ABSTRACT
The 5ʹ-UTR of the actin-related protein 2/3 complex subunit 2 (ARPC2) mRNA exists in two variants. Using a bicistronic reporter construct, the present study demonstrates that the longer variant of the 5ʹ-UTR harbours an internal ribosome entry site (IRES) which is lacking in the shorter one. Multiple control assays confirmed that only this variant promotes cap-independent translation. Furthermore, it includes a guanine-rich region that is capable of forming a guanine-quadruplex (G-quadruplex) structure which was found to contribute to the IRES activity. To investigate the cellular function of the IRES element, we determined the expression level of ARPC2 at various cell densities. At high cell density, the relative ARPC2 protein level increases, supporting the presumed function of IRES elements in driving the expression of certain genes under stressful conditions that compromise cap-dependent translation. Based on chemical probing experiments and computer-based predictions, we propose a structural model of the IRES element, which includes the G-quadruplex motif exposed from the central stem-loop element. Taken together, our study describes the functional relevance of two alternative 5ʹ-UTR splice variants of the ARPC2 mRNA, one of which contains an IRES element with a G-quadruplex as a central motif, promoting translation under stressful cellular conditions
Graphical abstract
Introduction
The actin-related protein 2/3 complex subunit 2 (ARPC2) gene encodes one of the seven subunits of the human actin-related protein 2/3 (ARP2/3) protein complex. This complex is evolutionally conserved and is involved in the control of actin polymerization and branching in cells [Citation1]. It thereby plays multiple roles in cellular processes such as cellular migration, adhesion, trafficking, endocytosis, and it is required to generate lamellipodia at the leading edge of cells [Citation2,Citation3]. The exact role of the protein encoded by ARPC2, the p34 subunit, has yet to be determined; however, the expression of ARPC2 was shown to be upregulated in tumour tissues such as gastric cancer [Citation3], and ARPC2 knockdown resulted in cellular growth arrest [Citation4]. Despite its obvious relevance in various physiological and pathophysiological processes, the regulation of ARPC2 expression is not yet fully understood.
We recently provided evidence that a guanosine-rich sequence in the 5ʹ-UTR of the ARPC2 mRNA is capable of forming a guanosine-quadruplex (G-quadruplex) structure [Citation5,Citation6]. G-quadruplexes are non-canonical secondary structures in DNA or RNA that are composed of planar quartets, each of which consists of four guanine bases that associate through Hoogsteen hydrogen bonds. Multiple π-π stacked G-quartet planar arrangements then form the three-dimensional G-quadruplex motif. The presence of a central monovalent cation, such as K+, plays a crucial role for the stability of the G-quadruplex. Emerging evidence suggests that G-quadruplexes have important roles in a variety of biological processes, including transcription, recombination, replication, splicing, polyadenylation, translation and chromosome stability [Citation7,Citation8]. Likewise, G-quadruplexes have been shown to be involved in numerous physiological and pathophysiological processes, including cancer [Citation9] and neurodegeneration [Citation10].
One of the most intensively investigated functions of G-quadruplex motifs in the 5ʹ-UTR of mRNAs is their capacity to repress translation. This phenomenon was first observed in vitro in a reticulocyte lysate for a G-quadruplex located for the 5ʹ-UTR of the neuroblastoma RAS viral oncogene homolog (NRAS) [Citation11]. Since then, numerous examples have been found in which G-quadruplex structures in mRNAs inhibit translation in living eukaryotic cells, e.g. in the genes of the zinc-finger protein of the cerebellum1 (zic-1) [Citation12], the B-cell lymphoma gene 2 (Bcl-2) [Citation13,Citation14], and the gene encoding the membrane type 3 metalloproteinase (MT3-MMP) [Citation15]. In most cases, the G-quadruplexes repress translation to a level of 30%–70% of the control [Citation16].
Unexpectedly, some cases were reported, in which G-quadruplexes in the 5ʹ-UTR of an mRNA augment translation. One example is the G-quadruplex in the mRNA of the transforming growth factor β2 (TGF-β2), which inhibits expression in reporter assays, but enhances gene expression in the context of the entire 5ʹ-UTR [Citation17]. Moreover, G-quadruplex structures were also reported to influence cap-independent translation through internal ribosome entry sites (IRES) [Citation18]. Such elements were first discovered in picornaviruses [Citation19,Citation20] and have since then been established as the main mechanism of translation initiation in numerous plus-stranded RNA viruses. Following their discovery in viral genomes, numerous cellular IRES Elements have been described, many of which are located in proto-oncogenes and other cancer-related genes [Citation21–Citation23]. IRES elements initiate protein synthesis by a non-canonical and 5ʹ-cap-independent manner through recruiting ribosomes, thereby bypassing the requirement of interaction with the cap-binding protein eIF4E [Citation24]. The presence of IRES elements in cellular mRNAs has been controversial for many years, but an increasing body of evidence supports their existence in certain 5ʹ-UTRs [Citation25,Citation26]. The exact biological function of this alternative mode of translational initiation still remains to be elucidated, but numerous studies suggest that IRES-mediated translation becomes relevant under certain stressful conditions, when cap-dependent translation is compromised [Citation25–Citation28]. In contrast to viral IRES elements, which can be classified into four groups according to their sequence and structure, no such common motifs have been identified that allow prediction of whether an mRNA sequence has IRES activity [Citation25,Citation29]. The function of some smaller IRES elements in eukaryotic cells was described as to mediate interactions with the 18S rRNA in a Shine-Dalgarno kind of manner, which is actually known from prokaryotes [Citation25].
Several IRES elements in 5ʹ-UTRs of cellular mRNAs have been reported to contain G-quadruplex motifs. One example is the IRES in the mRNA of the fibroblast growth factor 2 (FGF2), which harbours a G-quadruplex motif and is responsible for translation initiation at four alternative codons [Citation30]. Furthermore, a G-quadruplex was found to be essential for cap-independent initiation of translation by an IRES element in the mRNA of the vascular endothelial growth factor (VEGF) [Citation31]. A subsequent mechanistic study demonstrated that the G-quadruplex structure directly interacts with the 40S ribosomal subunit in the absence of other protein factors [Citation32]. Further studies led to the idea that specific ligands can be used to stabilize the VEGF G-quadruplex and thereby modulate its cap-independent translation [Citation33]. Another IRES element harbouring a G-quadruplex motif was found in the 5ʹ-UTR of the α-synuclein mRNA [Citation34]. Various conditions, such as plasma-membrane depolarization, serum starvation and oxidative stress stimulated α-synuclein protein translation via its 5ʹ-UTR and enhanced its IRES activity.
