ABSTRACT
As the adaptor that decodes mRNA sequence into protein, the basic aspects of tRNA structure and function are central to all studies of biology. Yet the complexities of their properties and cellular roles go beyond the view of tRNAs as static participants in protein synthesis. Detailed analyses through more than 60 years of study have revealed tRNAs to be a fascinatingly diverse group of molecules in form and function, impacting cell biology, physiology, disease and synthetic biology. This review analyzes tRNA structure, biosynthesis and function, and includes topics that demonstrate their diversity and growing importance.
Genetic information is passed from DNA to RNA to protein through the processes of transcription and translation. Central to the information flow from RNA to protein are transfer RNA (tRNA) molecules [Citation1]. tRNAs are the vehicle that bring amino acids to the growing polypeptide chain at the ribosome and read the three base codons that define protein sequence [Citation2–5]. At the textbook level, tRNA structure and function can be described quite simply. In reality, they are significantly more complex, with nuances highly relevant to disease, evolution and biotechnology. This review will focus on the molecular biology of tRNAs, assembling some of the information that is not discussed in depth when considering general genetic information flow. Topics central to all organisms will be discussed but there will be specific focus on eukaryotic cytosolic tRNAs. The breadth of the review has meant that we have been unable to reference all of the relevant literature; however, in each section, we refer to more detailed review articles.
Structure
Holley et al. [Citation6] determined the first tRNA sequence, a 77 base tRNAAla from yeast. Overall the length of canonical tRNAs varies from 76 to 90 bases [reviewed in Citation7]. This gives the impression that they are small molecules; however, their average mass is greater than 26,000 g/mol, the approximate mass of a 230 amino acid residue protein. Their structure is defined by internal base pairing that results in a stem-loop pattern when depicted in two dimensions (). From 5ʹ to 3ʹ, the stem-loop structures are the acceptor stem, the D-arm, anticodon stem, variable loop and T-arm. The D-loop is named after the modified dihydrouridine base present in the loop and is responsible for stabilizing the tRNA tertiary structure [Citation8]. The T-arm, also known as the TΨC arm, is named after the presence of the universally conserved modified bases thymidine, pseudouridine and cytidine [Citation6] and is responsible for facilitating interactions with the ribosome [Citation9]. As suggested by the name, the length of the variable arm is variable. In eukaryotes, it is most prominent for the serine (tRNASer), selenocysteine (tRNASec) and leucine (tRNALeu) tRNAs. Longer variable arms are also found in eubacterial and organellar tRNATyr and in a systematic search for other variable arm containing tRNAs, Hamashima et al. [Citation10] identified 253 additional non-serine or non-leucine tRNAs with an extended variable arm from a range of eukaryotic organisms.
Figure 1. tRNA structure. (A) Cloverleaf representation of a tRNA in 2D. From 5ʹ to 3ʹ the regions of the tRNA are: acceptor stem (green), D-arm (pink), anticodon arm (blue) containing the anticodon (darker blue), variable arm (orange) and T-arm (purple). The discriminator base (base 73) is the unpaired base at the 3ʹ end [yellow) before the terminal CCA residues. Aminoacyl-tRNA synthetases ligate an amino acid onto the terminal 3ʹ adenosine. Numbered positions are constant bases, as described in Citation32. (B) Three-dimensional tRNA structure of a yeast tRNAPhe [Citation14, PDB: 1EHZ]. Colour of the 2D elements is the same as in (A). Intramolecular interactions between the T- and D-arms (purple and pink respectively) in the elbow region facilitate tRNA folding. (C) A surface representation of tRNAPhe (left; PDB: 1EHZ) and the ribosome recycling factor (right; PDB: 1EH1) demonstrate how molecules that interact with the translational machinery often adopt tRNA-like structures
![Figure 1. tRNA structure. (A) Cloverleaf representation of a tRNA in 2D. From 5ʹ to 3ʹ the regions of the tRNA are: acceptor stem (green), D-arm (pink), anticodon arm (blue) containing the anticodon (darker blue), variable arm (orange) and T-arm (purple). The discriminator base (base 73) is the unpaired base at the 3ʹ end [yellow) before the terminal CCA residues. Aminoacyl-tRNA synthetases ligate an amino acid onto the terminal 3ʹ adenosine. Numbered positions are constant bases, as described in Citation32. (B) Three-dimensional tRNA structure of a yeast tRNAPhe [Citation14, PDB: 1EHZ]. Colour of the 2D elements is the same as in (A). Intramolecular interactions between the T- and D-arms (purple and pink respectively) in the elbow region facilitate tRNA folding. (C) A surface representation of tRNAPhe (left; PDB: 1EHZ) and the ribosome recycling factor (right; PDB: 1EH1) demonstrate how molecules that interact with the translational machinery often adopt tRNA-like structures](/cms/asset/3bdf4943-9e10-4e31-ad35-d3a2d925acad/krnb_a_1809197_f0001_c.jpg)
tRNAs have two key single-stranded regions required for translation: the three base anticodon determines translation specificity by base pairing with the mRNA codon and the 3ʹ acceptor CCA is the site of covalent amino acid attachment in an ester linkage. The enzymes responsible for aminoacylation, or charging, of an amino acid onto its cognate tRNAs are called aminoacyl-tRNA synthetases [aaRS]. The specificity of aminoacylation is critical because the ribosome does not read the amino acid on the tRNA. As demonstrated by Chapeville et al. [Citation11], the ribosome will incorporate a non-coded amino acid if the tRNA is incorrectly aminoacylated. The base 5ʹ to the terminal CCA is also not base paired. It is referred to as the discriminator base because of its importance in determining aminoacylation specificity [Citation12].
