ABSTRACT
The purpose of this review is to highlight several areas of lncRNA biology and cancer that we hope will provide some new insights for future research. These include the relationship of lncRNAs and the epithelial to mesenchymal transition (EMT) with a focus on transcriptional and alternative splicing mechanisms and mRNA stability through miRNAs. In addition, we highlight the potential role of enhancer e-lncRNAs, the importance of transposable elements in lncRNA biology, and finally the emerging area of using antisense oligonucleotides (ASOs) and small molecules to target lncRNAs and their therapeutic implications.
KEYWORDS:
Introduction
Two decades ago, our laboratory was interested in studying the protective effect of an early pregnancy on breast cancer. The best experimental model for these studies at that time involved treating rats with a carcinogen. To perform these studies, we used a now obsolete, but unbiased technique called subtractive suppressive hybridization. One of the genes isolated from these studies was a new unannotated noncoding RNA which we called G.B7 [Citation1]. At this time, there were only a few long noncoding RNAs (lncRNAs) described with known functions, the best described of which was Xist. With encouragement from a colleague, Dr. John Mattick, a pioneer in emphasizing the potential importance of lncRNAs [Citation2], a postdoctoral fellow, Dr. Melanie Ginger, along with a graduate student, Amy Shore, undertook the study of this lncRNA, which we designated PINC, for pregnancy induced noncoding RNA [Citation3–5]. PINC was subsequently shown to be a marker of mammary epithelial cell fate and to interact with the polycomb repressive complex 2 [Citation5]. More recently, lncRNAs have been shown to be epigenetically regulated in a tissue-specific manner and to be excellent biomarkers of cell identity [Citation6].
Fast forwarding to the beginning of 2020, there are now estimated to be approximately 60,000 human lncRNAs and almost a third as many publications with long noncoding RNAs listed in PubMed including many excellent recent reviews (e.g. see Yao et al. [Citation7]). There are also many databases documenting associations between lncRNAs and diseases, in particular focusing on cancer, including breast cancer (e.g. see Ma et al. [Citation8]. and Huang et al. [Citation9]). Accordingly, the challenge for this review was to highlight several areas of lncRNA biology that we hope will provide some new insights for future research. These include the relationship of lncRNAs and the epithelial to mesenchymal transition (EMT) with a focus on transcriptional and alternative splicing mechanisms and mRNA stability through miRNAs. In addition, we highlight the potential role of enhancer e-lncRNAs, the importance of transposable elements in lncRNA biology, and finally the emerging area of using antisense oligonucleotides (ASOs) and small molecules to target lncRNAs and their therapeutic implications.
Transcriptional regulation by lncRNAs
LncRNAs play a crucial role in tumour progression and metastasis mainly through transcriptional regulation. LncRNAs adopt different mechanisms to regulate transcription, including modifying chromatin architecture, binding to the DNA in a sequence specific manner and bringing transcriptional factors and complexes to activate or suppress transcription [Citation10,Citation11]. At the epigenetic level, hundreds of lncRNAs have been found to interact directly with chromatin remodelling complexes, controlling their localization and activity [Citation12] (). While many still need to be functionally validated, this suggests that epigenetic modification is likely a core function of lncRNAs. Among the extensively studied lncRNAs that form a scaffold for chromatin modifying enzymes is HOTAIR (HOX transcript antisense intergenic lncRNA). HOTAIR regulates gene expression by binding to Polycomb Repressive Complex 2 (PRC2) at its 5´domain and changing its genome-wide occupancy [Citation13]. PRC2 is a critical chromatin modifying complex consists of four main subunits (Enhancer of zeste homolog 2 [EZH2], embryonic ectoderm development [EED], suppressor of zeste 12 [SUZ12], and retinoblastoma suppressor associated protein 46/48 [RbAp46/48]) [Citation14]. Many other lncRNAs bind to different components of PRC2 to induce changes in gene expression. PRC2 induces gene silencing through histone H3 lysine 27 trimethylation (H3K27m3). HOTAIR also has multiple binding sites at its 3´domain that can bind to lysine-specific histone demethylase 1A (LSD1), restrictive element 1-silencing transcription factor (REST) and REST corepressor 1 (LSD1/coREST/REST) complex demethylating H3K4 [Citation15]. HOTAIR has been found to be associated with increased risk of metastasis in breast cancer [Citation16], and required for EMT and stemness of breast, colon and cervical cancers [Citation17,Citation18].
Figure 1. Mechanisms of transcriptional regulation by lncRNAs. (A). Recruitment of chromatin remodelling complex PRC2 by lncRNAs to induce epigenetic silencing. (B). Binding to DNA methyltransferase DNMT1 and inhibiting its epigenetic modification. (C). Formation of R-loops by binding of lncRNA to the DNA duplex allowing access of the transcription factor (NF-kB) to the promoter of target genes. (D). Recruitment of a transcription factor (TF) by lncRNA to the promoter of the target gene. (E). Suppression of transcription by lncRNA GAS5, which functions as a decoy glucocorticoid response element (GRE) for the glucocorticoid receptor (GR). (F). Binding of DHFR lncRNA to the transcription factor TFIIB to suppress its transcription activity.
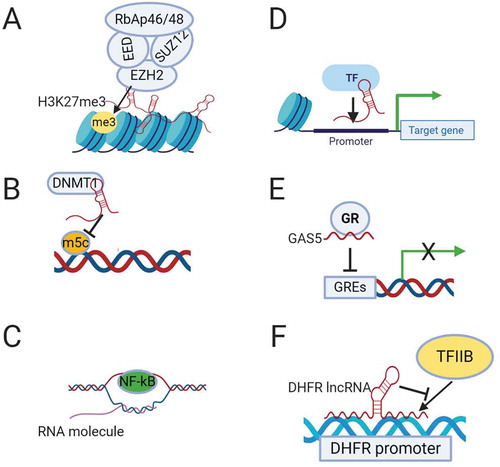
H19 is another lncRNA that binds EZH2 and promotes EMT in bladder cancer by downregulating E-cadherin [Citation19]. MALAT1 (Metastasis associated lung adenocarcinoma transcript 1), also known as nuclear-enriched abundant transcript 2 (NEAT2), is another extensively studied lncRNA that interacts with the chromatin remodelling complex PRC2 to regulate gene expression. MALAT1 is induced by TGF-β and promotes EMT by suppressing the expression of the epithelial marker E-cadherin through its binding to SUZ12 [Citation20]. NEAT1 (nuclear enriched abundant transcript1), another well-known lncRNA, is mainly localized to the nuclear paraspeckles and recruits the histone deacetylase SIN3A to the transcription suppressor FOXN3 to form a repressive complex that induces EMT and invasion in ER-positive breast cancer [Citation21].
To explain the mechanism by which lncRNAs direct PRC2 to target genes, Mondal et al. mapped the binding site of MEG3 lncRNA, which also binds to EZH2. They found that MEG3 forms RNA-DNA triplexes with GA-rich regions distal to the promoters of the target genes and directs the chromatin modifiers to these genes. Through this mechanism, MEG3 lncRNA regulates TGF-β pathway target genes [Citation22]. This may represent a general mechanism by which lncRNAs recruit different chromatin modifying enzymes to the target genes.
A second mechanism by which lncRNAs regulate transcription at the epigenetic level is through DNA methyltransferases (DNMTs) function (). LncRNA DBCCR1-003, which is derived from the locus of the tumour suppressor DBCCR1 (deleted in bladder cancer chromosome region 1), directly binds to DNMT1 and blocks its methylation of the promoter of DCCR1, subsequently suppressing the proliferation of bladder cancer cells [Citation23].
LncRNAs can regulate transcription through other mechanisms such as R-loop (DNA:RNA hybrid) formation, binding directly to the promoter or recruiting transcriptional activators or suppressors. The lncRNA VIM-AS1, which is transcribed head to head to VIM intermediate filament, is a good example. VIM-AS1 regulates the transcription of VIM by forming a stable R-loop that reduces chromatin condensation around the VIM promoter and makes it accessible for transcription factor NF-kB [Citation24] (). LncRNAs can also recruit transcription factors to the target genes (). For example, LncRNA-HIT (HOXA transcript induced by TGFβ), which has been found to increase invasion and migration in non-small cell lung cancer (NSCLC) [Citation25], regulates transcription by binding to the transcription factor E2F1 and regulating its occupancy on gene promoters [Citation26]. ELIT-1 (EMT associated lncRNA induced by TGFβ) is a newly discovered lncRNA that also works as a scaffold by binding to SMAD3 and facilitates SMAD3/4 complex binding to the promoters of target genes, promoting EMT in lung and gastric cancer [Citation27]. LncRNA BX-111, which is induced by HIF1-α, mediates hypoxia-induced EMT in pancreatic cancer by recruiting the transcription factor Y-box protein (YB1) to the ZEB1 promoter [Citation28].
A number of lncRNAs work as decoys for transcription factors and suppress their transcriptional activity. The best example is GAS5 (growth arrest specific 5), which works as a decoy for the glucocorticoid receptor (GR) [Citation29] (). GAS5 binds to DNA-binding domain of GR by mimicking the glucocorticoid receptor elements (GREs) and hence competes with DNA GREs, resulting in growth arrest during stress [Citation29]. GAS5 is considered a tumour suppressor and has been found to affect proliferation, apoptosis, EMT and metastasis in different types of cancer [Citation30,Citation31].
