ABSTRACT
PIWI homologs constitute a subclass of the Argonaute family. Traditionally, they have been shown to associate with a specific class of small RNAs, piRNAs, to suppress transposable elements and protect genomic integrity in germ cells. Recent studies imply that PIWI proteins may also exert important biological functions in somatic contexts, including the brain. However, their exact role in neural development remains unknown. Hence we investigated whether PIWI proteins are involved in neuronal differentiation. By using an established cell model for studying neurogenesis, NTera2/D1 (NT2) cells, we found that a particular PIWI homolog, PIWIL4 was increasingly upregulated throughout the course of all-trans retinoic acid (RA)-mediated neuronal differentiation. During this process, PIWIL4 knockdown led to partial recovery of embryonic stem cell markers, while suppressing RA-induced expression of neuronal markers. Consistently, PIWIL4 overexpression further elevated their expression levels. Furthermore, co-immunoprecipitation revealed an RA-induced interaction between PIWIL4 and the H3K27me3 demethylase UTX. Chromatin immunoprecipitation showed that this interaction could be essential for the removal of H3K27me3 from the promoters of RA-inducible genes. By a similar mechanism, PIWIL4 knockdown also suppressed the expression of PTN and NLGN3, two important neuronal factors secreted to regulate glioma activity. We further noted that the conditioned medium collected from PIWIL4-silenced NT2 cells significantly reduced the proliferation of glioma cells. Thus, our data suggest a novel somatic role of PIWIL4 in modulating the expression of neuronal genes that can be further characterized to promote neuronal differentiation and to modulate the activity of glioma cells.
KEYWORDS:
Introduction
PIWI (P-element induced wimpy testis) homologs belong to a subclade of Argonaute family of RNA-binding proteins. They can associate with a group of small non-coding RNAs called piRNAs, which are 24–31 nucleotide long and highly enriched in germline cells [Citation1–Citation3]. Previous studies have identified an indispensable role of the PIWI/piRNA complex in maintaining the genomic integrity in germline cells by suppressing the activity of transposable elements (TE) [Citation4–Citation7]. More recently, it has been suggested that the roles of PIWI are not just restricted to germline cells; instead they may have uncharacterized functions in somatic cells and contexts. PIWIL4 has been found to suppress gene expression by H3K9 methylation in human cancer cells [Citation8]. Surprisingly, in breast cancer cells, PIWI and piRNA can also recruit epigenetic factors, such as histone methyltransferase MLL3 and histone demethylase UTX, to facilitate gene activation [Citation9]. Our previous study has shown that oestrogen can elevate PIWIL4 expression, which subsequently modulates the migration and invasion of breast cancer cells [Citation10].
Interestingly, a potential role of PIWI homologs in the nervous system has started to emerge. In one study, the presence of PIWI and piRNA has been discovered in the neurons of Aplysia and shown to modulate long term memory at the epigenetic level [Citation11]. Additionally, silencing PIWIL1 expression in the cerebral cortex of rodent brain downregulates various genes related to cytoskeletal organization and affects radial migration of cortical neurons [Citation12]. However, the exact somatic role of PIWI homologs in neural development remains to be characterized. Hence, to characterize the potential involvement of PIWI homologs in modulating neuronal differentiation, we utilized an embryonal carcinoma (EC) model, Ntera2/D1 (NT2) cells. These cells can be induced by all-trans retinoic acid (RA) to differentiate along the neuronal lineage [Citation13–Citation15].
RA has been established to promote neural patterning, neuronal differentiation and axonal outgrowth [Citation16,Citation17]. It is also essential for neurogenesis in adult hippocampus [Citation18]. In vitro, RA has been widely used to differentiate embryonic stem cells (ESCs), EC cells, neural precursor cells from hippocampus and subventricular zone (SVZ) into neurons [Citation19–Citation24]. Once transplanted, the exogenously generated neurons have been shown to successfully replace degenerated neurons in the animal models of Parkinson’s disease (PD) [Citation25], Huntington’s disease (HD) [Citation26] and stroke [Citation27], suggesting that the cell-based therapy could be a promising approach to treat neurodegenerative diseases, such as PD [Citation28].
Paradoxically, neural precursor cells have been related to glioma cell proliferation and migration [Citation29–Citation33]. Pleiotrophin (PTN), a neurite outgrowth promoting factor secreted by precursor cells, can activate Rho/ROCK signalling in glioma and cause migration of tumour cells towards SVZ [Citation31]. Indeed, glioblastoma (GBM) recurrence has been positively correlated with the proximity to neural stem cell niche [Citation30]. In addition, neuronal activity in presynaptic neurons can induce the secretion of neuroligin-3 (NLGN3) which serves as a potent mitogen for high-grade glioma (HGG) cells [Citation32].
In this study, we found that a particular PIWI homolog, PIWIL4, was increasingly upregulated throughout the course of RA-mediated neuronal differentiation. Suppression of PIWIL4 upregulation significantly reduced the RA-induced expression of neuronal markers, while partially restored that of ESC markers, NANOG and OCT-4. Then we noted an interaction between PIWIL4 and the H3K27me3 demethylase UTX and showed that PIWIL4 was needed to efficiently remove the repressive histone mark at the promoters of neuronal genes, such as CYP26A1 and NEUROD1. We even noticed that silencing PIWIL4 decreased the expression of PTN and NLGN3, at least partially through maintaining the H3K27me3 modification at their promoters. Consistently, the conditioned medium collected from PIWIL4-depleted NT2 cells significantly reduced glioma cell proliferation. These observations suggest that PIWIL4 may regulate not only the neurogenic genes that are essential for normal neurogenesis but also those implicated in modulating glioma cell activity.
Materials and Methods
Cell culture
The human NTera2 clone D1 cell line (NT2) and 293 T cells were purchased from ATCC. The U87 MG cell line was a kind gift from A/Prof Lim Kah Leong (Department of Physiology, National University of Singapore). All the cell lines were cultured using Dulbecco’s Modified Eagle’s Medium with 10% FBS. They were maintained at 37°C in 5% CO2.
For NT2 cell differentiation, cells were treated for 28 days with 10 µM all-trans retinoic acid (Sigma-Aldrich) with the medium changed every 3 days.