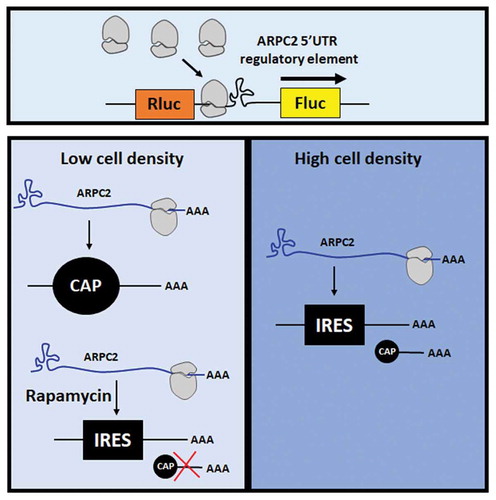
The present study investigates in depth the regulatory function of the ARPC2 5ʹ-UTR. As outlined for other examples above, the isolated G-quadruplex motif of the 5ʹ-UTR of the ARPC2 mRNA represses cap-dependent translation, but, when embedded in the environment of its full-length 5ʹ-UTR, it is part of an IRES element that mediates cap-independent translation. A special feature of the ARPC2 mRNA is the existence of two splice variants of the 5ʹ-UTR, one of which harbours the IRES element with the G-quadruplex, while the shorter variant lacks these elements. We show that under certain stressful conditions, such as high cell density, the IRES activity results in a relative increase of the ARPC2 expression. Based on structural probing experiments and bioinformatic prediction programs, we propose a model for the folding of the IRES element.
Materials and methods
Cell culture
Human Embryonic Kidney 293 (HEK293) cells were cultured in low glucose Dulbecco’s modified Eagle’s medium (PAA Laboratories GmbH, Pasching, Austria) containing 10% foetal bovine serum, 2 mM glutamine, non-essential amino acids and the antibiotics penicillin and streptomycin (50 I.U./ml and 50 μg/ml, respectively). The culture medium was additionally supplemented with 4.5 mg/ml glucose. MCF7 breast cancer cells were cultured in RPMI medium containing 10% foetal bovine serum, 2 mM glutamine, non-essential amino acids and the antibiotics penicillin and streptomycin (50 I.U./ml and 50 μg/ml, respectively). The culture medium was additionally supplemented with 100 mM pyruvate. Cells were grown at 37°C in a humidified atmosphere containing 5% CO2. HEK293 and MCF7 cells seeded for transfection were kept in medium without antibiotics.
Plasmid constructs
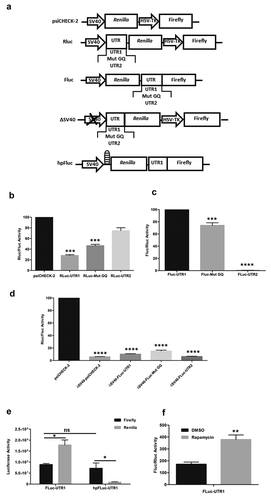
To study the influence of different human ARPC2 5ʹ-UTR sequences on translation, the dual luciferase reporter vector psiCHECK-2 was used (Promega, Madison, WI, USA). The G-quadruplex-containing 5ʹ-UTR1, a corresponding mutant sequence and the short variant 5ʹ-UTR2 were cloned into the unique NheI restriction site of Renilla luciferase 5ʹ-UTR. The resulting vectors were named Rluc-UTR1, Rluc-Mut GQ and Rluc-UTR2, respectively ()).
For insertions of different human ARPC2 5ʹ-UTR sequences upstream of firefly luciferase, unique MluI and XhoI restriction sites in the vector were used to excise the HSV-TK promoter region between the two luciferase cistrons, followed by blunting. Different human ARPC2 5ʹ-UTR sequences (synthesized by Life Technologies GmbH, Darmstadt, Germany) were digested with ApaI and SacI and the resulting fragments were then blunt end ligated into the blunted MluI and XhoI sites.
The following sequences of different ARPC2 5ʹ-UTRs were cloned:
ARPC2 UTR1:
5ʹ-GGCGCCGCGGGGCCGCGCGTCCAGGCGGCGAGCGGAAGTGGGTGTGAGAGCGGAAGTGGCCGGCTAGAGCCGGGGGCTGGGCGGGGACCGGGCTTGTCGGTGAAGCGGCAGTGGCGGCGGCGGCGGCGGCTCGGCAGGCGGGTTCAGGCTTCGGGGGCCAGCCGCCGCC-3ʹ
ARPC2 with a mutated G-quadruplex:
5ʹ-GGCGCCGCGGGGCCGCGCGTCCAGGCGGCGAGCGGAAGTGGGTGTGAGAGCGGAAGTGGCCGGCTAGAGCCGTAGACTGAGCGAAGACCGAGCTTGTCGGTGAAGCGGCAGTGGCGGCGGCGGCGGCGGCTCGGCAGGCGGGTTCAGGCTTCGGGGGCCAGCCGCCGCC-3ʹ
ARPC2 UTR2:
5ʹ-CTCCCTCCGTCCTTGCCTCCCTTACCCACCCTCACCGGCCCTTGTTTCTCCTTCCCCTGGGGGCAGCCGCCGCC-3ʹ
The resulting vectors were named Fluc-UTR1, Fluc-Mut GQ and Fluc-UTR2, respectively.
To generate promoter-less bicistronic constructs that allow analysis of promoter activity of the DNA insert in the intergenic region, the Simian virus 40 (SV40) promoter sequence was removed by BqlII and StuI digestion. The resulting vectors were named ΔSV40-psiCheck-1, ΔSV40-Rluc-UTR1, ΔSV40-Rluc-Mut GQ and ΔSV40-Rluc-UTR2, respectively
To exclude ribosomal readthrough, a sequence encoding a stable hairpin (ΔG −55 kcal/mol) that blocks translation [Citation23] was cloned upstream of the Renilla luciferase. The In-Fusion cloning technology (Clontech, Mountain View, CA, USA) was used to insert a 60 bp palindromic sequence upstream of the Renilla luciferase encoding sequence of the Fluc-UTR1 vector using the following oligonucleotides:
Oligonucleotide 1:
5ʹ-AGCCATGGTGGCTAGAGATCTGGTACCGAGCTCCCCGGGCTGCAGGATATCCTGCAGCCCGGGGAGCTCGGTACCAGATCTCTAGCCTATAGTGAG-3ʹ
Oligonucleotide 2:
5ʹ-CTCACTATAGGCTAGAGATCTGGTACCGAGCTCCCCGGGCTGCAGGATATCCTGCAGCCCGGGGAGCTCGGTACCAGATCTCTAGCCACCATGGCT-3ʹ
The oligonucleotides were first annealed, then phosphorylated using T4 PNK (New England Biolabs, Frankfurt, Germany) and the plasmids were linearized using NheI restriction enzyme, which was used to perform In-Fusion HD cloning based on the manufacturer’s instructions. The resulting vector was named hpFluc-UTR1.
The integrity of all plasmid constructs was confirmed by DNA sequencing.