In addition to all these mechanisms, a few lncRNAs directly interfere with the transcriptional machinery. The transcription of the DHFR (dihydrofolate reductase) gene is regulated by the DHFR lncRNA that is transcribed upstream to the DHFR minor promoter. The DHFR lncRNA binds to both the DHFR major promoter and the transcription factor TFIIB, preventing the formation of a pre-initiation complex and suppressing the transcription of DHFR in quiescent cells [Citation32,Citation33](). RNA polymerase II elongation is also regulated by non-coding RNAs such as 7SK snRNA that is transcribed by RNA polymerase III. 7SK suppresses the activity of positive transcription elongation factor b (P-TEFb) by forming a RNA-protein complex (7SK/HEXIM/P-TEFb) that can be disassembled by the helicase activity of DDX21 (DEAD-box RNA helicase) to release the active P-TEFb [Citation34].
These are key examples of the different mechanisms by which lncRNAs contribute to transcriptional regulation of EMT. There are many other lncRNAs that play a role in regulating EMT at the transcriptional level, but their mechanism of action remains to be elucidated.
Enhancer and enhancer-associated RNAs
Enhancer regions within the human genome are defined by an increased ratio of histone H3 lysine 4 monomethylation (H3K4me1) to trimethylation (H3K4me3), as well as the acetylation of histone H3 on lysine 27 (H3K27Ac) [Citation35–37]. Furthermore, enhancers have been shown to bind acetyltransferases and coactivators such as p300 and CREB (cyclic AMP response element binding) protein [Citation38]. Enhancers play a central role in the transcriptional regulation by recruiting transcription machinery to promoter regions within the genome. In addition, they themselves are transcribed. RNAs transcribed from enhancer sequences are named eRNAs, and, based on their length, many of them fall into the category of lncRNAs (>200 nucleotides). The presence of eRNAs correlates with enhancer activity [Citation38,Citation39]. Likewise, super-enhancers are classified as cluster of enhancers that are hyperacetylated and actively transcribed [Citation40]. They are often found near actively transcribed coding genes. These super-enhancer genomic regions also produce large numbers of eRNAs. Since the initial description of eRNAs as a subtype of lncRNAs, their function and what role they may play at a cellular level has been up for debate. Many questions have been raised regarding how they are controlled, and how they exert their effects on their target genes (whether in cis through looping with their targets or in trans), especially in disease progression.
What functions do enhancer-associated lncRNAs serve and their potential roles in cancer?
Contrasting to the other types of noncoding RNAs whose roles have been elucidated, the functional role of enhancer-associated lncRNAs (e-lncRNAs) is debated. e-lncRNAs could have a role in the recruitment of transcriptional machinery or just be a byproduct of enhancer transcription. The passive role of e-lncRNAs is broadly agreed upon, however, recent studies have illustrated they may be more than just byproducts. For example, Sigova et al. observe that these e-lncRNAs are involved in recruiting the transcription factor Yin-Yang. Furthermore, by knocking down the RNA exosome complex, the authors showed that e-lncRNAs are sensitive to activity by this protein complex as accumulation of e-lncRNAs was seen [Citation41]. In contrast, a decrease in the Yin-Yang transcription factor was observed, implying that nascent e-lncRNAs are necessary for the recruitment of the Yin-Yang factor to the enhancer. The RNA exosome protein complex allows the 3ʹ end of the e-lncRNA to pull away from RNA polymerase II and other transcription machinery. Consequently, the newly transcribed e-lncRNAs can interact with transcription factors and guide them to associated enhancer regions [Citation42]. As a result, e-lncRNAs have been indicated to play a vital role in engaging with transcription factors and localizing them to cognate enhancers. The 3′-end processing activity of the RNA exosome may be important for trimming the e-lncRNAs to appropriate sizes that support functionality; alternatively, the degradation activity may reduce the level of free e-lncRNAs.
In support of this, kinetic experiments assessing how cells respond to different stimuli have demonstrated the time dependent transcription of e-lncRNAs. For example, mammalian cells exposed to pathogens and different growth stimuli exhibit the transcription of e-lncRNAs first, then mRNAs that encode transcription factors, and ultimately, the transcription of the gene of interest [Citation43,Citation44]. This indicates that e-lncRNAs might be essential in recruiting transcription factors for target gene transcription under certain stimuli. These recent findings increase our understanding of what role e-lncRNAs may serve but also highlight why enhancers are usually found near genes they regulate. Within the past decade, work on lncRNAs have highlighted several potential candidates and their role in cancer metastasis. Li et al. [Citation45] described two lncRNAs, AC026904.1 and UCA1, which upregulate Slug expression at both transcriptional and post-transcriptional levels, exerting critical roles in TGF-β-induced EMT in breast cancer. However, a concerted effort is required to understand how enhancer associated lncRNAs facilitate gene expression in cancer progression. Whether an e-lncRNA acts in a cis or trans manner to regulate transcription of target genes, specifically genes crucial for epithelial to mesenchymal transition of cells, still remains to be studied.
Cis vs trans action of e-lncRNAs
e-lncRNAs are speculated to act either in cis or trans to regulate transcription of target genes. First, in the pure cis model, e-lncRNA are thought to be byproducts of the open chromatin and the transcription machinery present at the enhancer locus. Thus, the e-lncRNAs themselves do not play a role in recruiting transcription factors rather their transcription denotes how active the enhancer is. Current studies and models indicate that in a 3D environment, chromosomal motion is limited within a nucleus [Citation46]. Therefore, intrachromosomal interactions are more prevalent than interchromosomal interactions and suggest that the cis–trans model likely reflects nature of e-lncRNAs and their function.
An alternative to the purely cis model is the cis–trans model. In this model, the e-lncRNA guides the enhancer, from which it was transcribed, to a promoter region of a coding gene. This coding gene could again be on a different chromosome or elsewhere on the same chromosome. In other words, the e-lncRNA is a component that facilitates the interaction of the enhancer and the promoter. For example, HOTTIP (long intergenic noncoding RNA), which resides at the 5′ tip of the HOXA locus, regulates the transcription of various HOXA genes in vivo through chromosomal looping of its enhancer to the promoter region of these genes. The HOTTIP RNA interacts with WDR5 (WD repeat domain 5) directly and directs WDR5/MLL (mixed lineage leukaemia) complex to the HOXA locus. Ectopically expressed HOTTIP RNAs were unable to activate genes within the HOXA locus through similar function. Accordingly, HOTTIP would seem to function in a cis–trans manner [Citation47].
A purely trans acting e-lncRNA can regulate genes on different chromosomes or genes farther away on the same chromosome from which they were transcribed Identifying trans-acting e-lncRNAs has been a challenge; however, the best example of a trans-acting e-lncRNA known so far is the aforementioned HOTAIR. HOTAIR is a lncRNA 2.2-kb in length transcribed from the antisense strand of the HOXC cluster on human chromosome 12 [Citation48]. Knockdown of HOTAIR exhibited no effects to neighbouring genes, but led to transcriptional activation of HOXD genes (through the loss of H3K27me3 marks present at the HOXD locus) located on human chromosome 2. This result shows that HOTAIR can function in a trans-acting manner [Citation49].
Finally, super-enhancers display increased levels of chromatin interaction and chromatin organization. This high order of chromatin organization is necessary for recruitment of transcription machinery and regulating expression of important genes. However, what coordinates the local chromatin organization within the super-enhancers and what recruits the regulatory complexes to the super-enhancers are yet to be fully explored. Soibam et al. has described a set of lncRNAs (which they term super-lncRNAs) that interacts with these super-enhancer genomic regions via RNA:DNA:DNA triplex formation [Citation50]. These lncRNAs can recruit essential transcription facilitators to the super-enhancers and allow for organization of local chromatin in super-enhancer regions driving transcriptional activity.
LncRNA regulation of splicing
Newly transcribed RNA molecules have coding regions (exons) and non-coding regions (introns) that are spliced out during or immediately after transcription. RNA splicing is catalysed by spliceosomes, RNA-protein complexes composed of small nuclear RNAs (snRNAs) that are assembled with small nuclear proteins to form ribonucleoproteins (snRNPs). The splicing process can produce two or more transcripts from a single RNA molecule by a process called alternative splicing (AS) increasing the proteomic complexity. This process is regulated by cis-acting sequences (enhancer and silencer sites) and trans-acting proteins called splicing factors (SFs) [Citation51]. The most common splicing factors are the serine/arginine-rich proteins (SR proteins), which have one or more RNA recognition motifs (RRMs) [Citation52]. Abnormalities in alternative splicing have been linked to many human diseases including cancers [Citation53].
