ABSTRACT
LncRNA ANCR plays important roles in the modulation of epithelial mesenchymal transition (EMT) and tumour metastasis in many tumours. However, the role of ANCR in regulating hepatocellular carcinoma (HCC) metastasis is still not known. The current study aims to investigate the underlying mechanism for tumour oncogenesis of ANCR in HCC metastasis. HCC cell proliferation and migration/invasion were measured by MTT and Transwell assays. Xenograft model was established to determine the effect of ANCR on HCC growth and metastasis. ChIP assay was used to detect the H3 and H4 histone acetylation levels at the ANCR promoter region. RNA pull-down and RIP assay was performed to analyse the relationship between ANCR and heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1). Dual-luciferase reporter gene assay was conducted to determine the interaction between ANCR and miR-140-3p. The results indicated that ANCR was highly expressed in HCC tissues and cells, which promoted the proliferation and migration/invasion of HCC cells. In vivo experiments showed interfering ANCR suppressed the growth and metastasis of HCC. H3/H4 histone acetylation levels at the ANCR promoter region were elevated in HCC tissues and cells, and interfering histone deacetylases 3 (HDAC3) significantly up-regulated ANCR expression. ANCR could bind to HNRNPA1, and promoted the expression of HNRNPA1 through regulating its degradation. In addition, ANCR upregulated the expression of HNRNPA1 through sponging miR-140-3p. Finally, we found that ANCR promoted the EMT and invasion/migration of HCC cells through regulating HNRNPA1. In conclusion, ANCR promoted HCC metastasis by upregulating HNRNPA1, inhibiting HNRNPA1 degradation and sponging miR-140-3p.
Introduction
Hepatocellular carcinoma (HCC) is one of the common malignant tumours and the major cause of death in the world with an increasing number of new cases year by year [Citation1,Citation2]. Studies have highlighted that chronic hepatitis c virus infection, alcohol abuse, obesity and metabolic syndrome are risk factors for HCC. So far, great progress has been made in the study of the aetiology and prognosis of HCC. However, the 5-year survival rate of HCC patients after surgical intervention is still not high (approximately 46.8%), mainly due to frequent metastasis of HCC [Citation3]. Studies indicated that genes associated with invasion/metastasis of cancer cells are overexpressed in cancer tissues and cells [Citation4,Citation5]. The expression of heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) is elevated in HCC, especially in recurrent HCC tissues, and high expression of HNRNPA1 is associated with low survival rate of HCC patients [Citation6]. Besides, HNRNPA1 overexpression promotes the invasion of HCC, and can be used as an indicator of poor prognosis for HCC patients [Citation7]. However, the molecular mechanisms that control HNRNPA1 expression in HCC metastasis are incompletely understood.
Increasing evidences suggest that long non-coding RNAs (lncRNAs) are important modulators in tumour metastasis, and exert its control function through binding to proteins or sponging microRNAs (miRNAs) [Citation8]. Anti-differentiation noncoding RNA (ANCR), located on human chromosome 4, is first reported in regulating the progenitor differentiation and is 7.64 × 1090 times more likely to be a noncoding transcript than protein-coding [Citation9]. It has been proved that ANCR plays important roles in the modulation of cell proliferation, migration/invasion, epithelial mesenchymal transition (EMT) and tumour metastasis in various tumours [Citation10,Citation11]. In addition, ANCR can regulate EZH2 stability and its downstream molecules expression through directly binding with EZH2, thus to affect osteosarcoma and colorectal cancer cell growth, invasion and metastasis [Citation11,Citation12]. ANCR can also act as a sponge of miR-758 to regulate Notch2 expression in osteogenic differentiation of periodontal ligament stem cells [Citation13]. These studies indicate that ANCR has the ability to binding with protein to regulate protein level, and sponging miRNA to regulate the target gene expression. However, whether the dysregulation of ANCR occurs during HCC metastasis is still unknown.
In our preliminary experiments, RNA pull-down and Mass spectrometry assays show that interfering ANCR remarkably down-regulates the expression of HNRNPA1 in HCC cells, indicating ANCR can positively regulate HNRNPA1 expression. In this study, we mainly focus on the impacts of ANCR on HCC proliferation and migration/invasion, and whether ANCR regulates HNRNPA1 expression through regulating HNRNPA1 protein stability and sponging miRNA. Collectively, ANCR may serve as a potential target for preventing HCC metastasis.
Materials and methods
Patients and HCC tissues
HCC tissues and the paired normal tissues were collected from HCC patients who underwent surgery at the Second Affiliated Hospital of Nanchang University. All patients recruited in this study did not receive chemotherapy before surgery. All collected tissues were frozen with liquid nitrogen after surgery and stored at −80°C for further use. This study was approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University and the written informed consent was obtained from all patients. The hepatitis B surface antigen (HBsAg) and alpha fetoprotein (AFP) were detected by MEIA (Abbott AxSYM, USA), the tumour size and portal vein tumour thrombus were determined by surgical evaluation, and the tumour differentiation and tumour node metastasis (TNM) stage were confirmed by histopathology.
Cell culture and transfection
Human hepatocytes (LO2) and human HCC cell lines (Hep3B, HepG2 and Huh7) were purchased from American Type Culture Collection (ATCC, VA, USA) and cultured in Minimum Essential Medium (MEM; Gibco, CA, USA) supplemented with 10% foetal bovine serum (FBS; Gibco, CA, USA), 1% glutamax (Invitrogen, CA, USA), 1% non-essential amino acids (Invitrogen, CA, USA) and sodium pyruvate 100 mM solution (Gibco, CA, USA) at 37°C with 5% CO2 and 95% air.
