S7B). We over-expressed them in the BmF cells, and found that they did not change AS of Bm-dsx in the BmF cells with or without OE of BxRBP1 (Fig. S7C), unlike the previously described effects of BxRBP3-B. Taken together, these data implied that lack of an NLS, BxRBP2 might have a different or additional regulatory function for doublesex gene in the Lepidoptera.
In the BmM cells, knockdown of BxRBPs and Masc did not change AS of the Bm-dsx and the expression of BxRBP3, respectively (). These results imply that other factors may be involved in the AS regulation of Bm-dsx, although our insufficient efficiency of RNAi silencing could be another possibility. In addition, individual over-expression of BxRBP1 or BxRBP3 in the BmF cells is sufficient to skip exon 3 or 4 of the Bm-dsx mini-gene, which has intact exons but shorter introns ( & ), suggesting that the cooperative function between BxRBP1 and BxRBP3 is no-longer necessary. Taken together, there may be other factor(s), which could interact with intron sequence of Bm-dsx and play a key role in the regulation of Bm-dsx AS.
Materials and methods
Silkworm culture and sample collection
R01 and P50 strains were cultured by standard methods [Citation35]. Embryonic samples were collected from R01 strain. Larval, pupal, and adult samples were collected from P50 strain. Silkworm individuals and tissues were separated and collected by gender, frozen in liquid nitrogen, and stored at −70°C.
Cell line culture
To establish sex-specific embryonic cell lines, primary cell cultures were initiated from silkworm embryos after 5 days of the termination of diapauses using an R01 strain, in which the egg shells of females are dark blue and those of males are pink (Fig. S4A). Approximately 25 embryos without an eggshell of each gender were cultured at 27°C in a 25 cm2 flask with TNM-FH medium (Sigma) plus 10% fetal bovine serum (GIBCO) and antibiotics penicillin-streptomycin (GIBCO). To obtain enough cells for subculture, half of the primary culture medium was changed every two weeks. Cells for subcultures were dissociated by treatment with 0.25% Trypsin-EDTA solution (GIBCO) and transferred to a new flask at a split ratio of 1:2. This process was continued ~3 years to establish two stable cell lines, which were named BmF cells from the female embryos and BmM cells from the males. The doubling time of both cell lines is approximately 3 days. The BmN cell line was cultured by standard methods [Citation33], and the HEK293T cell line was obtained from ATCC and cultured in DMEM (GIBCO) with 10% fetal bovine serum at 37°C with 5% CO2.
Yeast three-hybrid screens and yeast two-hybrid (Y2H) assay
The yeast three-hybrid system, including YBZ-1 strain and a pIIIA/MS2-2 plasmid, was provided by Prof. Marvin Wickens at University of Wisconsin–Madison. cDNA libraries from the silkworm testis and R01 strain embryos were prepared in pDEST32 (Invitrogen) and pGADT7 vector (Oebiotech), respectively. All six yeast three-hybrid screens were performed according to the previously described protocol [Citation34] with the following modifications. The first-step selection was carried out on – His media with 0.5 mM 3-aminotriazole (AT) for embryo cDNA libraries and without 3-AT for testis cDNA library. After the second-step selection by assays for β-galactosidase activity, plasmids with MS2-ligated dsx exon 3 or 4 was replaced by control MS2-only plasmid using 0.1% 5-FOA media to exclude false-positive clones. Quantitative β-galactosidase assays were performed as described [Citation34]. For Y2H assay, coding sequences of RBPs were individually cloned into pGADT7 prey vector or pGBKT7 bait vector and then transformed into Y2HGold yeast strain. The Y2H assay was performed according to the manufacturer’s instructions (Matchmaker® Gold Yeast Two-Hybrid System, Clontech).
Total RNA isolation and RT-PCR
Total RNA was extracted using Trizol (ThermoFisher) and reverse transcriptions were performed using RevertAid First Strand cDNA Synthesis kit (ThermoFisher). PCRs were performed using 2× Hieff™ PCR Master Mix (Yeasen) with programs containing 21–35 cycles (58°C for annealing). Primers used for RT-PCR are listed in Table S1.
