ABSTRACT
The ribosomal RNA (rRNA) genes are generally organized as an operon and cotranscribed into a polycistronic precursor; therefore, processing and maturation of pre-rRNAs are essential for ribosome biogenesis. However, rRNA maturation pathways of archaea, particularly of methanoarchaea, are scarcely known. Here, we thoroughly elucidated the maturation pathway of the rRNA operon (16S-tRNAAla-23S-tRNACys-5S) in Methanolobus psychrophilus, one representative of methanoarchaea. Enzymatic assay demonstrated that EndA, a tRNA splicing endoribonuclease, cleaved bulge-helix-bulge (BHB) motifs buried in the processing stems of pre-16S and pre-23S rRNAs. Northern blot and quantitative PCR detected splicing-coupled circularization of pre-16S and pre-23S rRNAs, which accounted for 2% and 12% of the corresponding rRNAs, respectively. Importantly, endoribonuclease Nob1 was determined to linearize circular pre-16S rRNA at the mature 3′ end so to expose the anti-Shine–Dalgarno sequence, while circular pre-23S rRNA was linearized at the mature 5′ end by an unknown endoribonuclease. The resultant 5′ and 3′ extension in linearized pre-16S and pre-23S rRNAs were finally matured through 5′-3′ and 3′-5′ exoribonucleolytic trimming, respectively. Additionally, a novel processing pathway of endoribonucleolysis coupled with exoribonucleolysis was identified for the pre-5S rRNA maturation in this methanogen, which could be also conserved in most methanogenic euryarchaea. Based on evaluating the phylogenetic conservation of the key elements that are involved in circularization and linearization of pre-rRNA maturation, we predict that the rRNA maturation mode revealed here could be prevalent among archaea.
Introduction
The ribosome is the machinery for protein synthesis in all life, while functional ribosome biogenesis demands correct assembly of the ribosomal RNAs (rRNAs) and specific sets of ribosomal proteins [Citation1]. In most organisms, the rRNA genes are organized as an operon and co-transcribed as a polycistronic precursor (pre-rRNA) that comprises the mature small and large subunit rRNAs separated by internal spacers [Citation2–Citation4]. Therefore, the pre-rRNAs must be processed and matured for assembling a functional ribosome. Disruption of rRNA maturation will generate defective ribosomes and cause retarded growth or death of organisms [Citation5,Citation6]. Pre-rRNA processing and maturation is a multistep and coordinated process that requires endo- and exoribonucleolytic activities [Citation7–Citation9]. Pathways and ribonucleases involved in pre-rRNA processing are intensively studied in bacteria and eukaryotes. In bacteria, the leader and spacer of 16S and 23S rRNAs are annealed to form double-stranded processing stems, which are recognized and cleaved by RNase III [Citation10]. For further maturation, the released pre-16S, pre-23S, and pre-5S rRNAs will be subjected to endoribonucleolysis combined with 3′-5′ exoribonucleolytic processing in Gram-negative bacteria like enterobacteria and/or 5′-3′ exoribonucleolytic processing in Gram-positive bacteria like Firmicutes [Citation11–Citation13]. In eukaryotes, cleavage at the internal transcribed spacers (ITSs) releases pre-18S, 5.8S, and 25S/28S rRNAs, and the maturation process also undergoes a series of endo- and exoribonucleolytic events [Citation14,Citation15].
Nevertheless, detailed pre-rRNA maturation process in archaea remains largely undeciphered. Since 1980s, the archaeal pre-rRNA processing has been individually studied in halophilic and thermophilic archaea [Citation16–Citation22], which have found that the rRNA operons are organized as 16S-tRNAAla-23S-5S in most halophilic euryarchaea, but as 16S-23S in thermophilic euryarchaea and crenarchaea. The 16S-23S organization pattern is also found among the archaea in the TACK superphylum [Citation3,Citation9]. Except for the 16S rRNA of Thermoplasma acidophilum [Citation20], the regions flanking the mature 16S and 23S rRNAs in these archaea are generally folded into a long processing stem containing a bulge-helix-bulge (BHB) motif. The BHB motif, a conserved secondary structure presents in the exon-intron boundary in some intron-containing archaeal tRNAs and rRNAs, is specifically recognized and cleaved by the tRNA splicing endoribonuclease EndA [Citation23,Citation24]. Accompanying EndA cleavage, the 5′ and 3′ halves of the spliced tRNA were ligated into a mature tRNA, while the excised introns were self-ligated as stable circular molecules [Citation25]. Coincidentally, small RNA-omics and circular RNA sequencing detected the by-product and junction sequence of the predicted circular pre-rRNA processing intermediates [Citation26,Citation27], suggesting that the splicing mechanism may be involved in the archaeal pre-rRNA maturation. Recently, BHB motif was determined essential for the circularization of pre-rRNA intermediates and rRNA maturation in Haloferax volcanii [Citation28]. However, the intact entities and proportion of the circular pre-rRNA intermediates have not been confirmed and the subsequent processing steps are also not unravelled in archaea.
Compared with the halophilic and thermophilic euryarchaea (Halobacterium, Thermoplasma, Archaeoglobus) and also the thermophilic crenarchaea (Saccharolobus, Sulfolobus), knowledge of the pre-rRNA maturation process in methanogenic euryarchaea, the ubiquitously distributed and mostly cultured archaeal group, is scarce. In the present study, based on the pre-rRNA processing sites captured by 5′ monophosphate transcript sequencing (5′P-seq) [Citation29], and through molecular approaches and enzymatic assays, we comprehensively investigated the pre-rRNA maturation process in Methanolobus psychrophilus R15, one representative of methanogenic euryarchaea. We unravelled the detailed circularization and linearization for the maturation of pre-16S and pre-23S rRNA, and also identified a novel processing mode of ‘endoribonucleolysis plus exoribonucleolysis’ for pre-5S rRNA maturation. Surveying the wide distribution of the key elements involved in pre-rRNA maturation predicted the prevalence of the processing mode determined here among most archaea.
Results
5′P-seq captures multiple pre-rRNA processing sites
M. psychrophilus encodes three copies of ribosomal RNA (rrn) operons, which are identical in sequences, and each contains the three species of rRNAs and two interspersed tRNA genes (), but the fourth 5S rRNA gene is separately encoded. Previously, differential RNA-seq (dRNA-seq) identified two transcription start sites of each rRNA operon [Citation30], which transcribed a 338-nt and 235-nt 5′ external transcribed spacer (ETS), respectively (Fig. S1). Later, using 5′P-seq to capture the 5′ end of the processed transcripts harbouring 5′ monophosphate [Citation29], we obtained a dozen of processing sites in pre-rRNA (rrnB operon in ), and equivalent sites in rrnA and rrnC operons (Fig. S2). Positioning the processing sites at the predicted pre-rRNA secondary structures by reference to the Comparative RNA Web Site [Citation31] (), we identified some processing sites that are likely generated by known enzymes, including that of RNase P (nucleotides (nt) 1780288 and 1783516 at 5′ ends of tRNAAla and tRNACys, respectively), and of RNase Z (nt 1780362 at 3′ end of tRNAAla). Moreover, processing at nt 1780197, which is just downstream of the mature 3′ end of 16S rRNA, implies a possible endoribonucleolytic site of Nob1, which cleaves at 16 rRNA 3′ end in Pyrococcus horikoshii [Citation32]. These indicate that 5′P-seq is able to capture the processing events in pre-rRNA maturation, and could be used to explore the undescribed processing pathways. Examples as the two processing sites with high coverage at nts 1778514 and 1783600 that are located at the loop between helices A and B and the helix between tRNACys and 5S rRNA (, magenta arrows pointed), respectively. Analysing the flanking sequences of the two sites, we surprisingly found a 10-nt signature sequence identical to the mRNA processing motif (UNMND↓NUNAY) in M. psychrophilus [Citation29], hinting that the methanoarchaeon may use the same endoribonuclease to process both pre-rRNAs and mRNAs.
Figure 1. 5′P-seq detected processing sites in the polycistronic pre-rRNA of M. psychrophilus. (a) Schematic shows the 5′P-seq reads and genome location of the polycistronic pre-rRNA processing sites, in which the representative ones are noted in (b) with the same coloured arrows and letters. Arrows in brown and orange indicate the cleavage sites of ribonucleases RNase P and RNase Z. Arrows in red, magenta, and purple point the processing sites produced by unknown endoribonucleases, and arrow in cyan specifies the mature 5′ end of 5S rRNA. Blue arrows indicate the two transcription start sites (TSSs) of the rrn operon detected by dRNA-seq (Fig. S1). (B) RNA secondary structures of the polycistronic pre-rRNA include six helices (A, B, C, E, F, C′) and a processing stem each in pre-16S and pre-23S rRNAs, and two tRNAs and 5S rRNA.