ANCR overexpressing vector (ANCR) and vector, small interfering RNA ANCR-1 (si-ANCR-1, 5ʹ-UAUAUUGUAAGCCGGUCAU-3ʹ), si-ANCR-2 (5ʹ-UUGACUUAAGCCAACAUC-3ʹ), si-HADC1-1 (5ʹ-AACTATGGTCTCTACCGAAAA-3ʹ), si-HADC1-2 (5ʹ-AACCGGTCATGTCCAAAGTAA-3ʹ), si-HADC2-1 (5ʹ-ACTGCATATTAGTCCTTCATT-3ʹ), si-HADC2-2 (5ʹ-GUGUAAUGACGGUAUCAUU-3ʹ), si-HADC3-1 (5ʹ-AAGGAGCTTCCCTATAGTGAA-3ʹ), si-HADC3-2 (5ʹ-GAUCGAUUGGGCUGCUUUA-3ʹ), si-HADC4-1 (5ʹ-CGACTCATCTTGTAGCTTATT-3ʹ), si-HADC4-2 (5ʹ-GAATCTGAACCACTGCATTTC-3ʹ) and si-control (5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ), miR-140-3p mimic and pre-negative control (NC), pcDNA-HNRNPA1 and pcDNA-control were synthesized by RiboBio (Guangzhou, China) and transfected into the cells using transfection reagent (Invitrogen, CA, USA) according to the manufacturer’s instruction.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from HCC tissues or HCC cells using TRIzol Reagent (Invitrogen, CA, USA), and cDNA was synthesized using High-Capacity RNA-to-cDNA Kit (Applied Biosystems, CA, USA). Reverse transcription was conducted using the SuperScript III Reverse Transcriptase kit (Invitrogen/Thermo Fisher Scientific Inc., USA) using primers specific for miR-140-3p. Special miR-140-3p reverse transcriptional primer was: 5ʹ-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCGTTT-3ʹ. qRT-PCR was performed using SYBR Green Real-Time PCR Master Mixes (Applied Biosystems, CA, USA) and a standard protocol. GAPDH level was used as the internal control for ANCR, and U6 level was used as the internal control for miR-140-3p. The primers for ANCR and miR-140-3p were as follows: ANCR, F: 5ʹ-GACATTTCCTGAGTCGTCTTCGAACGGAC-3ʹ, R: 5ʹ-TAGTGCGATTTAGAGCTGTACAAGTTTC-3ʹ; miR-140-3p, F: 5ʹ-ACACTCCAGCTGGGAGGCGGGGCGCCGCGGGA-3ʹ; R: 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ.
Cell proliferation assay
HCC cells were seeded into a 96-well plate at a density of 2 × 103 cells and incubated for 0 h, 24 h, 48 h, 72 h and 96 h. MTT solution (10 μl, 5 mg/ml; Beyotime Biotechnology, Shanghai, China) was added into each well and cultured in an incubator for 4 h. Then, 100 μl Formazan solution was added into each well and cultured in an incubator for 3 h. The absorbance at the 490 nm was determined at the spectrophotometer (ThermoFisher Scientific, CA, USA).
Cell migration and invasion assay
For cell migration assay, HCC cells were digested with Tyrisin Protease (Pierce, CA, USA). Then, cells were centrifuged at 200 × g for 5 min, and re-suspended in DMEM medium (Gibco, CA, USA) at a density of 4 × 105 cells/ml. HCC cells (100 μl, 4 × 104 cells) were seeded on the upper chamber of Transwell containing a porous membrane (pore size 8 μm; Corning, New York, USA). The bottom chamber of Transwell was added with 700 μl serum-free DMEM medium (Gibco, CA, USA) supplemented with 10% FBS (Gibco, CA, USA). After incubation for 48 h at 37°C with 5% CO2 and 95% air, the remaining cells on the upper chamber were removed, and cells on the bottom chamber were fixed with 4% paraformaldehyde and stained with 0.1% Crystal Violet solution. Cell number was counted in three random fields by a light microscope (Leica; Frankfurt, Germany). For invasion assay, the Transwell was coated with Matrigel (final concentration of 250 μg/ml/well), and the other procedures were the same as cell migration assay.
In vivo animal experiments
Interfering ANCR-1 lentivirus plasmid (lenti-si-ANCR) and negative control (lenti-GFP) were purchased from Genechem (Shanghai, China). HepG2 cells were transfected with lenti-si-ANCR or lenti-GFP with 5 μg/ml of polybrene (Sigma, Wisconsin, USA). Forty-eight hours later, puromycin (1.5 μg/ml; Sinopharm Chemical Reagent, Shanghai, China) was used for the selection of stable cells. Lentivirus-infected HepG2 cells (1.5 × 107) in 0.2 ml PBS were injected subcutaneously into the right dorsal regions of 5-week-old female nude mice to establish xenograft model (n = 6). Mice were checked every four days for the measurement of tumour volume, and sacrificed 28 days later. The tumours were collected and weighed. Lentivirus-infected HepG2 cells (3 × 106) were injected into the mice by tail vein to determine the metastatic ability. Mice were sacrificed 40 days later, and the number of metastatic nodules in lung was counted. All animal experiments were approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was used to detect the H3 and H4 histone acetylation levels at the ANCR promoter region. HCC tissues were cut into pieces smaller than 1 mm3, and Tyrisin Protease (Pierce, CA, USA) was added to digest the pieces into single cells. HCC cells were fixed with 1% formaldehyde for 5 min and then lysed for 10 min using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) on the ice. The protein-DNA complexes of cells were cross-linked using formaldehyde for 10 min at 25°C. The genomic DNA of lysed cells was sheared to 200–1000 bp by sonication. The final cell lysates were incubated with acetyl-histone H3/H4 antibodies (Cell Signalling Technology, Massachusetts, USA) or normal rabbit IgG (Cell Signalling Technology, Massachusetts, USA) for 12 h at 4°C and precipitated with protein G magnetic beads. After washing three times, the DNA-protein complex was collected for reverse crosslink, and qRT-PCR was conducted for the detection of acetylation of ANCR promoter after ChIP assay. The comparative method 2−ΔΔCt was used to calculate the relative expressions of DNA-H3 complex level and DNA-H4 complex level.