Dual luciferase assay
HEK293 and MCF7 cells were seeded onto 24-well plates prior to transfection. Then, cells were transfected with 0.8 µg of the respective reporter plasmid using Lipofectamine 2000 (Life Technologies GmbH, Darmstadt, Germany) according to the manufacturer’s instructions. Cells were kept at 37°C in a humidified atmosphere containing 5% CO2. Activities of firefly and Renilla luciferase were measured 24 h after transfection with the Dual-Luciferase Assay kit (DLA) (Promega, Mannheim, Germany) using the TriStar2 Multimode Reader LB 942 Luminometer (Berthold Technologie GmbH & Co. KG, Bad Wildbad, Germany).
Western blotting
Western blotting was performed from cell monolayers lysed directly with SDS lysis buffer (3% 2-mercaptoethanol, 20% glycerine, 0.05% bromophenol blue, 3% SDS) as described previously [Citation35]. Briefly, cell lysates were harvested and boiled at 95°C for 10 min. Equal amounts of protein were separated using SDS-PAGE and immunoblotted for anti-ARPC2 (Millipore, Cat No. 07–227) and anti-actin (Sigma, Cat No. A2228). Goat anti-rabbit and goat anti-mouse IgG peroxidase conjugated secondary antibodies were purchased from Thermo Scientific. Images were visualized using ECL substrate (Pierce, Cat No. 32,109) and the ChemiDoc™ MP Imaging System (Bio-Rad Laboratories, CA, USA) followed by densitometric analysis using Image Lab Version 4.1 (Bio-Rad).
Treatment with rapamycin
Cells were treated with different concentrations of rapamycin (Tocris Bioscience, CAS 53123–88-9) for the indicated periods of time. To determine the effects of rapamycin on cell viability, supernatants were collected from the cells at the end of the treatment and the lactate dehydrogenase colorimetric assay (Thermo Scientific) was carried out according to manufacturer’s instructions. The absorbance at 490 nm was measured with the Sunrise microplate reader (Tecan, Männedorf, Switzerland).
DNA templates and RNA synthesis
For the generation of dsDNA templates for in vitro transcription, the plasmids Rluc-UTR1 and Rluc-Mut GQ were used. The dsDNA templates were amplified by PCR with the Q5-MasterMix (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocols. The forward primer included the promotor sequence for the T7 polymerase. The following primers were used:
T7 For: 5ʹ-TAATACGACT CACTATAGGC GCCGCGGGGC CGCGCGTC-3ʹ
Rev: 5ʹ-GGCGGCGGCT GGCCCCCGAAG-3ʹ
As a result, two dsDNAs encoding the desired RNA sequences were generated, containing the T7 promoter at the 5ʹ end. The reaction products were purified using the PCR/DNA Clean Up Kit (EURx, Gdansk, Poland) and the obtained dsDNA templates were dissolved in TE buffer.
In the next step, the 169-nucleotide 5ʹ-UTR1 of ARPC1 and its mutated variant Mut GQ, which is no longer capable of folding into a G-quadruplex, were transcribed. Transcription reactions were performed using the TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific Waltham, MA, USA) according to the manufacturer’s instructions. In case of transcripts to be used for 5ʹ-end labelling, a solution of 2 mM guanosine was added to the transcription reaction and incubated at 37°C for 3 h. The synthesized RNAs, 5ʹ-UTR1 and Mut GQ, were checked for size, integrity and homogeneity on a denaturing agarose gel, and purified with the GeneJet RNA Cleanup and Concentration Micro Kit (Thermo Fisher Scientific). In order to structurally characterize the 5ʹ portion of the RNAs, they were labelled with 32P at their 5ʹ end with (γ-32P) ATP (4600 Ci/mmol) from Hartmann Analytic (Braunschweig, Germany) and polynucleotide kinase (Thermo Fisher Scientific) according to standard procedures.
RNA structure probing in vitro
The in vitro SHAPE (Selective 2ʹ-Hydroxyl Acylation analysed by Primer Extension) reaction, DMS methylation and RNA cleavage in the presence of lead ions (Pb2+), were performed on the basis of protocols published elsewhere [Citation36,Citation37]. Briefly, after standard RNA renaturation in a buffer containing 130 mM potassium chloride (Sigma-Aldrich, St. Louis, United States), the SHAPE reagent 2-methylnicotinic acid imidazolide (NAI) (Merck KGaA, Darmstadt, Germany) was added to the sample at a final concentration of 200 mM. One control sample was treated with DMSO only (Sigma-Aldrich). After 10 min incubation at 37°C, the samples were subjected to RNA precipitation. For a second probing assay, 8.3% DMS (Sigma-Aldrich) solution in ethanol was added to two samples at a final concentration of 0.4%. The second control sample was treated with ethanol only. After 7 or 14 min incubation at 37°C, the samples were treated with ice-cold 100 mM dithiothreitol (Sigma-Aldrich) to stop the reaction, then subjected to RNA precipitation. Lead acetate (Sigma-Aldrich) was added to four samples at a final concentration of 0.25, 0.5, 1 and 2 mM. An additional control sample was treated with water only. After 5 min incubation at 37°C, the samples were treated with chilled 100 mM EDTA (Sigma-Aldrich) to stop the reaction and subjected to RNA precipitation. The obtained purified RNA was suspended in RNase-free water.
To determine sites in the RNA that were modified by DMS or NAI treatment or cleaved in the presence of 0.5 mM and 2 mM lead ions (Pb2+), reverse transcription reactions were performed with 0.2 μg of the RNA and a DNA primer (5ʹ-GGCGGCGGCT GGCCCCCGAAG-3ʹ) labelled with 32P at its 5ʹ-end. To assign the cleavage and chemical modification sites, products of the primer extension reaction were run on polyacrylamide gels along with dideoxy sequencing markers.
Dideoxy sequencing markers were generated for the RNAs 5ʹ-UTR1 and Mut GQ as described in [Citation36] with the only difference being the use of Superscript IV reverse transcriptase (Thermo Fisher Scientific) with 10 min incubation time and analysis with the FLA 5100 image analyser (Fujifilm, Tokyo, Japan). For the 5ʹ-labelled RNA monitoring, two ladders were generated: a formamide ladder and a guanine ladder using T1 ribonuclease (Thermo Fisher Scientific) as described elsewhere [Citation37].
Secondary structure modelling
Secondary structure models for the 5ʹ-UTR1 and the mutated variant Mut GQ were built with RNAstructure 6.0 [Citation38] using default options of the program and structural constraints taken from our experimental data. The nucleotides indicated as chemically modified or cleaved by at least two methods were used as structural constraints. This means, that they remain unpaired, are present at the terminal positions of double-stranded stems or are adjacent to any G-U wobble pair in the proposed structure model. For modelling of the 5ʹ-UTR1, the part of the sequence presumed to form a G-quadruplex (nucleotides 72–92) was treated as single-stranded.