Cells were transfected using Lipofectamine 3000 with siRNAs (100 nM). The MISSION® pre-designed siRNAs used for transfection were purchased from Sigma-Aldrich. The sequence information is as follows: siPL4-1 (SASI_Hs01_00210309), siPL4-2 (SASI_Hs02_00366170). The third sequence of siPL4-3 (GGACGGAACTTCTATAATC) adopted from a previous report [Citation9] was synthesized with IDT. The ON-TARGETplus SMARTpool siRNA targeting PIWIL4 (p-siPL4) was purchased from Dharmacon. The negative control siRNA was purchased from Qiagen.
The PIWIL4-pLX304 plasmid was obtained from DNASU Plasmid Repository. The PIWIL4 coding sequence was then cloned into pcDNA3.1-3xHA vector using the BamHI and XhoI sites. pCS2-UTX-F (plasmid #17438) and pCAG-HA-NLGN3 WT (plasmid #59318) were purchased from Addgene. Human PTN was cloned from the cDNA of NT2 cells as the template and inserted into pcDNA3.1(+)-3xFLAG vector using the BamHI and EcoRI sites. All the cloned sequences were validated by Sanger sequencing.
Western Blot
Culture medium was removed carefully, and cells washed twice with 1X PBS. Then 1X RIPA buffer (Pierce, ThermoFisher Scientific, Waltham, MA, USA) with protease (Roche, Sigma-Aldrich, USA), phosphatase inhibitors (Pierce, ThermoFisher Scientific, Waltham, MA, USA) were added to cells and incubated for 15 minutes on ice to lyse the cells. The lysate was collected using cell scrapper and centrifuged at 14,000 g for 15 minutes. The supernatant was collected and further used for protein concentration estimation using Bradford reagent (Bio-Rad, Hercules, CA, USA). 60 µg total proteins were loaded on SDS-PAGE gel and separated at 100 V for 2 hours. Then they were transferred onto PVDF membrane using semi-dry transfer cassette (Bio-Rad, Hercules, CA, USA). The blots were blocked with 5% skimmed milk for 1 hour at room temperature followed by incubation with the following primary antibodies overnight at 4°C: PIWIL1 (1:1000; ab12337, Abcam), PIWIL2 (1:1000; ab181340, Abcam), PIWIL3 (1:1000; SAB4200150, Sigma Aldrich), PIWIL4 (1:1000; ab87939, Abcam), OCT4 (1:1000; #2750, Cell Signalling Technology), NANOG (1:1000; #3580, Cell Signalling Technology), HOXA1(1:1000; ab168179, Abcam), MAP2 (1:1000; #4542, Cell Signalling Technology), TUBB3 (1:1000; #4466, Cell Signalling Technology), NLGN3 (1:400; NBP1-90,080, Novus biologicals), PTN (1:300; sc-74,443, Santa Cruz), UTX (1:1000; #33,510, Cell Signalling Technology), HA-Tag (1:1000; #3724, Cell Signalling Technology) and β-actin (1:2000; sc-47,778, Santa Cruz). The next day, after washing with 1X TBST, the blots were incubated with HRP-conjugated secondary antibodies (anti-mouse or anti-Rabbit, 1:3000, Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) for 1 hour at room temperature. After further washing with 1X TBST, the immunoreactive bands were detected using the chemiluminescence detection solution (Pierce, ThermoFisher Scientific, Waltham, MA, USA).
To detect PTN and NLGN3 in the conditioned medium of NT2 cells, the medium was collected and concentrated using the Amicon® Ultra-4 Centrifugal Filter Unit (10,000 MWCO, Cat# UFC801008, Merck Millipore, MA, USA). The concentrate was then subject to immunoblotting as described above.
Real-time PCR
RNA was extracted using Trizol reagent (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) as per manufacturer’s protocol. 2 µg of total RNAs were initially subjected to DNase treatment (TURBO DNase, Ambion) for 30 minutes at 37°C to get rid of traces of DNA contamination. The enzyme activity was inactivated by the treatment with EDTA at 70°C for 10 minutes, followed by cDNA conversion using the GoScript Reverse Transcription System (Promega). Real-time qPCR was performed using the GoTaq qPCR Master Mix (Promega) in the Real-Time PCR System (7900HT, Applied Biosystems, USA). The 10 µl reaction mixture that includes SYBR master mix, 1 µM forward and reverse primer, nuclease-free water and cDNA was added to each well of optical 96 well reaction plate. The reaction setup was done as per the recommendation by the instrument manufacturer. The data were analysed by the ΔΔCt approach with GAPDH as internal control. The primer sequences are given in .
Table 1. Primer sequences.
Immunocytochemistry
The coverslips were coated with Poly-L-ornithine at room temperature for 6 hours and then about 2 × 104 cells were seeded on coverslips in 24-well culture plates. After treatment, the cells were fixed with 4% paraformaldehyde for 15 minutes, washed and permeabilized with 1X PBS+0.1% Triton X-100. The cells were then blocked with 1% BSA for 30 minutes followed by incubation with PIWIL4 primary antibody (ab87939, Abcam) and H3K27me3 (#9733, Cell Signalling Technology) (1:100 in 0.2% BSA) for 90 minutes. After washing three times with PBS, the cells were then incubated with secondary goat anti-rabbit Alexa Fluor 555 (1:200 in 0.2% BSA) for 1 hour at room temperature. The cells were washed with 1X PBS thrice and counterstained with Hoechst 33342 (1: 5000) for 5 minutes. The coverslips were mounted using DAKO mounting medium and image captured using Olympus CKX53 microscope.
Co-Immunoprecipitation
The cells were washed with 1X ice cold PBS and lysed with 1 ml of lysis buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P40, 0.05% sodium deoxycholate,1X protease inhibitor) for 10 cm culture dish. The collected cells were kept on a rotor at 4°C for 20 minutes and centrifuged at 14,000 rpm for 15 minutes. The supernatant was collected and to reduce background due to non-specific absorption, the lysate was precleared with Protein G-plus agarose beads (Santa Cruz) for 1 hour at 4°C and 10% of the sample was stored as Input at −80°C. To the remaining sample, appropriate amount of control IgG or specific antibody was added and subjected to slow rotation at 4°C for 1 hour followed by adding 40 µl of Protein G-plus agarose beads. The incubation was performed overnight at 4°C. The next day, Protein G beads were sequentially washed with the lysis buffer, high salt buffer (50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 0.1% Nonidet 40) and low salt buffer (50 mM Tris-HCl (pH 7.5), 0.1% Nonidet P-40) for 10 minutes each. Then the samples were eluted by adding 1X LDS sample buffer with 5% β-mercaptoethanol and boiling the suspension to 100°C for 5 minutes, including the input samples. After centrifugation, the supernatant was collected and analysed by western blotting.