RNA pull-down
RNA pull-down assay was performed using the Magnetic RNA-Protein Pull-Down kit (Pierce, MA, USA) with cell lysates to analyse the relationship between ANCR and HNRNPA1. The biotinylated ANCR or antisense RNA probe was synthesized by Genepharm (Shanghai, China). Proteins (1 mg) extracted from HepG2 and Huh7 cells were mixed with 50 pmol biotinylated ANCR or antisense RNA probe, and incubated with streptavidin agarose beads (Thermo Scientific, CA, USA). Western blotting was used to measure proteins binding to the streptavidin-coupled beads.
RNA immunoprecipitation (RIP)
AGO2-RIP experiment was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Massachusetts, USA). Generally, HepG2 or Huh7 cells at 80-90% confluency was collected and were lysed using RIP lysis buffer. Then 100 µl cell extract was incubated with RIP buffer containing magnetic beads conjugated with human anti-Ago2 antibody (ab32381, Abcam, Cambridge, UK) or negative control Anti-IgG. The samples were incubated with Proteinase K to digest the protein and then the immunoprecipitated RNA was isolated. The purified RNA was further used for qRT-PCR analysis of ANCR and miR-140-3p. Specific primers were used for the detection of mature miR-140-3p: forward 5ʹ-ACACTCCAGCTGGGAGGCGGGGCGCCGCGGGA-3ʹ, reverse 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ. The products of qRT-PCR were collected and an agarose gel electrophoresis was performed to straightly see the existence of ANCR and miR-140-3p in the AGO2-complexes. To explore the binding between ANCR and HNRNPA1, the HNRNPA1 antibody (ab5832, Abcam) was used in the RIP experiment.
Western blotting
HCC cells were collected and lysed using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) with 1% PMSF and phosphatase inhibitors. The concentration of protein samples was measured by Pierce BCA Protein Assay Kit (Thermo Scientific, CA, USA). The samples were separated on 12% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membrane (Invitrogen, CA, USA). The membrane was incubated with primary antibodies against HNRNPA1 (1:1000; PA5-19,431, Invitrogen, CA, USA), E-cadherin (1:500; 13–1700, Invitrogen, CA, USA), ZO-1 (1:500; PA5-85,256, Invitrogen, CA, USA), N-cadherin (1:1000; 33–3900, Invitrogen, CA, USA), vimentin (1:1000; PA5-27,231, Invitrogen, CA, USA), Twist1 (1:2000; PA5-49,688, Invitrogen, CA, USA), Nanog (1:1000; PA5-85,110, Invitrogen, CA, USA) and β-actin (1:5000; MA5-15,739, Invitrogen, CA, USA) at 4°C overnight and subsequently incubated with horseradish peroxidase conjugated goat anti-rabbit IgG for 1.5 h at 25°C. Signals were detected using Pierce Western Blot Chemiluminescent Substrate (Thermo Scientific, CA, USA), and blots were visualized by ChemiDoc™ MP Imaging System (Bio-Rad, CA, USA).
Dual-luciferase reporter gene assay
Dual-luciferase reporter gene assay was conducted to determine the interaction between ANCR and miR-140-3p. The recombinant luciferase reporter vectors containing ANCR-wt (pGL3-ANCR-wt) or ANCR-mut (pGL3-ANCR-mut) were constructed by Genepharm (Shanghai, China). Then, 0.2 μg pGL3-ANCR-wt or 0.2 μg pGL3-ANCR-mut combined with 40 pmol miR-140-3p mimic or pre-NC were transfected into 293T cells using Lipofectamine 2000 reagent (ThermoFisher Scientific, CA, USA). 48 h after transfection, luciferase activity was detected using Dual Luciferase Assay System (Promega, Wisconsin, USA) in a TD-20/20 Luminometer (Turner BioSystems, CA, USA).
Statistical analysis
The data were presented as mean±standard deviation (SD), and were analysed by SPSS software (version 18.0; Chicago, Illinois, USA). The significance of difference was detected using t test or one-way analysis of variance (ANOVA), with p < 0.05 considered statistically significant.
Results
Lncrna ANCR was up-regulated in HCC tissues and cells
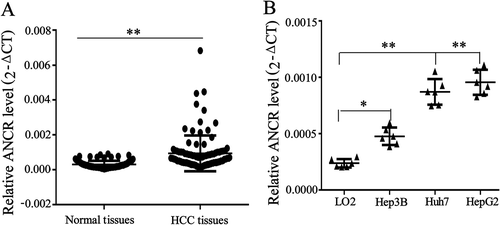
The expression of ANCR was measured by qRT-PCR in HCC tissues and paired normal tissues. As shown in , ANCR was significantly up-regulated in HCC tissues than that of normal tissues (p < 0.01). As shown in , the high level of ANCR was associated with tumour size (p = 0.009), tumour differentiation (p = 0.000), TNM stage (p = 0.000), and portal vein tumour thrombus (p = 0.000), and was irrelevant to age, gender and HBV infection, indicating that ANCR was associated with the proliferation and metastasis of HCC. Next, we measured the expression of ANCR in hepatocytes (LO2) and HCC cells (Hep3B, HepG2 and Huh7). ANCR was significantly up-regulated in HCC cell lines, including Hep3B (p < 0.05), HepG2 (p < 0.01), and Huh7 (p < 0.01), in comparison with that in LO2 cells ().
Table 1. Correlation between clinicopathological features and ANCR expression levels of HCC patients.
Figure 1. LncRNA ANCR was up-regulated in hepatocellular carcinoma (HCC) tissues and cells. A. ANCR was detected in HCC tissues and normal tissues using qRT-PCR. B. ANCR was detected in hepatocyte LO2 cells and HCC cell lines (Hep3B, HepG2, and Huh7). *p < 0.05, **p < 0.01. Statistical analysis of qRT-PCR was calculated by 2−△CT.
