ABSTRACT
Circular RNAs (circRNAs) have a critical role in the control of gene expression. Their function in spermatozoa (SPZ) is unknown to date. Twenty-eight genes, involved in SPZ/testicular and epididymal physiology, were given in circBase database to find which of them may generate circular transcripts. We focused on circNAPEPLDiso1, one of the circular RNA isoforms of NAPEPLD transcript, because expressed in human and murine SPZ. In order to functionally characterize circNAPEPLDiso1 as potential microRNA (miRNA) sponge, we performed circNAPEPLDiso1-miR-CATCH and then profiled the expression of 754 miRNAs, by using TaqMan® Low Density Arrays. Among them, miRNAs 146a-5p, 203a-3p, 302c-3p, 766-3p and 1260a (some of them previously shown to be expressed in the oocyte), resulted enriched in circNAPEPLDiso1-miR-CATCHed cell lysate: the network of interactions generated from their validated targets was centred on a core of genes involved in the control of cell cycle. Moreover, computational analysis of circNAPEPLDiso1 sequence also showed its potential translation in a short form of NAPEPLD protein. Interestingly, the expression analysis in murine-unfertilized oocytes revealed low and high levels of circNAPEPLDiso1 and circNAPEPLDiso2, respectively. After fertilization, circNAPEPLDiso1 expression significantly increased, instead circNAPEPLDiso2 expression appeared constant.
Based on these data, we suggest that SPZ-derived circNAPEPLDiso1 physically interacts with miRNAs primarily involved in the control of cell cycle; we hypothesize that it may represent a paternal cytoplasmic contribution to the zygote and function as a miRNA decoy inside the fertilized oocytes to regulate the first stages of embryo development. This role is proposed here for the first time.
Introduction
Endocrine, paracrine and autocrine communications along the hypothalamus-pituitary-gonad (HPG) axis orchestrate spermatogenesis [Citation1–Citation3], a succession of spermatogonia self-renewal, meiosis and post-meiotic maturation events (spermiogenesis), which allows the production of highly specialized cells such as spermatozoa (SPZ). SPZ are generally considered transcriptionally and translationally inert cells, which lose most of their cytoplasm and enclosed RNA molecules during spermiogenesis. SPZ contain a large repertoire of small non-coding RNAs (ncRNAs), such as transfer RNA-derived small RNAs (tsRNAs) and microRNAs (miRNAs), which have long been considered ‘junk’ molecules with no biological functions. Recent discoveries have suggested an important biomolecular role of SPZ-derived RNA cargo as means of conveying paternal features to the offspring, such as obesity, metabolic disorders, and stress, thus affecting its reproductive health and neurological or behavioural status [Citation4]. Such a mechanism is defined as a trans-generational epigenetic inheritance [Citation5,Citation6].
In the scenario of ncRNAs, notable attention has been gained by the competitive endogenous RNA (ceRNA) hypothesis, according to which RNAs, as long non-coding RNAs (lncRNAs) or circular RNAs (circRNAs), may function as decoys to sequester RNA binding proteins (RBPs) or miRNAs: this latter mechanism would repress miRNA activity, therefore allowing mRNA targets to escape degradation [Citation7–Citation14]. Accordingly, circRNAs may be considered an important group of post-transcriptional regulators. Interestingly, over two decades ago the first circRNA was discovered from the testis-determining gene Sry (sex-determining region Y) [Citation15]. Recently, about 16,000 circRNAs have been identified in human testis, making this organ only second to the brain in circRNA content [Citation16]: this is probably due to the need of accurate gene expression regulation during spermatogenesis. Testis-derived circRNAs have also been detected in human seminal plasma, and a fine analysis of circRNAs in isolated mouse spermatogenic cells has also been carried out [Citation17]. However, to date, neither indication about a possible SPZ-derived circRNA cargo has been provided in mammals, nor what may be the contribution of such circRNA cargo in fertilization and embryo development is known. An impressive work by Dang and co-workers [Citation18] paints a complete landscape of embryo transcriptome, including circRNAs, during human pre-implantation development. Interestingly, the comparison between human and murine embryos reveals both high conservation, but also a clear distinction between these two species, since human embryos generate more types of circRNAs than the murine ones [Citation18]. It is important to note that circRNA cargo in human embryos has mainly been ascribed to maternal gene contribution, since most of the circRNAs are present before fertilization and persist during pre-implantation development. What may be a paternal contribution in such a context is to date unexplored.
With this in mind, we investigated the presence of circRNAs in both human and mouse SPZ, starting from a list of 28 linear transcripts known to be involved in SPZ/testicular physiology or epididymal function. Among them, we focused on circNAPEPLDiso1 and explored its ability to tether miRNAs. Accordingly, we predicted the circRNA-associated ceRNA network (ceRNET) potentially regulated by SPZ circNAPEPLDiso1 through in silico analysis and speculated the possibility that circNAPEPLD may be transmitted from SPZ to oocytes during fertilization.