In three dimensions, tRNAs fold into an L-shaped structure, primarily through intramolecular tertiary interactions of the T- and D-arms (; [Citation13,Citation14]). At one end of the L is the amino acid acceptor branch and at the other is the anticodon branch. The amino acid acceptor branch is formed from interactions between the acceptor stem, generally seven base pairs, and T-stem, generally five base pairs. It is this 12 base pairs in a 7/5 configuration that is recognized by the elongation factor EF1A in eukaryotes and archaea and EF-Tu in bacteria [Citation15]. Theanticodon branch consists of the anticodon and D-arms. The region where the branches meet is referred to as the elbow [Citation16]. It should be noted that whereas the L-shaped structure of the tRNA is generally conserved across organisms, the exact structural details are not [reviewed in Citation17, Citation18]. For example, both bacterial and eukaryotic tRNASec have a 13 base-pair acceptor branch [Citation19,Citation20]. There are even functional mitochondrial tRNAs that lack one or both of the D- or T-arms [Citation21-24]. Other proteins and RNA molecules adopt the canonical L shape to interact with translational machinery in a process known as tRNA mimicry. For example, some RNA viruses have internal ribosome entry sites (IRES) to direct translation of overlapping open reading frames, where a pseudoknot RNA structure within the viral RNA transcript adopts a tRNA-like structure to recruit the host ribosome without the need for an initiator tRNAMet [reviewed in Citation25]. In other cases, proteins such as elongation factor G, responsible for translocation of the tRNA and mRNA through the ribosome, and the release factor proteins, responsible for translation termination, adopt tRNA-like structures to facilitate interactions with the ribosome (; reviewed in Citation26, Citation27).
tRNAs that accept the same amino acid are called isoacceptors. They differ in their anticodon sequence reflecting that all amino acids other than methionine and tryptophan are coded for by more than one codon. Generally, tRNA sequences in the same isoacceptor family are similar but not identical. There are 21 types of isoacceptors, one for each of the 20 standard amino acids and one for selenocysteine. The three stop codons are represented by release factor proteins that terminate translation [Citation28], rather than tRNAs; though it should be noted that stop codons have been reassigned to encode amino acids in some organisms [reviewed in Citation29]. Not all of the 61 amino acid encoding codons have a corresponding tRNA with a completely complementary anticodon sequence. Wobble at the third position of the codon allows for decoding of all codons (discussed below). tRNAs with the same anticodon and thus decode the same set of codons are called isodecoders. tRNAs in the same isodecoder family differ in their primary sequence at sites other than the anticodon.
The number of tRNAs found in different organisms varies widely; for example, Saccharomyces cerevisiae and humans contain 275 and 416 high confidence tRNA encoding genes, respectively [Citation30, http://gtrnadb.ucsc.edu]. Because of wobble, 32 tRNA species are sufficient to decode all 61 codons. There are, however, many different isodecoders per organism, each often having multiple genomic copies. The additional copies provide redundancy in case of a loss of function mutation and a buffer should a mutation result in noncognate decoding [Citation31]. These multiple copies provide the raw material for genetic code evolution.
tRNAs have a standard numbering ending with the terminal 3ʹ CCA being numbers 74, 75 and 76 (; [Citation32]). Base 1 in the acceptor arm is the 5ʹ base that pairs with base 72. tRNAHis has an additional 5ʹ base, which is added post-transcriptionally, numbered 0 but sometimes referred to as −1 (discussed below). Bases with conserved function or invariant nucleotides always have the same number. Most notable of these are the three anticodon positions, which are always numbered 34, 35 and 36. Other constant bases include two invariant guanine residues at positions 18 and 19 in the D-loop and TΨC residues at positions 54, 55 and 56. Additional bases found after a conserved position are designated with letters following the conserved position number (e.g. 20A and 20B). The discriminator base is numbered 73. The variable arm begins with bases 44 and 45 and ends with bases 46, 47 and 48. In tRNAs with extended variable arms, mainly tRNASer, tRNASec and tRNALeu, additional bases are numbered after position 45 and designated with an e. They are not numbered sequentially but rather numbered in a way that corresponds to their base-pairing partner. For example, position e11 would be the first extended variable arm base and base pair with the last extended variable arm base, position e21. The single-stranded loop region in an extended variable arm is numbered as single digits beginning with e1. Full tRNA numbering can be found in [Citation32].
tRNA biosynthesis
Transcription
In eukaryotes, tRNAs are transcribed by the relatively α-amanitin resistant RNA polymerase III [RNAPIII; Citation33–35, reviewed in Citation36]. RNAPIII is a 17-subunit enzyme with its two largest subunits resembling β and β’ from bacterial RNA polymerase [Citation37]. RNAPIII transcribes other small noncoding RNAs including 5S rRNA, U6 snRNA of the spliceosome and the RNA component of both RNase P and the signal recognition particle [reviewed in Citation38]. The tRNA genes are highly expressed at approximately 300,000 copies per cell and on par with ribosomal RNA genes [Citation39]. The elements required to transcribe tRNA genes, known as the A box (consensus sequence TRGYNNARNNG) and B box (consensus sequence RGTTCRANTCC), are internal to the tRNA gene within the sequences encoding the D- and T-arms [Citation40–42]. The spacing between the A and B box varies greatly depending on whether a tRNA encoding gene contains an intron and the length of the variable arm. Segall et al. [Citation43] were the first to characterize the factors required for RNAPIII transcription at tRNAs. They identified two column fractions they termed B and C that allowed in vitro transcription of tRNA genes. Further purification of the activities in these fractions led to the designations TFIIIB and TFIIIC. Transcription is initiated with the binding of TFIIIC to the A and B box (; [Citation44,Citation45]). Bound TFIIIC recruits TFIIIB to the 5ʹ side of the gene [Citation46,Citation47]. One of the three protein subunits of TFIIIB is the TATA-binding protein [Citation48,Citation49]. As part of the TFIIIC-TFIIIB-DNA ternary complex, TFIIIB recruits RNAPIII [Citation50] and participates in promoter melting with the C37 subunit of RNAPIII [Citation51,Citation52], whether TFIIIC is displaced during elongation is unclear. Turowski et al. [Citation53] suggest that it is displaced first from the A box then the B box as transcription proceeds. Transcriptional termination requires a stretch of five or more T residues on the non-template strand [Citation54] and three subunits of RNAPIIII [Citation55]. Transcriptional re-initiation, which allows enhanced transcription of tRNA genes, is thought to occur by TFIIIB maintaining contact with RNAPIII through the termination process [Citation56, reviewed in Citation57]. The exception to the basic mechanism described above is tRNASec. In most eukaryotes, tRNASec recruits RNAPIII through an internal B box and upstream elements also found in U6 snRNA genes [see, for example, Citation58]. The distinction is even clearer in trypanosomes where tRNASec is transcribed by RNA polymerase II [Citation59].