Lncrna ANCR promoted the migration and invasion of HCC cells
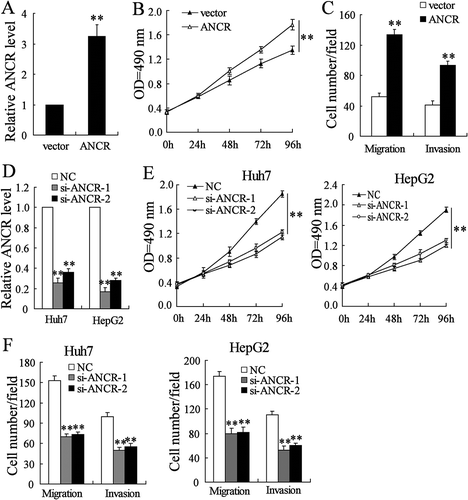
As ANCR was lower in Hep3B cells than in HepG2 and Huh7 cells, we overexpressed ANCR in Hep3B cells and interfered ANCR in HepG2 and Huh7 cells. We found that ANCR was significantly up-regulated in Hep3B cells after the transfection of ANCR overexpressing vector (ANCR) (, P < 0.01). Besides, the proliferation of Hep3B cells was significantly promoted after the transfection of ANCR (, P < 0.01), and the migration and invasion were significantly induced (, both p < 0.01). Next, we found that ANCR was significantly down-regulated in HepG2 and Huh7 cells after the transfection of si-ANCR-1 or si-ANCR-2 (, both p < 0.01). The proliferation of HepG2 and Huh7 cells was significantly suppressed after the transfection of si-ANCR-1 or si-ANCR-2 (, both p < 0.01), and the migration and invasion were also significantly suppressed (, both p < 0.01).
Figure 2. LncRNA ANCR promoted the migration and invasion of HCC cells. Hep3B cells were transfected with ANCR overexpressing vector (ANCR) or control vector (vector) for 48 h. A. ANCR was detected in Hep3B cells using qRT-PCR. B. The proliferation of Hep3B cells was detected using MTT assay. C. The migration and invasion of Hep3B cells were detected using Transwell assay. HepG2 and Huh7 cells were transfected with si-ANCR-1, si-ANCR-2 or negative control (NC) for 48 h. D. ANCR was detected in HepG2 and Huh7 cells using qRT-PCR. E. The proliferation of HepG2 and Huh7 cells was detected using MTT assay. F. The migration and invasion of HepG2 and Huh7 cells were detected using Transwell assay. **p < 0.01, compared with vector or NC. Statistical analysis of qRT-PCR was calculated by 2−△△CT.
Interfering ANCR in vivo suppressed the proliferation and metastasis of HCC
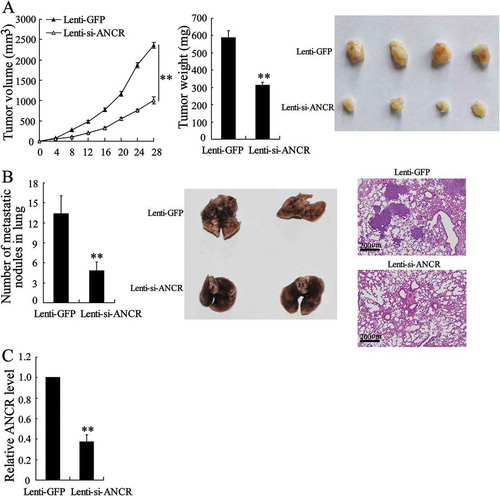
Since interfering ANCR in HepG2 and Huh7 cells suppressed the proliferation, migration and invasion of tumour cells, we interfered ANCR in vivo to observe the effect of ANCR on the proliferation and metastasis of HCC. HepG2 cells were transfected with Lenti-si-ANCR or Lenti-GFP, then lentivirus-infected HepG2 cells were injected subcutaneously into the right dorsal regions of 5-week-old female nude mice. Mice were sacrificed and tumours were collected 28 days later. As shown in , the tumour volume and tumour weight were significantly decreased in the Lenti-si-ANCR group than the Lenti-GFP group (p < 0.01), indicating that interfering ANCR suppressed the proliferation of HCC. Next, Lentivirus-infected HepG2 cells were injected into the mice by tail vein and lung nodules were counted to determine the anti-metastatic ability of interfering ANCR. We found that the number of metastatic nodules in lung was significantly reduced in the Lenti-si-ANCR group than the Lenti-GFP group (p < 0.01), and the H&E staining showed less tumour cells in nodule samples (), indicating that interfering ANCR suppressed lung metastasis of HCC. Also, the expression of ANCR was significantly down-regulated in HCC tumour of Lenti-si-ANCR group than Lenti-GFP group (, P < 0.01), suggesting the inhibition of proliferation and metastasis of HCC was associated with the down-regulation of ANCR.
Figure 3. Interfering lncRNA ANCR in vivo suppressed the proliferation and metastasis of HCC. A. HepG2 cells were transfected with Lenti-si-ANCR or Lenti-GFP. Lentivirus-infected HepG2 cells (1.5 × 107) in 0.2 ml PBS were injected subcutaneously into the right dorsal regions of 5-week-old female nude mice (n = 6 in each group). Mice were sacrificed and tumours were collected 28 days later. Tumour volume and tumour weight were detected. B. Lentivirus-infected HepG2 cells (3 × 106) were injected into the mice by tail vein. Mice were sacrificed 40 days later. The number of metastatic nodules in lung was counted and the nodule samples were stained by H&E. C. The expression of ANCR was detected in tumour tissues using qRT-PCR. **p < 0.01, compared with Lenti-GFP. Statistical analysis of qRT-PCR was calculated by 2−△△CT.