Results
Candidate circRNA expression in human and murine SPZ and tissues
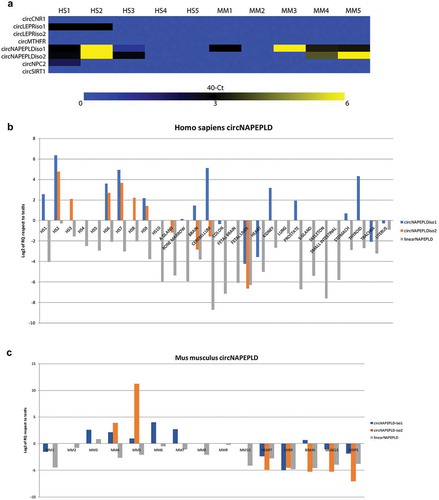
We profiled the expression of eight candidate circRNAs (see Materials and Methods, and ), generated from six linear transcripts (CNR1, LEPR, MTHFR, NAPEPLD, NPC2, SIRT1) in five RNA samples from human (HS) and murine (MM) SPZ, respectively, through qRT-PCR ()). Only the two circular isoforms of NAPEPLD (that we named circNAPEPLDiso1 and circNAPEPLDiso2) were found to be largely expressed in both HS and MM SPZ. In order to evaluate circNAPEPLDiso1 and circNAPEPLDiso2’s cell-type and tissue specificity, we assayed their expression in a panel of 19 and 6 fresh-frozen HS and MM tissues, respectively, plus 10 RNA samples from HS and MM SPZ (,c)). CircNAPEPLDiso1 and circNAPEPLDiso2 were expressed at low levels in testis and at high levels in SPZ; however, this expression trend was heterogeneous in the 10 samples analysed. circNAPEPLDiso1 was expressed only in few analysed HS tissues (i.e. brain, cerebellum, kidney, prostate, stomach and thyroid), whereas circNAPEPLDiso2 was not expressed or was expressed at very low levels in the analysed tissues (). CircNAPEPLDiso1 and circNAPEPLDiso2 were expressed at high levels in MM SPZ but, also in this case, we observed heterogeneity of expression among the 10 samples analysed. Both circular and linear isoforms were strongly downregulated in MM adipose, brain, heart, liver and muscle tissue samples (). This parallel expression analysis allowed us to conclude that both isoforms of circNAPEPLD showed similar expression levels in HS and MM SPZ and tissues. Furthermore, circNAPEPLDiso1 and circNAPEPLDiso2 expression appeared to be independent of the linear NAPEPLD, which was downregulated in all samples, compared to the HS testis, where this mRNA was highly expressed ().
Table 1. Linear transcripts and HS circRNAs retrieved from circBase. Linear transcript, circRNA circBase ID, circRNA genomic position, circRNA genomic length and circRNA spliced length are reported in columns in the table.
Table 2. Linear transcripts and MM circRNAs retrieved from circBase. Linear transcript, circRNA circBase ID, circRNA genomic position, circRNA genomic length and circRNA spliced length are reported in columns in the table.
Figure 1. Expression of candidate circRNAs (a), circNAPEPLD and linear NAPEPLD in HS (b) and MM (c) SPZ and tissues. (a) Heat Map showing the expression of the six candidate circRNAs. Data are shown as 40 – Ct. (b and c) Bar chart showing the expression values relating to NAPEPLD circular and linear isoforms in Homo sapiens and Mus musculus samples, respectively. RNA from testis was used as calibrator. Expression is shown as Log2 of RQ compared to testis.
circNAPEPLDiso1 physically interacts with miRNAs
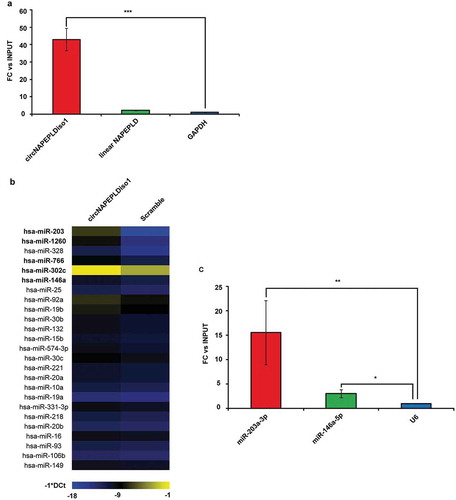
Following miR-CATCH experiments, we specifically pulled down circNAPEPLDiso1, as shown in . Mir-CATCH experiments (see Materials and Methods) followed by TaqMan® Low Density Array (TLDA) qRT-PCR analyses highlighted the enrichment (fold-changes more or equal than 2.5- see Materials and Methods) of five miRNAs (miR-203a-3p, miR-1260a, miR-766-3p, miR-302c-3p, miR-146a-5p) in circNAPEPLDiso1-miR-CATCHed with respect to scramble-miR-CATCHed cell lysates . Among them, miR-203a-3p and miR-146a-5p appeared to be the most and the least enriched miRNAs in circNAPEPLDiso1-miR-CATCHed samples, respectively. The enrichment of these two miRNAs was confirmed by single TaqMan® miRNA assays (). The number of miRNA responsive elements (MREs) within the circNAPEPLDiso1 sequence, retrieved by BLAST analysis for each candidate miRNA, is shown in .
Table 3. miRNAs interacting with circNAPEPLDiso1 as revealed by TLDA analysis. MiRNAs are ordered following decreasing LOG fold-change (FC) values (N/A = not annotated).
Figure 2. circNAPEPLDiso1 miR-CATCH. (a) Fold enrichment (FC) of circNAPEPLDiso1, linear NAPEPLD and U6 are shown as IPed samples/input. U6 FC was set to one and FC of circNAPEPLDiso1 and linear NAPEPLD mRNA were calculated accordingly (*** p-value, < 0.0001; N = 3, Student’s t-test). (b) Heat Map showing the expression of miRNAs enriched in circNAPEPLDiso1-miR-CATCHed samples as compared to negative control (Scramble-miR-CATCHED). Data are reported as −1*DCt and U6 was used as endogenous control. MiRNAs with FC ≥2.5 (TLDA data) and predicted to have at least one binding site within the circNAPEPLDiso1 sequence are shown in bold. (c) Fold enrichment (FC) of miR-203a-3p, miR-146a-5p and U6 are shown as IPed samples/input. U6 FC was set to one, and FC of miR-203a-3p and miR-146a-5p were calculated accordingly (**p-value, < 0.01; *p-value, < 0.05; N = 3, Student’s t-test).