Figure 2. tRNAs are transcribed by RNA polymerase III. The internal A- and B-box sequences are recognized by TFIIIC (i). Bound TFIIIC leads to the recruitment of TFIIIB which binds upstream of the tRNA gene and recruits RNAPIII (ii). TFIIIB and RNAPIII melt the promoter, leading to transcription of the tRNA (iii and iv). Termination is signalled by a string of thymine residues on the non-templated strand (v). Transcriptional re-initiation, where RNAPIII is recycled to the transcriptional start site, can enhance transcription of tRNA genes (iv)
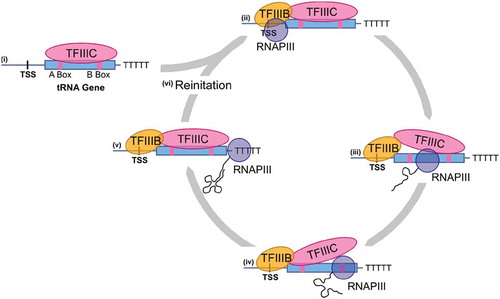
RNAPIII transcription is regulated by Maf1, which inhibits transcriptional re-initiation in response to nutrient stress [Citation60]. Maf1 is regulated by phosphorylation from a range of kinases including TORC1 [Citation61,Citation62]. Interestingly, Gerber et al. [Citation63] recently demonstrated that in HEK293 cells, RNA polymerase II transcription through a subset of tRNA encoding genes inhibits RNAPIII transcription in a Maf1 dependent fashion. Also in mammalian cells, RNAPIII is regulated by Myc [reviewed in Citation64], p53 [Citation65] and Rb [Citation66], emphasizing the importance of RNAPIII transcription in cell cycle control and disease processes.
tRNA processing
After transcription, pre-tRNA transcripts are processed (). Transcription results in a 5ʹ leader of approximately 10 bases that is removed by RNase P, the first transacting ribozyme identified [Citation67]. Though there are exceptions [Citation68], the canonical eubacterial, archaeal and eukaryotic RNase P is a ribonucleoprotein where the RNA plays a catalytic role [Citation69–71]. Cleavage of the scissile phosphodiester bond leaves the 5′ end of the RNA with a 3′-OH and the 3′ end of the RNA with a 5′-phosphate [Citation69, reviewed in Citation72]. In eukaryotes, cleavage of pre-tRNA leader sequences by RNase P requires base pairing at the first and second base pairs in the acceptor stem and generates an acceptor stem with the canonical seven base pairs [Citation73–75]. The 3ʹ ends of pre-tRNAs are a short U tract [reveiwed in Citation76]. This U tract is the binding site for the La protein, which protects the 3ʹ end of the transcript from exonuclease digestion and assists pre-tRNA folding [Citation77, reviewed in Citation78]. The 3ʹ endonuclease RNase Z cleaves after the discriminator nucleotide leaving a 3ʹ-OH [Citation79]. Both 5ʹ and 3ʹ cleavage occur in the nucleus. It should be noted that in eukaryotes, the 3ʹ exonucleases Rex1p and Rpr6p process pre-tRNA trailer sequences under some conditions [Citation80,Citation81], while processing for bacterial tRNAs with an encoded 3ʹ CCA uses a different mechanism. Following 3ʹ cleavage, the CCA adding enzyme [ATP (CTP): tRNA nucleotidyltransferase] catalyzes the addition of the CCA terminus or any nucleotides required to complete the CCA terminus, without the need of a template [Citation82]. In organisms where the CCA is encoded in the genome, the enzyme repairs 3ʹ termini [Citation83].
Figure 3. Lifecycle of a tRNA. (A) tRNAs are transcribed (i) with 5ʹ leader and 3ʹ trailer sequences that are cleaved by RNase P and RNase Z respectively (ii). After trimming, partial modification (iii), indicated by the red circles, and addition of the terminal CCA residues (iv) occur in the nucleus. In yeast, the tRNA is exported out of the cytoplasm by Los1 (v) and if the tRNA contains an intron, it is spliced on the mitochondrial surface (vi). In mammals, splicing occurs before nuclear export. In the cytoplasm, additional modifications may be added (viii). tRNAs can then be aminoacylated by their cognate aminoacyl-tRNA synthetase (viii) or be re-imported (ix) by Mtr10 into the nucleus for further modification (x) and quality control through nuclear aminoacylation (xi). Re-export of tRNAs is facilitated by both Los1 and an Msn5 dependent pathway that specifically recognizes mature tRNAs (xii). Once aminoacylated, tRNAs are used in translation (xiii). (B) Quality control mechanisms can lead to the degradation of tRNAs. After transcription, nuclear surveillance (i) monitors tRNAs for end maturation and specific modifications. Improper tRNAs are polyadenylated by the TRAMP complex and degraded by the nuclear exosome. After processing and export, tRNAs are monitored and degraded through the rapid tRNA pathway by the 5ʹ-3ʹ exonuclease Xrn1 in the cytoplasm (ii) and Rat1 in the nucleus (iii)
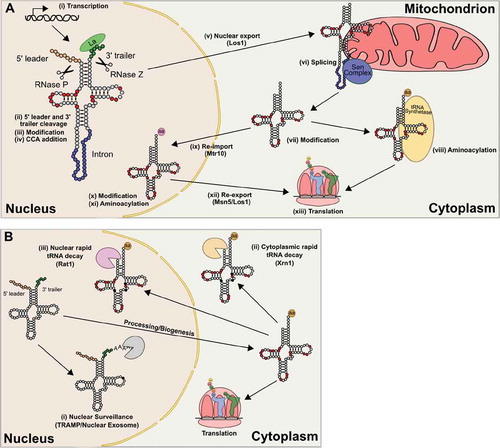
tRNAHis has an additional G nucleotide at its 5ʹ end that is not genomically encoded [reviewed in Citation84]. In eukaryotes, this is accomplished after processing by Thg1 enzymes, which use a 3′–5′ addition reaction to add the G residue [Citation85, Citation86, reviewed in Citation87]. The 3ʹ-5ʹ reaction is the reverse of other known DNA and RNA polymerases. The reaction occurs in a multistep process which begins with adenylation of the processed 5ʹ end in a high energy 5ʹ-5ʹ linkage [Citation86,Citation88]. The 3′-OH of GTP then attacks the pyrophosphate of the adenylylated tRNA, leaving a 5ʹ triphosphate, which Thg1 removes leaving a single 5ʹ phosphate. Thg1 specificity for tRNAHis occurs through recognition of the GUG anticodon [Citation89].