Chromatin Immunoprecipitation
The cells were crosslinked with 1% formaldehyde and the reaction quenched using 2.5 M glycine. The cells were collected in 1X ice cold PBS and the pellet was washed with buffer 1, pH 6.5 (0.25% Triton, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES) and buffer 2, pH 6.5 (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES). Then the pellet was resuspended with lysis buffer (0.1% SDS, 0.1% Sodium Deoxycholate, 10 mM EDTA, 50 mM Tris-HCl (pH 7.8–8.1), Protease Inhibitor) and sonicated with a Bioruptor (Diagenode) for 30 cycles at 4°C. After centrifugation, the supernatant was collected, and 10% Input was stored at −80°C. The remaining samples were diluted with dilution buffer (1% Triton, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl (pH 7.8–8.1), Protease inhibitor) and precleared using Protein G-plus agarose beads by rotating at 4°C for 1 hour. After preclearing, 2.5 µg of H3K27me3 antibody (#9733, Cell Signalling Technology) was added to the sample with normal Rabbit IgG (#2729, Cell Signalling Technology) as control and rotated overnight at 4°C. Then the samples were continued to rotate for 4 hours after adding 40 μl of Protein G-plus agarose beads. Then these beads were washed with TSE1 buffer, TSE2 buffer and buffer 3 at room temperature. After finally washing them with TE buffer, elution buffer was added to the beads along with the Input and rotated at room temperature for 15 minutes. 12.5 µl of 5 M NaCl was added to each tube and kept for de-crosslinking at 65°C overnight. Then further using QIAquick PCR purification kit, DNA was eluted and stored at −20°C. The primer sequences to amplify selected promoter region are given in .
Table 2. ChIP-qPCR primer sequences.
Cell proliferation assay
U87 MG glioma cells were seeded as 5,000 cells per well in 96-well plates. The conditioned medium collected from variously treated NT2 cells were mixed 1:1 with the fresh complete medium and added to glioma cells in 96-well plates. After 48 hours of treatment, 20 µl of MTS CellTiter 96®AQueousOne Solution (Cat #G3582, Promega, USA) was added to each well and the plate incubated at 37°C for 1–4 hours in a humidified, 5% CO2 Incubator. The absorbance was measured at 490 nm using microplate spectrophotometer and cell viability was plotted as a function of absorbance.
Results
Expression profiling of PIWI homologs during RA-induced neuronal differentiation
To explore the potential role of PIWI homologs in neuronal differentiation, we utilized a well-characterized cellular model, Ntera2/D1 (NT2) cells [Citation13,Citation15,Citation34]. They closely resemble pluripotent stem cells from early embryos such as human ESCs in terms of differentiation potentials. The two cell models are reported to have similar expression profile of genes and transcriptional factors in the regulatory network [Citation35,Citation36]. Upon exposure to RA, NT2 cells can be induced to express neuronal markers, and acquire similar morphological and electrophysiological properties of post-mitotic neurons of the central nervous system [Citation13–Citation15].
As confirmation, we treated NT2 cells with 10 µM all-trans RA over a course of 28 days as reported [Citation37] and the transcript levels of some typical neuronal genes were examined by qPCR at different time points. The data showed that the neural precursor cell marker, Nestin (NES) was upregulated at the initial phase of RA-induced differentiation followed by the expression of mature neuronal markers, including MAP2 and TUBB3 (), which is consistent with previous studies [Citation21].
Figure 1. PIWIL4 was upregulated with RA treatment in NT2 cells. (A) qPCR measurement of early neuronal differentiation marker, Nestin and terminal markers, MAP2 and TUBB3 at different time points of RA treatment in NT2 cells. Fold change is normalized to GAPDH as an internal reference. Expression of PIWI homologs, PIWIL1-4, was evaluated at the transcript level using qPCR (B) and at the protein level by immunoblotting (C). (D) Immunofluorescence staining of PIWIL4 showed nuclear localization in non-treated and RA-treated NT2 cells. 3D: 3-day treatment. 9D: 9-day treatment. Scale bar: 10 μm. The fluorescence intensity was measured by Image J. Bars: mean±sd; n = 9; **p < 0.01 by Student’s t-test.
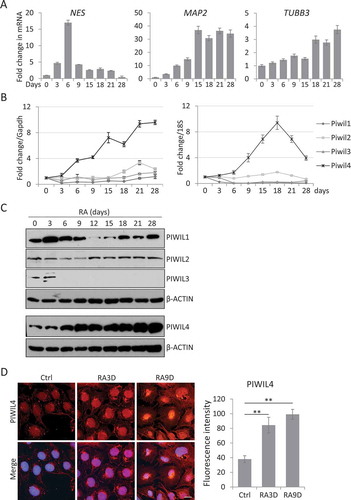
Subsequently, we followed the abovementioned treatment scheme and checked the transcript levels of the four PIWI homologs by qPCR. Remarkably, only PIWIL4 was consistently upregulated by RA over the entire period of RA treatment (). Immunoblotting confirmed that only PIWIL4 was increasingly upregulated by the RA treatment in a time-dependent manner, while PIWIL1-3 appeared to be either suppressed or minimally affected by the RA treatment (). The RA-induced expression of PIWIL4 was further validated by immunofluorescence analysis in NT2 cells (). As reported previously [Citation8,Citation38,Citation39], PIWIL4 was predominantly localized in the nucleus, implying some potential transcriptional or post-transcriptional role in neuronal differentiation. Taken together, we decided to focus on PIWIL4 in the following experiments to further pursue whether this homolog could actively regulate the process of neuronal differentiation.