Histone acetylation at LncRNA ANCR promoter in HCC tissues and cells up-regulated the expression of LncRNA ANCR
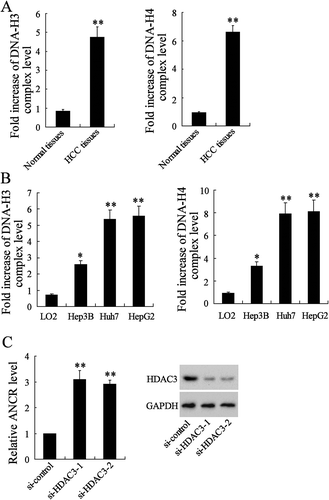
It has been reported that ANCR expression can be regulated by the histone acetylation at ANCR promoter in breast cancer [Citation10]. Therefore, we measured the H3 and H4 histone acetylation levels of ANCR promoter to figure out whether ANCR expression can be regulated by the histone acetylation at ANCR promoter in HCC. As shown in , H3 and H4 histone acetylation levels were significantly up-regulated in HCC tissues than normal tissues. Besides, H3 and H4 histone acetylation levels were significantly up-regulated in HCC cell lines, including Hep3B (p < 0.05), HepG2 (p < 0.01), and Huh7 (p < 0.01), compared with those in LO2 cells (). Moreover, interfering HDAC3 (si-HDAC3-1 and si-HDAC3-2) significantly up-regulated ANCR expression in Hep3B cells (, P < 0.01), whereas there was no significant difference in ANCR expression (p > 0.05) after the transfection of HDAC1 (si-HDAC1-1 and si-HDAC1-2), HDAC2 (si-HDAC2-1 and si-HDAC2-2), and HDAC4 (si-HDAC4-1 and si-HDAC4-2) (Fig. S1). The efficacy of HDAC3 knockdown was confirmed by western blot analysis (). These findings suggested that the up-regulation of ANCR in HCC was associated with increased H3 histone acetylation at ANCR promoter.
Figure 4. Histone acetylation at lncRNA ANCR promoter in HCC tissues and cells up-regulated the ANCR expression. A. H3 and H4 histone acetylation levels were detected in HCC tissues and normal tissues using ChIP assay. B. H3 and H4 histone acetylation levels were detected in LO2 cells and HCC cell lines (Hep3B, HepG2 and Huh7). C. Hep3B cells were transfected with si-HDAC3-1, si-HDAC3-2, or si-control for 48 h. ANCR expression was detected using qRT-PCR. HDAC3 protein expression was detected using western blot. *p < 0.05, **p < 0.01, compared with normal tissues, LO2 or si-control. Statistical analysis of qRT-PCR was calculated by 2−△△CT.
LncRNA ANCR affected the proteasome degradation of HNRNPA1 via binding with HNRNPA1
We found that HNRNPA1 was one of the most obvious molecules with elevated expression after overexpressing ANCR in Hep3B cells by LC-MS/MS analysis and RNA pull-down assay (Table S1). Therefore, we speculated that ANCR could interact with HNRNPA1. Biotinylated ANCR was incubated with protein lysates from HepG2 and Huh7 cells, and we found that biotinylated ANCR could bind to HNRNPA1, whereas antisense RNA probe could not bind to HNRNPA1 (). In addition, ANCR was accumulated after immunoprecipitation of HNRNPA1 antibody in HepG2 and Huh7 cells (). Next, truncated fragment of biotinylated ANCR was incubated with protein lysates from HepG2 cells, and we found that 3ʹ-end (605–855 nt) of ANCR was necessary for binding to HNRNPA1 (). In addition, HNRNPA1 expression was down-regulated in HepG2 and Huh7 cells after the transfection with si-ANCR-1 and si-ANCR-2 (), while it was up-regulated after the transfection with ANCR overexpressing vector (), indicating HNRNPA1 expression was positively related to ANCR. Moreover, ANCR overexpression restored the expression of HNRNPA1 after the treatment of cycloheximide (CHX) which is a protein synthesis inhibitor, and the proteasome inhibitor MG-132 did not affect the expression of HNRNPA1 with or without ANCR overexpression (), indicating that ANCR modulated the proteasome degradation pathway of HNRNPA1 without affecting its synthesis. Then we further investigated the effect of ANCR on the HNRNPA1 degradation. HepG2 and Huh7 cells were transfected with HA-Ub and si-ANCR-1. As shown in , si-ANCR significantly promoted the ubiquitination of HNRNPA1. Moreover, HepG2 and Huh7 cells transfected with si-ANCR or si-control were treated with CHX for 0 h, 2 h, 4 h and 6 h. We found that si-ANCR increased the degradation rate of HNRNPA1 after the CHX treatment (, P < 0.01). These findings indicated that ANCR inhibited HNRNPA1 degradation via suppressing its ubiquitination.
Figure 5. LncRNA ANCR affected the degradation of HNRNPA1. A. HepG2 and Huh7 cells were transfected with biotinylated ANCR or antisense RNA probe. RNA pull-down showed that biotinylated ANCR could bind to HNRNPA1, whereas antisense RNA probe could not bind to HNRNPA1. B. RIP assay showed that ANCR was accumulated after immunoprecipitation of HNRNPA1 antibody in HepG2 and Huh7 cells. C. HepG2 cells were transfected with truncated fragment of biotinylated ANCR. RNA pull-down showed that 3ʹ-end (605–855 nt) of ANCR was necessary for binding to HNRNPA1. D. HNRNPA1 expression was detected in HepG2 and Huh7 cells transfected with si-ANCR-1, si-ANCR-2, or si-control using western blot. E. HepG2 and Huh7 cells were transfected with ANCR overexpressing vector (ANCR) or control vector (vector). After the treatment of CHX (50 μg/ml) and MG132 (10 μl/ml, an inhibitor of the proteasome), the expression of HNRNPA1 was detected using western blot. **p < 0.01.