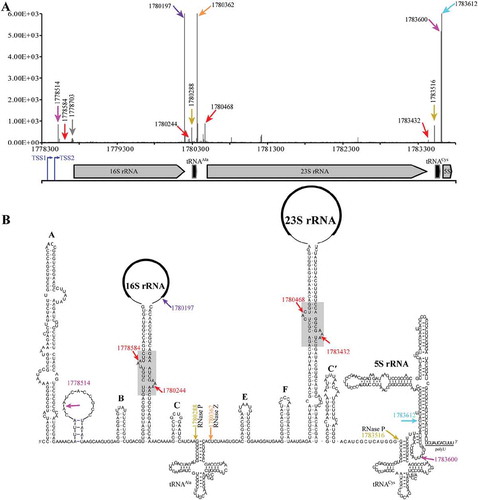
5′P-seq also captured four cleavage sites at the long processing stems of pre-16S and pre-23S rRNA (, red arrows pointed), which are located at the predicted BHB motifs (, grey boxed background). This suggests that the splicing endoribonuclease that specially cleaves at the BHB motif in intron-containing tRNA or rRNA is involved in cropping pre-16S and pre-23S rRNA from the polycistronic pre-rRNA.
EndA cleaves the BHB motifs at pre-16S and pre-23S rRNA processing stems
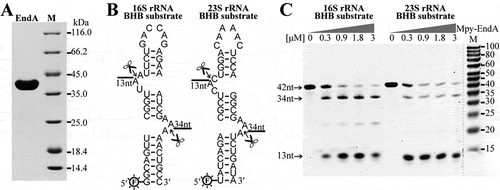
Although in silico analyses have predicted BHB motifs occurring in the flanking sequences of 16S and 23S rRNAs [Citation16–Citation22,Citation26-Citation28], whether they are cleaved by the splicing endoribonuclease has not been enzymatically confirmed. To evaluate this, a gene annotated as tRNA splicing endoribonuclease (EndA, Mpsy_0954) was heterogeneously expressed, and the purified recombinant protein () was used in enzymatic assay. RNA substrates in length of 42 nt, which contain the identified BHB motifs and partial processing stem sequences of the pre-16S and pre-23S rRNA, were synthesized (). Enzymatic assay showed that at a molecular ratio of 2:1 of protein to RNA, two fragments of 13 nt and 34 nt were generated from the 16S- and 23S-BHB substrates, respectively (), the expected products cleaved at the BHB motifs. This verified the function of the tRNA splicing endoribonuclease EndA in cropping pre-16S and 23S rRNAs from the polycistronic transcript, a key step for rRNA maturation.
Figure 2. Cleavage at the BHB motifs in the pre-16S and pre-23S rRNA processing stems by EndA. (a) The Mpy-EndA (Mpsy_0954) was overexpressed in E. coli and the purified ~40 kDa protein was detected by SDS-PAGE. (b) Two 42-nt RNA substrates embedded with the 16S and 23S rRNA BHB motifs were synthesized and labelled with 6-FAM at 5′ ends. The cleavage sites of EndA and the predicted product lengths are indicated. (c) Denaturing urea-PAGE analysing the cleavage products of the 16S and 23S rRNA BHB substrates by increased concentrations of Mpy-EndA. The substrates of 42 nt and the cleaved products with expected lengths of 34 nt and 13 nt are indicated by arrows. The gradient protein concentrations are indicated below the grey triangles and 0.15 μM RNA substrate was used. M, low molecular weight marker (10–100 nt).
Splicing-coupled formation of circular pre-16S and pre-23S rRNA intermediates
Referenced to that the processing of tRNA and rRNA introns by EndA is accompanied by an inter-molecularly ligation catalysed by the RtcB RNA ligase [Citation33], we tested the RtcB-ligated circular pre-rRNA intermediates upon splicing as depicted in in the growing cells of M. psychrophilus. A ligation-omitted circularized RT-PCR (cRT-PCR) was first performed to amplify the invert permuted cDNA fragments corresponding to the junctions of the 5′ leader and 3′ trailer of pre-16S and pre-23S rRNA. Given that EndA cleaves at the splice sites and a coupled ligation would occur between the upstream and downstream cleaved fragments, productions of 186-nt and 48-nt precursor sequences embedded in the circular pre-16S and pre-23S intermediates were predicted, respectively. Ligation-omitted cRT-PCR was performed by using the primer pairs as indicated in and reverse transcription generated cDNA from total RNA as template. Expectedly, DNAs in lengths of 450 nt and 300 nt that contain the precursor fragments were amplified using the primer pair of F1/R1 targeting pre-16S rRNA and F3/R2 targeting pre-23S rRNA, respectively ( upper panel). Addition of T4 RNA ligase did not enhance the permuted amplification products, supporting the authentic presence of circular rRNA entities in the growing cells. The sequencing of PCR product confirmed the junction sequences between the 3′ BHB and 5′ BHB halves at EndA cleavage sites ( lower panel), and also verified the circularization of pre-16S and pre-23S rRNAs. Next, using probes P1 and P3 that specifically complement the junction regions in circular pre-16S and pre-23S rRNAs, respectively, Northern blot detected the whole-length of the circular pre-rRNAs (,), left panels). Using probes P2 and P4 that respectively complement the mature 16S and 23S rRNA (,), right panels), the circular pre-16S and pre-23S rRNA intermediates were estimated as ~1% and >10% of the total corresponding rRNAs in the exponential cells, respectively.
Figure 3. Determination of the presence and percentages of circular pre-rRNA intermediates in total cellular rRNA pool. (a) Schematic shows the predicted circularization coupled with EndA cleaving at BHB motifs (grey shaded boxes, upper panel) to separate pre-16S and pre-23S rRNA from the polycistronic transcript; the spliced products are presumably ligated by RtcB as circular pre-rRNA intermediates (lower panel). Thick black lines indicate mature rRNAs. Thin dark cyan and red lines indicate the precursor sequences from BHB 5′ splice site to the mature 5′ end of 16S or 23S rRNA and that from the mature 3′ end of 16S or 23S rRNA to BHB 3′ splice site, respectively. Same labels are also used in the following figures. (b) Detection of the circular pre-16S and pre-23S rRNA intermediates using ligation-omitted cRT-PCR as described in Materials and methods. The PCR primer pairs are shown in (A), the nested PCR products on cDNA templates that are reverse-transcribed from total RNA with (+) or without (−) T4 RNA ligase pretreatment were detected on a 2% agarose gel (upper panel). Longer and shorter arrows specify the products of the first and second round of PCR, respectively. The lower panels show the DNA sequencing results of the PCR products with an inverted ligation between the 3′ half and 5′ half of 16S and 23S rRNA BHBs. (c–d) Northern blot detected the circular pre-16S (c) and pre-23S rRNA intermediate (d) (left panel) and total cellular 16S or 23S rRNA (right panel) using different probes that spanned the junction region or targeted the mature ends of rRNAs. Locations of the probes are shown in (A). Total RNA contents used are indicated at the top of panel. M1 and M2 indicated ssRNA size markers. (E–F) qRT-PCR determined the percentages of circular pre-rRNA intermediates in the total pool using primers matching the precursor and mature sequences. Percentages of the circular pre-rRNA intermediates in the total were determined at the early, middle, late and stationary cultures. All of the data were calculated from three independent measurements and standard deviations are shown.
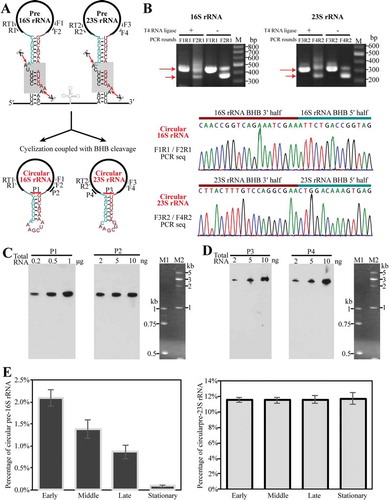
Using qRT-PCR, 2% and 12% circular pre-16S and pre-23S rRNAs were precisely determined in the earlier growing cells, respectively (). Interestingly, while the circular pre-16S rRNA contents gradually decreased during growth and to an undetectable level in the stationary phase, the circular pre-23S rRNA remained at similar percentages along the growth (). This implies that further processing of the circular pre-16S rRNA intermediates would be more rapid than that of circular pre-23S rRNA. Altogether, we verified the existence of the circular pre-16S and pre-23S rRNA intermediates and also quantified their proportions in M. psychrophilus.