PIWIL4 is involved in regulating RA-induced neuronal gene expression
To examine whether PIWIL4 was essential for NT2 differentiation, we suppressed the RA-induced expression of PIWIL4 with three individual siRNAs, siPL4-1/2/3 (). The immunoblotting showed that the suppression of PIWIL4 partially restored the expression of stemness factors, NANOG and OCT4, in NT2 cells treated with RA for three days (). On the other hand, although the canonical target of RA signalling, HOXA1 [Citation40], was upregulated by RA treatment, the induction was reduced by PIWIL4 depletion in NT2 cells (). Similarly, we observed that the RA-induced expression of some early neuronal factors, CYP26A1 [Citation41] and NEUROD1 [Citation42], was significantly decreased by knocking down PIWIL4 in NT2 cells (), suggesting a potential role of PIWIL4 in regulating neuronal gene expression. To confirm this notion and to minimize the off-target effect of individual siRNAs, we introduced the ON-TARGETplus siRNA pool targeting PIWIL4 (p-siPL4) into NT2 cells (, E), and observed a partial recovery in the expression of OCT4 and NANOG in such cells (). Moreover, a sustained knockdown of PIWIL4 by repeated transfection also suppressed the expression of terminal neuronal markers, MAP2 and TUBB3 [Citation21] (), indicating that PIWIL4 could regulate both the early and late neuronal marker genes.
Figure 2. PIWIL4 knockdown suppressed expression of neuronal markers. (A) The knockdown efficiency of three individual siRNAs of PIWIL4 (siPL4-1/2/3) was analysed by RT-PCR. The decimals below the gel strips represent the relative abundance of PIWIL4 to GAPDH as an internal reference. siCtrl: negative control siRNA. (B) NT2 cells were transfected with siPL4-1/2/3 and concurrently treated with RA for 3 days. Immunoblotting was then performed to assess the ESC markers, OCT4 and NANOG, and the early developmental gene HOXA1. (C) qPCR measurement of RA-induced expression of early neuronal genes, NEUROD1 and CYP26A1, in PIWIL4-depleted NT2 cells which were treated with RA for 3 days. (D) The knockdown efficiency of the ON-TARGETplus siRNA pool targeting PIWIL4 (p-siPL4) was assessed by qPCR at different concentrations. (E) Immunoblotting of PIWIL4 in p-siPL4-transfected NT2 cells. The decimals below the gel strips denote the relative abundance of PIWIL4 to β-ACTIN as an internal reference. (F) Immunoblotting of OCT4 and NANOG in RA-treated NT2 cells transfected with the indicated concentration of p-siPL4. (G) Immunoblotting of MAP2 and TUBB3 in p-siPL4-transfected NT2 cells which were treated with RA for 9 days. Bars: mean±sd; n = 4; *p < 0.05, **p < 0.01 by Student’s t-test.
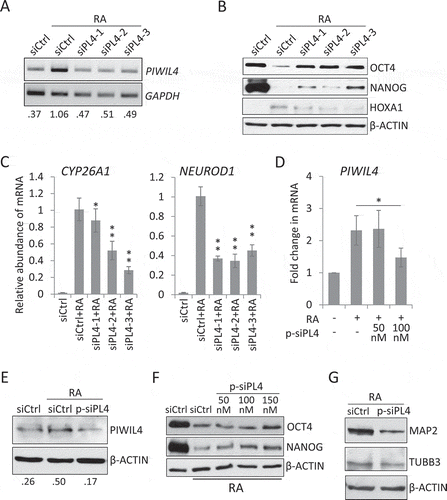
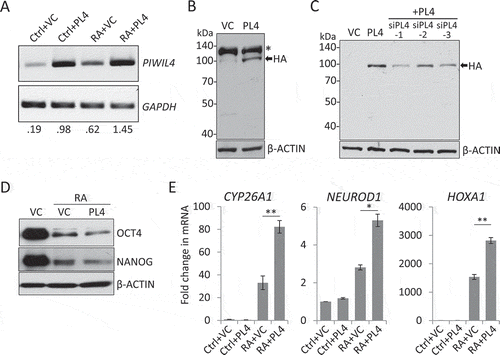
Reciprocally, we further examined the effect of PIWIL4 overexpression on stemness factors and neuronal genes in NT2 cells (-C). It was noted that while OCT4 and NANOG expression was reduced by PIWIL4 overexpression (), the RA-induced upregulation of CYP26A1, NEUROD1 and HOXA1 was significantly enhanced (), supporting the notion that PIWIL4 could be a positive regulator of the neuronal gene expression.
Figure 3. PIWIL4 overexpression increased expression of neuronal markers. The overexpression of PIWIL4 (PL4) was verified by RT-PCR (A). The decimals below the gel strips denote the relative abundance of PIWIL4 to GAPDH as an internal control. VC: vector control (pcDNA3.1-3xHA). The overexpression was confirmed by immunoblotting in NT2 (B) and 293 T cells (C). * in (B) denotes a non-specific immunoreactive band of the HA antibody, which was not seen in 293 T cells (C). (D) Immunoblotting of OCT4 and NANOG in PIWIL4-overexpressing NT2 cells treated with RA for 3 days. (E) Expression of early differentiation markers, HOXA1, NEUROD1 and CYP26A1, was evaluated by qPCR in PIWIL4-overexpressing NT2 cells treated with RA for 3 days. Bars: mean±sd; n = 4; *p < 0.05, **p < 0.01 by Student’s t-test.
PIWIL4 interacts with histone demethylase UTX
In both germline and somatic cells, PIWI homologs localized in nucleus have been found to have an important role in epigenetic regulation [Citation5,Citation6,Citation11]. For example, PIWIL4 was reported to be involved in regulating histone modification by interacting with relevant epigenetic factors [Citation8,Citation9]. In particular, UTX (KDM6A) is a di- or tri-methyl H3K27 demethylase [Citation43], and it has been reported to interact with PIWIL4 in the WDR5 complex to regulate gene expression by histone modification [Citation9]. Interestingly, our immunofluorescence study showed that PIWIL4 was more localized in the nucleus of NT2 cells (). Hence, we subsequently examined whether PIWIL4 could be involved in RA-mediated neuronal differentiation through interacting with epigenetic regulators.