LncRNAs play a role in the regulation of mRNA splicing. The first lncRNAs to be associated with RNA splicing are NEAT1 and MALAT1 (NEAT2), which have been found to be associated with SC35 SF-containing nuclear speckles [Citation54]. NEAT1 regulates AS of PPARγ during adipogenesis through its effect on the phosphorylation of the splicing factor SRp40 [Citation55]. MALAT1 is mainly localized to the nuclear speckles and interacts with SR splicing factors. Tripathi et al. found that MALAT1 modulates the nuclear localization and phosphorylation status of SR proteins (), which is required for AS regulation [Citation56]. MALAT1 depletion also has been found to change the pattern of AS for many genes [Citation56,Citation57]. The mechanism by which MALAT1 regulates the phosphorylation of SR proteins is suggested to be through its modulation of the localization and activity of kinases like SRPK1, or the phosphatases (PP1 or PP2A) that influence AS through modifying SR protein [Citation58]. Direct binding of MALAT1 to splicing factors may disrupt their interaction with other proteins and hence modulate their activity, e.g. NEAT2 binding to SFPQ (proline- and glutamine-rich SF). SFPQ works as a suppressor by binding to the proto-oncogene PTBP2 (polypyrimidine-tract-binding protein) that promotes tumour growth. MALAT1 has been found to bind competitively to SFPQ, releasing the proto-oncogene PTBP2 in colorectal cancer (). This mechanism may explain how MALAT1 induces tumour growth and metastasis [Citation59].
Figure 2. Regulation of splicing by lncRNAs. (A) MALAT1 modulates the localization and phosphorylation of serine arginine rich splicing factors (SR). (B) MALAT1 disrupts the interaction between the splicing factor SFPQ and the proto-oncogene PTBP, releasing PTBP from SFPQ. (C) LINC01133 functions as a decoy for the splicing factor SRSF6, blocking its effect on target mRNAs. (D) SAF NAT forms base-pairing with FAS pre-mRNA to recruit the splicing factor SPF45 in order to induce exon 6 exclusion. (E) ZEB2-AS1 blocks the splicing donor site on ZEB2 mRNA, leading to the inclusion of an IRES that promotes ZEB2 mRNA translation. (F) The lncRNA asFGFR2 recruits polycomb-group proteins and KDM2a to FGFR2 pre-mRNA, masking the binding site of the chromatin splicing complex and causing exon IIIb inclusion.
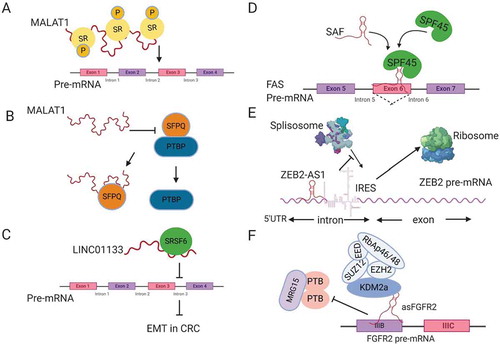
Beside its ability to shuttle SFs between the nuclear speckles and transcription start sites, MALAT1 increases the expression of the splicing factor SRSF1 in hepatocellular carcinoma. SRSF1 modulates the AS of a set of genes that play an important role in cancer progression and maintenance [Citation60]. MALAT1 also regulates the expression of another splicing factor called RBFOX2, which modulates AS of pro-apoptotic tumour suppressor KIF1B, promoting anchorage independent survival and EMT in ovarian cancer [Citation61].
Another lncRNA that binds to SF and affects colorectal cancer metastasis is LINC01133, which is downregulated by TGFβ. LINC01133 suppresses EMT by working as a target mimic for the splicing factor SRSF6, which has been found to induce EMT [Citation62]. By this mechanism, LINC01133 works as a decoy for SRSF6 titrating it away from its target mRNA (), thus suppressing EMT induction by SRSF6.
Another mechanism by which lncRNAs modulate AS involves a group of lncRNAs that are transcribed in the opposite direction to some mRNA called Natural Antisense Transcripts (NATs). NATs can base pair with their sense transcripts, forming RNA-RNA duplexes to regulate the stability, nuclear export and splicing of mRNA [Citation58,Citation63]. One of the known NATs that plays an important role in protecting cancer cells from programmed cell death, or apoptosis, is SAF lncRNA. SAF is transcribed anti-sense to the FAS ligand (FASL), which initiates apoptosis by binding to the FAS receptor at the cell membrane. SAF binds to FAS pre-mRNA overlapping exon 6 that codes for transmembrane sequences. At the same time, it binds to splicing factor 45 (SPF45), which facilitate AS of FASL pre-mRNA leading to exon 6 skipping (). The FAS transcript without exon 6 produces soluble FAS (sFAS) that cannot induce cell death. By this mechanism, SAF lncRNA protects cancer cells from apoptosis [Citation64]. SAF is an example of a NAT that mediates exon skipping. ZEB2NAT or ZEB2-AS1 (zing finger E-box- binding homeobox 2 antisense RNA 1) is another example for a NAT that mediates intron inclusion of the transcription factor ZEB2 inducing EMT [Citation65]. ZEB2NAT sequences are complementary to an intron on the 5´UTR of ZEB2 mRNA, which has an internal ribosomal entry site (IRES) that is normally removed by splicing. ZEB2NAT binds to ZEB2 pre-mRNA, covering a splicing donor site at that intron, leading to conservation of IRES and inducing ZEB2 translation [Citation65] ().
Antisense lncRNAs can also regulate AS through recruitment of chromatin modifiers to their sense locus. The best studied example is lncRNA asFGFR2 (antisense for fibroblast growth factor receptor 2, FGFR2). In epithelial cells, asFGFR2 binds to the FGFR2 locus and recruits Polycomb-group proteins and H3K36 demethylase KDM2a, masking the binding site for the chromatin-splicing complex. By this mechanism, asFGFR2 promotes inclusion of exon IIIb of FGFR2 in epithelial cells that is normally excluded in mesenchymal stem cells, which lack asFGFR2 transcript [Citation66] ().
From these examples, we can conclude that lncRNAs regulate alternative splicing through several mechanisms including: (1) Regulating the transcription of some SFs, (2) Interacting with SFs modulating their nuclear localization and activity, (3) Altering the phosphorylation status of SFs, (4) Working as a decoy for splicing factors and (5) Antisense lncRNAs (NATs) form RNA-RNA duplexes with sense RNAs to activate or suppress splicing site [Citation58].
LncRNA regulation of miRNAs
One of the best characterized functions of lncRNAs is by working as a competitive endogenous RNA (ceRNA), which is also one of the mechanisms by which lncRNAs regulate mRNA stability (see also section on Transposable Elements and ). These lncRNAs are also described as ‘molecular sponges’, where they compete with mRNAs for binding miRNAs [Citation67]. Among the extensively studied microRNAs that play a crucial role in cancer initiation and metastasis is the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429), which inhibits EMT by targeting ZEB1 and ZEB2 [Citation68–71]. A number of lncRNAs have been demonstrated to promote EMT by working as sponges for the miR-200 family. The most studied lncRNAs that function as ceRNAs for miR-200s are HULC (highly upregulated in liver cancer) [Citation72,Citation73], which induces EMT by competing with ZEB1 for miR-200a-3p [Citation74]; LncRNA-ATB (activated by TGFβ), which induces TWIST by targeting miR-200c [Citation75,Citation76]; and lncRNA-HIT, which is associated with tumour growth and metastasis in breast cancer [Citation77].
Other EMT-related miRNAs and mRNAs are also regulated by lncRNAs through the ceRNA mechanism. For instance, H19 binds to miR-200s and miR-138, de-repressing ZEB1, ZEB2 and vimentin expression in lung cancer and colorectal cancer [Citation78,Citation79]. LncRNA-PNUTS, derived from PNUTS (phosphatase 1 nuclear targeting subunit) pre-mRNA in the absence of the splicing silencer hnRNP E1 that is suppressed by TGFβ, induces EMT by competing with PNUTS mRNA for binding to miR-205 [Citation80]. Linc-ROR has been found to increase invasion and metastasis in triple negative breast cancer (TNBC) by working as ceRNA for ARF6, a small GTPase that facilitates cellular invasion, by binding to miR-145 [Citation81]. Linc-ROR also binds to other miRNAs such as miR-205, de-repressing ZEB2 expression to induce EMT [Citation82]. TUG1 (Taurine upregulated 1) induces EMT by serving as ceRNA for different miRNAs such as miR-145, which targets ZEB1 [Citation83].
A few other lncRNAs compete with Snail1 and Snail2 (Slug) for miRNA binding such as CAR10 (chromatin-associated RNA 10), which binds miR-30 and miR-203 and induces metastasis of lung adenocarcinoma [Citation84]. LncRNA TTN-AS1, associated with increased invasion and metastasis in oesophageal squamous cell carcinoma, induces EMT by competing for miR-133b binding. This stabilizes miR-133b target mRNAs such as Snail1, HuR and FSCN1 (actin-binding protein fascin homolog1) [Citation85]. FSCN1 is required for the migration and invasion of cancer cells and has been found to be associated with poor prognosis [Citation86]. Another lncRNA that competes with Snail1 and Snail2 for binding miR-203 is HCP5 (histocompatibility leukocyte antigen complex P5) lncRNA that is induced by TGFβ and promotes metastasis in lung adenocarcinoma [Citation87]. TINCR (terminal differentiation-induced noncoding RNA) induces EMT by titrating miR-125b away from Snail1 in Trastuzumab-resistant breast cancer [Citation88]. UCA1 lncRNA increases the expression of Slug in breast cancer by competing for binding to miR-1 and miR-203a [Citation45].