Endoribonuclease Nob1 linearizes the circular pre-16S rRNA intermediate at the mature 3′ end
The circular pre-rRNA intermediates have to be linearized through an endoribonucleolytic cleavage for further processing. The highly enriched 5′P site at nt 1780197 ()) is located downstream of the predicted mature 3′ end of 16S rRNA, suggesting that the endoribonuclease Nob1 homolog could cleave at the mature 3′ end of 16S rRNA in M. psychrophilus as that in P. horikoshii [Citation32], and this endoribonucleolytic event may linearize the circular pre-16S rRNA intermediate as well. To test this hypothesis, we first determined the mature 3′ end of the M. psychrophilus 16S rRNA by 3′-RACE, and identified a sequence of 5′-UCACCUCCU-3′ (), which exactly matches the predicted mature 3′ end of 16S rRNA. We then purified the recombinant Mpy-Nob1 and one catalytic inactive mutant D6N by substitution mutagenesis of Asp6 in the predicted active site. A linear RNA of 80 nt (L80), which consists of 54 nt of the 16S rRNA 3′ end and 26 nt of the downstream spacer sequence, was used as the substrate. At 1:1 and 2:1 of Mpy-Nob1 to RNA, two cleaved RNA fragments of 54 nt and 26 nt were generated (). Again, 3′-RACE determined the 3′ end of the 54-nt cleavage product as 5′-UCACCUCCU-3′. The mutant D6N was incapable of cleaving L80 RNA, confirming that Mpy-Nob1 possesses the intrinsic 16S rRNA 3′ end cleavage activity.
Figure 4. The endoribonuclease Nob1 mediates the linearization of circular pre-16S rRNA intermediate. (a) 3′-RACE determined the mature 3′ end of 16S rRNA. A diagram shows the circular pre-16S rRNA intermediate (left upper panel) with the precursor sequence in the same colour as in . Using a procedure depicted in the upper right panel, the nested PCR products of 3′-RACE were detected by agarose gel electrophoresis and sequenced (lower panel). (b–d) The endoribonucleolytic activity of Mpy-Nob1 was assayed on the RNA substrates of a linear RNA L80 (b), a circular-like RNA SL37 (c), and an actual circular RNA C251 (d). Diagrams display the three RNA substrates as described in Materials and methods. Incubation of gradient concentrations of Mpy-Nob1 as indicated with 0.2 μM L80 (B) and 0.2 μM SL37 substrates (C) respectively and the cleavage products (black arrows) were run on 10% urea-PAGE, while C251 (black circle, 3 nM) and the cleavage products were analysed on a 6% urea-PAGE with L251 substrate as control (D, lane L, black line). The D6N mutant was included in the three substrate cleavage assays. M, low molecular weight marker (10–100 nt). (e) Circular pre-16S rRNA in total RNA was linearized by Nob1. Total cellular RNA was incubated with the indicated concentrations of Mpy-Nob1. After incubation for 30 min, ligation-omitted cRT-PCR was performed using the same approach as in . Agarose gel displays the PCR products of circular pre-16S and pre-23S rRNAs. M in (A) and (E), 100 bp DNA ladder marker showing the migration of PCR products.
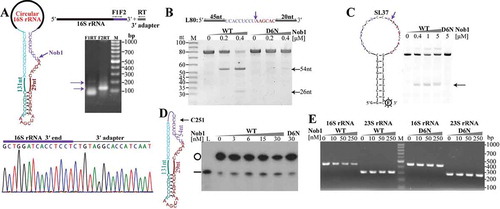
To test the activity of Nob1 in linearizing the circular pre-16S rRNA intermediate, two RNA substrates were synthesized. One is a circular-like substrate of SL37 RNA consisting of a 18-nt loop that embeds with the Mpy-Nob1 16S rRNA 3′ end cleavage site and a stem with 9-GC base pairs (), and the other is a linear 251-nt RNA (L251) for producing an actual circular RNA (C251), which contains 54-nt 3′ end and 11-nt 5′ end sequences of the mature 16S rRNA, and a 186-nt precursor sequence of the circular pre-16S rRNA intermediate (). Using a 3′ FAM-labelled SL37 as substrate, Mpy-Nob1 efficiently cleaved at the 16S mature 3′ end embedded in the loop of SL37 RNA and generated a product with expected length, but the D6N mutant did not (). By cyclizing the 5′-end [γ-32P] labelled L251 RNA to make the circular C251, we determined the capability of Mpy-Nob1 in linearizing the actual circular C251 and producing a linear product with the same migration as L251 (). These results experimentally demonstrated that Mpy-Nob1 cleaves at 3′ end of 16S rRNA not only for 3′ end maturation but also linearization of the circular pre-16S rRNA intermediates.
Furthermore, through incubation of Mpy-Nob1 with the total RNA pool of M. psychrophilus and using the ligation-omitted cRT-PCR, we determined a dose-dependent reduction of the circular pre-16S rRNA with gradient concentrations of Mpy-Nob1, but not with that of the D6N mutant (). However, such a reduction mode was not found for the circular pre-23S rRNA intermediate, even at the highest Mpy-Nob1 concentration (). This further verified that Mpy-Nob1 specifically linearizes the circular pre-16S rRNA.
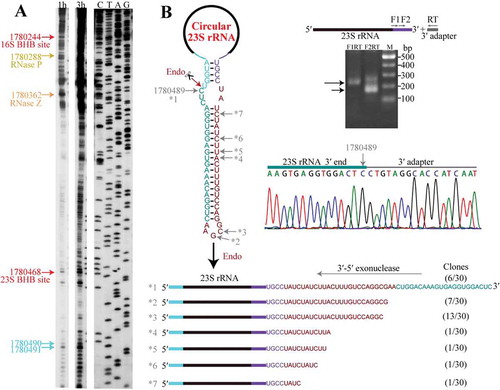
Circular pre-23S rRNA intermediate is linearized at the mature 5′ end
To explore how the circular pre-23S rRNA intermediate is linearized for further processing, we first evaluated the major processing sites of pre-23S rRNA and the mature 5′ and 3′ ends of 23S rRNA. Primer extension verified the processing sites that were determined by 5′P-seq, including 3′ and 5′ splice sites of EndA at pre-16S and pre-23S rRNA, respectively, and the cleavage sites of RNase P and RNase Z on tRNAAla upstream of 23S rRNA, as well as two main mature 5′ ends (nts 1780491 and 1780490, ) of 23S rRNA, which were verified by 5′-RACE (Fig. S3). Unexpectedly, 3′-RACE detected a highly accumulated 3′ end at nt 1780489, just 1 nt upstream the mature 5′ end of 23S rRNA (). This suggests an endoribonucleolytic cleavage between nts 1780489 and 1780490, i.e. at the mature 5′ end of 23S rRNA. As neither primer extension nor 5′P-seq detected other processing sites preceding the mature 5′ end of 23S rRNA, we predict that this endoribonucleolytic action could linearize the circular pre-23S rRNA intermediate. The cleavage should directly produce a mature 5′ end but leave a 48-nt 3′ extension to be further processed.
Figure 5. Linearization of the circular pre-23S rRNA intermediate followed by 3′-5′ exoribonucleolytic trimming. (a) Primer extension detected the major processing sites upstream of 23S rRNA. Cyan arrows indicate the two mature 5′ ends of 23S rRNA, and the upstream major processing sites are indicated by the same coloured arrows shown in . 1 h and 3 h at gel top indicate autographing times. (b) 3′-RACE assayed the 3′-5′ exoribonucleolytic processing trace in pre-23S rRNA maturation. A diagram of the circular pre-23S rRNA shows the complete precursor sequence (left upper panel) with the featured RNA fragments coloured same as in . An endoribonucleolytic processing event (dark red arrow) was predicted to linearize the circular pre-23S rRNA. 3′-RACE was performed as depicted in the right upper panel and the products were sequenced. The nested PCR products in 3′-RACE were analysed on 2% agarose gel (right upper panel) and the DNA sequencing results of nt 1780489 (right lower panel) are shown. The remaining six representative 3′ ends (*2 to *7) captured by 3′-RACE are shown as a diagram (lower panel), and the corresponding sites are indicated in the sequence at the upper panel. Numbers in parenthesis indicate the sequenced clones.
Linearized pre-16S and pre-23S rRNAs are matured by 5′-3′ and 3′-5′ exoribonucleolytic pathways, respectively
Furthermore, we performed 3′-RACE to capture the major processing sites in trimming the 48-nt 3′ extension of the linearized pre-23S rRNA intermediate. Notably, a series of 23S rRNA 3′ ends upstream the endoribonucleolytic site of 1780489 were identified (), exposing a 3′-5′ exoribonucleolytic trace, i.e. the 3′ extension of linearized pre-23S rRNA is trimmed by a 3′-5′ exoribonuclease. While, 3′-RACE did not capture the mature 3′ end of 23S rRNA, which could be attributed to a four-base-pair stem formed between the mature 5′ and 3′ ends that limited the adapter ligation.