5′- and 3′-RACE
The 5′ and 3′ terminal sequences of BxRBPs in silkworm samples were obtained using 5′-Full RACE Kit and 3′-Full RACE Core Set Ver.2.0 (TaKaRa), respectively. Primers used are listed in Table S1. Nested PCR programs for RACE were performed in 25 cycles with outer primers, followed by 32 cycles with inner primers using KOD-Plus-Neo DNA polymerase (TOYOBO) with additional 5% dimethyl sulfoxide. PCR products were then purified by DNA gel extraction kit (Axygen) and cloned into pMD18-T simple vector (TaKaRa) for sequencing. Obtained sequences were compared by BLAST against the SilkDB (Xia et al, 2009) to confirm their novelties. Encoded proteins and homologs were aligned and analyzed by DNAman (LynnonBiosoft). Novel sequences in this study have been deposited to GenBank under accession numbers from MH745573 to MH745580.
Over-expression and RNAi knockdown
For over-expression of proteins containing V5 and 6xHis C-terminal tags, full-length coding sequences of BxRBP1, BxRBP2, BxRBP3, and Fem piRNA-resistant Masc [Citation11] were cloned into the pIZT/V5-his vector (Invitrogen). For RNAi knockdown of each target gene, two different short interfering RNAs (siRNAs) were used (sequences listed in Table S1). siRNAs were synthesized by GenePharma Corp and dissolved in RNase-free water (GIBCO) for storage at −80°C. Transfections were performed using TransIT-Insect Transfection Reagent (Mirus) according to the manufacturer’s instructions. Cells were then collected at 72 hr after transfection for further analyses. Protein signals were then detected by western blotting using anti-Masc polyclonal antibody (Angobiotech)
Subcellular localization
For detecting the subcellular localization of the RBPs, EGFP was fused to the C-terminus of RBPs in a GFP-minus pIZT/V5-his vector. BmF cells were transferred onto coverslips after 2 days of transfection and fixed for 15 min with 4% paraformaldehyde in PBS, and permeabilized for 20 min in 0.1% Triton X-100 in PBS. Nuclei then were counterstained with DAPI (ThermoFisher). Images were captured at room temperature using a Nikon ECLIPSE Ni-U microscope with NIS-Elements Documentation software.
Recombinant protein purification
Coding sequences were individually cloned into pGEX-4T-1 (GST tag) or pET-28a vector (6xHis tag). Recombinant proteins were induced by 1.0 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and expressed in E. coli BL21 for 24 hr at 16°C and then purified by either glutathione-Sepharose (GE Healthcare) or Ni agarose (Qiagen) chromatography under standard conditions followed by dialysis against buffer D (20 mM Tris-HCl at pH 7.9, 0.2 mM EDTA, 100 mM KCl, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 20% glycerol).
In vitro protein–protein interaction
Purified 6xHis-tagged protein (10 pmol) and GST-tagged protein (40 pmol) were incubated together in 600 µL with binding buffer (20 mM Tris–HCl at pH 7.9, 150 mM NaCl, 1mM EDTA, 0.2% Triton X-100, 0.5 mM PMSF) for 4 hr at 4°C with glutathione-Sepharose. Bead pellets were washed five times with wash buffer (50 mM Tris-HCl at pH 7.9, 140 mM NaCl, 1 mM EDTA, 0.1% Triton X-100) and then resuspended in 50 µL of sample loading buffer for SDS-PAGE. For detecting the interaction between BxRBP3-B and PSI, 1 × 107 BmM cells were treated by lysis buffer (10 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.2 mM sodium ortho-vanadate, 0.2 mM PMSF, 1% Triton X-100, 0.5% NP-40), then incubate with 10 µg GST-tagged BxRBP3-B protein for 4 hr at 4°C with glutathione-Sepharose. Protein signals were then detected by western blotting using anti-GST (GST-2) monoclonal antibody (Sigma), anti-BxRBP1 and anti-PSI polyclonal antibody (Angobiotech).
Mini-gene assay
The Bm-dsx mini-gene, the same structure as previous described [Citation33], was cloned into pIZT/V5-his vector. For a higher expression level in HEK293T cell line, Masc was cloned into pcDNA3 vector. Mini-gene and RBPs or Masc were co-transfected into HEK293T cells by using Attractene Transfection Reagent (Qiagen) according to the manufacturer’s instructions. Cells were then collected at 48 hr after transfection. Protein signals were then detected by western blotting using anti-V5-HRP antibody or anti-tubulin (Sigma).