Statistical analysis
Data were analysed using non-parametric one-way ANOVA or Student’s t-test (GraphPad Prism 6, GraphPad Software, Inc., La Jolla CA, USA). Data are represented as mean ± SD, p-values are considered significant by *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
Results
The ARPC2 mRNA exists in multiple variants, according to the NCBI Nucleotide Database. Transcript variant 1 (NM_152862.2) has a comparatively longer 5ʹ-UTR of 169 nucleotides, which contains a G-rich sequence. In our previous publication [Citation6], we have shown by bioinformatic analysis as well as by CD spectroscopy and UV melting analysis that this G-rich sequence can adopt a G-quadruplex structure. Furthermore, this motif was found to repress translation in a dual luciferase reporter assay. According to the database, a second, shorter transcript exists (NM_005731.3), denoted further on as 5ʹ-UTR2, which is devoid of the G-rich sequence (for a schematic representation of the two RNA variants, see Fig. S1A). We confirmed by specific RT-PCR analysis that both variants are expressed in HEK293 cells (Fig. S1B).
To analyse the influence of both variants on translation, we cloned the full length 5ʹ-UTR1 (RLuc-UTR1), a variant with a mutated G-quadruplex (RLuc-Mut GQ) and the shorter 5ʹ-UTR2 (RLuc-UTR2) upstream of the Renilla luciferase in a psiCHECK-2 vector for dual luciferase assays. Schematic representations of all constructs used in the present study are summarized in ). In line with our previously published results [Citation6], the full-length 5ʹ-UTR1 repressed translation, an effect which is significantly reduced when perturbing the G-quadruplex by introducing mutations ()). The shorter 5ʹ-UTR2 lacking the G-rich sequence does not have a substantial effect on translation at all.
Figure 1. Dual Luciferase Assays for functional analysis of the ARPC2 5‘-UTR variants. a) Schematic representation of the vectors used in the present study. b) Influence of different 5ʹ-UTR variants positioned upstream of Renilla luciferase. The ratio of Renilla to firefly luciferase is shown. c) Activity of the different 5ʹ-UTRs on the second, firefly luciferase of a bicistronic vector. The ratio of firefly to Renilla luciferase is shown. d) Luciferase activity of various constructs lacking the SV40 promotor. The ratio of Renilla to firefly luciferase is shown. e) Activity of the individual luciferases in the bicistronic vector in the absence or presence of a hairpin upstream of the Renilla luciferase. Activities of the individual luciferases are shown. f) Effect of rapamycin, an inhibitor of cap-dependent translation, on the luciferase expression in the bicistronic vector. The ratio of firefly to Renilla luciferase is shown. All data represent averages ± SD of three independent experiments. Further details are given in the text.
The 5ʹ-UTR1 of ARPC2 harbours an IRES element
As previous studies have shown that G-quadruplex motifs may not only repress translation, but may also be part of an IRES element that mediates cap-independent translation [Citation31], and our previous interaction study found several ribosomal proteins to interact with the G-quadruplex of the ARPC2 mRNA [Citation6], we reasoned that the 5ʹ-UTR1 of ARPC2 harbouring the G-quadruplex may function as an IRES element. We therefore designed a bicistronic construct, in which the HSV-TK promotor driving the second, firefly luciferase was replaced by either of the 5ʹ-UTR variants. The resulting plasmids were named: Fluc-UTR1, Fluc-mut GQ, Fluc-UTR2. As can be seen in ), the second, firefly luciferase of the bicistronic construct was actively translated in the presence of the 5ʹ-UTR1. The activity was slightly lower when the G-quadruplex was disrupted. Although this reduction was less than 30%, it was statistically highly significant. Even more importantly, the activity of the second, firefly luciferase was completely shut down in the presence of the short 5ʹ-UTR2. These data can be regarded as a first indication that the 5ʹ-UTR1 harbours an IRES element.
In the experiments described above, we observed substantial translation of the downstream cistron of the bicistronic mRNA containing the 5ʹ-UTR1 of ARPC2, an activity that is absent in the short 5ʹ-UTR2. However, this translational activity might result from various mechanisms. It is not necessarily a result of an IRES element, but the 5ʹ-UTR1 may also contain a cryptic promoter or an element which stimulates readthrough to the firefly luciferase cistron. To exclude the presence of a cryptic promoter in the 5ʹ-UTR1 of ARPC2, the SV40 promoter upstream of the Renilla luciferase was excised from the original psiCHECK-2 vector and from psiCHECK-2 vectors containing different 5ʹ-UTRs as described above. The remaining activity of both luciferase was determined in HEK293 cells 24 h post transfection using dual luciferase assays. Our findings indicated that neither of the constructs lacking the SV40 promotor exhibited substantial promoter activity compared to intact psiCHECK-2 reporter ()), and thus, these results prove that the ARPC2 G-quadruplex-UTR is devoid of cryptic promoter activity.
Further experiments were carried out to exclude that firefly luciferase activity of the bicistronic vector resulted from ribosomal readthrough. To this end, a palindromic sequence was cloned upstream of the Renilla luciferase coding sequence in the bicistronic vector. When transcribed, this sequence forms a stable hairpin (hp) and the vector was named hpFluc-UTR1. This hairpin can be expected to diminish cap-dependent translation of the Renilla cistron, whereas IRES-mediated, cap-independent translation of the firefly luciferase should remain unaffected. ) shows the values of each of the luciferases in the presence or absence of the hairpin. While the original bicistronic vector Fluc-UTR1 without the hairpin-forming sequence produces substantial levels of both luciferases, the hairpin represses translation of the Renilla luciferase. In contrast, activity of the firefly luciferase remains basically unchanged. This finding further supports its translation by IRES activity of the 5ʹ-UTR1.
In a final confirmatory experiment, the effect of rapamycin was studied, a well-established inhibitor of cap-dependent translation. As can be seen in the Fig. S2, treatment of the cells with rapamycin did not exert cytotoxic effects. For the analysis of its effect on translation, cells were transfected with the vector Fluc-UTR1 and treated with DMSO and 20 µM rapamycin 3 h post-transfection, respectively. Following an incubation period of 21 h, cells were analysed further in dual luciferase assays. Treatment with rapamycin significantly increased firefly luciferase activity relative to the Renilla luciferase activity FigureF, which is in line with the assumption that rapamycin selectively repressed cap-dependent translation while the IRES-induced translation remained unaffected.
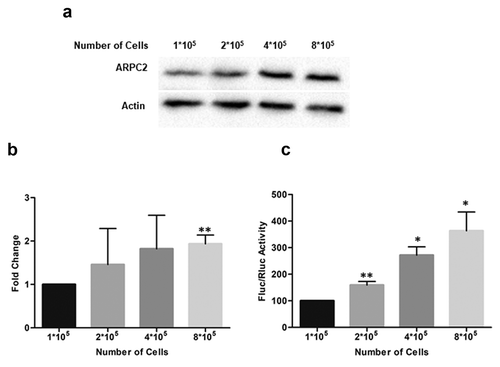
Figure 2. ARPC2 protein level at different cell densities. Increasing numbers of MCF7 cells (1*105 to 8*105) were seeded in 6-well plates. Twenty-four hours later, cells were harvested and boiled for protein analysis. a) Western blotting of ARPC2 and actin at increasing cell densities. b) The band densities were quantified from (a) and normalized to the β-actin control. Alterations in expression levels compared to sub-confluent (1*105) controls are represented as fold change ± SD from three independent experiments. c) Effect of different cell densities on the luciferase expression in the bicistronic vector. The ratio of firefly to Renilla luciferase is shown. All Data represent averages ± SD of three independent experiments.