In addition to regulating miRNAs function by sponging, lncRNAs can also regulate the expression of miRNAs at the epigenetic level. HOTAIR induces EMT in gastric cancer by suppressing the expression of miR-34a, subsequently de-repressing the expression of its target c-Met and Snail [Citation89]. MEG8 induces EMT by recruiting EZH2 to the regulatory region of miR-34a and miR-203, resulting in upregulation of Snail1 and Snail2 transcription factors that suppress the expression of E-cadherin in lung and pancreatic cancers [Citation90]. MEG3 recruits JARID2 and EZH2 to the promoters of E-cadherin and miR-200 family genes, silencing them and mediating EMT induction by TGF-β in lung cancer [Citation91]. The lncRNA B3GALT5-AS1 suppresses the expression of miR-203 by directly binding to its promoter, subsequently upregulating ZEB2 and Snail2 to induce EMT in colon cancer [Citation92].
Collectively, these examples illustrate that the interplay between lncRNAs and miRNAs is crucial for fine-tuning the level of expression of EMT markers and hence regulating metastasis in different types of cancers.
LncRNA regulation of mRNA stability
Regulation of mRNA stability shares a similar feature to AS regulation, being mediated by cis-acting sequences like AU-rich elements (AREs) and trans-acting proteins such as RNA binding proteins (RBPs), including stabilizing and destabilizing factors. LncRNAs have also been found to play a vital role in regulating mRNA stability by different mechanisms:
The first mechanism is by recruiting different stabilizing and destabilizing RBPs to the target mRNA [Citation52]. Linc-ROR has been found to bind both stabilizing and destabilizing factors to regulate c-Myc mRNA stability. On one hand, linc-ROR interacts with hnRNP1 to facilitate its binding to c-Myc mRNA and hence stabilizes c-Myc mRNA (). On the other hand, linc-ROR binds AUF1 (AU-rich element RNA-binding protein 1), which is known to destabilize mRNAs, and inhibits its binding to c-Myc mRNA [Citation93]. Furthermore, Linc-ROR suppresses p53 induction by DNA damage by titrating hnRNP1 away from p53, since hnRNP1 binding to p53 increases its translation [Citation94]. Another example of a lncRNA that binds to mRNA-stabilizing proteins is LAST. LAST (lncRNA-assisted stabilization of transcripts) is an oncogenic lncRNA, induced by c-Myc, that promotes cell cycle progression by stabilizing CCND1 (cyclin D1) mRNA through its interaction with CNBP, a RNA binding protein [Citation95].
Figure 3. LncRNA regulation of mRNA stability. (A) Linc-ROR mediates binding of hnRNP I to c-Myc mRNA and prevents AUF1 from binding to c-Myc. (B) LncRNA PDCD4-AS1 forms base-pairing with PDCD4 mRNA, blocking HuR-binding site. (C) LncRNA MACC1-AS1 directly binds MACC1 mRNA and stabilizes the mRNA by activating AMPK and inducing lin28 binding to MACC1 mRNA. (D) LncRNA OCC-1 binds HuR RBP and enhances its interaction with E3 ubiquitin ligase β-TrCP1 for proteasomal degradation. (E) LncRNA lnc-ASNR, sequesters AUF1 in the nucleus and reduces its cytoplasmic pool, leading to an increase in the stability of AUF1 target mRNAs. (F) ½ sbsRNA lncRNA forms base-pairing with an Alu element at the 3´UTR of SMD target mRNAs to generate STAU1-binding site (SBS). STAU1 recognizes SBS and binds the ½ sbsRNA/mRNA duplex to initiate SMD.
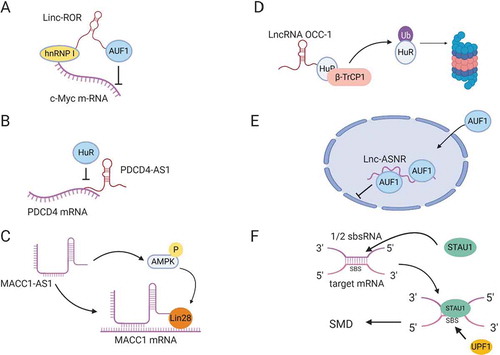
The second mechanism that lncRNAs adopt to regulate mRNA stability is by directly binding to mRNAs. One example is LncRNA AC132217.4 that is induced by KLF8 (Krueppel like factor 8) and promotes EMT in oral squamous cell carcinoma by binding to the 3´UTR of IGF2 (Insulin-like growth factor 2) mRNA in order to increase its stability [Citation96]. NATs can also regulate the stability of their sense mRNAs. PDCD4-AS1 is a lncRNA that is complementary to PDCD4 (programmed cell death 4) mRNA, a tumour suppressor gene suppressed in TNBC and metastatic diseases. PDCD4-AS1 binds to PDCD4 mRNA and inhibits HuR binding, thus stabilizing PDCD4 mRNA in epithelial cells () [Citation97]. Another example of NAT regulation is MACC1-AS1 (metastasis associated in colon cancer protein1-antisense 1) lncRNA, which enhances metabolic plasticity in gastric cancer by stabilizing MACC1 mRNA mediated by AMPK/lin28 pathway () [Citation98].
Third, some lncRNAs regulate the stability of mRNAs indirectly by modulating the stability of RBPs required for mRNA stability. LncRNA OCC-1 (over expressed in colorectal cancer) has been found to reduce cell death in colon cancer. LncRNA OCC-1 binds directly to HuR, which stabilizes thousands of mRNAs, and targets it for ubiquitination by enhancing its interaction with E3 ubiquitin ligase β-TrCP1 (). By this mechanism, lncRNA OCC-1 destabilizes mRNAs that are required for cell growth through the promotion of HuR degradation [Citation99].
Fourth, lncRNAs can function as a decoy for RBPs that mediate mRNA degradation [Citation7]. Lnc-ASNR (apoptosis suppressing-noncoding RNA) is a nuclear localized lncRNA that is upregulated in cancer cells. Lnc-ASNR interacts with AUF1, which is known to induce degradation of Bcl2, an apoptotic suppressor, mRNA, and reduces its cytoplasmic pool, ultimately suppressing apoptosis () [Citation100].
Lastly, lncRNAs can target mRNAs for decay through processes like staufen mediated decay (SMD). Staufen (STAU1) is a double-stranded RNA (dsRNA) binding protein that binds intramolecular or intermolecular dsRNAs to initiate SMD. A number of lncRNAs have been found to form partial base-pairing with the 3´UTR of target mRNAs through their Alu elements, resulting in the formation of STAU1-binding sites (SBSs) that recruit STAU1 to initiate SMD () (see also section on Transposable Elements). These lncRNAs are called half-STAU1-binding site RNAs (1/2-sbsRNAs) and have been shown to target the SERPINE1 and SOWAHC mRNAs for degradation [Citation101].
Transposable elements: a new paradigm of LncRNA function
Transposable elements (TEs) are well recognized to be pervasive in the mammalian genome yet their roles in gene regulation still remain elusive. TEs comprise 50% of the human genome [Citation102] and up to 95% of the genome in some plants [Citation103]. TEs have been the driving force of major genome expansions and evolution of eukaryotes through their capability to multiply within the genome as found in maize by Barbara McClintock’s pioneering work [Citation104]. In cancer, LINE-1 transposition occurs somatically in 53% of tumours and contributes to tumour evolution and metastasis [Citation105]. TEs that are either transposition incompetent or inactive are located within lncRNAs, UTRs or introns of mRNAs as well as intergenic regions. A number of these TEs have the potential to act in cis or in trans to influence lncRNA’s localization, stability, splicing, transcription, and the lncRNA’s capacity to regulate metabolism of other transcripts particularly mRNAs [Citation101,Citation106,Citation107]. In this section, we are going to discuss in detail molecular functions of TEs that are part of lncRNAs and how they may contribute to cancer development.
Eighty three percent of lncRNAs contain TEs, which comprise 42% of lncRNA sequences, whereas only 6.2% of protein-coding genes contain TEs, which comprise only 0.32% of their nucleotides [Citation108]. This large contribution of TEs to lncRNA composition is possibly due to the fact that lncRNAs are more evolutionarily malleable since their sequences can tolerate insertion or deletion driven by TE-mediated transposition or recombination due to TE’s high sequence homology among its many copies across the genome. In contrast, protein-coding genes are more likely to suffer a loss of function due to such mutations [Citation109].
There is substantial evidence supporting a role for TEs as functional domains of lncRNAs that can mediate the involvement of lncRNAs in the aetiology of cancer as both cis and trans regulators. In terms of cis regulation, TEs, such as Alu, that are embedded in lncRNAs can influence the localization of lncRNAs by mediating their interactions with compartment-specific RNA-binding proteins (RBPs) such as HNRNPK [Citation110] (). We have recently demonstrated that a SINEB1 of MALAT1, a well-known metastasis-associated lncRNA that is frequently mutated in endocrine-resistant breast cancers, mediates MALAT1's nuclear retention [Citation111]. Deletion of the SINEB1 in the endogenous Malat1 with CRISPR-Cas9 in a mammary epithelial cell line causes MALAT1 to dissociate from HNRNPK, mislocalize to the cytoplasm and subsequently induce apoptosis and DNA damage as a result of a loss of function of nuclear MALAT1 [Citation111]. In addition to MALAT1, lincRNA-p21, another well-known cancer-associated lncRNA involved in the p53-mediated stress response, has inverted repeat Alu elements (IRAlus) that regulate lincRNA-p21’s localization to paraspeckles [Citation112]. These studies indicate that TEs that regulate the localization of cancer-associated lncRNAs can potentially impact the ability of the lncRNA to influence cancer development.