For this purpose, HA-tagged PIWIL4 and Myc-tagged UTX were co-expressed in NT2 cells. The reciprocal immunoprecipitation assays revealed an interaction between PIWIL4 and UTX (). Further experiment confirmed that UTX could pull down endogenous PIWIL4 in an RA-dependent manner in NT2 cells (), implying a functional relevance of PIWIL4 in regulating neuronal gene expression epigenetically.
Figure 4. PIWIL4 was associated with UTX and involved in histone modification. (A) pcDNA-PIWIL4-HA and pCS2-UTX-F were overexpressed in NT2 cells and immunoprecipitated (IP) with α-HA or α-Myc antibody. The precipitates were immunoblotted (IB) with the indicated antibodies. (B) The endogenous interaction between PIWIL4 and UTX was examined in RA-treated NT2 cells by immunoprecipitation with α-UTX antibody, followed by immunoblotting for PIWIL4. (C) Expression of UTX during RA-mediated neuronal differentiation of NT2 cells was profiled by immunoblotting. The decimals below the gel strips represent the relative abundance of PIWIL4 to β-ACTIN as an internal reference. (D) Immunofluorescence staining of H3K27me3 in RA-treated NT2 cells with or without PIWIL4 knockdown (KD). Scale bar: 10 μm. (E) NT2 cells were transfected with p-siPL4 and treated with RA for 3 days. The protein level of UTX was then determined by immunoblotting. (F) ChIP-qPCR measurement of H3K27me3 in the promoter regions of CYP26A1, NEUROD1, NANOG and OCT4 in NT2 cells treated as indicated. The chromatin binding of H3K27me3 was normalized to IgG. pr: promoter. Bars: mean±sd; n = 4; **p < 0.01 by Student’s t-test.
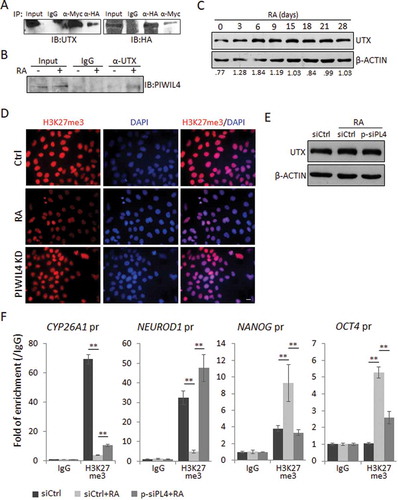
UTX (KDM6A) is known to be required for the activation of many RA responsive genes during the neuronal differentiation of NT2 [Citation43,Citation44]. Indeed, immunoblotting showed that the UTX expression was elevated by the RA treatment in NT2 cells, especially during the early phase of neuronal differentiation ().
PIWIL4 is involved in regulating H3K27me3 modification
To test whether UTX could efficiently remove the transcriptionally repressive histone mark, H3K27me3 during NT2 differentiation, and whether this process involves PIWIL4, we first immunostained the cells for H3K27me3 and strong labelling was observed in the nuclei of mock-treated cells (). As expected, RA dramatically reduced the immunolabelling, suggesting a global activation of gene transcription during neuronal differentiation (). Remarkably, PIWIL4 knockdown partially restored the immunolabelling of H3K27me3 (), without affecting the protein level of UTX (), implying that PIWIL4 could be required for UTX-mediated H3K27me3 demethylation.
H3K27me3 is a critical transcriptionally repressive mark in ESCs [Citation45]. Previously, it has been reported that the RA treatment of NT2 cells could increase UTX occupancy at the promoters of RA-inducible genes, such as HOXA1, hence removing the repressive H3K27me3 mark to activate gene expression [Citation43]. Indeed, the chromatin immunoprecipitation (ChIP) and subsequent qPCR confirmed that in NT2 cells, the RA treatment resulted in an almost complete loss of H3K27me3 at the promoters of CYP26A1 and NEUROD1 (). Remarkably, the suppression of PIWIL4 expression significantly restored the H3K27me3 enrichment at these loci (), which may explain the decreased expression of neuronal markers in these cells (). Consistently, H3K27me3 was recruited to the promoters of NANOG and OCT4 upon the RA treatment, which was significantly reduced by PIWIL4 depletion ().
PIWIL4 regulates the neuronal genes promoting glioma cell proliferation
Recent studies have suggested that neuronal differentiation and neuronal activity could have profound effect on glioma progression. In particular, Neuroligin 3 (NLGN3) and pleiotrophin (PTN), two well defined microenvironment determinants, once secreted from neuronal cells, could modify the microenvironment to potently drive the proliferation of glioma cells [Citation31,Citation32]. Interestingly, PTN is found to be highly expressed in neural precursor cells [Citation31]. Indeed, we verified that during the RA-induced differentiation of NT2 cells, the expression of PTN was gradually upregulated as reported [Citation36] whereas NLGN3 was found to be most increased during the first 15 days of the RA treatment (, B). We then inquired whether PIWIL4 was also involved in regulating these genes. For this purpose, we suppressed the RA-induced PIWIL4 expression with p-siPL4 and subsequently noticed decreased expression of NLGN3 and PTN ().
Figure 5. PIWIL4 depletion decreased the expression of critical factors for glioma. Expression of PTN and NLGN3 during RA-mediated neuronal differentiation was profiled at the transcript level by qPCR (A) and at the protein level by immunoblotting (B). dRA: days of RA treatment. (C) Immunoblotting of PTN and NLGN3 in PIWIL4-suppressed NT2 cells which were treated with RA for 9 days. The occupancy of H3K27me3 (H3) at the promoters of PTN and NLGN3 were assessed by ChIP assays followed by PCR amplification (D) and (E) qPCR measurement. In: Input; Ig: IgG in (D). The chromatin binding of H3K27me3 was normalized to Input. pr: promoter. Bars: mean±sd; n = 4; **p < 0.01 by Student’s t-test.