tRNAs are highly modified, with approximately 12% of nucleotides bearing a modification (; reviewed in Citation90). Currently, 93 unique tRNA modifications have been identified from more than 700 diverse tRNA sequences [https://mods.rna.albany.edu/; Citation91]. Many of these modifications are broadly conserved in bacteria, eukaryotes and archaea; for example, the ribothymidine in the T-stem and dihydrouridine in the D-loop. Saccharomyces cerevisiae tRNAs contain 25 unique modifications that occur at 36 positions (). These modifications include the addition of methyl groups to the ribose 2ʹ-OH or nucleobases, acetylation of nucleobases, modification of uridine to pseudouridine and dihydrouridine, deamination of adenine to inosine and addition of threonylcarbamonyl and isopentenyl groups to adenine, among others. Modification occurs at many stages of tRNA biosynthesis [reviewed in Citation92], including prior to processing in the nucleus [Citation93–95]. As such, some of the modifying enzymes are located in the nucleus, others in the cytoplasm and some have isozymic forms located in both [Citation96]. A detailed discussion of the functions of tRNA modifications is presented below.
Figure 4. tRNAs are highly post-transcriptionally modified. (A) Heat map showing modification frequency for each position in the tRNA on the 2D (left) and 3D (right) tRNA structures. Modification data for 715 unique tRNA sequences from archaea, bacteria and eukaryotes accessed from the MODOMICS database [http://genesilico.pl/modomics/; [Citation282]]. Circled bases are not modified in any of the 715 unique tRNA sequences in the database. (B) tRNA modifications found across all S. cerevisiae tRNAs [pseudouridine (Ψ); 2ʹ-O-methylcytidine (Cm); 2ʹ-O-methyladenosine (Am); 1-methyl-guanosine (m1G); N2-methylguanosine (m2G); N4-acetylcytidine (ac4C); dihydrouridine (D); 2ʹ-O-methylguanosine (Gm); N2,N2-dimethylguanosine (m2,2G); 3-methylcytidine (m3C); inosine (I); 5-methylcytidine (m5C); 5-methoxycarbonylmethyluridine (mcm5U); 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U); 5-carbamoylmethyluridine (ncm5U); 5-carbamoylmethyl-2ʹ-O-methyluridine (ncm5Um); 1-methylinosine (m1I); N6-isopentenyladenosine (i6A); wybutosine (yW); N6-threonylcarbamoyladenosine (t6A); 2ʹ-O-methyluridine (Um); 7-methylguanosine (m7G); ribothymidine (rT); 1-methyladenosine (m1A); 2ʹ-O-ribosyladenosine phosphate (Ar(p))]. The nucleobases of select-modified nucleotides are shown on the right
![Figure 4. tRNAs are highly post-transcriptionally modified. (A) Heat map showing modification frequency for each position in the tRNA on the 2D (left) and 3D (right) tRNA structures. Modification data for 715 unique tRNA sequences from archaea, bacteria and eukaryotes accessed from the MODOMICS database [http://genesilico.pl/modomics/; [Citation282]]. Circled bases are not modified in any of the 715 unique tRNA sequences in the database. (B) tRNA modifications found across all S. cerevisiae tRNAs [pseudouridine (Ψ); 2ʹ-O-methylcytidine (Cm); 2ʹ-O-methyladenosine (Am); 1-methyl-guanosine (m1G); N2-methylguanosine (m2G); N4-acetylcytidine (ac4C); dihydrouridine (D); 2ʹ-O-methylguanosine (Gm); N2,N2-dimethylguanosine (m2,2G); 3-methylcytidine (m3C); inosine (I); 5-methylcytidine (m5C); 5-methoxycarbonylmethyluridine (mcm5U); 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U); 5-carbamoylmethyluridine (ncm5U); 5-carbamoylmethyl-2ʹ-O-methyluridine (ncm5Um); 1-methylinosine (m1I); N6-isopentenyladenosine (i6A); wybutosine (yW); N6-threonylcarbamoyladenosine (t6A); 2ʹ-O-methyluridine (Um); 7-methylguanosine (m7G); ribothymidine (rT); 1-methyladenosine (m1A); 2ʹ-O-ribosyladenosine phosphate (Ar(p))]. The nucleobases of select-modified nucleotides are shown on the right](/cms/asset/38199a52-b1a3-44c2-907b-9fcb650882bd/krnb_a_1809197_f0004_c.jpg)
Transport in and out of the nucleus
Eukaryotic tRNAs are shuttled in and out of the nucleus through the nuclear pore complex [Citation97, reviewed in Citation98]. Primary export after synthesis requires a dedicated member of the β-importin family, Los1 in yeast and Xpot in vertebrates [Citation99–101]. Los1/Exportin-t directly binds tRNAs in a Ran-GTP-dependent fashion [Citation102–106]. Xpot is regulated by the small GTPase Ran and binds tRNAs in the nucleus in its GTP-bound form [reviewed in Citation107]. Binding requires a mature tRNA backbone and aminoacylation stem with processed 5ʹ and 3ʹ ends [Citation103] but does not require intron removal [Citation101,Citation102,Citation108]. Upon translocation through the nuclear pore, GTP hydrolysis results in tRNA release into the cytoplasm. Crm1 and Mex67-Mtr2 were identified in a genome-wide screen in yeast for other genes involved in primary tRNA nuclear export and participate in parallel pathways [Citation109,Citation110].