Figure 4. TE-mediated regulation of lncRNA metabolism and function in both cis and trans manners that can influence cancer development. (A) TEs can modulate lncRNA localization and stability by functioning as binding sites for RBP complexes. (B) TEs can function as miRNA binding sites on lncRNAs to control lncRNA stability and may sequester miRNAs from their active binding sites on other transcripts. (C) TEs may function as splice-sites that can be recognized by splicing regulators such as HNRNPC to regulate maturation of pre-lncRNAs. (D) TEs can from PAS to promote APA and formation of short lncRNA transcripts. (E) TEs promote formation of transcription factor binding sites to regulate lncRNA transcription. (F) TEs mediate lncRNA binding to mRNA through partial complementary base pairing in order to promote SMD. (G) TEs mediate antisense lncRNA’s ability to promote translation of sense mRNA. (H) TEs can also mediate lncRNA’s recruitment of chromatin remodelling factors to promoter regions of certain gene loci through formation of dsDNA-RNA triplexes in order to regulate mRNA transcription. (I) TEs embedded in lncRNA can direct RBP localization, aggregation/folding and function.
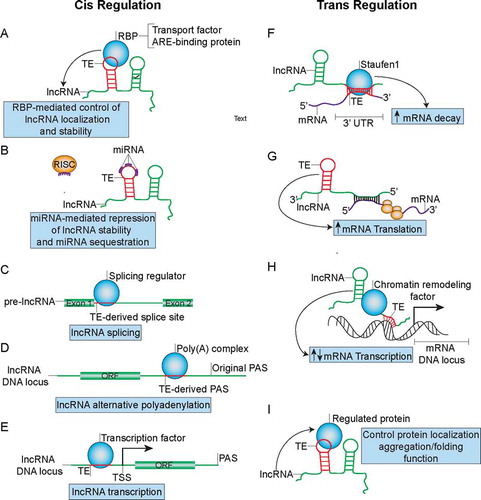
LncRNA TEs can regulate the stability or splicing of the host lncRNAs (–C) as well as the lncRNA’s ability to regulate other transcripts by functioning as binding sites for regulating RBPs such as HuR [Citation113] (). Genome-wide analysis of CLIP (crosslinking immunoprecipitation)-Seq experiments has identified a diversity of TE-specific RBP binding motifs on lncRNAs [Citation113]. In fact, poly(U) sequences in the antisense Alu, which are derived from poly(A) stretches in the sense Alu, contribute to the formation of approximately 40% of AU-rich elements (AREs) in the 3ʹ UTR of mRNAs [Citation114]. These Alu-derived elements can be bound by ARE-binding proteins such as TTP, AUF1 and HuR to either stabilize or destabilize TE-embedded lncRNAs (). The ability of lncRNAs to promote tumour development has been demonstrated to be regulated by ARE-binding proteins. For instance, NEAT1’s stability in ovarian cancer is regulated by HuR [Citation115]. Almost 50% of NEAT1’s sequence is made up of TEs, many of which are Alu, which likely function as AREs for HuR binding. In terms of splicing, intronic Alu elements, containing 5ʹ and 3ʹ splice sites, have been estimated to contribute to 5% of alternatively spliced exons in human [Citation116] and can be bound by RBPs involved in regulation of alternative splicing such as HNRNPC [Citation117] (). Since the majority of lncRNAs need to be spliced into mature lncRNAs like mRNAs, the intronic Alu elements likely regulate splicing of a fraction of lncRNA exons, a number of which may belong to cancer-associated lncRNAs.
LncRNA-embedded TEs have also been suggested to function as binding sites for microRNAs (miRNAs) to influence lncRNA’s stability or to titrate miRNAs away from binding sites on other transcripts [Citation118] (as mentioned above and ). For example, approximately 50% of the sequence of PTENP1, the pseudogene of PTEN, is comprised of TEs, predominantly LINEs. PTENP1, selectively lost in human cancer, functions as a ceRNA to titrate different miRNAs away from the tumour suppressor PTEN and has been implicated in a variety of cancers including breast cancer [Citation119,Citation120]. Thus, it is likely that the TEs may serve as miRNA binding sites on PTENP1 in order to influence cancer development. A similar mechanism may apply to the aforementioned lncRNA HULC, which regulates liver cancer tumorigenesis by potentially functioning as an endogenous ‘sponge’ for multiple miRNAs [Citation121] and contains many TEs within its sequence.
Alu embedded at the 3ʹ end of a lncRNA may function as a polyadenylation signal (PAS) () since many Alu TEs contain the canonical AAUAAA PAS sequence [Citation122] that can be bound by PABP, resulting in generation of transcripts with different lengths. Moreover, approximately 70% of human and mouse genes go through alternative polyadenylation (APA), many of which are lncRNAs [Citation123], suggesting the possibility that TE-derived PAS may drive APA in cancer-associated lncRNAs. An example of a cancer-associated lncRNA that undergoes APA is the lncRNA CCAT1, which is known to regulate long-range chromatin interactions at the MYC locus in colon cancer [Citation124].
TEs in the promoter regions of lncRNAs have also been associated with the tissue specificity of lncRNA expression and can function as binding sites for transcription factors [Citation125] (). These transcription factors may either drive tissue-specific expression of lncRNAs or alternatively be titrated away from other active binding sites. For instance, the LINE L1PA2 subfamily was reported to potentially function as a promoter for placenta-specific lncRNAs [Citation125]. The FANTOM4 project identified TE-derived transcription start sties (TSSs) in 6–30% of 5ʹ-capped transcripts in human and mouse, many of which are likely lncRNAs [Citation126], implicating the prevalence of promoter TEs that drive lncRNA expression. Another analysis of FANTOM data identified a promoter TE, a LTR7 C element, in the cancer-associated lncRNA UCA1, known to promote breast, bladder and pancreatic cancer. This is thought to contribute to the formation of breast cancer-associated transcription factor binding sites that drive lncRNA UCA1 transcription [Citation127]. Additionally, these promoter TEs may induce lncRNA transcription at alternative transcription start sties.
In terms of trans regulation, TEs can mediate intermolecular lncRNA-mRNA interactions to regulate the stability of cancer-associated mRNAs (). Alu or SINE elements in human and mouse lncRNAs, respectively, upon forming double-stranded RNA (dsRNA) duplexes with Alu or SINE in the 3ʹ UTRs of mRNAs through partial complementary base pairing can be bound by the dsRNA-binding protein Staufen1, which mediates degradation of the mRNA [Citation101]. For instance, the aforementioned lncRNA_AF087999 (1/2-sbsRNA1) has been demonstrated to regulate SERPINE1 and SOWAHC through SMD [Citation101]. SERPINE1 has been associated with poor prognosis in various cancers [Citation128]. This mode of regulation by lncRNA TEs may influence the stability of a wide range of cancer-associated genes, since Alu exists in a majority of lncRNAs and in at least 5% of human mRNA 3ʹ UTRs [Citation126]. However, the presence of a 3ʹUTR Alu does not guarantee that the mRNA will undergo SMD and hence it remains to be determined which subsets of 3ʹUTR Alu elements dictate the process.
TEs that are embedded in lncRNAs can also mediate the ability of lncRNAs to regulate translation of cancer-associated mRNAs (). For example, The SINEB2 repeat embedded in the antisense lncRNA UCHL1AS promotes translation of the sense mRNA Uchl1 through an unknown mechanism [Citation106], potentially by recruiting translation initiation factors to the 5ʹ UTR of the mRNA UCHL1. The lncRNA Uchl1AS forms complementary base pairing with the mRNA Uchl1 in a 5ʹ head-to-head fashion instead of through the SINEB2 repeat. Further bioinformatic analysis has identified 59 other similar antisense lncRNAs with embedded SINEB2 elements that may function in a similar manner to promote translation of sense mRNAs [Citation106]. A number of these regulated mRNAs have been associated with cancer progression including UCHL1 [Citation129] and UXT [Citation130]. The UXT-AS1 lncRNA shown to promote translation of UXT [Citation106] has also been associated with tumour progression [Citation131].
Transcription of mRNAs can be influenced by lncRNA TEs (). The TEs can promote direct binding between lncRNAs and promoters of specific genes through complementary base pairing for subsequent recruitment of transcription regulators to the promoter region. A LINE L1 embedded in the lncRNA FENDRR functions as a DNA-binding domain to promote recruitment of PRC2 to promoters of Foxf1 and Pitx2 through complementary base-pairing and dsDNA/RNA triplex formation [Citation132]. Likewise, the lncRNA ANRIL can recruit PRC1 and PRC2 complexes to the chromatin through Alu-mediated complementary base pairing to influence expression of genes that drive cell proliferation and reduce apoptosis [Citation133]. Both FENDRR and ANRIL have been associated with cancer progression and EMT [Citation134,Citation135].