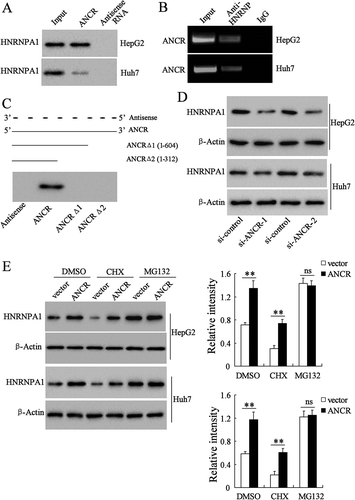
Figure 6. LncRNA ANCR inhibited the degradation of HNRNPA1 via suppressing its ubiquitination. A. HepG2 and Huh7 cells were transfected with HA-Ub and si-ANCR-1. HNRNPA1 specific antibody was used for immunoprecipitation, and then HA labelled antibody was used for immunoblotting. B. HepG2 and Huh7 cells were transfected with si-ANCR or si-control for 48 h and then treated with CHX for 0 h, 2 h, 4 h and 6 h. The expression of HNRNPA1 was detected in using western blot. **p < 0.01, compared with si-control.
LncRNA ANCR regulated the expression of HNRNPA1 through sponging miR-140-3p
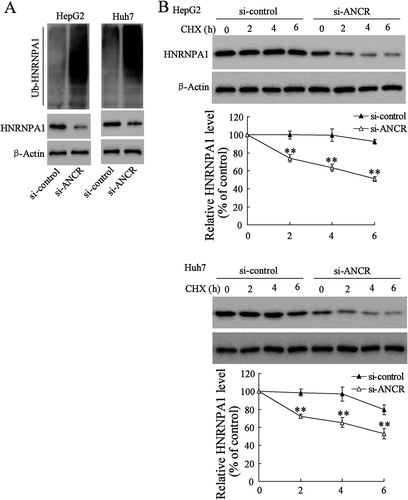
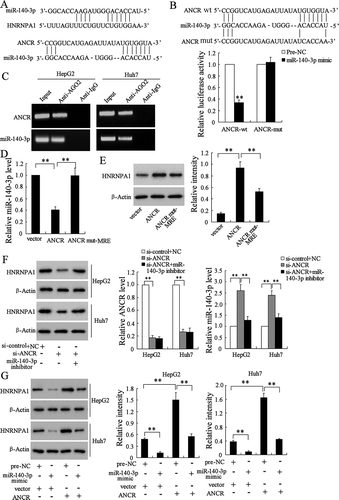
According to the prediction of bioinformatics software Targetscan and DIANA tools, we found that there were binding sites between miR-140-3p and HNRNPA1 3ʹ-UTR (), as well as between miR-140-3p and ANCR (). Dual-luciferase reporter assay showed that miR-140-3p mimic significantly down-regulated the activity of ANCR-wt (p < 0.01), whereas there was no significant difference in the activity of ANCR-mut (). In addition, AGO2 antibody could bind to ANCR and miR-140-3p in HepG2 and Huh7 cells, as ANCR and miR-140-3p were existed in the AGO2-IP products (). ANCR overexpression down-regulated the expression of miR-140-3p in HepG2 cells (). These findings suggested that ANCR could bind to miR-140-3p, and negatively regulated the expression of miR-140-3p. We further investigated the effect of ANCR on the expression of HNRNPA1, and found that ANCR overexpression up-regulated the expression of HNRNPA1 in HepG2 cells (). Moreover, si-ANCR down-regulated the expression of HNRNPA1 in HepG2 and Huh7 cells, whereas miR-140-3p inhibitor abolished the inhibition effect of si-ANCR (). Meanwhile, si-ANCR significantly down-regulated ANCR expression in HepG2 and Huh7 cells (, P < 0.01), and si-ANCR up-regulated miR-140-3p expression, whereas miR-140-3p inhibitor abolished the promotion effect of si-ANCR (, P < 0.01). Overexpressing miR-140-3p significantly down-regulated HNRNPA1 expression in HepG2 and Huh7 cells, and ANCR overexpressing abolished the inhibition effect of miR-140-3p (). We also found that overexpression of ANCR alone up-regulated the expression of HNRNPA1 (lane 3 compared to lane 1). These findings indicated that ANCR promoted the expression of HNRNPA1 via sponging miR-140-3p.
Figure 7. LncRNA ANCR regulated the expression of HNRNPA1 via sponging miR-140-3p. A. The binding sites between miR-140-3p and HNRNPA1 3ʹ-UTR, and the binding sites between miR-140-3p and lncRNA ANCR. B. 293T cells were transfected with pGL3-ANCR-wt or pGL3-ANCR-mut, miR-140-3p mimic or Pre-NC (negative control). The luciferase activities were detected using dual luciferase reporter assay. C. RIP assay showed AGO2 antibody could bind to ANCR and miR-140-3p in HepG2 and Huh7 cells. D. miR-140-3p expression was detected in HepG2 cells after transfected with vector, ANCR, or ANCR mut-MRE using qRT-PCR. E. The expression of HNRNPA1 was detected in HepG2 cells using western blot. F. The expressions of HNRNPA1, ANCR, and miR-140-3p were detected in HepG2 and Huh7 cells after transfected with si-control+NC, si-ANCR, and si-ANCR+miR-140-3p inhibitor. G. HNRNPA1 expression was detected in HepG2 and Huh7 cells after transfected with pre-NC+vector, miR-140-3p mimic+vector, pre-NC+ANCR, and miR-140-3p mimic+ANCR, and the relative intensity of HNRNPA1 protein expression was analysed. **p < 0.01, compared with Pre-NC, vector or ANCR. Statistical analysis of qRT-PCR was calculated by 2−△△CT.