Retrograde transport of mature tRNAs back into the nucleus occurs constitutively in some organisms and in response to stress and nutrient deprivation [Citation111–116, reviewed in Citation117]. The β-importin family member Mtr10 and interestingly the heat shock protein Hsp70 have been implicated in the process [Citation118,Citation119]. Constitutive retrograde transport is necessary for S. cerevisiae because the intron-containing tRNAPhe is spliced on the surface of mitochondria (see below) and subsequently subject to G37 methylation in the nucleus [Citation120]. The intron-containing tRNAPhe is not a substrate for modification. Similarly, in Trypanosoma brucei, tRNATyr modification by tRNA-guanine transglycosylase occurs after retrograde transport of the spliced tRNA [Citation121]. Schwenzer et al. [Citation122] recently demonstrated that in HeLa cells, tRNASec, which is integral to redox control because of its requirement in the synthesis of selenoproteins, is subject to retrograde transport regulated by the integrated stress response pathway.
Re-export of tRNAs to the cytoplasm involves three pathways [reviewed in Citation123]. Two of these are shared with the export of primary tRNAs, the Los1/Xpot pathway and the Mex67-Mtr2 pathway. The third, an Msn5 dependent pathway, appears specialized for re-export of mature tRNAs since it is unable to transport unspliced tRNA transcripts [Citation118]. Re-export is also partially dependent on nuclear aminoacylation of tRNAs [Citation108,Citation124,Citation125]. Conditional mutants of aaRSs or the CCA adding enzyme or drugs that inhibit aminoacylation [see, for example, Citation126] lead to mature tRNAs accumulating in the nucleus. This likely functions as a quality control mechanism to ensure the export of functional tRNAs.
Splicing
A subset of tRNA genes in all three kingdoms of life contain introns [Citation127]. In eukaryotes, the intron is positioned primarily following base 37 and does not affect the overal structure of the tRNA [reviewed in Citation128]. Splicing requires a tRNA-splicing endonuclease known as the Sen complex [Citation129,Citation130] and a tRNA ligase [Citation131,Citation132]. To allow flexible sequence selection, the Sen complex measures the distance between the body of tRNA and the splice sites [Citation133,Citation134]. In vertebrates, splicing occurs in the nucleus [Citation129, Citation135]; whereas in yeast the splicing endonuclease is located on the surface of the mitochondrion [Citation136]. Cleavage by the Sen complex results in a 2′,3′ cyclic phosphate on the 5ʹ exon and a 5′-OH on the 3ʹ exon [Citation137,Citation138]. How these are treated depends on the organism. In yeast, Trl1 ligates the two exons together by opening the 2ʹ,3ʹ cyclic phosphate, phosphorylating the 5ʹ terminus of the 3ʹ exon with GTP, activating the 5ʹ terminus with AMP and finally joining the two exons together [Citation131,Citation132]. Trl1 is also required to splice the HAC1 transcript in the unfolded protein response [Citation139]. In vertebrates, tRNA exons are ligated directly by 3ʹ-5ʹ ligation catalysed by a complex containing HSPC117 [Citation140].
tRNA turnover
Mature tRNAs are stable, with half-lives around 2 to 3 days in eukaryotes [Citation141–143]. Nonetheless, two main tRNA degradation pathways remove partially processed, hypomodified or misfolded tRNAs (; reviewed in Citation144). In the first pathway, the TRAMP (Trf4 Air2 Mtr4 polyadenylation) complex monitors pre-tRNAs and hypomodified tRNAs in the nucleus and targets them for degradation by the nuclear exosome. The pathway was recognized by Anderson et al. [Citation145], who demonstrated that initiator tRNAMet lacking methylation at A58 is unstable. The components required were identified genetically [Citation146, Citation147] when the temperature-sensitive lethality of a trm6 mutant, encoding the tRNA 1-methyladenosine methyltransferase responsible for initiator tRNAMet modification, was suppressed by deleting the nuclease subunit of the nuclear exosome or the poly(A) polymerase of the TRAMP complex. Hypomodified tRNAs are 3ʹ polyadenylated, which signals degradation by the nuclear exosome in a 3ʹ to 5ʹ manner. This pathway also monitors the 3ʹ ends of pre-tRNAs, where the loss of accurate 3ʹ end maturation leads to polyadenylation of select tRNA substrates and degradation by the nuclear exosome [Citation80,Citation81]. A large-scale study of nuclear exosome catalytic mutants performed by Gudipati et al. [Citation148] suggests that up to 50% of pre-tRNAs are degraded by this pathway.
The second tRNA decay pathway, the rapid tRNA decay (RTD) pathway, monitors specific mature tRNAs and degrades hypomodified or unstable tRNAs in a 5ʹ to 3ʹ manner. tRNAs targeted by the RTD pathway include hypomodified tRNAValAAC lacking 7-methylguanosine and 5-methylcytidine [Citation149] and tRNASerUGA and tRNASerCGA lacking 2ʹ-O-methyluridine and 4-acetylcytidine or 5-methylcytidine and 2ʹ-O-methylguanosine [Citation150–152]. In addition, tRNAs that are structurally unstable are targeted by this pathway [Citation153–156]. Degradation requires the 5ʹ-3ʹ exonucleases Rat1 or Xrn1 [Citation150].
tRNAs can also be degraded through endonucleolytic cleavage. This often occurs in response to stress and involves cleavage near or within the anticodon loop to generate tRNA half molecules that can be signalling molecules (discussed further below: Other functions of tRNAs).
Aminoacylation
Aminoacyl-tRNA synthetases (aaRS) are the enzymes that couple amino acids to tRNAs and thus allow tRNA molecules to serve as adaptors in the central dogma [Citation157]. Each tRNA isoacceptor family is aminoacylated by a specific aaRS [reviewed in Citation158]. The reaction requires specific recognition of an amino acid and a tRNA. In the first step of the two-step reaction, the amino acid is condensed with ATP to form an aminoacyl adenylate. Interestingly, the aaRSs were first identified for this step in the reaction [e.g. Citation159, see Citation2]. In the second step, the amino acid is transferred in a high energy ester linkage to the 2ʹ- or 3ʹ-OH of the terminal adenosine ribose [reviewed in Citation160]. In solution, the 2ʹ- and 3ʹ-linked amino acid transacylates to the neighbouring -OH [Citation161] with translation elongation factor EF-Tu/EF1A binding stabilizing the preferred 3ʹ-linked form [Citation162].