The 5ʹ-UTR1 of ARPC2 promotes translation in response to stress conditions
We next aimed at elucidating the biological function of the two alternatively spliced variants of the 5ʹ-UTR of ARPC2. As outlined above, IRES-dependent translation is assumed to gain importance during various stress conditions, when cap-dependent translation is attenuated. The expression of several genes has been reported to be rapidly downregulated upon contact inhibition, when cells reach confluence [Citation39–Citation41]. Examples are of FOXM1, PLK1 and cyclin A.
We therefore investigated the ARPC2 protein level at different cell densities. We seeded 1*105 to 8*105 cells in a 6-well plate and cultured the cells for 24 hours. Cells were lysed and the ARPC2 protein level was analysed by Western blotting ()). Densitometric analysis of the band intensities revealed a significant, approximately 2-fold increase of the ARPC2 protein level relative to the actin level at high cell density ()). Furthermore, we transfected different cell densities (1*105 to 8*105 cells) with Fluc-UTR1 to specifically test whether cell-cell contact inhibits the expression of firefly luciferase which is under control of the IRES activity. Our results show that firefly luciferase expression is significantly higher at high cell densities compared to low density cell monolayer ()). These data further support the model of confluent cell-cell contact promoting relative translation of ARPC2.
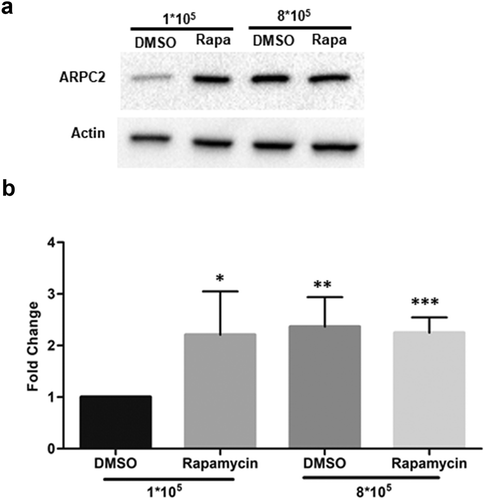
We next tested the effects of rapamycin, which we used in reporter assays above, on the cellular expression of ARPC2 in MCF7 cells. Sub-confluent (1*108 cells in a 6-well plate) and confluent (8*108 cells in a 6-well plate) cultures of MCF7 cells were cultivated for 16 h and then treated 10 μM rapamycin or DMSO. Following another incubation period of 8 h, cells were harvested for immunoblotting analysis. As can be seen in , treatment with rapamycin resulted in a highly significant, approximately 3-fold increase in ARPC2 expression relative to the actin level at low cell density. At higher cell density, the relative ARPC2 level was also increased in the DMSO-treated cells, which is in line with the data above (). Together, these cellular data support the hypothesis that conditions favouring IRES-mediated translation (i.e. high cell density stress or pharmacological inhibition of cap-dependent translation) increase the relative expression level of ARPC2 due to the IRES element located in the 5ʹ-UTR1 variant.
Figure 3. Rapamycin treatment of MCF7 cells. MCF7 cells (1*105 and 8*105 cells per well) were seeded in 6-well plates, cultured for 16 h, treated with 10 µM rapamycin and harvested 8 h thereafter. a) Western blotting of ARPC2 compared to actin. b) Quantification of band intensities. Values for ARPC2 are relative to those of the β-actin and were normalized to the DMSO control. Alterations in expression levels are represented as fold change ± SD of three independent experiments.
Secondary structure probing of the ARPC2 mRNA
The 5ʹ-UTR1 of ARPC2 is composed of 169 nucleotides, and has a high GC content, indicating that it is likely to fold into elaborate RNA secondary structures. We therefore carried out chemical in vitro structure probing experiments and used the RNAstructure 6.0 program to predict the secondary structure of the 5ʹ-UTR1 and its mutated control, which cannot form a G-quadruplex motif.
Three methods were used for structural probing: 1) SHAPE, using the NAI reagent, indicates single-stranded or highly flexible RNA regions without nucleotide character bias. 2) The use of DMS reveals single-stranded A and C (often weaker) residues located in terminal base-pairs or in base pairs adjacent to GU pairs [Citation42]. 3) The Pb2+-induced RNA cleavage method can detect single-stranded or highly flexible RNA regions, to some extent with sequence-structure context specificity [Citation43]. Following chemical treatment, reverse-transcription analysis was carried out to identify modified or cleaved sites and thereby detect single-stranded or flexible regions in the RNA under investigation. For better characterization of the 5ʹ portion of the RNAs, they were radioactively labelled on their 5ʹ-ends and analysed once again with the Pb2+-induced RNA cleavage method. The gels are shown in Supplementary Figure S3.
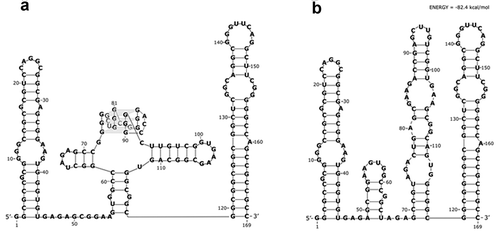
In the structure probing experiments, a set of positions were identified, which were cleaved in the presence of Pb2+ ions, NAI-adducted or DMS-methylated. The next step was to use the experimental data to propose a secondary structure model by bioinformatic means. For the most accurate RNA structure prediction, 15 nucleotide positions that were indicated by at least two methods, were used as structural constraints. These were: A36, A37, A47, A54, C61, G63, U65, A66, U94, U101, G102, A103, G105, G111, A136 (and additionally A74, A80, A84 for mutated variant). As the RNAstructure 6.0 program cannot predict higher ordered RNA structures, the G-rich part that can form a G-quadruplex structure was treated as single-stranded during bioinformatic structure analysis. Such an approach has been used by others before [Citation44]. In the end, the analysis confirmed that the longer 5ʹ-UTR of ARPC2 (5ʹ-UTR1) forms a very stable secondary structure. The secondary structure model consists of three parts ()): a 5ʹ hairpin, which is comparatively labile, the middle part harbouring the G-quadruplex structure and adjacent minor hairpins, and the highly stable 3ʹ hairpin with an unusually G-C-rich double-stranded stem.
We also made a prediction for the secondary structure of the mutated variant of the 5ʹ-UTR, which can no longer form a G-quadruplex structure, by chemical probing and bioinformatic analysis. Interestingly, no substantial differences in the structure probing reaction patterns were observed in the studied RNAs, with exception of the predicted G-quadruplex region of the wild-type sequence (nucleotide positions 72–92) and adjacent positions (Fig. S3). This observation indicates that both RNAs fold into very similar secondary structures outside of the G-quadruplex-forming stretch ()).