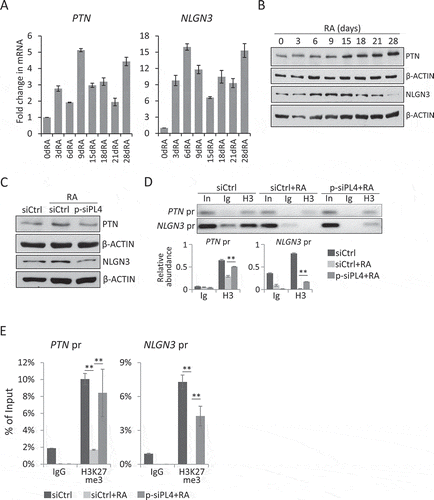
Interestingly, the Polycomb group of protein complex has been found to bind to the promoter of PTN, thus altering the H3K27me3 status and leading to gene activation in lung cancer cells [Citation46]. Hence, we analysed by ChIP assays whether PIWIL4 was involved in regulating H3K27me3 modification at the promoter of PTN and NLGN3 in RA-treated NT2 cells. PCR amplification of the precipitated DNA showed that at both promoters, the H3K27me3 level was reduced by the RA treatment, and the knockdown of PIWIL4 significantly restored the transcriptionally repressive mark (). This was then confirmed by qPCR of the precipitated chromatin (). Given the similar observations at the promoters of CYP26A1 and NEUROD1 (), it is conceivable that in the absence of PIWIL4, UTX might fail to remove the repressive histone mark efficiently during the RA-induced neuronal differentiation. Indeed, a previous study on breast cancer cells has reported a similar dependence of UTX on PIWIL1/4 in the context of H3K27me3 demethylation [Citation9].
PIWIL4 knockdown decreases glioma cell viability
These observations raised an interesting notion that PIWIL4 not only plays an essential role in RA-induced neuronal differentiation, but also has implication in regulating glioma cell viability via some microenvironment-determining neuronal proteins.
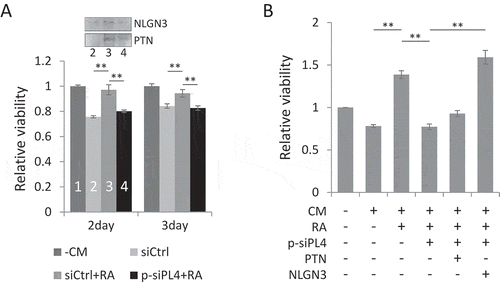
To test this hypothesis, we collected the conditioned medium (CM) from NT2 cells to treat U87 MG glioblastoma cells, and assessed their viability in MTS assays. We noticed that at both time points tested, the CM from RA-treated NT2 cells could significantly increase the cell viability of U87 MG, which may be partially attributed to the increased expression of PTN and NLGN3 (). Consistently, when the RA-induced expression of PIWIL4 was suppressed in NT2 cells, the CM showed significantly less potent effect on enhancing the viability of U87 MG (). We further observed that overexpression of NLGN3 in PIWIL4-depleted NT2 cells could restore the growth promoting effect of the CM on U87 MG cells (). These observations support an essential role of PIWIL4 in regulating the expression of some neuronal genes that could be further characterized and targeted to suppress glioma growth.
Figure 6. PIWIL4 depletion reduced the proliferation of glioma cells. (A) U87 MG glioma cells were treated for either 2 days or 3 days with the conditioned medium (CM) collected from indicated NT2 cells and subject to MTS viability assays. The gel strips above the bar graph indicate the immunoblotting of NLGN3 and PTN in the CM collected from the numbered samples. – CM: no CM used. (B) NT2 cells were treated with RA and transfected with p-siPL4 together with the plasmid overexpressing either PTN or NLGN3. The CM was then collected and used to treat U87 MG cells for 2 days. The proliferation was assessed by MTS viability assays. Bars: mean±sd; n = 3; **p < 0.01 by Student’s t-test.
Discussion
Traditionally, PIWI homologs have been found to be indispensable for epigenetic regulation of retrotransposons in germline cells [Citation2,Citation47,Citation48]. Interestingly, in Drosophila testis, Piwi proteins are responsible not only for germ stem cell maintenance, but also for the maintenance and differentiation of somatic cyst stem cell [Citation49], suggesting that the PIWI subclade of Argonaute family also contributes significantly to normal functions of somatic cells [Citation50–Citation52]. Recent studies have further revealed the presence of PIWI proteins and associated small RNAs in the brain [Citation53,Citation54] and showed that they are essential for dendritic spine development [Citation54] and organization of cytoskeleton [Citation12] in neurons. However, the exact function of PIWI homologs in neuronal development remains to be characterized. In this study, a specific PIWI homolog, PIWIL4, is found to be increasingly upregulated by RA throughout the course of neuronal differentiation ().
Silencing PIWIL4 in NT2 cells showed that the RA-induced expression of early differentiation markers such as HOXA1, NEUROD1 and CYP26A1 was universally suppressed along with the partial recovery in the expression of ESC markers such as OCT4 and NANOG (). This was later confirmed with the reciprocal experiment of PIWIL4 overexpression in RA-treated NT2 cells (). Similarly, PIWIL4 knockdown also suppressed the expression of terminal differentiation markers such as MAP2 and TUBB3 (). These observations support the notion that RA-induced PIWIL4 may facilitate the exit from stemness state and subsequently exert a neurogenic effect on NT2 cells.
A recent report has shown that PIWIL1/4 can regulate gene transcription by recruiting histone modifying proteins [Citation9]. H3K27me3 is a critical repressive mark found in ESCs and its demethylation plays an important role in the transcription activation of developmental genes. JMJD3 and UTX are two major H3K27me3 demethylases that can remove the repressive mark and promote developmental gene activation in RA signalling pathway [Citation55]. Previously it has been reported that RA treatment can increase UTX occupancy at the promoter of HOX genes, causing demethylation of H3K27me3 and transcription activation in NT2 cells [Citation43,Citation44]. Furthermore, removal of this mark at the promoters of other RA-inducible genes, such as CYP26A1 and NEUROD1, also leads to gene activation [Citation42,Citation56,Citation57].
Significantly, we noted an interaction between PIWIL4 and UTX in RA-treated NT2 cells (, B), raising an intriguing possibility that the interaction could be essential for the recruitment of UTX to remove the repressive histone mark. Indeed, our experiments showed that PIWIL4 knockdown could affect the UTX-mediated H3K27me3 demethylation at the promoters of CYP26A1 and NEUROD1 (), which could explain the decreased expression of these RA-inducible genes when PIWIL4 was knocked down (). Hence, as with the previous report [Citation9], our study supports an essential role of PIWIL4 in UTX-mediated demethylation during RA-induced neuronal differentiation.