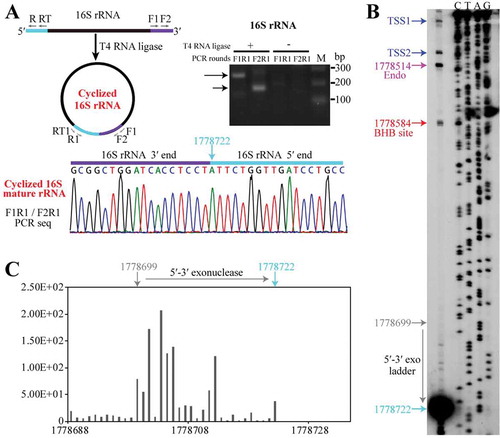
On the contrary, linearization of circular pre-16S rRNA at the mature 3′ end would produce a 186-nt 5′ extension. To dissect the further processing of the linearized pre-16S rRNA, we first determined the two mature ends of 16S rRNA using cRT-PCR (, left panel), and verified the mature 3′ end detected above by 3′-RACE (). cRT-PCR also identified the mature 5′ end of 16S rRNA at nt 1778722 (), which was confirmed by 5′-RACE (Fig. S3). Primer extension not only verified the mature 5′ end but also the two TSSs of the rrn operon, in addition of the newly identified endoribonucleolytic site at nt 1778514 between helices A and B, and the 5′ splice site of EndA (). Noticeably, an obvious cDNA ladder proximal the mature 5′ end was observed between the 5′ splice site and mature 5′ end of 16S rRNA (), revealing a 5′-3′ exoribonucleolytic trace. Consistently, a similar 5′-3′ exoribonucleolytic ladder was observed in 5′P-seq (). These indicate a 5′-3′ exoribonucleolytic action in the 5′ end maturation of pre-16S rRNA.
Figure 6. 5′-3′ exoribonucleolytic processing finalizes the 16S rRNA maturation. (a) cRT-PCR determined the mature 5′ and 3′ ends of 16S rRNA as depicted in upper left panel. The nested PCR products were amplified from cDNA templates generated from a reverse-transcription of the total RNA that were pretreated with (+) or without (−) T4 RNA ligase and analysed on an agarose gel (right upper panel) and sequenced (lower panel). Nucleotide 1778722 points the junction of 3′ and 5′ ends of 16S rRNA. (b) Primer extension analysis showing the processing sites at 5′ end of 16S rRNA. Two transcription start sites (TSS1 and TSS2, blue arrows), an endoribonucleolytic site (magenta arrow), 5′ splice site generated by EndA (red arrow) and the mature 5′ end of 16S rRNA (cyan arrow) are pointed. (c) A series of 1-nt 5′P-seq reads between the mature 5′ end (nt 1778722) and upstream nt 1778699 of 16S rRNA exposes a 5′-3′ exoribonucleolytic pattern.
Collectively, these results indicate that although linearization happens in both the circular pre-16S and pre-23S rRNA intermediates, the endoribonucleolytic action occurs at the mature 3′ and 5′ ends, respectively. Thus, the resultant 5′ and 3′ extensions are trimmed by 5′-3′ and 3′-5′ exoribonucleolytic pathways for final maturation, respectively.
Endoribonucleolysis coupled exoribonucleolysis on both ends mediates 5S rRNA maturation
Next, pre-5S rRNA maturation mode of M. psychrophilus was probed. 5′P-seq captured two main processing sites, nts 1783600 and 1783612, between tRNACys and 5S rRNA (). Consistently, primer extension verified the two sites upstream of 5S rRNA (Fig. S4A). The higher abundant site at nt 1783612 was noted as the mature 5′ end of 5S rRNA, which was confirmed by 5′-RACE (Fig. S4B). As mentioned above, processing site at nt 1783600 () perfectly conforms to the 10-nt motif (UNMND↓NUNAY) identified in mRNA processing sites of M. psychrophilus [Citation29], implying a same endonuclease acting on them. The endoribonucleolytic event at nt 1783600 could separate pre-5S rRNA from the upstream tRNACys as well as mediate the 3′ end maturation of tRNACys, because neither primer extension nor 5′P-seq detected the RNase Z cleavage site downstream tRNACys. However, the endoribonucleolysis at nt 1783600 would leave a 12-nt 5′ extension in pre-5S rRNA. By revisiting the 5′P-seq data, a 5′-3′ exoribonucleolytic ladder was found between nts 1783600 and 1783612 (Fig. S5), suggesting a 5′-3′ exoribonucleolytic trimming of the 12-nt 5′ extension for 5′ end maturation. On the other side, 3′-RACE determined the mature 3′ end of 5S rRNA being 15 nt distant to the predicted transcription end site (TES) of the polycistronic pre-rRNA (Fig. S4 C), and also detected several sites in between. This suggested a 3′-5′ exoribonucleolytic action for trimming the 15-nt 3′ trailer. Altogether, we speculated that an endoribonucleolysis coupled exoribonucleolysis mode mediates the 5S rRNA maturation in M. psychrophilus.
Circularization and linearization could be a conserved mode in archaeal rRNA maturation
To interrogate the prevalence of the circularization and linearization mode in pre-rRNA maturation found here among archaea, we chose 30 representative species from the four defined archaeal superphyla (Euryarchaeota, TACK, Asgard and DPANN) for investigation. The key elements involved in the process were surveyed, including the rRNA operon patterns (), the rRNA BHB motifs () and enzymes of EndA, RtcB, Nob1 and the 3′ end sequence of 16S rRNA recognized by Nob1 () and rRNA maturation mode (). Three major types of rRNA operons are found (), type I (most euryarchaea) contains either the three rRNA species with or without two tRNAs interspersed, type II (most other phyla) possesses 16S and 23S rRNAs but no 5S rRNA, and type III encodes the three rRNAs separately (a few thermophiles and uncultured species). Intensive in silico search found that the BHB motif occurs in most pre-16S and pre-23S rRNAs, except for Thermoplasmatales, Methanomassiliicoccales, and a few metagenome-assembled archaeal genomes ( and Fig. S6). EndA and RtcB, which are responsible for splicing and circularization, are highly conserved in all the investigated species (). Thereby, splicing-mediated circularization of pre-16S and pre-23S rRNAs is common among archaea.
Figure 7. The conserved rRNA maturation modes in archaea. By searching the available archaeal genomes in KEGG database, key elements involved in pre-rRNA processing and maturation were analysed in 30 representative species affiliating to the four superphyla of archaea. These include three organization patterns of rRNA operons (a), 16S and 23S rRNA BHB motifs (b), 16S 3′ end sequences cleaved by Nob1 (c), three maturation modes of 5S rRNA (d), and the presence of EndA, RtcB and Nob1 (e). The grey shadowed BHB motif in (B) consists of two bulges (red spheres) and an in-between helix (blue spheres). A conserved 3′ end sequence of 16S rRNA noted as anti-SD in (C) is found in almost all of the investigated archaea, and perfectly complementary to the M. psychrophilus SD sequence (black dot line framed). Three maturation modes of 5S rRNA are summarized in the 30 investigated archaeal species (d,e): (i) transcribed as matured ‘leaderless’ 5S rRNA prevalent in TACK, Asgard and DPANN, (ii) RNase Z-dependent maturation on a tRNA-like structure widely distributed in haloarchaea, and (iii) ‘endo+exo’ mode employed in methanoarchaea. A representative phylogenetic tree (e) is constructed on the homology analysis of the protein concatenation of EndA and Nob1 from the 30 investigated archaeal species. The tree was constructed using MEGA7.0, and the bar represents 10% sequence difference. E, Euryarchaeota (green background); T, Thaumarchaeota (brick red background); A, Aigarchaeota (blue background); C, Crenarchaeota (pink background); K, Korarchaeota (magenta background); N/A, Nanoarchaeota and ARMAN (yellow background). Distributions of the key elements involved in rRNA maturation are shown at the right side. Filled and empty symbols represent the presence and absence of items in corresponding archaeal species. Grey filled squares indicate incomplete or inaccurate 16S or 23S rRNA sequences that do not allow a prediction of BHB motif, and NA means no available 16S or 23S rRNA sequence in the database. Grey shaded letters in 3′ end of 16S rRNA indicate mutated sites by comparing with the conserved mature 3′ end sequence shown in (C).