Figure 4. Secondary structure model of the 169 nucleotide-long 5′-UTR1 of ARPC2 mRNA. Two RNA molecules were analysed: a) the wild type sequence with a G-rich sequence capable of forming a G-quadruplex structure and b) the mutated sequence, which can no longer form the G-quadruplex structure. The models were generated by the RNAstructure 6.0 computer program with the use of experimentally determined constraints.
Discussion
Alternative splicing is a cellular mechanism to increase the diversity of protein isoforms derived from a limited set of genes. The most common mode of action is the use of alternative splice sites in the coding region of an mRNA. However, in many cases alternative splicing occurs in untranslated regions and thus has no effect on the polypeptide sequence, but rather influences mRNA stability, translational activity or subcellular localization [Citation45]. For example, two splice variants of a proinsulin gene are expressed in human pancreatic islets and metabolic changes modulate the expression of the 5ʹ-UTR splice variants [Citation46]. The longer 5ʹ-UTR1 was found to increase translational activity compared to the native proinsulin mRNA. Another example is the human toll-like receptor 5 (TLR5) gene, for which splice variants in the 5ʹ-UTR were identified; however, the functional relevance of this finding remained speculative. In contrast, the biological role of distinct 5ʹ-UTRs in mRNA of the X-chromosome linked inhibitor of apoptosis (XIAP) could be clarified [Citation47]. The shorter 5ʹ-UTR promoted basal level XIAP expression under normal growth conditions, while the longer 5ʹ-UTR was found to contain IRES activity and support cap-independent translation during stress.
The present study investigates the function of the two 5ʹ-UTR variants of ARPC2 found in human cells. ARPC2 is a key component of Arp2/3 complex, that controls actin polymerization and branching and has a crucial role in regulating cellular migration in health and disease [Citation1–Citation4]. Although the biochemistry and cell biology of the Arp2/3 complex have been well studied, far less is known about the regulation of its cellular level. The present study thus aims at investigating the relevance of the two splice variants for the expression level of ARPC2. The longer variant, named 5ʹ-UTR1, contains a G-rich sequence, which we recently reported to be capable of forming a G-quadruplex structure and to decrease regular translational activity [Citation5,Citation6]. Repression of translation is a mechanism commonly attributed to G-quadruplex structures in 5ʹ-UTRs [Citation16]. As interaction studies identified various ribosomal proteins as binding partners [Citation5,Citation6], it is reasonable to assume that G-quadruplex motifs can cause stalling of the ribosome and thereby reduce translational efficiency.
In addition to their function as translational repressors, G-quadruplex structures were found to contribute to IRES-mediated initiation of translation [Citation18]. A mechanistic study revealed direct interaction between the G-quadruplex motif in the VEGF IRES element and the 40S ribosomal subunit in the absence of other protein factors [Citation32]. Similarly, an IRES element harbouring a G-quadruplex motif was found in the 5ʹ-UTR of the human α-synuclein mRNA [Citation34]. This IRES element significantly induced expression of α-synuclein as well as a luciferase reporter without influencing the mRNA levels. When cap-dependent translation was attenuated by rapamycin, the 5ʹ-UTR enhanced protein synthesis in a cap-independent mechanism. Just like the ARPC2 IRES described in the present study, the G-quadruplex in the α-synuclein IRES was not absolutely required for translational activity, but increased its efficiency. Taken together, these examples show that G-quadruplex motifs may not only inhibit cap-dependent translation, but may also recruit the ribosome for cap-independent translation in IRES elements. In fact, when we placed the 5ʹ-UTR1 of ARPC2 upstream of the second reporter in a bicistronic construct, it mediated cap-independent translation, which did not occur with the shorter 5ʹ-UTR2.
We carried out a set of control experiments to confirm the IRES activity of the ARPC2 5ʹ-UTR1. For example, careful inspection had shown that presumed IRES activities might be due to cryptic promotor activity in the DNA encoding the 5ʹ-UTR [Citation48]. We therefore deleted the promotor, which drives the transcription of the ARPC2 5ʹ-UTR and a downstream reporter luciferase. As a consequence, luciferase activity was completely abrogated, excluding the possibility that the IRES-encoding DNA sequence contains a measurable cryptic promotor. For a second confirmatory experiment, we placed a stable hairpin upstream of the first reporter in a bicistronic construct that contains the 5ʹ-UTR1 upstream of the second reporter. As expected, the stem-loop structure substantially repressed translation of the first reporter, while initiation of translation of the second reporter by the IRES element remained unaffected. This test ruled out the possibility that the presumed IRES activity resulted from ribosomal readthrough.
A further confirmatory experiment used rapamycin, a macrolide compound targeting mTOR, the mechanistic (or mammalian) target of rapamycin. This central regulator of a signalling pathways controls initiation of cap-dependent translation [Citation49]. Rapamycin can thus be used to block cap-dependent translation, while cap-independent translation remains unaffected. In fact, in the presence of rapamycin, translation of the bicistronic construct shifted towards the second reporter, further confirming its translation by a cap-independent mechanism.
While these experiments demonstrated the IRES activity of the 5ʹ-UTR1, another aspect of interest was the function of the G-quadruplex which is located in the centre of the sequence. Disruption of the G-quadruplex structure by the introduction of mutations significantly decreased translational efficiency by approximately 30%. Interestingly, this effect is less pronounced than reported for the IRES element in the 5ʹ-UTR of VEGF. Morris et al. found the IRES activity to decrease gradually, depending on the number and position of mutations introduced. However, when the capacity to form a G-quadruplex was completely disrupted by the introduction of four mutations, the IRES activity was found to be completely abolished [Citation31]. This indicates that for the VEGF IRES, the flanking secondary structure elements support optimal interaction between the G-quadruplex containing domain and the 40S ribosomal subunit; however, without the G-quadruplex structure interaction with the ribosome was disturbed. In their follow-up paper investigating the structure-function relationship in more detail, the authors describe G-quadruplex structure of the IRES to ‘single-handedly’ interact with the 40S ribosomal subunit, a feature which is typical for type 3 and type 4 viral IRES elements [Citation32]. This is in contrast to the IRES element described in the present study, in which disruption of the G-quadruplex structure reduced translational activity by 30% only, indicating that the flanking regions can interact, directly or indirectly via ITAFs, with the ribosome. However, the possibility cannot be ruled out the introduced mutations in the G-quadruplex sequence may have resulted in the creation of additional binding sites for ITAFs, like hnRNA A1.