In silico prediction of pathways regulated by circNAPEPLDiso1’s interacting miRNAs
The gene interaction network generated on the basis of the validated targets of the five miRNAs tethering circNAPEPLDiso1 was made of 1659 nodes and 1788 edges. This network was significantly enriched in genes involved in the pathway of cancer (p-value = 0.00000596) (). Centrality analysis revealed that this network was centred on 131 nodes linked through 260 edges (betweenness cut-off = 5). Hub genes were significantly associated with cell cycle (p-value = 0.05) ().
Table 4. Top 10 pathways enriched within miRNA targets interaction network.
Table 5. Top 10 pathways enriched within miRNA targets interaction network filtered for betweenness centrality (cut-off = 5).
circNAPEPLDiso1 and circNAPEPLDiso2 expression in murine oocytes and one-cell zygotes
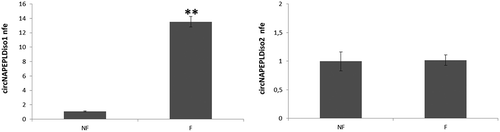
With the aim to hypothesize a paternal contribution in circNAPEPLD transmission to oocytes we analysed the expression level of both circular isoforms in murine-unfertilized oocytes (NF) and in one-cell zygotes (F), making sure that male and female pronuclei were not fused. Interestingly, the expression level of circNAPEPLDiso1 was very low in NF and significantly increased after fertilization (p-value <0.01, ). Instead, circNAPEPLDiso2 was expressed at a higher level in NF and remained constant in F ().
Figure 3. Expression analysis of circNAPEPLDiso1 (a) and circNAPEPLDiso2 (b) in murine-unfertilized oocytes (NF) and in one-cell zygotes (F) by qRT-PCR. N = 5 pools each containing 10 NF and N = 5 pools each containing 10 F were used for the expression analysis. Data are expressed as normalized fold expression (nfe) using Actb as endogenous control (**p-value, < 0.01 Student’s t-test).
circNAPEPLDiso1 is predicted to interact with the Eukaryotic Translation Initiation Factor 4A3 (EIF4A3) and to be translated into a functional protein
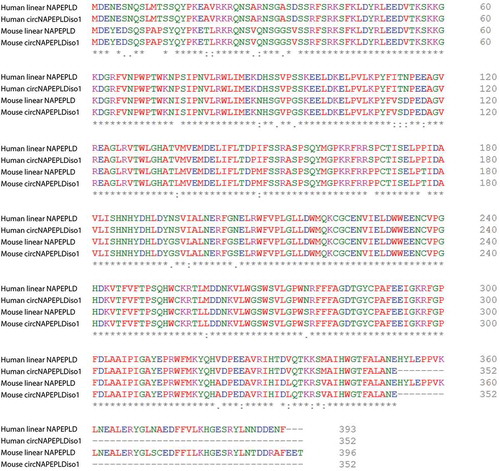
Querying circInteractome database revealed that circNAPEPLDiso1 has 11 binding sites for EIF4A3. In order to evaluate circNAPEPLDiso1’s potential translatability into a protein, we checked for predicted Internal Ribosomes Entry Sites (IRESs) within its sequence by querying circRNADb database (http://202.195.183.4:8000/circrnadb/circRNADb.php) [Citation19]. Through this analysis, we identified two IRESs (at positions: 294–436 and 892–1039) and a predicted ORF (Open Reading Frame) within the sequence of HS and MM circNAPEPLDiso1. We compared the protein sequences (352 amino acids/aa) predicted to be synthesized by the HS and MM ORFs with those encoded by the linear transcripts (393 and 396 aa, in Homo sapiens and Mus musculus, respectively) by using Clustal Omega (). We found that these sequences differ only for C-terminus length, since the predicted ORF within circNAPEPLDiso1 encodes for a shorter peptide (length 352 aa). In particular, there are 41 and 44 aa lacking at C-terminus of the protein encoded by HS and MM circNAPEPLDiso1, respectively (). However, these differences did not alter the functional domains of NAPEPLD original structure, as predicted by SMART database (http://smart.embl-heidelberg.de/).
Discussion
Since their discovery more than 30 years ago [Citation20], circRNAs have been characterized in several mammalian cells, and their biomolecular functions are being characterized in an increasing number of biological systems [Citation21]. The main functions described to date comprise: (1) sponging activity for miRNAs and other RNAs; (2) sequestering RBPs; (3) ability to encode for a protein through a cap-independent and IRES-dependent mechanisms; (4) competition with linear mRNA counterparts through autoregulatory loops. Even though many aspects of circRNAs’ biology (i.e. developmental stage-specific expression, subcellular localization, turnover, degradation, contribution to trans-generational epigenetic inheritance) have been described, a full comprehension of their biomolecular functions remains to be gathered [Citation22]. New computational strategies allowed a more detailed exploration of high-throughput sequencing data and paved the way to new perspectives in the study of circRNAs [Citation23]. Currently, circRNAs represent the largest RNA group within the human transcriptome, especially in brain and testis within which researchers have identified 65,731 [Citation24] and 15,996 circRNAs [Citation16], respectively. Their discovery in different species, their stability and resistance to exonucleases, both in cells and in serum exosomes, reliably suggest important biological functions of circRNAs.
CircRNAs have also been identified in individual human oocytes and pre-implantation embryos, implying their potential functions in oogenesis and reproduction [Citation18]. Interestingly, the contribution of SPZ to the circRNAs’ landscape is still an open question: only in the nematode Caenorhabditis elegans, they have been characterized in oocytes, SPZ, and all embryonic developmental stages [Citation25]. Based on this observation, we investigated the presence of circRNAs in both HS and MM SPZ. Among candidate circRNAs, only circNAPEPLDiso1 and circNAPEPLDiso2 were found to be largely expressed in both HS and MM SPZ.