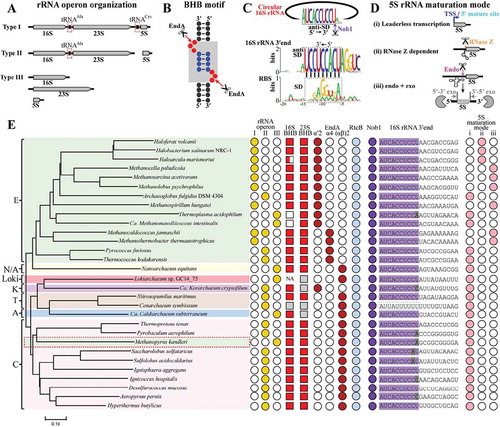
Sequence alignment revealed that the mature 3′ end (5′-AUCACCUCCU-3′) of 16S rRNA harbouring the anti-SD sequence is also highly conserved among the investigated species (,). However, a single base mutation occurred in some members of the TACK superphylum, and a distinctive mature 3′ end sequence was found in Cenarchaeum symbiosum. Remarkably, the ubiquitous distribution of Nob1 and the conserved 3′ end of mature 16S rRNA together suggest that Nob1-mediated linearization of the circular pre-16S rRNA could be a general strategy utilized among archaea.
Moreover, a phylogenetic tree based on a concatenation of EndA and Nob1 is almost topologically identical to that of the 45 concatenated conserved proteins [Citation34] (). This suggests that the two proteins could be vertically inherited along the evolution of archaeal species. Taken together, the circularization and linearization of the processing intermediates during rRNA maturation, which was experimentally demonstrated in a methanogenic archaeon in this study, is a conserved rRNA processing mode employed by most archaea.
Discussion
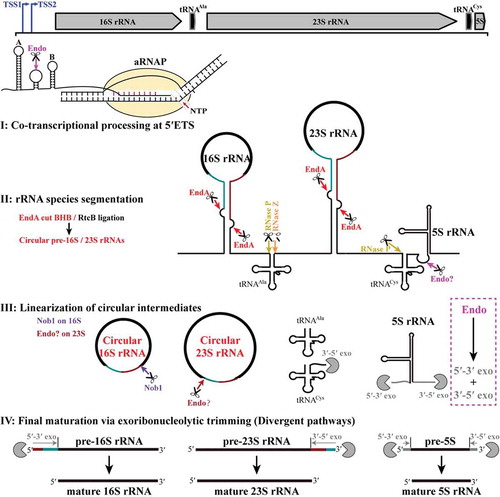
Ribosomal RNA maturation is a key biological process for ribosome biogenesis [Citation5,Citation15]. However, compared to our comprehensive knowledge of the rRNA maturation pathways in eukaryotes and bacteria, understanding of this important process in archaea is just sporadic, and particularly scarce in methanogenic archaea. Mainly omics-based approaches implied that maturation of the pre-rRNAs in euryarchaea (Archaeoglobus and Halobacterium) and crenarchaea (Saccharolobus and Sulfolobus) appeared to be subjected to a circularization process [Citation26,Citation27]; yet, the intact circular pre-rRNA entities have not been demonstrated and how the circular intermediates linearized remains unknown. In the present study, via combination of the 5′P-seq data with biochemical and molecular biological experiments, we comprehensively revealed the maturation steps of pre-rRNAs in M. psychrophilus, one representative methanogenic archaeon. The maturation mode depicted in includes an initial co-transcriptional processing within the 5′ ETS (Step I), splicing-mediated rRNA segmentation and circularization (Step II), linearization of circular pre-16S and pre-23S rRNA intermediates (Step III) and divergent exoribonucleolytic trimming pathways of pre-16S, 23S and 5S rRNAs for final maturation (Step IV). The comprehensive map not only elucidated the pre-rRNA maturation pathway in methanogen but also could expose a conserved circularization and linearization strategy widely utilized for rRNA maturation among most archaea.
Figure 8. The pre-rRNA processing and maturation mode in M. psychrophilus. The rRNA operon (16S-tRNAAla-23S-tRNACys-5S) is transcribed as a polycistronic RNA from two transcription start sites (TSS1 and TSS2). A co-transcriptional endoribonucleolytic cleavage (Endo) at 5′ ETS that removes helix A initiates the processing (Step I). Following pre-rRNA transcription and folding, rRNAs and tRNAs are segmented (Step II) by the corresponding endoribonucleases. Splicing endonuclease EndA cleaves at BHB motifs buried in the processing stems to excise pre-16S and pre-23S rRNAs and RNA ligase RtcB catalyzes the coupled circularization. Cleavages of RNase P and RNase Z release tRNAAla, while cleavages of RNase P and an unknown endoribonuclease separate tRNACys and 5S rRNA. The circular pre-16S and pre-23S rRNA intermediates are linearized (Step III). Nob1 linearizes the circular 16S rRNA at the mature 3′ end and generates a long 5′ extension. In contrast, an unknown endoribonuclease linearizes the circular 23S rRNA at the mature 5′ end and generates a short 3′ extension. Finally, linearized pre-16S, 23S, and 5S rRNAs are trimmed through different exoribonucleolytic pathways (Step IV). The 5′ and 3′ extensions of pre-16S and pre-23S rRNAs are finally matured by 5′-3′ and 3′-5′ exoribonucleolytic trimming, respectively, while exoribonucleolytic trimming of both ends are involved in 5S rRNA maturation. The scissor and Pacman icons represent an endoribonuclease and exoribonuclease, respectively. Dark cyan and red lines at pre-16S and pre-23S rRNAs show the precursor regions in circular intermediates. Magenta dotted line frames a postulated pre-5S rRNA maturation mode by a combination of endoribonucleolysis and exoribonucleolysis.
Early endoribonucleolytic processing in 5′ ETS of pre-rRNA could be a shared step by Euryarchaeota and Crenarchaeota
By reference to the processing at site A0 in the pre-rRNA 5′ ETS of S. cerevisiae as an initial event [Citation35,Citation36], processing at nt 1778514 in the pre-rRNA 5′ ETS of M. psychrophilus could represent an early processing action as well. This initial step could remove the bulky secondary structure of helix A, so to facilitate the subsequent downstream splicing cleavage of pre-16S rRNA. Previous studies also identified similar cleavage sites between helices A and B in the 5′ ETS, which were named site 0 in S. solfataricus or site 1 in S. acidocaldarius [Citation22,Citation37,Citation38]. Thus, the early processing in 5′ EST could be a common step in Crenarchaeota and Euryarchaeota. Interestingly, processing at nt 1778514 in M. psychrophilus, site 0 in S. solfataricus and site 1 in S. acidocaldarius all share an identical processing sequence identical to the 10-nt mRNA processing motif (UNMND↓NUNAY) identified in M. psychrophilus [Citation29]. This suggests that a conserved sequence-specific endoribonuclease could be responsible for the early processing of pre-rRNA and processing of mRNAs in archaea.
The tRNA splicing endoribonuclease EndA cleaves the rRNA BHB motifs for isolation of 16S and 23S rRNAs from the polycistronic pre-rRNA
The tRNA splicing endoribonuclease EndA orthologs are universally distributed in eukaryotes and archaea. Although firstly reported as a tRNA intron splicing endoribonuclease [Citation23,Citation24], EndA also scissors the introns of 16S and 23S rRNAs [Citation39–Citation42], and an intron in the cbf5 pre-mRNA of Crenarchaeota [Citation43]. However, introns are infrequently present in the prokaryotic genes, except in a few archaeal rRNAs, tRNAs, and mRNAs. Recently, a signal recognition particle (SRP) RNA of Thermoproteus tenax was also identified as a substrate of EndA [Citation44]. These observations inspired us to query the biological functions of the highly conserved EndA in archaea. An in vitro biochemical assay demonstrated that EndA cleaves the BHB motifs embedded in the processing stem of pre-16S and pre-23S rRNA () so to release the two rRNAs from the polycistronic precursor. Based on the ubiquitous distribution of rRNA BHB motifs among archaea (), we predict that EndA-mediated splicing of pre-16S and pre-23S rRNAs is a common pathway in archaeal rRNA maturation. Although the rRNA BHB motif was missing in a few archaeal species presumably due to the incomplete genomes through metagenomic assembly, the EndA orthologs occur in all investigated species of the four archaeal superphyla (). Phylogenetic analysis established EndA as a good phylogenetic marker as well (), with the α′2 and α4 types occurring in Euryarchaeota and Korarchaeota, and the (αβ)2 type in other archaeal phyla [Citation45]. The (αβ)2 heterotetrameric form EndA had the ability of cleaving relaxed BHB motif [Citation46,Citation47], exampled as that of Nanoarchaeum equitans (Fig. S6).
Different percentages of the circular pre-16S and pre-23S rRNAs, divergent linearization and the final maturation modes
Since the products of splicing cleavage at the BHB motif are ligated by RtcB RNA ligase [Citation33], circularized pre-16S and pre-23S rRNAs are expected. This work not only identified the circular intermediate entities, but also found that the cellular circular pre-16 S rRNA intermediate was at a relatively lower percentage (~2%) than that of the pre-23S rRNA (~12%). However, Northern blot did not detect the corresponding by-products of splicing, 16S-D and 23S-D in M. psychrophilus, indicating that they are more labile compared to the counterparts of A. fulgidus and S. solfataricus [Citation26], and also warning that detection of by-products is not reliable in predicting the existence of circular pre-16S and pre-23S rRNA intermediates.