It is currently a matter of debate, to what extent G-quadruplex structures occur in mammalian cells, a controversy that was initiated by the report of Guo and Bartel, according to which G-quadruplex structures are globally unfolded in eukaryotic cells [Citation50]. The DEAH-box helicase Rhau was reported to represent the major RNA G-quadruplex helicase activity in HeLa cells [Citation51]. However, while we found Rhau to have a preference for mRNAs possessing a G-quadruplex motif, only a small fraction (1.9%) of all proteins were substantially downregulated following RNAi-mediated silencing of Rhau [Citation52]. It is thus likely that additional helicases are involved in the regulation of G-quadruplex structure folding and unfolding [Citation53]. Altogether, this debate indicated that G-quadruplex structures might occur in living eukaryotic cells to a far lesser extent than originally assumed [Citation8]. It is conceivable that they only form temporarily under certain conditions. This mode of action compatible with the activity of IRES elements, which are thought to promote translation under stressful cellular conditions [Citation25–Citation28].
To get a better understanding of the biological function of the IRES element, we analysed the endogenous ARPC2 protein level in MCF7 breast cancer cells under stressful cellular conditions. At high cell density, the ratio of the ARPC2 to actin protein level increased statistically significantly by approximately 2-fold. This finding is in line with the hypothesis that IRES elements maintain efficient translation under stressful conditions, when regular cap-dependent translation is compromised [Citation25–Citation28,Citation54]. The IRES activity was further strengthened when we used rapamycin to modulate endogenous translation. Consistent with the above discussed reporter assays, treatment of MCF7 cells with rapamycin increased the relative ARPC2 level, even at low cell density. This can be interpreted as a repression of general cap-dependent translation through rapamycin, while IRES-mediated translation of ARPC2 remained unaffected. At higher cell density, the ratio of the ARPC2 to actin protein level is increased, even in the absence of rapamycin, as the mode of protein synthesis is shifted from cap-dependent to cap-independent translation due to the cell-cell contact inhibition. Consequently, the addition of rapamycin does not induce further effects.
Following the functional characterization of the IRES element in the 5ʹ-UTR1 of the ARPC2 mRNA, we propose a structural model based on chemical probing and bioinformatic prediction by the RNAstructure 6.0 program. Multiple approaches were taken for the in vitro probing, including SHAPE with the NAI reagent, DMS and Pb2+-induced cleavage. Sites that were identified in the chemical probing experiments were fixed as constraints in the following bioinformatic analysis, which revealed that the 5ʹ-UTR1 is highly structured. According to the model, the RNA folds into three hairpins, the central of which contains the G-quadruplex motif exposed on top of the stem-loop structure (). To date, no structural elements have been identified that are common to cellular IRES elements [Citation25,Citation29]. However, a recent mechanistic study demonstrated by in vitro RNA footprinting that the G-quadruplex located in the IRES element of the VEGF mRNA interacts directly with the ribosomal 40S subunit [Citation32]. In addition, several IRES transacting factors (ITAFs) are usually recruited onto the mRNA in order to render the 5ʹ-cap and some of the initiation factors nonessential [Citation25,Citation55,Citation56]. One example is the La antigen, which binds purine-rich stretches like GAGA GNRA, GAAG or GNRR (with N being any nucleotide and R being a purine), preferentially located in apical loops [Citation57]. Interestingly, the 5ʹ-UTR1 under investigation here contains three such motifs, one in an apical loop and two in a single-stranded linker. The mutated variant (Mut GQ) lacks not only the G-quadruplex motif, but also has only two of the above mentioned sequence motifs, one in an apical loop and one in an internal loop. In case the La antigen is one of ITAFs for ARPC2, these differences may account for the decreased translational activity induced by the mutated 5ʹ-UTR.
In their analysis of the FGF-2 IRES, Bonnal et al. also presented a structural model, which was based on chemical probing and bioinformatic predictions [Citation30]. They found a very stable stem-loop at the 3ʹ-end of the sequence, like in the structure under investigation here. In addition, the FGF-2 structure also contains single-stranded GAGA and GAAG motifs and is thought to contain the G-quadruplex structure between two large stem-loops, like in the model described here. It was shown that the FGF-2 IRES interacts with hnRNP A1, which stimulates translation of that mRNA [Citation58]. The nucleotide sequence recognized by hnRNP A1 consists of one or two 5ʹ-AG-3ʹ motifs [Citation59]. Such motifs are also present multiple times in the single-stranded regions of the 5ʹUTR of the ARPC2 mRNA. It is furthermore worth mentioning that cytoplasmic localization of hnRNP A1 as an ITAF substantially increases during hypoxia stress [Citation60] and could be at least partly responsible for translational activation of some particular IRES elements like the one present in ARPC2 5ʹUTR. This possibility needs to be confirmed yet.
In summary, the present study provides evidence that the 5ʹ-UTR1 of ARPC2 has IRES activity and regulates translation during stressful cellular conditions such as high cell density. We propose a structural model based on chemical probing experiments and bioinformatic predictions that places a G-quadruplex motif in the apical loop of a central hairpin structure. Interestingly, an alternative splice variant of the 5ʹ-UTR exists which does not harbour the G-quadruplex structure and does not promote cap-independent translation. As ARPC2 has an important role in cell migration, tumour invasiveness and endocytosis, and it is important for the branching and nucleation of the actin cytoskeleton, it is desirable to understand how the IRES activity in the 5ʹ-UTR contributes to the regulation of the ARPC2 expression. In the long run, this may have implications for the development of anti-cancer compounds that target the RNA structures.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Supplemental Material
Download MS Word (1.4 MB)Acknowledgments
The authors are thankful to Marlena Waliszewska for excellent technical help during RNA biochemical synthesis and RNA structure probing. Furthermore, we are thankful to Erik Wade for carefully proofreading the manuscript and valuable comments.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726.
- Rauhala HE, Teppo S, Niemela S, et al. Silencing of the ARP2/3 complex disturbs pancreatic cancer cell migration. Anticancer Res. 2013;33:45–52.
- Zhang Y, Shen H, Liu H, et al. Arp2/3 complex controls T cell homeostasis by maintaining surface TCR levels via regulating TCR(+) endosome trafficking. Sci Rep. 2017;7:8952.
- Melboucy-Belkhir S, Pradere P, Tadbiri S, et al. Forkhead Box F1 represses cell growth and inhibits COL1 and ARPC2 expression in lung fibroblasts in vitro. Am J Physiol Lung Cell Mol Physiol. 2014;307:L838–847.
- Serikawa T, Spanos C, von Hacht A, et al. Comprehensive identification of proteins binding to RNA G-quadruplex motifs in the 5ʹ UTR of tumor-associated mRNAs. Biochimie. 2018;144:169–184.
- von Hacht A, Seifert O, Menger M, et al. Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res. 2014;42:6630–6644.
- Dolinnaya NG, Ogloblina AM, Yakubovskaya MG. Structure, properties, and biological relevance of the DNA and RNA G-Quadruplexes: overview 50 years after their discovery. Biochemistry (Mosc). 2016;81:1602–1649.
- Al-Zeer MA, Kurreck J. Deciphering the enigmatic biological functions of RNA guanine-quadruplex motifs in higher eukaryotes. Biochemistry. 2019;59:305–311.
- Wolfe AL, Singh K, Zhong Y, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70.