NAPEPLD is one of the main actors of the endocannabinoid system, involved in the biosynthesis of the endocannabinoid anandamide (AEA) [Citation26], with important roles in reproduction [Citation27–Citation31]. AEA levels are spatio-temporally regulated in the uterus during early pregnancy, so that they may mediate reciprocal interaction between blastocysts and uterus – with lower levels in the receptive uterus and at the implantation site [Citation32,Citation33].
Compared to linearNAPEPLD, which is highly expressed in testis, HS and MM circNAPEPLDiso1 and circNAPEPLDiso2 appeared highly expressed in SPZ with respect to the panel of tissues assayed, even if with a heterogeneous trend. This result suggests that it may exist an individually variable transmission of epigenetic traits, as it is known for genetic traits.
CircRNA sponge activity has been demonstrated in mammalian testis, since the testis-specific circRNA SRY is able to bind to mir-138 in mice [Citation8]. In line with these results, we investigated the possibility that circNAPEPLDiso1 may act as sponge by retrieving miRNAs physically interacting with it through a miR-CATCH approach (see Materials and Methods). Interestingly, we identified five miRNAs physically interacting with circNAPEPLDiso1, many of which are known to be expressed in HS oocytes, blastocysts and blastocoel fluid (see ) [Citation34–Citation43] and whose targets are involved in pathways related to cancer and cell cycle (see and ). Therefore, it is conceivable that SPZ-derived circNAPEPLD may function as a decoy to inhibit the anti-proliferative activity of some oocyte-derived miRNAs, thus allowing cell proliferation, a massive event during the first stages of embryo development. Of course, more experimental work is needed to further explore this hypothesis.
Overall, these data confirm the hypothesis of an active function performed by the circNAPEPLDiso1/miR-203a-3p, miR-1260a, miR-766-3p, miR-302c-3p, miR-146a-5p/miRNA targets axis, in the control of cell cycle and, probably, in the first phases of embryogenesis. Of course, this hypothesis needs further in vivo and in vitro studies in order to be validated. However, the first speculation of a paternal contribution in circNAPEPLD transmission has been here suggested. Interestingly, while circNAPEPLDiso2 was already expressed in NF at high levels and did not undergo a significant change after fertilization, circNAPEPLDiso1, instead, showed a scanty expression in NF and it significantly increased in F.
Intriguingly, circRNAs also regulate RBP functions by acting as competing elements, with a mechanism similar to their modulation of miRNAs [Citation14,Citation44]. CircRNAs bind to Argonaute [Citation25], MBL [Citation45], QKI [Citation46] and FUS [Citation47], and compete with full-length linear mRNA production through their association with RNA polymerase II machinery [Citation48]. Our data suggest that circNAPEPLDiso1 is also able to act as a decoy for eIF4A3 and therefore modulate gene expression in a post-transcriptional manner.
Unlike the general idea of circRNAs as non-coding RNAs, untranslatable because they are not associated with ribosomes, compelling evidence indicates that circRNAs can be translated with a cap-independent, IRES-dependent mechanism to recruit ribosomes [Citation49]. CircZNF609 is an example of a circRNA that is able to produce a nuclear-localized protein involved in myogenesis [Citation50]. Interestingly, N6-methyladenosine residues, enriched in circRNAs, are an input to drive circRNA translation [Citation51]. Our computational data suggest that both HS and MM circNAPEPLDiso1 are potentially translatable, with a predicted ORF coding for a 352 aa protein showing a shorter C-terminus, as compared to the protein encoded by the linear transcript. The biological functions of this predicted protein will be a target of future investigations. Moreover, similarly to our findings, a dual role was already reported for circSHPRH, which can act as miRNA sponge but also it encodes a novel short form of SHPRH protein (SHPRH-146 aa) [Citation52,Citation53].
CircRNAs’ history is still in its infancy. We hypothesize that these molecules may take part in the SPZ-derived RNA repertoire and mediate trans-generational epigenetic inheritance, together with several other non-coding RNAs. The inherent structure and stability of circRNAs make them good candidates to transmit paternal features to offspring and also make them potential diagnostic biomarkers to be incorporated into clinical practice.
Materials and methods
Cell culture
Human embryonic kidney cell line HEK293 was purchased from American Type Culture Collection (ATCC) and cultured at 37°C in a humidified CO2 incubator in complete high-glucose DMEM medium supplemented with 4 mM L-glutamine. The medium was renewed 2 times a week.
Human semen samples and their purification by Percoll density gradient centrifugation
Semen samples (n = 10) were gathered from healthy normozoospermic volunteer donors. Informed consent was obtained from each subject. All these experiments were performed in accordance with the relevant guidelines and regulations. After 3 days of sexual abstinence, semen samples were produced by masturbation and collected in sterile sample containers, which were delivered to the laboratory within 1 h of ejaculation. The sperm samples were allowed to liquefy for 30 min at 37°C and were then purified on a Percoll density gradient.
Purification of HS SPZ was achieved using a 40% and 80% discontinuous Percoll (GE Healthcare, Castle Hill, Australia) centrifugation gradient. For this procedure, Percoll (90 ml) was supplemented with 10 ml of Dulbecco phosphate-buffered saline (PBS) 10-fold concentrated solution (Lonza, Basel, Switzerland). The resulting solution (considered to be 100% Percoll) was further diluted with PBS 1x to give 40% and 80% Percoll solutions. The pH was equilibrated to 7.4. The gradient was prepared by placing 1 ml of each of the two solutions in a conical plastic tube 30 mm in diameter. The human semen sample (1 ml) was loaded at the top of the gradient and centrifuged at 300 g for 20 min. Following centrifugation, the seminal plasma and Percoll were removed and discarded. Purified SPZ were recovered from the base of the 80% Percoll fraction and washed once with 5 ml of PBS to remove the Percoll, followed by centrifugation at 500 g for a further 15 min and finally frozen at −80°C prior to processing for total RNA isolation. The different Percoll fractions were evaluated under light microscopy, in particular, the pellet containing SPZ with good morphology and motility, without other cellular contaminants.