Circularized RNAs would be more resistant to exoribonucleases [Citation44] and could also serve as an rRNA scaffold in recruiting the early processing factors and assembling ribosomal proteins. Interestingly, the circular intermediates of pre-16S rRNA gradually decreased along with cell growth, but that of pre-23S rRNA retained at almost constant levels. This suggests that 16 S rRNA processing could be more important in ribosome biogenesis as well as in physiology. Supportively, a recent in vivo study indicates that circularization of pre-16S rRNA is required for the formation of circular pre-23S and the mature 16S/23S rRNAs in H. volcanii [Citation28]. Otherwise, linearization of the circular pre-23S rRNA may not be of the same importance as the circular pre-16S rRNA. This could be because the shorter insert (48 nt) in the circular pre-23S rRNA may less interfere the mature 5′ and 3′ ends aligning at close proximity inside ribosome [Citation1], or even the circular 23S rRNA could be assembled into the functional 50S ribosome. However, the longer insert (186 nt) in the circular pre-16S rRNA must be removed to expose the anti-SD sequence and warrant translation initiation. Consistently, generally shorter precursor sequences were found in circular pre-23S rRNAs than in circular pre-16S rRNAs among the investigated species from the four archaeal superphyla (Table S1).
Linearization of circular pre-16S and pre-23S rRNAs respectively occurs at the mature 3′ and 5′ ends, and the resultant 5′ and 3′ extensions were trimmed by 5′-3′ and 3′-5′ exoribonucleases, respectively. A handful of 5′-3′ exoribonucleases have been identified in archaea, such as the widely distributed β-CASP superfamily proteins aRNase J and aCPSF [Citation9,Citation48,Citation49], while exosome and RNase R exhibit 3′-5′ exoribonucleolytic activity in archaea [Citation50]. These enzymes could play roles in trimming the unnecessary extensions in pre-16S and pre-23S rRNA for the final maturation.
Three maturation modes of the archaeal 5S rRNA
Pre-rRNA processing pathways and related ribonucleases appear to have coevolved with the pre-rRNA sequences and structures, which are exemplified by the pre-5S rRNA processing patterns among various archaea phyla. The ‘leaderless’ 5S rRNAs are prevalent in the superphyla of TACK, Asgard and DPANN (,)), i.e. the primary transcripts already possess the mature 5′ ends and no further processing is required. This may represent the first type of maturation mode (mode i) of archaeal 5S rRNA. While in Halobacteriales, the upstream sequence of 5S rRNA folds into a tRNA-like structure and an RNase Z-mediated cleavage at its 3′ end directly generates the mature 5′ end (,) , mode ii) [Citation51]. Unlike the aforementioned two modes, isolation of the pre-5S rRNA of M. psychrophilus is subjected to an endoribonucleolytic cleavage at the upstream ITS, which leaves a 5′ leader for further exoribonucleolytic trimming, therefore corresponding to the newly identified ‘endo+exo’ processing mode (mode iii). Interestingly, the same 10-nt processing motif (UNMND↓NUNAY) was found upstream 5S rRNA in most investigated orders of methanogens (), including Methanomicrobiales, Methanocellales, Methanosarcinales, Methanococcales, and Methanobacteriales, implying that the ‘endo+exo’ maturation of 5S rRNA could be the methanogen-specific mode.
In conclusion, this study reveals a comprehensive rRNA maturation map based on analysing the detailed rRNA processing steps and pathways in a methanogen, which includes formation and linearization of circular pre-rRNA intermediates and followed by 5′-3′ or 3′-5′ exoribonucleolysis for maturation. The work suggests a conserved circularization and linearization mode for pre-rRNA maturation in archaea, and would shed lights on understanding the overall perspective of the rRNA maturation process in archaea and the evolution of rRNA maturation mode in three domains of life.
Materials and methods
Identification of the processing sites in pre-rRNAs
The majority of 16S and 23S rRNAs were depleted in the previous differential RNA sequencing (dRNA-seq) [Citation30] and 5′ monophosphate transcript sequencing (5′P-seq) [Citation29], which were performed to identify genome-wide transcription start sites (TSSs) and RNA processing site in M. psychrophilus, respectively. Nevertheless, the residual reads remaining in these samples were sufficient for analysing the processing sites of pre-rRNAs. Sites enriched in the dRNA-seq (+) library were considered as the transcription start sites, while those enriched in the 5′P-seq library were defined as processing sites. Sites with low read counts were all abandoned as they could be RNA degradation products. The two types of sites were all enriched in the dRNA-seq (−) library. Promoter motifs and processing motifs were searched using the MEME algorithm [Citation52] and sequence logos were generated by WebLogo [Citation53]. RNA structures pertinent to various types of the processing sites were predicted by Mfold software [Citation54] and fine-tuned according to the standard structures in the comparative RNA web (CRW) site [Citation31]. Secondary structure displays of the RNA molecules were plotted by RnaViz program [Citation55].
BHB motif searching
The BHB motif in RNAs that are recognized by splicing endonucleases is generally buried in a long inverted repeat sequence [Citation56]. To find the BHB motif of pre-rRNA, we aligned the reverse complementary downstream sequences of mature 16S or 23S rRNA with the upstream sequences of rRNA, and found a 7-nt mismatch sequence flanked by two aligned base-pairings each in the two rRNAs, which were defined as candidate BHB motifs. RNA structures that carry candidate BHB motifs were predicted by the Mfold software and fine-tuned according to the classic BHB motif. To find the possible rRNA BHB motif in the four superphyla of archaea (Euryarchaeota, TACK, Asgard and DPANN), the sequences flanking 16S and 23S of representative strains were collected from the KEGG database [Citation57] and analysed.
Overexpression and purification of Mpy-EndA and Mpy-Nob1
The M. psychrophilus gene Mpsy_0954, which encodes a putative tRNA splicing endonuclease, was cloned into the NcoI/XhoI sites of pET28a (Novagen) to generate pET28a-0954 and then transformed into E. coli BL21(DE3)pLysS to overexpress the recombinant Mpy-EndA with a C-terminal 6× His tag. Similarly, the M. psychrophilus Mpsy_1826, a gene encoding Nob1, was cloned into NdeI/NotI sites of pET23b (Novagen) to generate pET23b-1826. Substitution mutagenesis of Asp6 (D6N), one of the catalytic inactive mutants of Nob1, was constructed using a site-directed mutagenesis kit (Stratagene) with pET23b-1826 as template. The recombinant proteins, Mpy-EndA, and Mpy-Nob1 and its catalytic mutant D6N, were induced and purified using the methods described previously [Citation58]. Briefly, cells with pET28a-0954 or pET23b-1826 were cultured and resuspended in buffer A (20 mM Tris, pH 8.0, 250 mM NaCl, 20 mM imidazole and 5% (w/v) glycerol), sonicated and centrifugation (10,000 × g, 30 min, 4°C). The recombinant Mpy-EndA or Mpy-Nob1 or D6N mutant in the supernatant was purified by HisTrap HP column, HiTrap Q HP column, HiTrap desalting column (GE Healthcare), and concentrated using Amicon Ultra-15 30 K centrifugal filter device (Millipore), flash frozen and stored at −80°C for biochemical assays.
Nuclease assay of EndA
For the RNA cleavage assay of Mpy-EndA, the BHB substrates of 16S and 23S rRNAs () were synthesized and 5′ end labelled with 6-carboxyfluorescein (6-FAM) fluorescent dye (GenScript). The RNA cleavage mixture in a final volume of 10 μl contained 150 nM 6-FAM labelled BHB RNA substrates, 0.3–3 μM Mpy-EndA, 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM DTT, 25 mM KCl, 5 mM MgCl2, and 5% (w/v) glycerol. The cleavage reaction was initiated by addition of the enzyme, then it was incubated at 30°C for 30 min, and stopped by adding formamide loading buffer and denatured at 95°C for 3 min. The cleaved products were loaded onto a 20% denaturing polyacrylamide gel that was run in 1× TBE, 20 mA for 1 h next to a low molecular weight marker (10–100 nt, Affymetrix). The cleavage products were visualized by Typhoon FLA 9500 (GE Healthcare).