Understanding the underlying mechanism of neuronal differentiation has great therapeutic significance in the treatment of neurodegenerative diseases, such as PD, Alzheimer’s disease (AD), HD and amyotrophic lateral sclerosis (ALS), which are characterized by massive cell death of neurons. In fact, the stem cell-derived dopaminergic neurons have undergone successful preclinical testing in PD and AD models [Citation28,Citation58,Citation59]. However, it remains a major challenge of how to consistently generate specific types of neurons in sufficient quantity from various sources of stem cells, including ESCs, iPSCs, MSCs and neural precursor cells. Hence, it is of interest that our experiments have revealed a somatic role of PIWIL4 in regulating neurogenesis from NT2 cells. Since they share similar gene expression profiles and differentiation potentials with ESCs [Citation35,Citation36], it is conceivable that PIWIL4 may play similar roles in those pluripotent cells, which will be examined in future studies.
In addition, PIWIL4 silencing downregulates the expression of PTN and NLGN3 (). The former is a chemoattractant secreted by neural precursor cells in SVZ and is shown to promote the invasion of glioma cells [Citation31], and the latter is secreted by presynaptic neurons and functions as a potent mitogen for glioma. In fact, neural precursor cells have been implicated in the recurrence of HGGs [Citation30] and neuronal activity is also found to influence many forms of HGGs [Citation32,Citation33]. Given that HGGs are the major cause of brain tumour related death in both children and adults [Citation60–Citation62], our observations suggest that we may target PIWIL4 to regulate the gene expression in stem cells and the derived neuronal cells to modulate glioma cell activity. Indeed, we observed in MTS assays that the conditioned medium collected from PIWIL4-depleted NT2 cells was less potent in driving the proliferation of glioma cells ().
Interestingly, PIWIL4 has also been found to have a direct oncogenic role in glioma cells [Citation63]. In addition, PIWIL1 has also been correlated with glioma tumour grades in patients [Citation64], and silencing PIWIL1 directly in human glioblastoma inhibits cell proliferation and migration accompanied with an increase in apoptosis of tumour cells [Citation65]. These studies clearly propose the potential involvement of PIWI homologs in glioma.
In summary, our study has identified a novel somatic role of PIWIL4 in neuronal differentiation, which can be further explored to not only enhance the efficiency of generating functional neurons from stem cells to ultimately improve the efficacy of cell-based therapy for neurodegenerative diseases, but also modulate the activity of glioma cells.
Author Contributions
C.S.S. performed a majority of the experiments in the study. Q.C., C.W., Z.S.L.H., and Z.Z. provided technical assistance in cell culture, Western blotting and imaging. Q.H. started and supervised the project. C.S.S. and Q.H. analyzed the data and wrote the manuscript.
Acknowledgments
We would like to thank Mr. Lal Zo Thanga for administrative assistance, and A/Prof Lim Kah Leong (Department of Physiology, National University of Singapore) for the kind gift of U87MG cells. This study was supported by the NUS Start-up Grant R-181-000-155-133 and MOE Tier 1 Grant R-181-000-179-114. C.S.S. and C.W. were supported by the NUS Postgraduate Scholarships.
Disclosure statement
The authors declare no competing interests.
Additional information
Funding
References
- Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207.
- Carmell MA, Girard A, van de Kant HJ, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12(4):503–514.
- Girard A, Sachidanandam R, Hannon GJ, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202.
- Aravin AA, Sachidanandam R, Bourc’his D, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799.
- Huang XA, Yin H, Sweeney S, et al. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 2013;24(5):502–516.
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–917.
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27(4):400–412.
- Sugimoto K, Kage H, Aki N, et al. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem Biophys Res Commun. 2007;359(3):497–502.
- He X, Chen X, Zhang X, et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015;43(7):3712–3725.
- Heng ZSL, Lee JY, Subhramanyam CS, et al. The role of 17β estradiol induced upregulation of Piwi like 4 in modulating gene expression and motility in breast cancer cells. Oncol Rep. 2018;40(5):2525–2535.
- Rajasethupathy P, Antonov I, Sheridan R, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149(3):693–707.
- Zhao PP, Yao MJ, Chang SY, et al. Novel function of PIWIL1 in neuronal polarization and migration via regulation of microtubule-associated proteins. Mol Brain. 2015;8(1):39.
- Andrews PW. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984;103(2):285–293.
- Hill EJ, Jimenez-Gonzalez C, Tarczyluk M, et al. NT2 derived neuronal and astrocytic network signalling. PLoS One. 2012;7(5):e36098.
- Pleasure SJ, Lee VM. NTera 2 cells: a human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J Neurosci Res. 1993;35(6):585–602.
- Dhara SK, Stice SL. Neural differentiation of human embryonic stem cells. J Cell Biochem. 2008;105(3):633–640.
- Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8(10):755–765.
- Jacobs S, Lie DC, DeCicco KL, et al. Retinoic acid is required early during adult neurogenesis in the dentate gyrus. Proc Natl Acad Sci U S A. 2006;103(10):3902–3907.
- Haile Y, Fu W, Shi B, et al. Characterization of the NT2-derived neuronal and astrocytic cell lines as alternative in vitro models for primary human neurons and astrocytes. J Neurosci Res. 2014;92(9):1187–1198.
- Okada Y, Shimazaki T, Sobue G, et al. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev Biol. 2004;275(1):124–142.
- Przyborski SA, Morton IE, Wood A, et al. Developmental regulation of neurogenesis in the pluripotent human embryonal carcinoma cell line NTERA-2. Eur J Neurosci. 2000;12(10):3521–3528.
- Takahashi J, Palmer TD, Gage FH. Retinoic acid and neurotrophins collaborate to regulate neurogenesis in adult-derived neural stem cell cultures. J Neurobiol. 1999;38(1):65–81.
- Wichterle H, Lieberam I, Porter JA, et al. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110(3):385–397.
- Wohl CA, Weiss S. Retinoic acid enhances neuronal proliferation and astroglial differentiation in cultures of CNS stem cell-derived precursors. J Neurobiol. [1998];37(2):281–290.