LncRNA TEs can regulate not only mRNA expression, but also protein localization, metabolism and function (). We have demonstrated that the SINEB1 of MALAT1, is required for proteostasis of TDP-43, a RBP that is prone to forming cytotoxic aggregates [Citation111]. MALAT1 without the SINEB1 loses its ability to scaffold a number of RBPs including HNRNPK, but binds stronger to TDP-43 and hijacks TDP-43 to the cytoplasm for subsequent TDP-43 aggregation and activation of apoptosis and unfolded protein response. An independent study reported the role of a L1 transcript, expressed in 25% of hepatocellular carcinomas examined, that functions as a lncRNA to induce nuclear translocation of β-catenin and activate WNT signaling[Citation136]. The Wnt pathway is known to promote cancer invasion and synergizes with different cancer pathways such as FGF to accelerate tumorigenesis [Citation137]. LncRNAs generated from Alu and SINEB2 elements can directly bind RNA polymerase II (Pol II) to form stable complexes at promoter DNA of specific genes and blocks their transcription during the heat shock response [Citation138]. One of the repressed genes is GLUD-1, which has been implicated to drive growth of IDH1-mutant glioma tumours [Citation139]. It remains to be determined whether these heat shock responsive Alu-SINEB2-derived lncRNAs have roles in cancer adaptation and evolution.
Collectively, mutations in the TEs that weaken or strengthen their abilities to bind RNA, DNA or RBP can contribute to cancer development. Particularly, ADAR’s A-to-I editing, more than 90% of which occur in Alu elements [Citation140,Citation141], can disrupt TE-mediated gene regulation in cancer, which commonly overexpress ADAR1 [Citation142]. The study on the role of the SINEB1 element in mouse MALAT1 also suggests an involvement of lncRNA TEs in potentially influencing the cancer immune microenvironment, since a global upregulation of dsRNAs and activation of PKR, a key regulator of innate immunity, upon deletion of the MALAT1 SINEB1 was observed. It is likely that not all lncRNA TEs are important in regulating lncRNA metabolism and function. Therefore, it will be important to identify those TEs that represent functional domains of lncRNAs in order to further our understanding of their role in cancer development and guide the development of emerging cancer therapeutics targeting RNAs.
Targeting lncRNAs with ASOs and small molecules
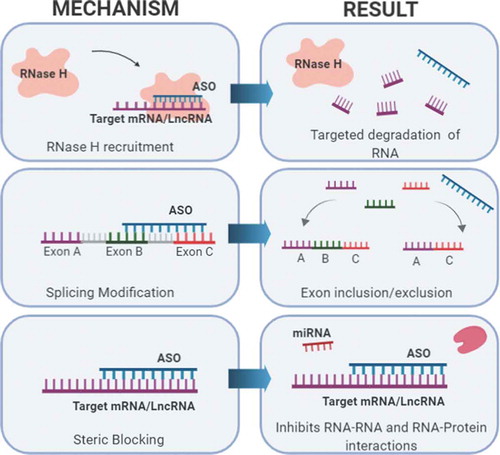
Targeting lncRNAs that play a role in cancer pathogenesis will help expand therapeutic options for cancers that are difficult to treat. Though there are multiple approaches to target lncRNAs, the use of antisense oligonucleotides (ASOs) has potential clinical applications. Advances in ASO chemistry has increased potency, safety, and tissue distribution of the oligonucleotides, making them a promising drug candidate. ASOs bind to the targeted RNA through Watson and Crick base pairing and can alter RNA function using multiple mechanisms such as steric hindrance, splicing alterations, and RNA degradation via the endogenous enzyme RNase H [Citation143] ().
Figure 5. Targeting lncRNAs with ASOs. When administered, ASOs bind to target RNAs with base pair complementarity and can exert various effects. Three mechanisms commonly used in preclinical models and human clinical trial development are shown. These mechanisms include: 1) Targeted degradation of mRNA or lncRNA via recruitment of RNase H, 2) Alternative splicing modification to include or exclude exons, and 3) Causing a steric block that inhibits RNA or protein interactions.
In order for antisense drugs to overcome the instability of RNAs in the body, modifications to the oligonucleotide are required. First generation ASOs were created with a modification in the backbone, replacing one of the non-bridging oxygen atoms in the molecule with a sulphur atom. This modification, known as a phosphorothiate bond (PS), increases the resistance to nucleotlytic degradation, allowing a systematic administration of the drug while also helping to facilitate drug uptake and RNase H cleavage of the target RNA [Citation144,Citation145]. Further modifications to the 2ʹ sugar moiety (2ʹ-O-methoxyethyl) have enhanced ASO drug-like capabilities by greatly increasing RNA binding affinity and improving tissue half-life. Known as second generation ASOs, the modifications made to the sugar backbone of these oligonucleotides cause minimal RNAse H activity [Citation146]. Though these ASOs are not ideal for RNA knockdown, they are effective as splice-switching ASOs and can be used to alter splicing patterns of respective RNA targets by blocking splicing enhancers or repressor binding sites [Citation147]. In order to overcome the reduced RNase H activity made with this modification, gapmer ASOs were created. Gapmer ASOs are chimeric RNA–DNA–RNA hybrids, where RNA residues still contain a 2-O-methoxyl modified sugar backbone [Citation146]. ASO chemistry has advanced further with the introduction of the S-constrained ethyl (cEt) modifications that once again improved potency of the drug, while limiting toxicity found in the earlier ASO generations [Citation147].
The increased potency and tolerability of second generation ASO has translated to a potential clinical benefit, and ASOs targeting different mRNAs have entered clinical trials for multiple diseases including cancer. Custersin is an ASO that targets the gene clusterin, an inhibitor of apoptosis, and has been well tolerated and shown promise when used as a single agent or in combination with chemotherapy in Phase I and II trials for the treatment of prostate cancer [Citation148]. Danvatirsen, a cET gapmer ASO targeting signal transducer and activator of transcription 3 (STAT3), has shown promising results as a single agent in a Phase II clinical trial against STAT3-enriched cancers with results in diffuse large B cell lymphoma being the most noteworthy [Citation149]. Another cET gapmer in Phase II clinical trials targets the androgen receptor and has delayed chemo resistant tumour growth in prostate cancer preclinical models [Citation150].
Targeting lncRNAs with ASO technology is still a relatively new endeavour, however there has been promising preclinical results. A large amount of lncRNAs reside in the nucleus and since ASO mediated knockdown in the nucleus can be achieved due to the abundance of RNase H within, ASOs are an ideal approach for targeted lncRNA knockdown. Identifying oncogenic lncRNAs and creating oligonucleotides for targeted inhibition may improve clinical outcome when used as a single agent or in combination with other therapeutics. The Spector laboratory has shown a proof of concept with the targeting of the nuclear enriched lncRNA, MALAT1. Subcutaneous delivery of MALAT1 ASO in the MMTV–PyMT mouse model of luminal B breast cancer caused differentiation of primary tumours as well as nearly 80% reduction in metastasis relative to non-specific ASO-treated control mice [Citation151]. Furthermore, ASO mediated MALAT1 knockdown reduced branching morphogenesis in a 3D organoid model derived from MMTV–PyMT tumours and a HER2 amplified mouse mammary tumour model [Citation151]. These results suggest that MALAT1 ASOs may emerge as a potential therapeutic for metastatic disease in several cancer types, but further assessment into the mechanisms behind this discovery is needed [Citation152]. As more lncRNAs responsible for promoting cancer dissemination become known, ASO therapies provide a way to combat metastasis and hopefully improve clinical outcome.
An alternative approach to the use of ASOs to target lncRNAs is the design and validation of small molecules to target RNA based upon RNA structure (see review in [Citation153]). Small molecules have been developed that can cross-link and cleave their RNA targets, and even compete for protein binding to the RNA. While there are still many hurdles to overcome, small molecules may ultimately be preferable as therapeutics. The clinical efficacy of both ASOs and small molecules to target lncRNAs for cancer treatment remains to be demonstrated.
EMT and RNA Modifications
EMT involves both inactivation of epithelial genes and activation of mesenchymal genes. Accomplishment of this fundamental state change includes epigenetic mechanisms (DNA methylation and histone marks) [Citation154,Citation155], alternative splicing [Citation156,Citation157], specific miRNA expression [Citation155,Citation158]and lncRNAs; the subject of this review. There are also specific RNA modifications that are enriched or repressed in the EMT state that play a role in this process. Epitranscriptomics, the study of RNA modifications, represents an exciting new frontier for identifying the importance of RNA modifications as a code in RNA’s control of gene expression. Post-transcriptionally modified nucleosides in RNA are critical for many aspects of biology including germline development, cellular signalling, neurodegeneration, circadian rhythm, and cancer [Citation159–164]. These modifications are pervasive across different RNA types including messenger RNA (mRNA) [Citation165,Citation166], transfer RNA (tRNA) [Citation167], ribosomal RNA (rRNA) [Citation168], small nuclear RNA (snRNA) [Citation169], and microRNA (miRNA) [Citation170]. In fact, nucleoside modifications are one of the most evolutionarily conserved properties of RNAs, with the sites of modification being under strong selective pressure. Many of these modifications, over 110 types currently documented (http://mods.rna.albany.edu/), as well as their prevalence and impact, have only been recently discovered [Citation171,Citation172]. During EMT, m6A modifications in mRNA, the most abundant modification found in eukaryotic mRNA [Citation173], significantly increase in cancer cells [Citation174]. The increased m6A in the coding sequence of the Snail mRNA has been shown to trigger polysome-mediated translation of Snail through the m6A binding protein or ‘reader’ YTHDF1 [Citation174]. Snail recruits chromatin modifiers which lead to repression of the CDH1 promoter. Methyltransferase-like 3 (METTL3)-mediated m6A modification has been shown to be important for EMT in both gastric and liver cancers [Citation174,Citation175]. Although currently unexplored, other RNA modifications as well as RNA editing are likely to play additional roles that are critical in regulating gene expression during the EMT process.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We apologize to those authors whose work we were unable to cite due to space limitations.