Animals
CD1 adult male mice were purchased by Charles River (Charles River Laboratories, Lecco, Italy) and maintained under constant temperature (22°C) and lighting (12L:12LD), with food and water provided ad libitum. Before the sacrifice, food was removed from the cage at 5 p.m. and animals were killed the day after, between 9 and 11:30 a.m., under ether anaesthesia by cervical dislocation. Brain, testis, muscle, adipose tissue, liver and heart were collected.
Experiments were approved by the Italian Ministry of Education and the Italian Ministry of Health. Procedures involving animal care were carried out in accordance with the National Research Council’s publication Guide for Care and Use of Laboratory Animals (National Institutes of Health Guide).
Mouse semen samples and their purification by Percoll density gradient centrifugation
CD1 adult male mice (n = 10) were euthanized, their vasculature was perfused with pre-warmed PBS to minimize the possibility of blood contamination. The epididymes were then removed, separated from their fat and overlying connective tissue and carefully dissected. SPZ were recovered by placing the tissues in 6 ml PBS and cutting into a few pieces to let SPZ flow out from the ducts. Then, SPZ samples were filtered and the eluted solution was recovered from this buffer by Percoll density gradient centrifugation; 1 ml of the sperm cell suspension was layered above 3 ml of 27% Percoll (GE Healthcare), prepared by Percoll 100 and diluted with PBS 1x to obtain a 27% solution. The gradient was centrifuged at 700 g for 30 min at 25°C and SPZ collected from the pellet were washed by gentle centrifugation, 400 g for 20 min, and frozen at −80°C prior to processing for total RNA isolation. Quality control of MM purified SPZ samples was carried out by analysing possible non-sperm cell markers contaminants, such as CD4 (biomarker of leukocytes) and E-cadherin (biomarker of epithelial cells) [Citation54] (data not shown).
Identification of candidate SPZ-related circRNAs in Homo sapiens and Mus musculus
Based on the hypothesis that linear transcripts and their circular counterparts could have potential biological and molecular relationships [Citation55–Citation57], we chose genes having an important function in SPZ biology, whose linear transcripts are abundant in SPZ or testis, as hosts for candidate circRNAs. More specifically, we focused on components of the endocannabinoid system (CNR1, CNR2, FAAH, NAPEPLD) [Citation58–Citation62], modulators of protein folding (DNAJB1, DNAJB3 [Citation2,Citation63], HSP90AA1 [Citation64]; HSPA1B, HSPD1 [Citation65]), intragonadic regulators (AR, ESR1, ESR2, GNRH1, GNRHR, GPER1, KISS1, KISS1R) [Citation2,Citation3,Citation66,Citation67]; (LEP, LEPR) [Citation68]; MTHFR [Citation69], PIWIL1 [Citation70]; RARA [Citation71]; RXFP1 [Citation72]; SIRT1 [Citation73]] and epididymal markers (ADAM7 [Citation74],GLIPR1L1 [Citation75], NPC2 [Citation76], SPAM1 [Citation77]). Querying circBase database (http://www.circbase.org/), we found that only six of the previously selected linear RNAs had HS and MM homologous circular isoform counterparts. All the selected HS and MM circRNAs are listed in and , respectively.
RNA isolation
Total RNA was extracted from HS and MM SPZ, as well as from MM tissues, using Trizol® Reagent (Invitrogen Life Technologies, Paisley, UK), according to the manufacturer’s instructions. Total RNA was quantified by GenQuantPro spectrophotometer (Biochrom). To remove potential contamination of genomic DNA, RNA aliquots (10 µg) were treated with DNaseI (RNase-free DNase I, Ambion, Thermo Fisher Scientific, Massachusetts, USA) according to the manufacturer’s recommendations. A commercially available panel of total RNA from 18 HS tissues [Total RNA Master Panel II (BD Biotech Clontech, Palo Alto, CA)] was also used to assay the expression of candidate circRNAs.
PCR primer and oligoprobe design
Primers were designed through Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), and specific primers spanning the back-splicing junction were designed to amplify circRNAs. Primer sequences are shown in .
DNA antisense oligoprobe against circNAPEPLDiso1 (TTGGTGAAGAACTCATTTGCTA) spanned the backsplice junction, while a scrambled DNA oligoprobe (GTGAGGCGTTGTAAGAGTGGTTAAG) was used as not-targeting negative control (NC), as reported by De Santi C and Coll [Citation78]. All the oligos were modified with a 5ʹ biotin-TEG (triethyleneglycol) tail.
miRNA expression profile
TaqMan® Human MicroRNA Arrays v. 3.0 A and B (ThermoFisher Scientific, Waltham, MA USA) were used to amplify 754 miRNAs as previously described [Citation79]. Briefly, 160 ng of total RNA were reverse transcribed through Megaplex™ RT Primers, Human Pool Set v3.0 (ThermoFisher Scientific) and then pre-amplified through Megaplex™ PreAmp Primers, Human Pool Set v. 3.0 (ThermoFisher Scientific), according to manufacturer’s instructions. Diluted pre-amplification products were loaded into TaqMan® Human MicroRNA Arrays, according to manufacturer’s instructions. Only amplification curves with cycle thresholds (Ct) ≤33 were considered for downstream analyses. U6 was used as endogenous control, data were analysed through SDS v. 2.3 and RQ Manager v. 1.2, and reported as LOG 2−ΔΔCt: values ≥0.39 were considered as representative of a real enrichment of a specific miRNA in circNAPEPLDiso1-miR-CATCHed samples as compared to negative controls. Validation of data obtained by using TLDAs was performed through single TaqMan® assays. qRT-PCR reactions were run on a 7900HT Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA) by using TaqMan® Universal Master mix II (ThermoFisher Scientific).