Circularized RT-PCR
Circularized RT-PCR (cRT-PCR), which was performed according to the protocols described by Slomovic et al. [Citation59] with some modifications, was used to identify the 5′ and 3′ termini of rRNA simultaneously. The 5′ and 3′ ends of 5 μg total RNA were self-ligated by 1-h incubation with 5 U T4 RNA ligase (Ambion) in the presence of 20 U RNasin (Promega) at 37°C. Circularized RNA was precipitated by addition of an equal volume of ice-cold isopropanol. A T4 RNA ligase absent circularization reaction was used as a negative control. Notably, for detecting the natural circular pre-16S and pre-23S rRNA intermediates occurring in total RNAs, the T4 RNA ligase was also omitted from the reaction (ligation-omitted cRT-PCR). Subsequently, an RT-PCR protocol was performed to generate cDNA comprising the adjoined 5′ and 3′ ends using a reverse transcriptase and a gene-specific primer complementary to the 5′ ends of rRNAs. Each 20 μl RT reaction contains: 4 μl 5× First-Strand Buffer (Invitrogen), 1 μl 10 mM dNTP Mix, 1 μl 0.1 M DTT, 0.5 μl 40 U/μl RNasin (Promega), and 1 μl 200 U/μl SuperScript III reverse transcriptase (Invitrogen). Two rounds of PCR were conducted with reverse primer and nested forward primers (Table S2, Sangon Biotech). PCR products were run on a 2% agarose gel for purification, and the purified products were cloned into the pMD19-T vector (TaKaRa) for sequencing. Sequences around the circularization junction were aligned with genomic DNA.
Northern blot
Total RNA of M. psychrophilus was extracted and separated on 4% polyacrylamide gels containing 7.6 M urea. After heating at 65°C for 10 min and chilling on ice, RNA was loaded to the gels in 50% formamide. RNA Century™-Plus Markers (Ambion) and ssRNA Ladder (New England Biolabs) were used as ssRNA size markers in denaturing gel electrophoresis. Gels were electroblotted in 0.5× TBE onto a Hybond N+ membrane (GE Healthcare) and then fixed by ultraviolet irradiation for 2 min. Hybridization between DNA probes and mature rRNAs or circular intermediates was performed at 42°C in 50% formamide, 5× SSC, 5× Denhardt’s reagent, 0.5% SDS, and 200 μg/ml denatured salmon sperm DNA. The DNA probes were 5′ end labelled with biotin and complementary to the mature end of rRNAs. For circular RNA processing intermediates, the probes (Sangon Biotech) were complementary to the circularization junctions of the precursor sequences of rRNAs. Membranes were washed for 10 min each in 1×, 0.2×, and 0.1× SSC–0.1% SDS solutions. Relative quantities of the targeting RNAs on Northern blots were determined using a chemiluminescent nucleic acid detection module (Thermo Scientific) according to the manufacturer’s instructions.
Quantitative RT-PCR (qRT-PCR)
qRT-PCR was performed for quantifying the ratios of circular pre-16S or pre-23S rRNA intermediates to the corresponding mature rRNAs during growth. Total RNAs were extracted from the M. psychrophilus cells at early, middle, and late-exponential and stationary phases and purified using TRIzol reagent (Invitrogen). cDNAs were then generated from 2 μg of total RNAs with random primers using Moloney murine leukaemia virus reverse transcriptase (Promega). qPCR amplifications were performed at Mastercycler ep realplex2 (Eppendorf) using the corresponding primers (Table S2, Sangon Biotech) specific for amplification of the circular pre-16S or pre-23S rRNA intermediates and the mature rRNAs. To estimate the copy numbers of the tested rRNA species, standard curves of the corresponding primers were generated using 10-fold serially diluted PCR product as templates. Then, the ratios of the circular pre-rRNA intermediates to the mature rRNAs of 16S and 23S were calculated.
Enzymatic assays of Nob1
To assay Mpy-Nob1 in processing the mature 3′ end of 16S rRNA, a linear RNA of L80, which contains the 54 nt of mature 16S rRNA 3′ end sequence and the flanking 26 nt precursor sequence (), was used. To assay the potential of Nob1 to linearize the circular 16S rRNA intermediate, a circular-like stem-loop RNA of SL37 () and an artificial circular rRNA of C251 () were used. The SL37 substrate, which was synthesized and labelled by 6-FAM at the 3′ end by GenScript company, has a 9-bp stem and an 18-nt loop containing the 11-nt mature 16S rRNA 3′ end sequence and a 7-nt downstream spacer sequence. The C251 substrate was circularized from a radiolabeled linear L251 RNA, which contains all of the precursor sequence, an 11-nt and a 54-nt mature 5′ end and 3′ end sequence of 16S rRNA. The linear L80 substrate and L251 RNA were synthesized via an in vitro transcription using MEGAshortscript T7 Transcription Kit (Ambion). After purification, L251 was 5′ end radiolabeled with [γ-32P]ATP (PerkinElmer) using T4 polynucleotide kinase (Thermo Scientific) at 37°C for 1 h, and then circularized by T4 RNA ligase to produce the C251. The circularized C251 product was first separated on 6% PAGE gel, and sliced for purification using ZR small-RNATM PAGE Recovery Kit (Zymo Research). Gradient concentrations of the purified recombinant Mpy-Nob1 () were incubated respectively with L80 and SL37 (0.2 μM), or C251 (3 nM) in a buffer (25 mM Tris-HCl, pH 8.0, 75 mM NaCl, 2 mM DTT, 100 μg/ml BSA, 0.8 U/μl RNasin, 5 mM MnCl2 and 5% (w/v) glycerol) at 30°C for 60 min. Cleavage reaction was stopped by addition of formamide loading buffer and denaturation at 95°C for 3 min, and the cleavage products of L80 and SL37 were loaded onto 10% urea-PAGE or products of C251 was on 6% urea-PAGE next to the radiolabeled L251 RNA as a migration marker. Cleavage products were visualized by fluorescence imaging directly (for SL37 substrate) or after SYBR Gold staining (for L80 substrate) with Storm Phosphor/fluorescence Imager (GE Healthcare), or by autoradiography (for C251 substrate) with X-ray film (Kodak).
Primer extension
Primer extension was performed to validate the pre-rRNA processing sites that were captured by 5′P-seq previously [Citation29]. Briefly, DNA fragments complementary to the downstream sequences proximal to the predicted mature 5′ ends of 16S, 23S and 5S rRNAs were used as the corresponding primers (Table S2, Sangon Biotech) for reverse transcription and sequencing reactions. Primers were 5′ end radiolabeled in a reaction containing T4 polynucleotide kinase (Thermo Scientific) and [γ-32P]ATP (PerkinElmer) that was incubated at 37°C for 1 h. 5 μg of total RNA extracted from the mid-log M. psychrophilus cells was used in RT reaction. cDNA was synthesized by incubation at 55°C for 1 h using SuperScript III reverse transcriptase (Invitrogen). Sequencing ladders were generated by using a Thermo Sequenase Cycle Sequencing Kit (USB). The products of reverse transcription and DNA sequencing reactions were separated on a 6% polyacrylamide sequencing gel with 7 M urea, and visualized by autoradiography on an X-ray film.
5′- and 3′-RACE analysis
5′ and 3′ rapid amplification of cDNA ends (5′- and 3′-RACE) were performed to identify the 5′ and 3′ termini of RNA, respectively. Based on the fact that the 5′ ends of the processed transcripts are 5′P or 5′OH, the pretreatment by tobacco acid pyrophosphatase of the standard 5′-RACE protocol was omitted. 0.5 nmol of a 5′ RNA adapter (5′-CAGACUGGAUCCGUCGUC-3′; Integrated DNA Technologies) was ligated to 10 μg of total RNA by incubation at 17°C for 16 h with 40 U of T4 RNA ligase (Ambion). The adapter-ligated RNA was recovered by isopropanol precipitation and one aliquot of 2 μg was then used in reverse transcription (RT). The RT reaction was carried out with a gene-specific primer (Table S2, Sangon Biotech) using 200 U of SuperScript III reverse transcriptase (Invitrogen). After RT, nested PCR was conducted to obtain gene-specific products. PCR products were separated on a 2% agarose gel and bands of interest were excised and cloned into pMD19-T vector (TaKaRa). The 5′ terminal nucleotide immediately downstream the adapter sequence was considered as a candidate processing site. Similarly, 3′-RACE was used to identify the mature 3′ end of rRNA or the upstream fragment of an endoribonucleolytic event following published methods [Citation60]. Briefly, 10 μg of total RNA was ligated with 100 pmol of a 3′ RNA adapter (5′-App-CUGUAGGCACCAUCAAU-ddC-3′; Integrated DNA Technologies), and the subsequent operation was the same as the 5′-RACE except for the primers used in RT and PCR reactions. The first nucleotide upstream of the 3′ RNA adapter was assigned to the 3′ end of RNA.
Supplemental Material
Download MS Word (1.4 MB)Acknowledgments
We thank Huanmin Liu and Lei Song for experimental work with radioisotope. All members of the Anaerobes Research Group are acknowledged for helpful discussions.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Ferreira-Cerca S. Life and death of ribosomes in archaea. In: Clouet-d’Orval B, editor. RNA metabolism and gene expression in archaea. Cham: Springer International Publishing AG; 2017. p. 129–158.