Additional information
Funding
References
- Ginger MR, Gonzalez-Rimbau MF, Gay JP, et al. Persistent changes in gene expression induced by estrogen and progesterone in the rat mammary gland. Mol Endocrinol. 2001;15(11):1993–2009.
- Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2(11):986–991.
- Ginger MR, Rosen JM. Pregnancy-induced changes in cell-fate in the mammary gland. Breast Cancer Res. 2003;5(4):192–197.
- Ginger MR, Shore AN, Contreras A, et al. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci U S A. 2006;103(15):5781–5786.
- Shore AN, Kabotyanski EB, Roarty K, et al. Pregnancy-induced noncoding RNA (PINC) associates with polycomb repressive complex 2 and regulates mammary epithelial differentiation. PLoS Genet. 2012;8(7):e1002840.
- Amin V, Harris RA, Onuchic V, et al. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nat Commun. 2015;6:6370.
- Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21(5):542–551.
- Ma L, Cao J, Liu L, et al. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019;47(D1):D128–D134.
- Huang QY, Liu GF, Qian XL, et al. Long non-coding RNA: dual effects on breast cancer metastasis and clinical applications. Cancers (Basel). 2019;11(11). DOI: 10.3390/cancers11111802.
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789.
- Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407.
- Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106(28):11667–11672.
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076.
- Shi Y, Wang XX, Zhuang YW, et al. Structure of the PRC2 complex and application to drug discovery. Acta Pharmacol Sin. 2017;38(7):963–976.
- Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693.
- Sorensen KP, Thomassen M, Tan Q, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142(3):529–536.
- Padua Alves C, Fonseca AS, Muys BR, et al. Brief report: the lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31(12):2827–2832.
- Kim HJ, Lee DW, Yim GW, et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46(2):521–530.
- Luo M, Li Z, Wang W, et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221.
- Fan Y, Shen B, Tan M, et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20(6):1531–1541.
- Li W, Zhang Z, Liu X, et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J Clin Invest. 2017;127(9):3421–3440.
- Mondal T, Subhash S, Vaid R, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743.
- Qi D, Li J, Que B, et al. Long non-coding RNA DBCCR1-003 regulate the expression of DBCCR1 via DNMT1 in bladder cancer. Cancer Cell Int. 2016;16:81.
- Boque-Sastre R, Soler M, Oliveira-Mateos C, et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci U S A. 2015;112(18):5785–5790.
- Jia X, Wang Z, Qiu L, et al. Upregulation of LncRNA-HIT promotes migration and invasion of non-small cell lung cancer cells by association with ZEB1. Cancer Med. 2016;5(12):3555–3563.
- Yu L, Fang F, Lu S, et al. lncRNA-HIT promotes cell proliferation of non-small cell lung cancer by association with E2F1. Cancer Gene Ther. 2017;24(5):221–226.
- Sakai S, Ohhata T, Kitagawa K, et al. Long noncoding RNA ELIT-1 acts as a Smad3 cofactor to facilitate TGF-β/Smad signaling and promote epithelial-mesenchymal transition. Cancer Res. 2019;79(11):2821–2838.
- Deng SJ, Chen HY, Ye Z, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37(44):5811–5828.
- Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8.
- Han L, Ma P, Liu SM, et al. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumour Biol. 2016;37(5):6847–6854.
- Ji J, Dai X, Yeung SJ, et al. The role of long non-coding RNA GAS5 in cancers. Cancer Manag Res. 2019;11:2729–2737.
- Martianov I, Ramadass A, Serra Barros A, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670.
- Nie L, Wu HJ, Hsu JM, et al. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4(2):127–150.
- Calo E, Flynn RA, Martin L, et al. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015;518(7538):249–253.
- Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–461.
- Pefanis E, Wang J, Rothschild G, et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514(7522):389–393.
- Pefanis E, Wang J, Rothschild G, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161(4):774–789.
- Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17(4):207–223.
- De Santa F, Barozzi I, Mietton F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):e1000384.
- Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319.
- Sigova AA, Abraham BJ, Ji X, et al. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350(6263):978–981.
- Lemay JF, Larochelle M, Marguerat S, et al. The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat Struct Mol Biol. 2014;21(10):919–926.
- Arner E, Daub CO, Vitting-Seerup K, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347(6225):1010–1014.
- Kim YJ, Xie P, Cao L, et al. Global transcriptional activity dynamics reveal functional enhancer RNAs. Genome Res. 2018;28(12):1799–1811.
- Li G-Y, Wang W, Sun J-Y, et al. Long non-coding RNAs AC026904.1 and UCA1: a “one-two punch” for TGF-β-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics. 2018;8(10):2846–2861.
- Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293.
- Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151–164.
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323.
- Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124.
- Soibam B. Super-lncRNAs: identification of lncRNAs that target super-enhancers via RNA:DNA:DNA triplex formation. RNA. 2017;23(11):1729–1742.
- Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15(2):108–121.
- He RZ, Luo DX, Mo YY. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6(1):6–15.
- Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009;1792(1):14–26.
- Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39.
- Cooper DR, Carter G, Li P, et al. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARgamma2 splicing during adipogenesis in 3T3-L1 Cells. Genes (Basel). 2014;5(4):1050–1063.
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938.
- Bernard D, Prasanth KV, Tripathi V, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. Embo J. 2010;29(18):3082–3093.
- Romero-Barrios N, Legascue MF, Benhamed M, et al. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018;46(5):2169–2184.
- Ji Q, Zhang L, Liu X, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111(4):736–748.
- Malakar P, Shilo A, Mogilevsky A, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017;77(5):1155–1167.
- Gordon MA, Babbs B, Cochrane DR, et al. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog. 2019;58(2):196–205.
- Kong J, Sun W, Li C, et al. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380(2):476–484.
- Khorkova O, Myers AJ, Hsiao J, et al. Natural antisense transcripts. Hum Mol Genet. 2014;23(R1):R54–63.
- Villamizar O, Chambers CB, Riberdy JM, et al. Long noncoding RNA Saf and splicing factor 45 increase soluble Fas and resistance to apoptosis. Oncotarget. 2016;7(12):13810–13826.
- Beltran M, Puig I, Pena C, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22(6):756–769.
- Gonzalez I, Munita R, Agirre E, et al. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22(5):370–376.
- Xu Q, Deng F, Qin Y, et al. Long non-coding RNA regulation of epithelial-mesenchymal transition in cancer metastasis. Cell Death Dis. 2016;7(6):e2254.
- Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6(9):6472–6498.
- Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601.
- Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(7):894–907.
- Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–2151.
- Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106.
- Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342.
- Li SP, Xu HX, Yu Y, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7(27):42431–42446.
- Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681.
- Li RH, Chen M, Liu J, et al. Long noncoding RNA ATB promotes the epithelial-mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018;9(12):1171.
- Richards EJ, Zhang G, Li ZP, et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) beta: lncRNA-hit-mediated TGFbeta-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290(11):6857–6867.
- Zhao Y, Feng C, Li Y, et al. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem. 2019;460(1–2):1–8.
- Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525.
- Grelet S, Link LA, Howley B, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19(9):1105–1115.
- Eades G, Wolfson B, Zhang Y, et al. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13(2):330–338.
- Hou P, Zhao Y, Li Z, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287.
- Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin (Shanghai). 2017;49(7):588–597.
- Ge X, Li GY, Jiang L, et al. Long noncoding RNA CAR10 promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis. Oncogene. 2019;38(16):3061–3076.
- Lin C, Zhang S, Wang Y, et al. Functional role of a novel long noncoding RNA TTN-AS1 in esophageal squamous cell carcinoma progression and metastasis. Clin Cancer Res. 2018;24(2):486–498.
- Chen Y, Tian T, Li ZY, et al. FSCN1 is an effective marker of poor prognosis and a potential therapeutic target in human tongue squamous cell carcinoma. Cell Death Dis. 2019;10(5):356.
- Jiang L, Wang R, Fang L, et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics. 2019;9(9):2460–2474.
- Dong H, Hu J, Zou K, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol Cancer. 2019;18(1):3.
- Liu YW, Sun M, Xia R, et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802.
- Terashima M, Ishimura A, Wanna-Udom S, et al. MEG8 long noncoding RNA contributes to epigenetic progression of the epithelial-mesenchymal transition of lung and pancreatic cancer cells. J Biol Chem. 2018;293(47):18016–18030.
- Terashima M, Tange S, Ishimura A, et al. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. 2017;292(1):82–99.
- Wang L, Wei Z, Wu K, et al. Long noncoding RNA B3GALT5-AS1 suppresses colon cancer liver metastasis via repressing microRNA-203. Aging (Albany NY). 2018;10(12):3662–3682.
- Huang J, Zhang A, Ho TT, et al. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44(7):3059–3069.
- Zhang A, Zhou N, Huang J, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23(3):340–350.