Linear and circular RNA expression profile by qRT-PCR
The expression of candidate linear and circRNAs were assayed through qRT-PCR by using Power SYBR Green RNA-to-CT™ 1-Step kit (Applied Biosystems), according to the manufacturer’s instructions. All qRT-PCR reactions were performed by using 50 ng of total RNA on a 7900HT Fast Real Time PCR System (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and actin beta (Actb) were used as endogenous control for HS and MM samples, respectively. Expression fold changes were calculated according to the 2−∆∆Ct method [Citation80]. Testis was used as calibrator tissue, both for HS and MM samples.
miR-CATCH protocol
MiR-CATCH protocol was performed and data were analysed according to Hassan T and Coll. and to Vencken S and Coll [Citation81,Citation82]. with some modifications. Briefly, a pellet of 5x107HEK293 cells was lysed in lysis buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton, 0.1% Sodium Deoxycholate), supplemented with protease inhibitors (Sigma-Aldrich, Saint Louis, MO, USA), Phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich) and RNasin® Ribonuclease Inhibitors (Promega Italia, Milano, Italy), without any previous fixation in formaldehyde. Ten per cent of the cell lysate was used as input; the remaining volume was divided into two halves: each of them was incubated at 37°C for 90 min with DNA antisense oligoprobe against circNAPEPLDiso1 and scrambled DNA oligoprobe, previously immobilized to Dynabeads™ MyOne™ Streptavidin C1 (ThermoFisher Scientific), respectively. Finally, the beads were treated with proteinase K (Sigma-Aldrich) at 37°C for 1 h, incubated at 95°C for 10 min to reverse the interaction between the biotin-labelled DNA oligoprobe:circRNA:miRNA complexes and the magnetic beads. Samples were chilled on ice and RNA was purified by using Trizol, as previously described.
Vasectomy procedure
Male animals (N = 5) selected for the vasectomy procedure were weighed and immediately anaesthetized; a ventral laparotomy was performed, at the Alba Line, making a cut necessary for the exposure of a testis with an adjoining deferent duct. Subsequently, the deferent ducts were cauterized with a disposable thermocautery; once the organs were repositioned, the peritoneum and skin were sutured. At the end of the vasectomy procedure, the animals were kept on the plate at 37°C and monitored until complete awakening.
Zygotes collection. RNA isolation and qRT-PCR analysis
The day before the collection, the CD1 females (N = 10) in the oestrus phase were selected by direct observation and mated with fertile (N = 5) or vasectomized/sterile (N = 5) CD1 males following the scheme: 1 male with 1 female. The following morning the CD1 females with positive vaginal plug were enrolled for the experiment. Pregnant female mice at 0.5 dpc were humanely euthanized by cervical dislocation, placed on its back on absorbent paper and the abdomen was generously sprayed with 70% EtOH for disinfection. The uterus were drawn from one side by tweezing the cervix and cutting at the level of the caudal end of the cervix itself, from the other by cutting at the end of the uterine horn (infundibular portion). Then, a uterine horn was laid in a drop of Hyaluronidase from the bovine test (H4272 Sigma-Aldrich) in M16 medium (M7292 Sigma-Aldrich) 1 mg/ml, and the fertilization ampoule was broken with a needle so that the embryos and the cells surrounding them could easily get out. Then, oocytes were collected in a drop of PBS to wash them from mucous residues and blood. The fertilized oocytes or one-cell zygotes (F) were selected by observation to the stereomicroscope and those not fertilized were eliminated. Five pools (1 pool/female) each containing 10 F were used for the expression analysis. The oocytes from the CD1 females mated with vasectomized/sterile males were taken with the same procedure and observed at the stereomicroscope to ensure that the fertilization had not actually occurred. Ten unfertilized oocytes (NF) for each CD1 female were collected in five pools (1 pool/female) and used for the expression analysis. Each pool of 10 F and 10 NF was placed in 10 µl of RNase-free water. According to a previously published protocol, RNA from murine oocytes was extracted by thermolysis: the samples were incubated for 1 min at 100°C in order to release nucleic acids [Citation83], put in ice for 1 min and vortexed for 30 sec. Every sample was directly reverse transcribed, without prior RNA purification, and utilized for qRT-PCR analysis as previously described.
In silico analysis
NCBI BLAST
(https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=blast2seq&LINK_LOC=align2seq) was used to predict the miRNA responsive elements within the circNAPEPLDiso1 sequence: a word size of 7 was set as a cut-off.
The gene interaction network was generated through miRNet [Citation84], giving official candidate miRNA names as input. Betweenness centrality was set at 5, and the subsequent network was generated accordingly. KEGG pathway functional enrichment was evaluated through the same online tool: more in details, q-values were automatically calculated by miRNet for each biological function, through hypergeometric test after adjustment for false discovery rate (FDR), with an α value set to 0.05.
RBP binding sites and IRESs within circRNA sequence were predicted through CircInteractome (https://circinteractome.nia.nih.gov/index.html) [Citation85] and circRNADb [Citation19] (version1.0.0) (http://202.195.183.4:8000/circrnadb/circRNADb.php), respectively. Multiple alignment of translated ORFs from linear and circular RNAs was performed by using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Statistical analysis
All the statistical tests used in this study are described throughout the text and in figure legends. We defined results as significant by default where α < 0.05. More in details, p-values were calculated according to Student’s t-test and q-values were generated through a hypergeometric test after adjustment for false discovery rate (FDR) (see the previous paragraph), as specified in the text.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download MS Word (16 KB)Acknowledgments
We thank Prof. Catherine M Greene, Dr. Sebastian Vencken and Dr. Chiara De Santi for their kind suggestions on the development of miR-CATCH protocol. We thank Dr. Angela Caponnetto for her collaboration to miR-CATCH experiments. We wish to thank the Scientific Bureau of the University of Catania for language support.