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Supplemental Material
Download MS Word (2.3 MB)Acknowledgments
We thank C. Query at Albert Einstein College of Medicine for critical reading and discussion of the manuscript, and other members in Xu lab for data entries and discussions. We thank Prof. Marvin Wickens at University of Wisconsin–Madison for providing the yeast three-hybrid system.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bachtrog D, Mank JE, Peichel CL, et al. Sex determination: why so many ways of doing it? PLoS Biol. 2014;12:e1001899.
- Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7:42–54.
- Salz HK. Sex determination in insects: a binary decision based on alternative splicing. Curr Opin Genet Dev. 2011;21:395–400.
- Shukla JN, Nagaraju J. Doublesex: a conserved downstream gene controlled by diverse upstream regulators. J Genet. 2010;89:341–356.
- Rao SR, Ali S. Insect sex chromosomes. VI. A presumptive hyperactivation of the male X chromosome in Acheta domesticus (L.). Chromosoma. 1982;86:325–339.
- Erickson JW, Quintero JJ. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 2007;5:e332.
- Lucchesi JC, Kuroda MI. Dosage compensation in Drosophila. Cold Spring Harbor Perspect Biol. 2015;7:a019398.
- Hall AB, Basu S, Jiang X, et al. Sex determination. A male-determining factor in the mosquito Aedes aegypti. Science. 2015;348:1268–1270.
- Sharma A, Heinze SD, Wu Y, et al. Male sex in houseflies is determined by Mdmd, a paralog of the generic splice factor gene CWC22. Science. 2017;356:642–645.
- Beye M, Hasselmann M, Fondrk MK, et al. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114:419–429.
- Kiuchi T, Koga H, Kawamoto M, et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature. 2014;509:633–636.
- Bopp D, Bell LR, Cline TW, et al. Developmental distribution of female-specific sex-lethal proteins in Drosophila melanogaster. Genes Dev. 1991;5:403–415.
- Inoue K, Hoshijima K, Sakamoto H, et al. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990;344:461–463.
- Lynch KW, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101.
- Saccone G, Salvemini M, Polito LC. The transformer gene of Ceratitis capitata: a paradigm for a conserved epigenetic master regulator of sex determination in insects. Genetica. 2011;139:99–111.
- Shukla JN, Palli SR. Sex determination in beetles: production of all male progeny by parental RNAi knockdown of transformer. Sci Rep. 2012;2:602.
- Hasselmann M, Gempe T, Schiott M, et al. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature. 2008;454:519–522.
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010.
- Fu G, Condon KC, Epton MJ, et al. Female-specific insect lethality engineered using alternative splicing. Nat Biotechnol. 2007;25:353–357.
- Jin L, Walker AS, Fu G, et al. Engineered female-specific lethality for control of pest Lepidoptera. ACS Synth Biol. 2013;2:160–166.
- Abe H, Fujii T, Tanaka N, et al. Identification of the female-determining region of the W chromosome in Bombyx mori. Genetica. 2008;133:269–282.
- Fujii T, Shimada T. Sex determination in the silkworm, Bombyx mori: a female determinant on the W chromosome and the sex-determining gene cascade. Semin Cell Dev Biol. 2007;18:379–388.
- Ohbayashi F, Suzuki MG, Mita K, et al. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Comp Biochem Physiol. 2001;128:145–158.
- Goldsmith MR, Shimada T, Abe H. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol. 2005;50:71–100.
- Suzuki MG, Imanishi S, Dohmae N, et al. Establishment of a novel in vivo sex-specific splicing assay system to identify a trans-acting factor that negatively regulates splicing of Bombyx mori dsx female exons. Mol Cell Biol. 2008;28:333–343.
- Suzuki MG, Imanishi S, Dohmae N, et al. Identification of a male-specific RNA binding protein that regulates sex-specific splicing of Bmdsx by increasing RNA binding activity of BmPSI. Mol Cell Biol. 2010;30:5776–5786.
- Marec F. Developmental genetics: female silkworms have the sex factor. Nature. 2014;509:570–571.
- Katsuma S, Sugano Y, Kiuchi T, et al. Two conserved cysteine residues are required for the masculinizing activity of the silkworm Masc protein. J Biol Chem. 2015;290:26114–26124.