In general, each aaRS is specific for one amino acid and one tRNA isoacceptor group, but there are exceptions. SerRSserylates the different tRNASer isoacceptors and tRNASec with serine [Citation163,Citation164]. Serylated tRNASec is then converted to Sec-tRNASec enzymatically [Citation165–168]. tRNASec has a UCA anticodon and is selectively inserted at UGA stop codons. Bacteria have a relatively common mechanism that differs from that found in archaea and eukaryotes [reviewed in Citation169]. Another example of an aaRS charging multiple isoacceptor groups occurs in several eubacteria and archaea that lack GlnRS and AsnRS [Citation170]. In these cases, tRNAGln and tRNAAsn are charged by GluRS and AspRS, respectively, with glutamate and aspartate [Citation171]. A glutamine-dependent amidotransferase (AdT) then converts Glu-tRNAGln and Asp-tRNAAsn to Gln-tRNAGln and Asn-tRNAAsn, respectively [Citation172]. Sauerwald et al. [Citation173] described an unusual circumstance in methanogenic archaea where Cys-tRNACys is synthesized in a two-step pathway that does not involve an aaRS specific for cysteine. Rather, O-phosphoserine (Sep) is first transferred to tRNACys by SepRS. Sep-tRNACys is then converted to Cys-tRNACys by Sep-tRNA:Cys-tRNA synthase (SepCysS). This pathway is likely more broadly conserved as bioinformatic analysis has revealed SepRS and SepCysS in other archaea and bacteria [Citation174].
The sequences that allow tRNA recognition by a specific aaRS are called identity elements. Identity elements consist of single nucleotides, nucleotide pairs and structural motifs [Citation175,Citation176]. For many tRNAs, the anticodon is a principal identity element making direct contact with the aaRS [e.g. Citation177]. This recognition provides a direct link between the anticodon and its amino acid assignment [Citation178]. However, other specific nucleotides in the acceptor stem provide essential structural characteristics for aaRS recognition and aminoacylation [Citation179–181]. In particular, base 73, the discriminator base positioned directly 5ʹ of the terminal CCA, is a common point of recognition. The additional G at −1 of tRNAHis is essential for charging by HisRS [Citation182,Citation183].
For tRNAAla, tRNASer and tRNASec the anticodon plays no role in recognition by their cognate aaRSs. In some organisms (e.g. S. cerevisiae and humans) this applies to a lesser extent for tRNALeu as well. For tRNAAla, the principal identity element is a G3:U70 base pair in the acceptor stem [Citation184–187]. In fact, including a G3:U70 base pair in non-alanine isoacceptors is sufficient for their charging by AlaRS [e.g. Citation188, Citation189]. This may explain the relative absence of G3:C70 or A3:T70 base pairs in non-alanine tRNAs, as a single mutation would change coding specificity [Citation190]. For SerRS, the major feature recognized in both tRNASer and tRNASec is the variable arm, with both the length and composition being important [Citation191–194]. For these tRNAs, a change in the anticodon to a sequence designating another amino acid will lead to a coding error (discussed below: tRNA-dependent mistranslation).
Ten aaRSs have pre- and post-transfer editing mechanisms to distinguish between similar amino acids and correct aminoacylation errors [IleRS, PheRS, ValRS, LeuRS, MetRS, ThrRS, AlaRS, ProRS, LysRS and SerRS; reviewed in Citation195]. Baldwin et al. [Citation196] discovered the first example of editing when they observed that IleRS can form Val-AMP, but addition of tRNAIle results in its hydrolysis. Further studies from Eldred and Schimmel [Citation197] and Yarus [Citation198] demonstrated post-transfer editing where Val-tRNAIle and Ile-tRNAPhe were hydrolysed by IleRS and PheRS, respectively. Pre-transfer editing was later observed by Fersht [Citation199] for activated Val-AMP by IleRS using fast-kinetic studies.
Structural studies by Nureki et al. [Citation200] revealed that editing occurs in a separate domain of the aaRS, supporting the double-sieving model proposed by Fersht [Citation199]. The first sieve is the aaRS active site, which activates only cognate, isosteric or smaller amino acids. The second sieve is the editing site, which rejects cognate amino acids based on hydrophobicity, but hydrolyzes mis-activated or mischarged amino acids. Similar to aaRSs, mischarged tRNAs are recognized through identity elements within the tRNA. For example, LeuRS and AlaRS recognize the elbow region of tRNALeu and the G3:U70 identity element of tRNAAla, respectively, for both aminoacylation and post-transfer editing [Citation201–203]. In other cases, the elements for editing are distinct from those for aminoacylation. For example, the D-loop of tRNAIle is essential for editing of mischarged Val-tRNAIle but mutation of the D-loop does not affect aminoacylation [Citation204,Citation205].
In addition to aaRS editing domains, separate trans-acting editing proteins exist. These include D-Tyr-tRNATyr (DTD) that hydrolyzes D-Tyr and other D-amino acids in many species [Citation206–208]. Other trans-editing proteins target mischarged L-amino acids; for example, bacterial YbaK hydrolyzes mischarged tRNAPro [Citation209,Citation210]. Other similar free-standing enzymes edit tRNAAla and tRNAThr [Citation211,Citation212].
It should be noted that aaRSs act in other key cell signalling pathways beyond protein synthesis [reviewed in Citation213]. The multiple functions of aaRSs are perhaps not surprising because they were evolutionarily among the first group of enzymes to acquire specificity for amino acid binding. The synthetases also interact with the principal cell energy source, ATP, and specifically recognize a charged or uncharged isoacceptor group of tRNAs. Aminoacyl-tRNA synthetases are thus in a unique position to monitor cellular nutritional and energy status.
Decoding at the ribosome
After aminoacylation, elongator aminoacyl-tRNAs (aa-tRNA) are recognized by GTP-bound EF-Tu in bacteria or EF1A in archaea and eukaryotes and brought to the ribosome for amino acid incorporation during the elongation stage of translation (; [Citation214–216, Citation9, Citation15, Citation217]). At the ribosome, speed and accuracy in protein synthesis are achieved by rapidly sampling many aa-tRNAs and assessing the stability of each codon–anticodon interaction.