Supplementary material
The supplementary material for this article can be accessed here
Additional information
Funding
References
- Pierantoni R, Cobellis G, Meccariello R, et al. Evolutionary aspects of cellular communication in the vertebrate hypothalamo-hypophysio-gonadal axis. Int Rev Cytol. 2002;218:69–141.
- Meccariello R, Chianese R, Chioccarelli T, et al. Intra-testicular signals regulate germ cell progression and production of qualitatively mature spermatozoa in vertebrates. Front Endocrinol (Lausanne). 2014;5:69.
- Chianese R, Cobellis G, Chioccarelli T, et al. Kisspeptins, estrogens and male fertility. Curr Med Chem. 2016;23:4070–4091.
- Fang P, Zeng P, Wang Z, et al. Estimated diversity of messenger RNAs in each murine spermatozoa and their potential function during early zygotic development. Biol Reprod. 2014;90:94.
- Grandjean V, Fourre S, De Abreu DA, et al. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci Rep. 2015;5:18193.
- Chen Q, Yan M, Cao Z, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Ragusa M, Barbagallo C, Statello L, et al. Non-coding landscapes of colorectal cancer. World J Gastroenterol. 2015;21:11709–11739.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283.
- Barbagallo D, Condorelli A, Ragusa M, et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7:4746–4759.
- Barbagallo D, Caponnetto A, Cirnigliaro M, et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19.
- Barbagallo C, Brex D, Caponnetto A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA Interactions. Mol Ther Nucleic Acids. 2018;12:229–241.
- Barbagallo D, Caponnetto A, Brex D, et al. CircSMARCA5 Regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers (Basel). 2019;11:194
- Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030.
- Dong WW, Li HM, Qing XR, et al. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci Rep. 2016;6:39080.
- Lin X, Han M, Cheng L, et al. Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells. RNA Biol. 2016;13:1011–1024.
- Dang Y, Yan L, Hu B, et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130.
- Chen X, Han P, Zhou T, et al. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985.
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340.
- Cortes-Lopez M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89:527–537.
- Li HM, Ma XL, Li HG. Intriguing circles: conflicts and controversies in circular RNA research. Wiley Interdiscip Rev RNA. 2019;e1538.
- Gao Y, Zhao F. Computational strategies for exploring circular RNAs. Trends Genet. 2018;34:389–400.
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338.
- Okamoto Y, Morishita J, Tsuboi K, et al. Molecular characterization of a phospholipase D generating anandamide and its congeners. J Biol Chem. 2004;279:5298–5305.
- Fasano S, Meccariello R, Cobellis G, et al. The endocannabinoid system: an ancient signaling involved in the control of male fertility. Ann N Y Acad Sci. 2009;1163:112–124.
- Grimaldi P, Di Giacomo D, Geremia R. The endocannabinoid system and spermatogenesis. Front Endocrinol (Lausanne). 2013;4:192.
- Bovolin P, Cottone E, Pomatto V, et al. Endocannabinoids are involved in male vertebrate reproduction: regulatory mechanisms at central and gonadal level. Front Endocrinol (Lausanne). 2014;5:54.
- Meccariello R, Battista N, Bradshaw HB, et al. Endocannabinoids and reproduction. Int J Endocrinol. 2014;2014:378069.
- Chianese R, Ciaramella V, Scarpa D, et al. Anandamide regulates the expression of GnRH1, GnRH2, and GnRH-Rs in frog testis. Am J Physiol Endocrinol Metab. 2012;303:E475–87.
- Guo Y, Wang H, Okamoto Y, et al. N-acylphosphatidylethanolamine-hydrolyzing phospholipase D is an important determinant of uterine anandamide levels during implantation. J Biol Chem. 2005;280:23429–23432.
- Wang H, Xie H, Sun X, et al. Differential regulation of endocannabinoid synthesis and degradation in the uterus during embryo implantation. Prostaglandins Other Lipid Mediat. 2007;83:62–74.
- Capalbo A, Ubaldi FM, Cimadomo D, et al. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil Steril. 2016;105:225–35 e1-3.
- Battaglia R, Palini S, Vento ME, et al. Identification of extracellular vesicles and characterization of miRNA expression profiles in human blastocoel fluid. Sci Rep. 2019;9:84.
- Battaglia R, Vento ME, Ragusa M, et al. MicroRNAs are stored in human MII oocyte and their expression profile changes in reproductive aging. Biol Reprod. 2016;95:131.
- Rosenbluth EM, Shelton DN, Sparks AE, et al. MicroRNA expression in the human blastocyst. Fertil Steril. 2013;99:855–61 e3.
- Nissan X, Denis JA, Saidani M, et al. miR-203 modulates epithelial differentiation of human embryonic stem cells towards epidermal stratification. Dev Biol. 2011;356:506–515.
- Machtinger R, Rodosthenous RS, Adir M, et al. Extracellular microRNAs in follicular fluid and their potential association with oocyte fertilization and embryo quality: an exploratory study. J Assist Reprod Genet. 2017;34:525–533.
- Parchem RJ, Moore N, Fish JL, et al. miR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep. 2015;12:760–773.
- Card DA, Hebbar PB, Li L, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438.
- Tian Y, Zhang Y, Hurd L, et al. Regulation of lung endoderm progenitor cell behavior by miR302/367. Development. 2011;138:1235–1245.
- Nguyen LS, Fregeac J, Bole-Feysot C, et al. Role of miR-146a in neural stem cell differentiation and neural lineage determination: relevance for neurodevelopmental disorders. Mol Autism. 2018;9:38.
- Qin M, Wei G, Sun X. Circ-UBR5: an exonic circular RNA and novel small nuclear RNA involved in RNA splicing. Biochem Biophys Res Commun. 2018;503:1027–1034.