- Haeusler AR, Donnelly CJ, Periz G, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200.
- Kumari S, Bugaut A, Huppert JL, et al. An RNA G-quadruplex in the 5ʹ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3:218–221.
- Arora A, Dutkiewicz M, Scaria V, et al. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296.
- Shahid R, Bugaut A, Balasubramanian S. The BCL-2 5ʹ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry. 2010;49:8300–8306.
- Serikawa T, Eberle J, Kurreck J. Effects of genomic disruption of a guanine quadruplex in the 5ʹ UTR of the Bcl-2 mRNA in melanoma cells. FEBS Lett. 2017;591:3649–3659.
- Morris MJ, Basu S. An unusually stable G-Quadruplex within the 5ʹ-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry. 2009;48:5313–5319.
- Bugaut A, Balasubramanian S. 5ʹ-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741.
- Agarwala P, Pandey S, Mapa K, et al. The G-Quadruplex augments translation in the 5ʹ untranslated region of transforming growth factor beta2. Biochemistry. 2013;52:1528–1538.
- Song J, Perreault JP, Topisirovic I, et al. RNA G-quadruplexes and their potential regulatory roles in translation. Translation (Austin). 2016;4:e1244031.
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325.
- Jang SK, Krausslich HG, Nicklin MJ, et al. A segment of the 5ʹ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643.
- Akiri G, Nahari D, Finkelstein Y, et al. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236.
- Coldwell MJ, Mitchell SA, Stoneley M, et al. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905.
- Stoneley M, Paulin FE, Le Quesne JP, et al. C-Myc 5ʹ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428.
- Terenin IM, Akulich KA, Andreev DE, et al. Sliding of a 43S ribosomal complex from the recognized AUG codon triggered by a delay in eIF2-bound GTP hydrolysis. Nucleic Acids Res. 2016;44:1882–1893.
- Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240.
- Jackson RJ. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol. 2013;5:a011569.
- Sherrill KW, Byrd MP, Van Eden ME, et al. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem. 2004;279:29066–29074.
- Li W, Thakor N, Xu EY, et al. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 2010;38:778–788.
- Yamamoto H, Unbehaun A, Spahn CMT. Ribosomal chamber music: toward an understanding of IRES mechanisms. Trends Biochem Sci. 2017;42:655–668.
- Bonnal S, Schaeffer C, Creancier L, et al. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J Biol Chem. 2003;278:39330–39336.
- Morris MJ, Negishi Y, Pazsint C, et al. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc. 2010;132:17831–17839.
- Bhattacharyya D, Diamond P, Basu S. An Independently folding RNA G-quadruplex domain directly recruits the 40S ribosomal subunit. Biochemistry. 2015;54:1879–1885.
- Cammas A, Dubrac A, Morel B, et al. Stabilization of the G-quadruplex at the VEGF IRES represses cap-independent translation. RNA Biol. 2015;12:320–329.
- Koukouraki P, Doxakis E. Constitutive translation of human alpha-synuclein is mediated by the 5ʹ-untranslated region. Open Biol. 2016;6:160022.
- Al-Zeer MA, Xavier A, Abu Lubad M, et al. Chlamydia trachomatis prevents apoptosis via activation of PDPK1-MYC and enhanced mitochondrial binding of hexokinase II. EBioMedicine. 2017;23:100–110.
- Gorska A, Blaszczyk L, Dutkiewicz M, et al. Length variants of the 5ʹ untranslated region of p53 mRNA and their impact on the efficiency of translation initiation of p53 and its N-truncated isoform DeltaNp53. RNA Biol. 2013;10:1726–1740.
- Dutkiewicz M, Ciesiolka J. Structural characterization of the highly conserved 98-base sequence at the 3ʹ end of HCV RNA genome and the complementary sequence located at the 5ʹ end of the replicative viral strand. Nucleic Acids Res. 2005;33:693–703.
- Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129.
- Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83.
- Faust D, Al-Butmeh F, Linz B, et al. Involvement of the transcription factor FoxM1 in contact inhibition. Biochem Biophys Res Commun. 2012;426:659–663.
- Kuppers M, Ittrich C, Faust D, et al. The transcriptional programme of contact-inhibition. J Cell Biochem. 2010;110:1234–1243.
- Mathews DH, Disney MD, Childs JL, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 2004;101:7287–7292.
- Ciesiolka J, Michalowski D, Wrzesinski J, et al. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs. J Mol Biol. 1998;275:211–220.
- Kwok CK, Ding Y, Shahid S, et al. A stable RNA G-quadruplex within the 5ʹ-UTR of Arabidopsis thaliana ATR mRNA inhibits translation. Biochem J. 2015;467:91–102.
- Mockenhaupt S, Makeyev EV. Non-coding functions of alternative pre-mRNA splicing in development. Semin Cell Dev Biol. 2015;47–48:32–39.
- Shalev A, Blair PJ, Hoffmann SC, et al. A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology. 2002;143:2541–2547.
- Riley A, Jordan LE, Holcik M. Distinct 5ʹ UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Res. 2010;38:4665–4674.
- Dumas E, Staedel C, Colombat M, et al. A promoter activity is present in the DNA sequence corresponding to the hepatitis C virus 5ʹ UTR. Nucleic Acids Res. 2003;31:1275–1281.
- Nandagopal N, Roux PP. Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation (Austin). 2015;3:e983402.
- Guo JU, Bartel DP. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016;353:aaf5371.
- Creacy SD, Routh ED, Iwamoto F, et al. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J Biol Chem. 2008;283:34626–34634.
- Vester K, Eravci M, Serikawa T, et al. RNAi-mediated knockdown of the Rhau helicase preferentially depletes proteins with a Guanine-quadruplex motif in the 5ʹ-UTR of their mRNA. Biochem Biophys Res Commun. 2019;508:756–761.
- Sauer M, Paeschke K. G-quadruplex unwinding helicases and their function in vivo. Biochem Soc Trans. 2017;45:1173–1182.
- Ozretic P, Bisio A, Musani V, et al. Regulation of human PTCH1b expression by different 5ʹ untranslated region cis-regulatory elements. RNA Biol. 2015;12:290–304.
- Faye MD, Holcik M. The role of IRES trans-acting factors in carcinogenesis. Biochim Biophys Acta. 2015;1849:887–897.
- Sharathchandra A, Lal R, Khan D, et al. Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol. 2012;9:1429–1439.
- Gao W, Li Q, Zhu R, et al. La autoantigen induces ribosome binding protein 1 (RRBP1) expression through internal ribosome entry site (IRES)-mediated translation during cellular stress condition. Int J Mol Sci. 2016;17.
- Bonnal S, Pileur F, Orsini C, et al. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J Biol Chem. 2005;280:4144–4153.
- Jain N, Lin HC, Morgan CE, et al. Rules of RNA specificity of hnRNP A1 revealed by global and quantitative analysis of its affinity distribution. Proc Natl Acad Sci U S A. 2017;114:2206–2211.
- Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035.