Figure 5. Aminoacyl-tRNA at the ribosome. Aminoacyl-tRNAs are recognized by GTP-bound EF-Tu in bacteria or EF1A in eukaryotes and recruited to the elongating ribosome in an mRNA-independent manner. During initial selection (i), codon–anticodon interactions are monitored by the ribosome and non-cognate tRNAs are rejected. A cognate codon–anticodon interaction triggers GTP hydrolysis by EF-Tu/EF1A and a conformational change that puts the aminoacyl branch of the tRNA into the peptidyl transferase centre. Proofreading (ii) occurs as a second check to ensure proper codon–anticodon pairing. Here, near-cognate matches that may have made it past initial selection are rejected. A cognate codon–anticodon interaction triggers peptide bond formation (iii) and the amino acid is added to the growing polypeptide chain. Translocation (iv) moves the tRNA containing the polypeptide chain into the P site and the cycle repeats
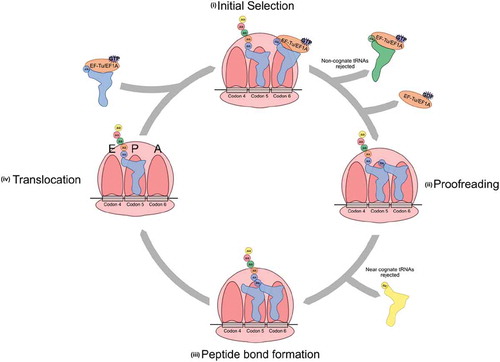
Elongation begins with the mRNA-independent binding of the aa-tRNA to the ribosome with EF-Tu and GTP [Citation218]. Ternary complex interactions are mediated through EF-Tu and ribosomal protein L7/L12 [Citation219]. The accuracy of the codon–anticodon interaction is assessed during two steps in the decoding centre. During the initial selection step, three universally conserved bases within the 16S rRNA, or 18S rRNA in eukaryotes, monitor the codon–anticodon interaction through the minor groove of the codon–anticodon helix [Citation220–223]. EF-Tu also interacts with the tRNA acceptor stem and amino acid during these steps to protect the labile aminoacyl ester bond and prevent hydrolysis [Citation224]. The correct codon–anticodon interaction, particularly at the first two positions of the codon, provides the free energy for conformational changes in the small ribosomal subunit, known as domain closure [Citation225,Citation226]. Domain closure activates EF-Tu GTPase activity, leading to GTP hydrolysis [Citation227]. Near-cognate codon–anticodon pairings, with G-U mismatches in the first or second codon position, adopt base pairs similar to Watson-Crick G-C pairs and induce domain closure similarly to cognate interactions [Citation228,Citation229].
After domain closure and GTP hydrolysis, GDP bound EF-Tu is released from the aa-tRNA, freeing the aminoacyl group for interactions with the peptidyl transferase centre of the ribosome [Citation230]. During the second specificity step, known as proofreading, near-cognate tRNAs disassociate from the ribosome [Citation231]. After EF-Tu release, the tRNA is less tightly bound to the ribosome and codon–anticodon interactions influence tRNA accommodation into the peptidyl transferase centre. Both the anticodon loop and the elbow region of the tRNA are monitored by either rRNA or a combination of rRNA and protein, respectively, to determine accommodation [Citation232]. Once the 3ʹ end of the tRNA moves into the peptidyl transferase centre, peptide bond formation rapidly occurs [Citation227].
Traditional Watson-Crick base pairing is not required at all three codon positions for an amino acid to be incorporated into a growing polypeptide chain [reviewed in Citation233]. Crick first identified that G-U wobble base pairing occurs in the third codon position and that only 32 tRNA species were required to decode 61 sense codons [Citation234]. Early experimental evidence demonstrated codon misreading and subsequent missense errors due to G-U wobble interactions [Citation235–237]. In bacteria and eukaryotes, G-U codon–anticodon mismatches at the middle position are the most frequent errors causing amino acid misincorporation [Citation238–240]. In vitro studies of G-U mismatches demonstrate that they are less destabilizing to the codon–anticodon helix than other non-Watson-Crick mismatches [Citation241,Citation242]. Pyrimidine–pyrimidine mismatches, such as U-U codon–anticodon pairing at the first codon position and third position, or C-U codon–anticodon pairing at the third position, also cause errors in translation [Citation239,Citation240,Citation243]. Recently, Pernod et al. [Citation283] investigated codon–anticodon interactions that are efficiently accepted by the eukaryotic ribosome in a high-throughput screening experiment using an IRES from the Cricket Paralysis Virus. This virus contains a pseudoknot that structurally and functionally mimics the codon–anticodon helix in the ribosome. They identified 59 near-cognate codon–anticodon interactions with one or two mismatches that permitted translational elongation. In their setup using unmodified RNA, the ribosome accepted G/U, G/A, C/A, C/U, A/A, C/C and G/G mismatches, with the majority occurring at the third position of the codon.
Superwobble, where an unmodified U in the wobble anticodon position reads all four nucleotides in the third codon position, also occurs in bacterial and organellar genomes that do not encode the full set of tRNAs required to read all codons. Rogalski et al. [Citation244]., demonstrated that of the two tRNA species that decode all four GGN glycine codons in chloroplasts, only tRNAGlyUCC was required. In other cases, modifications to nucleotides in the anticodon, as discussed above, can alter decoding. For example, in S. cerevisiae a currently unknown modification of tRNAProUGG allows it to decode all four CCN proline codons [Citation31].
Impact of tRNA modifications
Modifications play roles in all aspects of tRNA function, from structure and stability to aminoacylation and decoding at the ribosome. Modifications that affect tRNA stability or folding tend to occur in the body of the tRNA, outside of the anticodon stem-loop. As compared to unmodified tRNAs transcribed in vitro, native modified tRNAs show increased melting temperature, stabilized tertiary interactions and increased overall stability [Citation245-252]. A role for modifications in stabilizing tertiary interactions is suggested by findings that the number of modifications positively correlates with growth temperature [Citation111,Citation253–255]. Methylation of nucleobases at the junctions between the acceptor stem and D-arm or between the D-arm and anticodon stem-loop interrupt RNA duplex formation and facilitate the three-dimensional L-shape tRNA fold, as the methyl group on the Watson-Crick face of the nucleobase prevents base pairing [reviewed in Citation256]. Other studies show that pseudouridine stabilizes base-stacking interactions [Citation257–259], while dihydrouridine in the D-arm enhances tRNA flexibility for organisms that grow optimally at low temperature [Citation260–262]. Modifications also stabilize the secondary structure. For example, Nobles et al. [Citation263] found that adding any one of the highly conserved T-arm modifications (U54 modified to ribothymidine, C49 modified to 5-methylcytidine or U55 modified to pseudouridine) to an otherwise unmodified transcript significantly increased the interaction between the T-arm and D-arm.