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134.
- Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741.
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264.
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. Rna. 2015;21:172–179.
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular rna that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37 e9.
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641.
- Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814.
- Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun. 2018;503:1503–1509.
- Zhang X, Gao F, Fu J, et al. Systematic identification and characterization of long non-coding RNAs in mouse mature sperm. PLoS One. 2017;12:e0173402.
- Chen S, Huang V, Xu X, et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176:831–43 e22.
- Enuka Y, Lauriola M, Feldman ME, et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383.
- You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610.
- Pierantoni R, Cobellis G, Meccariello R, et al. Testicular gonadotropin-releasing hormone activity, progression of spermatogenesis, and sperm transport in vertebrates. Ann N Y Acad Sci. 2009;1163:279–291.
- Chianese R, Chioccarelli T, Cacciola G, et al. The contribution of lower vertebrate animal models in human reproduction research. Gen Comp Endocrinol. 2011;171:17–27.
- Cobellis G, Meccariello R, Chianese R, et al. Effects of neuroendocrine CB1 activity on adult leydig cells. Front Endocrinol (Lausanne). 2016;7:47.
- Ciaramella V, Meccariello R, Chioccarelli T, et al. Anandamide acts via kisspeptin in the regulation of testicular activity of the frog, Pelophylax esculentus. Mol Cell Endocrinol. 2016;420:75–84.
- Migliaccio M, Ricci G, Suglia A, et al. Analysis of endocannabinoid system in rat testis during the first spermatogenetic wave. Front Endocrinol (Lausanne). 2018;9:269.
- Meccariello R, Franzoni MF, Chianese R, et al. Interplay between the endocannabinoid system and GnRH-I in the forebrain of the anuran amphibian Rana esculenta. Endocrinology. 2008;149:2149–2158.
- Esakky P, Hansen DA, Drury AM, et al. Molecular analysis of cell type-specific gene expression profile during mouse spermatogenesis by laser microdissection and qRT-PCR. Reprod Sci. 2013;20:238–252.
- Scieglinska D, Vydra N, Krawczyk Z, et al. Location of promoter elements necessary and sufficient to direct testis-specific expression of the Hst70/Hsp70.2 gene. Biochem J. 2004;379:739–747.
- Chianese R, Colledge WH, Fasano S, et al. Editorial: the multiple facets of kisspeptin activity in biological systems. Front Endocrinol (Lausanne). 2018;9:727.
- Chimento A, Sirianni R, Casaburi I, et al. Role of estrogen receptors and g protein-coupled estrogen receptor in regulation of hypothalamus-pituitary-testis axis and spermatogenesis. Front Endocrinol (Lausanne). 2014;5:1.
- Tena-Sempere M, Barreiro ML. Leptin in male reproduction: the testis paradigm. Mol Cell Endocrinol. 2002;188:9–13.
- Khazamipour N, Noruzinia M, Fatehmanesh P, et al. MTHFR promoter hypermethylation in testicular biopsies of patients with non-obstructive azoospermia: the role of epigenetics in male infertility. Hum Reprod. 2009;24:2361–2364.
- Lin ZY, Hirano T, Shibata S, et al. Gene expression ontogeny of spermatogenesis in the marmoset uncovers primate characteristics during testicular development. Dev Biol. 2015;400:43–58.
- Chung SS, Wolgemuth DJ. Role of retinoid signaling in the regulation of spermatogenesis. Cytogenet Genome Res. 2004;105:189–202.
- Pimenta MT, Francisco RA, Silva RP, et al. Relaxin affects cell organization and early and late stages of spermatogenesis in a coculture of rat testicular cells. Andrology. 2015;3:772–786.
- Bell EL, Nagamori I, Williams EO, et al. SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development. 2014;141:3495–3504.
- Choi H, Han C, Jin S, et al. Reduced fertility and altered epididymal and sperm integrity in mice lacking ADAM7. Biol Reprod. 2015;93:70.
- Caballero J, Frenette G, D’Amours O, et al. Bovine sperm raft membrane associated glioma pathogenesis-related 1-like protein 1 (GliPr1L1) is modified during the epididymal transit and is potentially involved in sperm binding to the zona pellucida. J Cell Physiol. 2012;227:3876–3886.
- Vanier MT, Millat G. Structure and function of the NPC2 protein. Biochim Biophys Acta. 2004;1685:14–21.
- Martin-DeLeon PA. Epididymal SPAM1 and its impact on sperm function. Mol Cell Endocrinol. 2006;250:114–121.
- De Santi C, Vencken S, Blake J, et al. Identification of MiR-21-5p as a functional regulator of mesothelin expression using microRNA capture affinity coupled with next generation sequencing. PLoS One. 2017;12:e0170999.
- Barbagallo C, Passanisi R, Mirabella F, et al. Upregulated microRNAs in membranous glomerulonephropathy are associated with significant downregulation of IL6 and MYC mRNAs. J Cell Physiol. 2019.234(8):12625–12636.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408.
- Hassan T, Smith SG, Gaughan K, et al. Isolation and identification of cell-specific microRNAs targeting a messenger RNA using a biotinylated anti-sense oligonucleotide capture affinity technique. Nucleic Acids Res. 2013;41:e71.
- Vencken S, Hassan T, McElvaney NG, et al. miR-CATCH: microRNA capture affinity technology. Methods Mol Biol. 2015;1218:365–373.
- Di Pietro C, Vento M, Ragusa M, et al. Expression analysis of TFIID in single human oocytes: new potential molecular markers of oocyte quality. Reprod Biomed Online. 2008;17:338–349.
- Fan Y, Xia J. miRNet-functional analysis and visual exploration of miRNA-Target interactions in a network context. Methods Mol Biol. 2018;1819:215–233.
- Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.