- Cao L, Zhang P, Li J, et al. LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. Elife. 2017;pii:e30433.
- Li X, Ma C, Zhang L, et al. LncRNAAC132217.4, a KLF8-regulated long non-coding RNA, facilitates oral squamous cell carcinoma metastasis by upregulating IGF2 expression. Cancer Lett. 2017;407:45–56.
- Jadaliha M, Gholamalamdari O, Tang W, et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018;14(11):e1007802.
- Zhao Y, Liu Y, Lin L, et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17(1):69.
- Lan Y, Xiao X, He Z, et al. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46(11):5809–5821.
- Chen J, Liu L, Wei G, et al. The long noncoding RNA ASNR regulates degradation of Bcl-2 mRNA through its interaction with AUF1. Sci Rep. 2016;6:32189.
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3ʹ UTRs via Alu elements. Nature. 2011;470(7333):284–288.
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 2005;6(9):699–708.
- Goerner-Potvin P, Bourque G. Computational tools to unmask transposable elements. Nat Rev Genet. 2018;19(11):688–704.
- Fedoroff NV. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 2012;338(6108):758–767.
- Tubio JMC, Li Y, Ju YS, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345(6196):1251343.
- Carrieri C, Cimatti L, Biagioli M, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457.
- Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science. 2016;351(6274):aac7247.
- Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13(11):R107.
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5ʹ-untranslated regions of eukaryotic mRNAs. Science. 2016;352(6292):1413–1416.
- Lubelsky Y, Ulitsky I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555(7694):107–111.
- Nguyen TM, Kabotyanski EB, Reineke LC, et al. The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res. 2020;48(5):2621–2642.
- Chillon I, Pyle AM. Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res. 2016;44(19):9462–9471.
- Kelley DR, Hendrickson DG, Tenen D, et al. Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol. 2014;15(12):537.
- An HJ, Lee D, Lee KH, et al. The association of Alu repeats with the generation of potential AU-rich elements (ARE) at 3ʹ untranslated regions. BMC Genomics. 2004;5(1):97.
- Chai Y, Liu J, Zhang Z, et al. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016;5(7):1588–1598.
- Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12(7):1060–1067.
- Zarnack K, Konig J, Tajnik M, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152(3):453–466.
- Lehnert S, Van Loo P, Thilakarathne PJ, et al. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS One. 2009;4(2):e4456.
- Li RK, Gao J, Guo LH, et al. PTENP1 acts as a ceRNA to regulate PTEN by sponging miR-19b and explores the biological role of PTENP1 in breast cancer. Cancer Gene Ther. 2017;24(7):309–315.
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038.
- Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383.
- Lee JY, Ji Z, Tian B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3ʹ-end of genes. Nucleic Acids Res. 2008;36(17):5581–5590.
- Ziegler C, Kretz M. The more the merrier-complexity in long non-coding RNA loci. Front Endocrinol (Lausanne). 2017;8:90.
- Xiang JF, Yin QF, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24(5):513–531.
- Chishima T, Iwakiri J, Hamada M. Identification of transposable elements contributing to tissue-specific expression of long non-coding RNAs. Genes (Basel). 2018;9(1). DOI:10.3390/genes9010023
- Faulkner GJ, Kimura Y, Daub CO, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41(5):563–571.
- Jiang JC, Upton KR. Human transposons are an abundant supply of transcription factor binding sites and promoter activities in breast cancer cell lines. Mob DNA. 2019;10:16.
- Andreasen PA, Kjoller L, Christensen L, et al. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72(1):1–22.
- Hussain S, Bedekovics T, Chesi M, et al. UCHL1 is a biomarker of aggressive multiple myeloma required for disease progression. Oncotarget. 2015;6(38):40704–40718.
- Zeng J, Xiang W, Zhang Y, et al. Ubiquitous expressed transcript promotes tumorigenesis by acting as a positive modulator of the polycomb repressive complex 2 in clear cell renal cell carcinoma. BMC Cancer. 2019;19(1):874.
- Yin J, Luo W, Zeng X, et al. UXT-AS1-induced alternative splicing of UXT is associated with tumor progression in colorectal cancer. Am J Cancer Res. 2017;7(3):462–472.
- Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214.
- Holdt LM, Hoffmann S, Sass K, et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9(7):e1003588.
- Hao YR, Zhang DJ, Fu ZM, et al. Long non-coding RNA ANRIL promotes proliferation, clonogenicity, invasion and migration of laryngeal squamous cell carcinoma by regulating miR-181a/Snai2 axis. Regen Ther. 2019;11:282–289.
- Miao L, Huang Z, Zengli Z, et al. Loss of long noncoding RNA FOXF1-AS1 regulates epithelial-mesenchymal transition, stemness and metastasis of non-small cell lung cancer cells. Oncotarget. 2016;7(42):68339–68349.
- Lau CC, Sun T, Ching AK, et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25(3):335–349.
- Nguyen TM, Kabotyanski EB, Dou Y, et al. FGFR1-activated translation of WNT pathway components with structured 5ʹ UTRs is vulnerable to inhibition of EIF4A-dependent translation initiation. Cancer Res. 2018;78(15):4229–4240.
- Mariner PD, Walters RD, Espinoza CA, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29(4):499–509.
- Chen R, Nishimura MC, Kharbanda S, et al. Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc Natl Acad Sci U S A. 2014;111(39):14217–14222.
- Kim DD, Kim TT, Walsh T, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14(9):1719–1725.
- Levanon EY, Eisenberg E, Yelin R, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22(8):1001–1005.
- Xu LD, Ohman M. ADAR1 editing and its role in cancer. Genes (Basel). 2018;10(1). DOI:10.3390/genes10010012
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293.
- Bennett CF, Baker BF, Pham N, et al. Pharmacology of Antisense Drugs. Annu Rev Pharmacol Toxicol. 2017;57:81–105.
- Stein CA, Subasinghe C, Shinozuka K, et al. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 1988;16(8):3209–3221.
- Monia BP, Lesnik EA, Gonzalez C, et al. Evaluation of 2ʹ-modified oligonucleotides containing 2ʹ-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. 1993;268(19):14514–14522.
- Burel SA, Han SR, Lee HS, et al. Preclinical evaluation of the toxicological effects of a novel constrained ethyl modified antisense compound targeting signal transducer and activator of transcription 3 in mice and cynomolgus monkeys. Nucleic Acid Ther. 2013;23(3):213–227.
- Zielinski R, Chi KN. Custirsen (OGX-011): a second-generation antisense inhibitor of clusterin in development for the treatment of prostate cancer. Future Oncol. 2012;8(10):1239–1251.
- Hong D, Kurzrock R, Kim Y, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med. 2015;7(314):314ra185.
- Yamamoto Y, Loriot Y, Beraldi E, et al. Generation 2.5 antisense oligonucleotides targeting the androgen receptor and its splice variants suppress enzalutamide-resistant prostate cancer cell growth. Clin Cancer Res. 2015;21(7):1675–1687.
- Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51.
- Mendell JT. Targeting a long noncoding RNA in breast cancer. N Engl J Med. 2016;374(23):2287–2289.
- Disney MD. Targeting RNA with small molecules to capture opportunities at the intersection of chemistry, biology, and medicine. J Am Chem Soc. 2019;141(17):6776–6790.
- Malouf GG, Taube JH, Lu Y, et al. Architecture of epigenetic reprogramming following Twist1-mediated epithelial-mesenchymal transition. Genome Biol. 2013;14(12):R144.
- Taube JH, Malouf GG, Lu E, et al. Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Sci Rep. 2013;3:2687.
- Shapiro IM, Cheng AW, Flytzanis NC, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7(8):e1002218.
- Warzecha CC, Jiang P, Amirikian K, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. Embo J. 2010;29(19):3286–3300.
- Taube JH, Herschkowitz JI, Komurov K, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107(35):15449–15454.
- Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806.
- Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347(6225):1002–1006.
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature Chem Biol. 2011;7(12):885–887.
- Lin S, Choe J, Du P, et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345.
- Saletore Y, Meyer K, Korlach J, et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13(10):175.
- Satterlee JS, Basanta-Sanchez M, Blanco S, et al. Novel RNA modifications in the nervous system: form and function. J Neurosci. 2014;34(46):15170–15177.
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71(10):3971–3975.
- Wei CM, Gershowitz A, Moss B. 5ʹ-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. 1976;15(2):397–401.
- Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129.
- Iwanami Y, Brown GM. Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch Biochem Biophys. 1968;126(1):8–15.
- Bringmann P, Luhrmann R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 1987;213(2):309–315.
- Kaur S, Lotsari-Salomaa JE, Seppanen-Kaijansinkko R, et al. MicroRNA methylation in colorectal cancer. Adv Exp Med Biol. 2016;937:109–122.
- Cantara WA, Crain PF, Rozenski J, et al. The RNA modification database, RNAMDB: 2011 update. Nucl Acids Res. 2011;39(Database issue):D195–201.
- Machnicka MA, Milanowska K, Osman Oglou O, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucl Acids Res. 2013;41(Database issue):D262–267.
- Sibbritt T, Patel HR, Preiss T. Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA. 2013;4(4):397–422.
- Lin X, Chai G, Wu Y, et al. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of snail. Nat Commun. 2019;10(1):2065.
- Yue B, Song C, Yang L, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142.