LncRNA ANCR promoted the EMT and invasion/migration of HCC cells via up-regulating HNRNPA1 expression
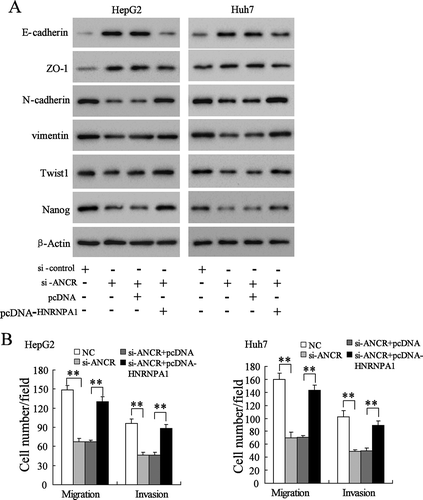
To further verify whether ANCR was involved in the EMT and invasion/migration of HCC cells via modulating HNRNPA1, we transfected si-ANCR and pcDNA-HNRNPA1 into HepG2 and Huh7 cells. As shown in , si-ANCR up-regulated the expressions of epithelial marker E-cadherin and tight junction protein ZO-1, down-regulated the expressions of mesenchymal markers N-cadherin, vimentin, Twist1 (an important transcription factors of EMT) and Nanog in HepG2 and Huh7 cells, whereas HNRNPA1 overexpression abolished these effects. Meanwhile, si-ANCR inhibited the invasion and migration of HepG2 and Huh7 cells, whereas HNRNPA1 overexpression abolished these effects (, P < 0.01). These findings indicated that ANCR promoted the EMT and invasion/migration of HCC cells via regulating HNRNPA1.
Figure 8. LncRNA ANCR promoted the EMT and invasion/migration of HCC cells via regulating HNRNPA1 expression. A. HepG2 and Huh7 cells were transfected with si-control, si-ANCR, si-ANCR+pcDNA, or si-ANCR+pcDNA-HNRNPA1 for 48 h. The expressions of E-cadherin, ZO-1, N-cadherin, vimentin, Twist1 and Nanog were detected using western blot. B. The invasion and migration of HepG2 and Huh7 cells were detected using Transwell assay. **p < 0.01, compared with NC or si-ANCR+pcDNA.
Discussion
Increasing evidences have proved that lncRNAs were abnormally expressed in cancers and play vital roles in the metastasis of HCC [Citation14,Citation15]. To the best of our knowledge, this is the first study to report the dysregulation of ANCR in HCC metastasis. We found that ANCR was highly expressed in HCC tissues and HCC cells, and positively correlated with tumour size, tumour differentiation, TNM stage and portal vein tumour thrombus. Moreover, in vitro experiments showed that ANCR overexpression promoted the proliferation, migration and invasion of HCC cells, whereas interfering ANCR produced the opposite results. In vivo experiments showed that interfering ANCR remarkably decreased tumour volume, tumour weight and number of metastatic nodules in lung, indicating that ANCR interfering could suppress the growth and metastasis of HCC.
More and more studies have shown that epigenetic regulatory factors, such as histone acetylation, can regulate the expressions of lncRNAs in HCC, thereby to regulate HCC metastasis [Citation16–Citation19]. A recent report found H3/H4 histone acetylation-dependent regulation of lncRNA MIAT may promote the proliferation and invasion of HCC [Citation20]. Increased H3 histone acetylation level at CCAL promoter region could promote the expression of lncRNA CCAL in HCC cells, thereby to accelerating tumour metastasis [Citation16]. LncRNA LET expression was inhibited by HDAC3 through decreasing the histone acetylation-mediated modulation of lncRNA LET promoter region, which result in the promotion of HCC cell invasion and metastasis [Citation17]. Our results found that H3/H4 histone acetylation level at ANCR promoter was up-regulated in HCC tissues and cells than normal tissues and hepatocytes. Moreover, silencing HDAC3 significantly up-regulated ANCR expression level in Hep3B cells, indicating histone acetylation is the key factor controlling the dysregulation of ANCR in HCC. These results showed the correlation between epigenetic regulation and lncRNAs, and provided evidences for epigenetic-related dysregulation of lncRNAs in HCC.
It is already known that lncRNAs can interact with proteins or sponging microRNA to regulate the expression of target molecules, thus to involve in the metastasis of HCC [Citation21,Citation22]. Importantly, previous studies also reported ANCR could be important regulators for the stability of their binding proteins through interacting with these proteins [Citation23]. However, the regulation mechanism of ANCR in HCC metastasis remains largely unknown. Our results demonstrated that ANCR could bind to HNRNPA1, and the protein level of HNRNPA1 was positively regulated by ANCR. Furthermore, we found that ANCR could bind to miR-140-3p, and negatively regulated the expression of miR-140-3p. Interestingly, HNRNPA1 was a target of miR-140-3p, and could be negatively regulated by miR-140-3p. To the best of our knowledge, this is the first study to report the effects of ANCR on the expression of target molecule through binding to the protein and sponging miRNA in HCC metastasis, which will provide reference value for other lncRNAs likely in HCC metastasis.
In Hep3B cells, the overexpression of ANCR reduced miR-140-3p expression and increased HNRNPA1 expression (,), while the knockdown of ANCR did not affect the expressions of miR-140-3p and HNRNPA1 (). In HepG2 and Huh7 cells, the knockdown of ANCR increased miR-140-3p expression and reduced HNRNPA1 expression (), while the overexpression of ANCR did not affect the expressions of miR-140-3p and HNRNPA1 in (Supplemental Figure 2B). In LO2 cells, the overexpression or knockdown of ANCR did not significantly affect the expressions of miR-140-3p and HNRNPA1, although ANCR knockdown slightly up-regulated miR-140-3p expression (p < 0.05) (Supplemental Figure 2C). These data indicated the ANCR-dependent regulatory network with HNRNPA1 and miR-140-3p in HCC cell lines, while the expressions of HNRNPA1 and miR-140-3p may not be regulated by ANCR in normal hepatocytes. In addition, in Hep3B cells, the proliferation, migration, and invasion were promoted by ANCR overexpression (–), while they were not significantly affected by ANCR knockdown (Fig. S2). In HepG2 and Huh7 cells, the proliferation, migration, and invasion were inhibited by ANCR knockdown (–), while they were not significantly affected by ANCR overexpression (Fig. S2B). In LO2 cells, cell proliferation was not affected by ANCR knockdown or overexpression (Fig. S2C). These data suggested that the HCC cellular ANCR needed to be elevated to a specific dosage compared to normal hepatocytes to mediate the up-regulation of HNRNPA1 and the pro-metastatic effect in HCC, although more evidence was still needed.