- Sugano Y, Kokusho R, Ueda M, et al. Identification of a bipartite nuclear localization signal in the silkworm Masc protein. FEBS Lett. 2016;590:2256–2261.
- Katsuma S, Kiuchi T, Kawamoto M, et al. Unique sex determination system in the silkworm, Bombyx mori: current status and beyond. Proc Jpn Acad Ser B Phys Biol Sci. 2018;94:205–216.
- Suzuki MG, Ohbayashi F, Mita K, et al. The mechanism of sex-specific splicing at the doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem Mol Biol. 2001;31:1201–1211.
- Baker BS. Sex in flies: the splice of life. Nature. 1989;340:521–524.
- Wang XY, Zheng ZZ, Song HS, et al. Conserved RNA cis-elements regulate alternative splicing of Lepidopteran doublesex. Insect Biochem Mol Biol. 2014;44:1–11.
- Stumpf CR, Opperman L, Wickens M. Chapter 14. Analysis of RNA-protein interactions using a yeast three-hybrid system. Methods Enzymol. 2008;449:295–315.
- Shao W, Zhao QY, Wang XY, et al. Alternative splicing and trans-splicing events revealed by analysis of the Bombyx mori transcriptome. Rna. 2012;18:1395–1407.
- Wang ZL, Li J, Xia QY, et al. Identification and expression pattern of Bmlark, a homolog of the Drosophila gene lark in Bombyx mori. DNA Sequence. 2005;16:224–229.
- Suetsugu Y, Futahashi R, Kanamori H, et al. Large scale full-length cDNA sequencing reveals a unique genomic landscape in a lepidopteran model insect, Bombyx mori. G3. 2013;3:1481–1492.
- Xia Q, Guo Y, Zhang Z, et al. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science. 2009;326:433–436.
- Sakurai H, Izumi S, Tomino S. In vitro transcription of the plasma protein genes of Bombyx mori. Biochim Biophys Acta. 1990;1087:18–24.
- Su CH, Hung KY, Hung SC, et al. RBM4 regulates neuronal differentiation of mesenchymal stem cells by modulating alternative splicing of pyruvate kinase M. Mol Cell Biol. 2017;37:e00466–16.
- Lin JC, Tarn WY. RBM4 down-regulates PTB and antagonizes its activity in muscle cell-specific alternative splicing. J Cell Biol. 2011;193:509–520.
- Lin JC, Tarn WY. Exon selection in alpha-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol Cell Biol. 2005;25:10111–10121.
- Dhananjaya D, Hung KY, Tarn WY. RBM4 modulates radial migration via alternative splicing of Dab1 during cortex development. Mol Cell Biol. 2018;38:e00007-18.
- Ayala YM, Pagani F, Baralle FE. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 2006;580:1339–1344.
- Dujardin G, Buratti E, Charlet-Berguerand N, et al. CELF proteins regulate CFTR pre-mRNA splicing: essential role of the divergent domain of ETR-3. Nucleic Acids Res. 2010;38:7273–7285.
- Buratti E, Brindisi A, Pagani F, et al. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325.
- Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887.
- Lukavsky PJ, Daujotyte D, Tollervey JR, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20:1443–1449.
- Oas ST, Bryantsev AL, Cripps RM. Arrest is a regulator of fiber-specific alternative splicing in the indirect flight muscles of Drosophila. J Cell Biol. 2014;206:895–908.
- Spletter ML, Barz C, Yeroslaviz A, et al. The RNA-binding protein Arrest (Bruno) regulates alternative splicing to enable myofibril maturation in Drosophila flight muscle. EMBO Rep. 2015;16:178–191.
- Fu XD, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701.
- Salz HK, Erickson JW. Sex determination in Drosophila: the view from the top. Fly (Austin). 2010;4:60–70.
- Verhulst EC, van de Zande L, Beukeboom LW. Insect sex determination: it all evolves around transformer. Curr Opin Genet Dev. 2010;20:376–383.
- Suzuki MG, Suzuki K, Aoki F, et al. Effect of RNAi-mediated knockdown of the Bombyx mori transformer-2 gene on the sex-specific splicing of Bmdsx pre-mRNA. Int J Dev Biol. 2012;56:693–699.