- Hage AE, Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1(1):10–15.
- Yip WS, Vincent NG, Baserga SJ. Ribonucleoproteins in archaeal pre-rRNA processing and modification. Archaea. 2013;2013:614735.
- Henras AK, Plisson-Chastang C, O’Donohue MF, et al. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6(2):225–242.
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526.
- Martin R, Straub AU, Doebele C, et al. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 2013;10(1):4–18.
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–391.
- Deutscher MP. Twenty years of bacterial RNases and RNA processing: how we′ve matured. RNA. 2015;21(4):597–600.
- Clouet-d’Orval B, Batista M, Bouvier M, et al. Insights into RNA-processing pathways and associated RNA-degrading enzymes in Archaea. FEMS Microbiol Rev. 2018;42:579–613.
- Young RA, Steitz JA. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci USA. 1978;75(8):3593–3597.
- Mackie GA, RNase E. at the interface of bacterial RNA processing and decay. Nat Rev Microbiol. 2013;11(1):45–57.
- Mohanty BK, Kushner SR. Enzymes involved in posttranscriptional RNA metabolism in Gram-negative bacteria. Microbiol Spectr. 2018;6(2):RWR-0011-2017.
- Baumgardt K, Gilet L, Figaro S, et al. The essential nature of YqfG, a YbeY homologue required for 3′ maturation of Bacillus subtilis 16S ribosomal RNA is suppressed by deletion of RNase R. Nucleic Acids Res. 2018;46(16):8605–8615.
- Woolford JL Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195(3):643–681.
- Pena C, Hurt E, Panse VG. Eukaryotic ribosome assembly, transport and quality control. Nat Struct Mol Biol. 2017;24(9):689–699.
- Chant J, Dennis P. Archaebacteria: transcription and processing of ribosomal RNA sequences in Halobacterium cutirubrum. EMBO J. 1986;5(5):1091–1097.
- Dennis PP, Ziesche S, Mylvaganam S. Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J Bacteriol. 1998;180(18):4804–4813.
- Kjems J, Garrett RA. Novel expression of the ribosomal RNA genes in the extreme thermophile and archaebacterium Desulfurococcus mobilis. EMBO J. 1987;6(11):3521–3530.
- Kjems J, Leffers H, Garrett RA, et al. Gene organization, transcription signals and processing of the single ribosomal RNA operon of the archaebacterium Thermoproteus tenax. Nucleic Acids Res. 1987;15(12):4821–4835.
- Ree HK, Zimmermann RA. Organization and expression of the 16S, 23S and 5S ribosomal RNA genes from the archaebacterium Thermoplasma acidophilum. Nucleic Acids Res. 1990;18(15):4471–4478.
- Durovic P, Dennis PP. Separate pathways for excision and processing of 16S and 23S rRNA from the primary rRNA operon transcript from the hyperthermophilic archaebacterium Sulfolobus acldocaldarius: similarities to eukaryotic rRNA processing. Mol Microbiol. 1994;13(2):229–242.
- Ciammaruconi A, Londei P. In vitro processing of the 16S rRNA of the thermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2001;183(13):3866–3874.
- Thompson LD, Daniels CJ. Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J Biol Chem. 1990;265(30):18104–18111.
- Kleman-Leyer K, Armbruster DW, Daniels CJ. Properties of H. volcanii tRNA intron endoribonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell. 1997;89:839–847.
- Salgia SR, Singh SK, Gurha P, et al. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA. 2003;9(3):319–330.
- Tang TH, Rozhdestvensky TS, d’Orval BC, et al. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 2002;30(4):921–930.
- Danan M, Schwartz S, Edelheit S, et al. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40(7):3131–3142.
- Jüttner M, Weiß M, Ostheimer N, et al. A versatile cis-acting element reporter system to study the function, maturation and stability of ribosomal RNA mutants in archaea. Nucleic Acids Res. 2020;48(4):2073–2090.
- Qi L, Yue L, Feng D, et al. Genome-wide mRNA processing in methanogenic archaea reveals post-transcriptional regulation of ribosomal protein synthesis. Nucleic Acids Res. 2017;45(12):7285–7298.
- Li J, Qi L, Guo Y, et al. Global mapping transcriptional start sites revealed both transcriptional and post-transcriptional regulation of cold adaptation in the methanogenic archaeon Methanolobus psychrophilus. Sci Rep. 2015;5:9209.
- Cannone JJ, Subramanian S, Schnare MN, et al. The comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2.
- Veith T, Martin R, Wurm JP, et al. Structural and functional analysis of the archaeal endoribonuclease Nob1. Nucleic Acids Res. 2012;40(7):3259–3274.
- Englert M, Sheppard K, Aslanian A, et al. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci USA. 2011;108(4):1290–1295.
- Williams TA, Szollosi GJ, Spang A, et al. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc Natl Acad Sci USA. 2017;114(23):E4602–E4611.
- Hughes JM, Ares M Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10(13):4231–4239.
- Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22(23):5139–5147.
- Ruggero D, Ciammaruconi A, Londei P. The chaperonin of the archaeon Sulfolobus solfataricus is an RNA-binding protein that participates in ribosomal RNA processing. EMBO J. 1998;17(12):3471–3477.
- Russell AG, Ebhardt H, Dennis PP. Substrate requirements for a novel archaeal endonuclease that cleaves within the 5′ external transcribed spacer of Sulfolobus acidocaldarius precursor rRNA. Genetics. 1999;152(4):1373–1385.
- Kjems J, Garrett RA. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988;54(5):693–703.
- Kjems J, Garrett RA. Ribosomal RNA introns in archaea and evidence for RNA conformational changes associated with splicing. Proc Natl Acad Sci USA. 1991;88(2):439–443.
- Burggraf S, Larsen N, Woese CR, et al. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc Natl Acad Sci USA. 1993;90(6):2547–2550.
- Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Evolution of introns in the archaeal world. Proc Natl Acad Sci USA. 2011;108(12):4782–4787.
- Yoshinari S, Itoh T, Hallam SJ, et al. Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endoribonuclease. Biochem Biophys Res Commun. 2006;346(3):1024–1032.
- Plagens A, Daume M, Wiegel J, et al. Circularization restores signal recognition particle RNA functionality in Thermoproteus. Elife. 2015;4:e11623.
- Kaneta A, Fujishima K, Morikazu W, et al. The RNA-splicing endonuclease from the euryarchaeaon Methanopyrus kandleri is a heterotetramer with constrained substrate specificity. Nucleic Acids Res. 2018;46(4):1958–1972.
- Marck C, Grosjean H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA. 2003;9(12):1516–1531.
- Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc Natl Acad Sci USA. 2005;102(43):15418–15422.
- Dominski Z, Carpousis AJ, Clouet-d’Orval B. Emergence of the β-CASP ribonucleases: highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim Biophys Acta. 2013;1829(6–7):532–551.
- Clouet-d’Orval B, Phung DK, Langendijk-Genevaux PS, et al. Universal RNA-degrading enzymes in Archaea: prevalence, activities and functions of β-CASP ribonucleases. Biochimie. 2015;118:278–285.
- Evguenieva-Hackenberg E, Gauernack S, Klug G. The archaeal exosome: degradation and tailing at the 3′-end of RNA. In: Clouet-d’Orval B, editor. RNA metabolism and gene expression in archaea. Cham: Springer International Publishing AG; 2017. p. 115–128.
- Hölzle A, Fischer S, Heyer R, et al. Maturation of the 5S rRNA 5′ end is catalyzed in vitro by the endoribonuclease tRNase Z in the archaeon H. volcanii. RNA. 2008;14(5):928–937.
- Bailey TL, Johnson J, Grant CE, et al. The MEME Suite. Nucleic Acids Res. 2015;43(W1):W39–W49.
- Crooks GE, Hon G, Chandonia JM, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190.
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415.
- De Rijk P, Wuyts J, De Wachter R. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19(2):299–300.
- Kjems J, Garrett RA. Secondary structural elements exclusive to the sequences flanking ribosomal RNAs lend support to the monophyletic nature of the archaebacteria. J Mol Evol. 1990;31(1):25–32.
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361.
- Zheng X, Feng N, Li DF, et al. New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family. Mol Microbiol. 2017;106(3):351–366.
- Slomovic S, Schuster G. Circularized RT-PCR (cRT-PCR): analysis of the 5′ ends, 3′ ends, and poly(A) tails of RNA. Methods Enzymol. 2013;530:227–251.
- Zhang J, Olsen GJ. Messenger RNA processing in Methanocaldococcus (Methanococcus) jannaschii. RNA. 2009;15(10):1909–1916.