Previous reports have demonstrated that HNRNPA1 can be degraded by ubiquitination pathway. For example, the interaction of ubiquilin-2 and HNRNPA1 enhances the degradation of HNRNPA1 [Citation24]. Also, HNRNPA1 is a direct substrate of TRAF6, and can be directly ubiquitinated by TRAF6 [Citation25]. However, the exact role of lncRNA in the regulation of HNRNPA1 degradation is not known. In this study, immunoprecipitation using HNRNPA1 specific antibody found that interfering ANCR enhanced the ubiquitination of HNRNPA1. After the treatment of CHX, HNRNPA1 protein level was down-regulated, and MG132 could abolish the increase of HNRNPA1 protein level induced by ANCR overexpression, which indicated that ANCR regulated HNRNPA1 degradation. To the best of our knowledge, no studies focused on the regulation of lncRNA in the degradation of HNRNPA1, our study will enrich the literature and provide directions for the regulation role of ANCR on protein degradation.
In conclusion, this study proposed a novel function of ANCR in the promotion of HCC metastasis by upregulating HNRNPA1 through inhibiting HNRNPA1 degradation and sponging miR-140-3p, which provided new insights into therapeutic strategies to treat HCC metastasis.
Supplemental Material
Download Zip (1.4 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Wallace MC, Preen D, Jeffrey GP, et al. The evolving epidemiology of hepatocellular carcinoma: a global perspective. Expert Rev Gastroenterol Hepatol. 2015;9:765–779.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
- Li X, Huang L, Leng X. Analysis of prognostic factors of more/equal to10 years of survival for liver cancer patients after liver transplantation. J Cancer Res Clin Oncol. 2018;144:2465–2474.
- Chen Y, Liu J, Wang W, et al. High expression of hnRNPA1 promotes cell invasion by inducing EMT in gastric cancer. Oncol Rep. 2018;39:1693–1701.
- Chaudhury A, Hussey GS, Ray PS, et al. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293.
- Chettouh H, Fartoux L, Aoudjehane L, et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res. 2013;73:3974.
- Zhou ZJ, Dai Z, Zhou SL, et al. Overexpression of HnRNP A1 promotes tumor invasion through regulating CD44v6 and indicates poor prognosis for hepatocellular carcinoma. Int J Cancer. 2013;132:1080–1089.
- Grelet S, Link LA, Howley B, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–1115.
- Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343.
- Li Z, Dong M, Fan D, et al. LncRNA ANCR down-regulation promotes TGF-β-induced EMT and metastasis in breast cancer. Oncotarget. 2017;8:67329–67343.
- Yang ZY, Yang F, Zhang YL, et al. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark. 2017;18:95–104.
- Zhang F, Peng H. LncRNA-ANCR regulates the cell growth of osteosarcoma by interacting with EZH2 and affecting the expression of p21 and p27. J Orthop Surg Res. 2017;12:103.
- Peng W, Deng W, Zhang J, et al. Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miRNA-758. Biochem Biophys Res Commun. 2018;503:815–821.
- Ni W, Zhang Y, Zhan Z, et al. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017;10:91.
- Guo D, Li Y, Chen Y, et al. DANCR promotes HCC progression and regulates EMT by sponging miR-27a-3p via ROCK1/LIMK1/COFILIN1 pathway. Cell Prolif. 2019;e12628.
- Liu Y, Yang Y, Wang T, et al. Long non-coding RNA CCAL promotes hepatocellular carcinoma progression by regulating AP-2α and Wnt/β-catenin pathway. Int J Biol Macromol. 2018;109:424–434.
- Yang F, Huo XS, Yuan SX, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096.
- Cheng CC, Liu YH, Lai YC, et al. Hypoacetylation in association with histone 3 modulation in human hepatocellular carcinoma. In Vivo. 2015;29:237–242.
- de Conti A, Dreval K, Tryndyak V, et al. Inhibition of the cell death pathway in nonalcoholic steatohepatitis (NASH)-related hepatocarcinogenesis is associated with histone H4 lysine 16 deacetylation. Mol Cancer Res. 2017;15:1163–1172.
- Huang X, Gao Y, Qin J, et al. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am J Physiol Gastrointest Liver Physiol. 2018;314:G559–g565.
- Chen J, Wu D, Zhang Y, et al. LncRNA DCST1-AS1 functions as a competing endogenous RNA to regulate FAIM2 expression by sponging miR-1254 in hepatocellular carcinoma. Clin Sci (Lond). 2019;133(2):367–379.
- Huang M, Wang H, Hu X, et al. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. Oncoimmunology. 2019;8:e1518628.
- Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59–71.
- Gilpin KM, Chang L, Monteiro MJ. ALS-linked mutations in ubiquilin-2 or hnRNPA1 reduce interaction between ubiquilin-2 and hnRNPA1. Hum Mol Genet. 2015;24:2565–2577.
- Koo JH, Lee HJ, Kim W, et al. Endoplasmic reticulum stress in hepatic stellate cells promotes liver fibrosis via PERK-mediated degradation of HNRNPA1 and up-regulation of SMAD2. Gastroenterology. 2016;150:181–193.e188.