- Dziewczapolski G, Lie DC, Ray J, et al. Survival and differentiation of adult rat-derived neural progenitor cells transplanted to the striatum of hemiparkinsonian rats. Exp Neurol. 2003;183(2):653–664.
- Bosch M, Pineda JR, Sunol C, et al. Induction of GABAergic phenotype in a neural stem cell line for transplantation in an excitotoxic model of Huntington’s disease. Exp Neurol. 2004;190(1):42–58.
- Ikeda R, Kurokawa MS, Chiba S, et al. Transplantation of neural cells derived from retinoic acid-treated cynomolgus monkey embryonic stem cells successfully improved motor function of hemiplegic mice with experimental brain injury. Neurobiol Dis. 2015;20(1):38–48.
- Barker RA, Drouin-Ouellet J, Parmar M. Cell-based therapies for Parkinson disease-past insights and future potential. Nat Rev Neurol. 2015;11(9):492–503.
- Adeberg S, Konig L, Bostel T, et al. Glioblastoma recurrence patterns after radiation therapy with regard to the subventricular zone. Int J Radiat Oncol Biol Phys. 2014;90(4):886–893.
- Chen L, Chaichana KL, Kleinberg L, et al. Glioblastoma recurrence patterns near neural stem cell regions. Radiother Oncol. 2015;116(2):294–300.
- Qin EY, Cooper DD, Abbott KL, et al. Neural Precursor-Derived Pleiotrophin Mediates Subventricular Zone Invasion by Glioma. Cell. 2017;170(5):845–859. e19.
- Venkatesh HS, Johung TB, Caretti V, et al. Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell. 2015;161(4):803–816.
- Venkatesh HS, Tam LT, Woo PJ, et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature. 2017;549(7673):533–537.
- Hu Q, Tanasa B, Trabucchi M, et al. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat Struct Mol Biol. 2012;19(11):1168–1175.
- Abu Dawud R, Schreiber K, Schomburg D, et al. Human embryonic stem cells and embryonal carcinoma cells have overlapping and distinct metabolic signatures. PLoS One. 2012;7(6):e39896.
- Przyborski SA, Smith S, Wood A. Transcriptional profiling of neuronal differentiation by human embryonal carcinoma stem cells in vitro. Stem Cells. 2003;21(4):459–471.
- Pleasure SJ, Page C, Lee VM. Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J Neurosci. 1992;12(5):1802–1815.
- Plestilova L, Neidhart M, Russo G, et al. Expression and Regulation of PIWIL-Proteins and PIWI-Interacting RNAs in Rheumatoid Arthritis. PLoS One. 2016;11(11):e0166920.
- Zhong F, Zhou N, Wu K, et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015;43(21):10474–10491.
- Martinez-Ceballos E, Gudas LJ. Hoxa1 is required for the retinoic acid-induced differentiation of embryonic stem cells into neurons. J Neurosci Res. 2008;86(13):2809–2819.
- Sonneveld E, van den Brink CE, Tertoolen LG, et al. Retinoic acid hydroxylase (CYP26) is a key enzyme in neuronal differentiation of embryonal carcinoma cells. Dev Biol. 1999;213(2):390–404.
- Pataskar A, Jung J, Smialowski P, et al. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. Embo J. 2016;35(1):24–45.
- Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450.
- Dhar SS, Lee SH, Chen K, et al. An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 2016;44(8):3659–3674.
- Moody JD, Levy S, Mathieu J, et al. First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor. Proc Natl Acad Sci U S A. 2017;114(38):10125–10130.
- Gao SB, Feng ZJ, Xu B, et al. Suppression of lung adenocarcinoma through menin and polycomb gene-mediated repression of growth factor pleiotrophin. Oncogene. 2009;28(46):4095–4104.
- Brower-Toland B, Findley SD, Jiang L, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21(18):2300–2311.
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450(7167):304–308.
- Gonzalez J, Qi H, Liu N, et al. Piwi Is a Key Regulator of Both Somatic and Germline Stem Cells in the Drosophila Testis. Cell Rep. 2015;12(1):150–161.
- Cox DN, Chao A, Baker J, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12(23):3715–3727.
- Mani SR, Megosh H, Lin H. PIWI proteins are essential for early Drosophila embryogenesis. Dev Biol. 2014;385(2):340–349.
- Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505(7483):353–359.
- Ghosheh Y, Seridi L, Ryu T, et al. Characterization of piRNAs across postnatal development in mouse brain. Sci Rep. 2016;6(1):25039.
- Lee EJ, Banerjee S, Zhou H, et al. Identification of piRNAs in the central nervous system. RNA. 2011;17(6):1090–1099.
- Kang SC, Kim SK, Chai JC, et al. Transcriptomic Profiling and H3K27me3 Distribution Reveal Both Demethylase-Dependent and Independent Regulation of Developmental Gene Transcription in Cell Differentiation. PLoS One. 2015;10(8):e0135276.
- Gillespie RF, Gudas LJ. Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. J Mol Biol. 2007;372(2):298–316.
- Martins-Taylor K, Schroeder DI, LaSalle JM, et al. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics. 2012;7(1):71–82.
- Kikuchi T, Morizane A, Doi D, et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature. 2017;548(7669):592–596.
- Tong LM, Fong H, Huang Y. Stem cell therapy for Alzheimer’s disease and related disorders: current status and future perspectives. Exp Mol Med. 2015;47(3):e151.
- Donaldson SS, Laningham F, Fisher PG. Advances toward an understanding of brainstem gliomas. J Clin Oncol. 2006;24(8):1266–1272.
- Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710.
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820.
- Zheng J, Liu X, Wang P, et al. CRNDE Promotes Malignant Progression of Glioma by Attenuating miR-384/PIWIL4/STAT3 Axis. Mol Ther. 2016;24(7):1199–1215.
- Sun G, Wang Y, Sun L, et al. Clinical significance of Hiwi gene expression in gliomas. Brain Res. 2011;1373:183–188.
- Wang X, Tong X, Gao H, et al. Silencing HIWI suppresses the growth, invasion and migration of glioma cells. Int J Oncol. 2014;45(6):2385–2392.