Interestingly, deleting genes encoding the enzymes responsible for many of the modifications outside of the anticodon loop of the tRNA does not lead to abnormal growth phenotypes, likely because of redundancy among the modifications that stabilize tRNA structure [reviewed in Citation90]. Only the removal of multiple genes leads to growth defects. For example, deleting either yeast TRM8 or TRM82, which methylate G46 [Citation264], results in no obvious growth defect. When removed in combination with genes that encode the enzymes that methylate cytosine or guanine [TRM4 or TRM10; Citation265, Citation266] or that modify uracil to pseudouridine or dihydrouridine [PUS7, DUS1, DUS2, DUS3 or DUS4; Citation267–269], synthetic slow or synthetic lethal interactions are observed [Citation149]. Whipple et al. [Citation156], discovered that the absence of 7-methylguanine and 5-methylcytidine or 2ʹ-O-methyluridine and N4-acetylcytidine modifications in the tRNA body triggers tRNA degradation by the RTD pathway due to decreased stability in the acceptor and T stems.
The highest density of tRNA modifications occurs in the anticodon stem-loop (ASL). Positions 32, 34, 37 and 38 are all frequently modified and have a major impact on growth, due to their role in translation and decoding at the ribosome. Modifications at position 34, the wobble position, can expand or restrict decoding potential [reviewed in Citation270]. Structural studies have shown that U34 modification, with moieties such as 5-oxyacetic acid, 5-methylaminomethyl or 2-thio, stabilize and pre-structure the ASL by facilitating intramolecular hydrogen bonds that decrease the entropic cost of codon–anticodon binding and allow U34 pairing with codons ending in C or U [Citation271,Citation272]. Adenine at position 34 is nearly always modified to inosine, which expands decoding to A, U or C [reviewed in Citation273]. Other modifications at position 34 restrict decoding to prevent mistranslation. For example, some bacteria decode AUA codons with tRNAIleCAU. Lysidine modification of cytidine at position 34 changes the base-pairing preference from G to A [Citation274]. This prevents misrecognition of the near-cognate AUG codon which codes for methionine. Modifications of uridine to 2-thiouridine at position 34 similarly restrict codon reading by altering the pucker of the ribose sugar and glycosidic angle leading to preferential binding of A rather than G [Citation275–277]. Interestingly, Gupta et al. [Citation278] have linked the thiolation of U34, which is seen in tRNAGlu, tRNAGln and tRNALys in yeast, to broad aspects of cell metabolism that control growth and cell cycle progression. They demonstrated that U34 thiolation diverts carbon metabolism from the pentose-phosphate pathway and nucleotide synthesis towards storing carbohydrates trehalose and glycogen in response to low methionine levels. The effect occurs with only small changes in translation but is achieved through limiting intracellular phosphate metabolism. Modification of U34 also maintains proteome integrity by optimizing codon translation speed and regulating co-translation protein folding [Citation279].
Modifications of the universal purine at position 37, 3ʹ of the anticodon, modulate the ASL structure by interfering with intra-ASL hydrogen bonding and also stabilize anticodon interactions through base stacking [Citation280,Citation281]. N6-isopentenyladenosine and N6-threonylcarbamoyladenosine often occur at position 37 for tRNAs that decode codons beginning in U or A, respectively [Citation282]. These modifications stabilize A-U Watson-Crick pairs in the first codon position and prevent hydrogen bonding between U33 and A37, which would disrupt the anticodon stem [Citation284, Citation285, reviewed in Citation286]. In tRNAPheGAA position 37 is similarly modified to wybutosine [Citation287]. Wyosine derivatives at position 37 enhance base stacking with adjacent bases to restrict flexibility of the anticodon loop, prevent frameshifting and enhance translational fidelity through stabilization of codon–anticodon interactions [Citation288–290].
Other modified positions in the ASL synergistically act to alter decoding. For example, inosine at position 34 should allow decoding of A, U and C, but for bacterial tRNAArgICG modification of 2-thiocytidine at position 32 prevents I-A base pairing at the third codon position [Citation291]. Pseudouridine at positions 32 and 39 stabilizes the open-loop structure required for decoding at the ribosomal A-site [Citation259,Citation292]. Modifications in the ASL also direct other modifications [reviewed in Citation293]. For example, in human and yeast tRNAPhe, wybutosine formation at position 37 is stimulated by 2ʹ-O-methylation of C32 and G34 [Citation294–296].
Some modifications act as identity elements for recognition and aminoacylation by an aaRS [Citation176]. For example, inosine at position 34 of yeast tRNAIle enhances aminoacylation by IleRS compared to an unmodified adenosine [Citation297]. In other cases, such as for yeast tRNAAsp, methylation of G37 blocks aminoacylation by ArgRS to prevent mis-charging [Citation298]. Other modifications influence tRNA interactions with the translational machinery; for example, ribosylation of A64 in initiator tRNAMet prevents its interaction with EF1A, restricting function to only the initiation AUG codon [Citation299].
tRNA modifications can be dynamic and regulate cellular processes in response to stimuli or changing cellular conditions. The role of U34 thiolation in the regulation of cell metabolism in response to starvation as discussed above is one example [Citation278]. Another is the increase of 2ʹ-O-methylcytidine, 5-methylcytidine and N2,N2-dimethylguanosine in response to hydrogen peroxide treatment [Citation300]. Yeast lacking the enzymes required for these modifications are more sensitive to a variety of stresses, including hydrogen peroxide, sodium arsenite and the DNA damaging agent methyl methanesulfonate [Citation300,Citation301]. Begley et al. [Citation301] found that Trm9 methylation of U34 in tRNAArgUCU and tRNAGluUUC allows for efficient translation of genes involved in the DNA damage response. Similarly, upon exposure to oxidative stress, Trm4 methylates tRNALeuCAA at the wobble position to enhance translation of Rpl22a, which is enriched in UUG codons [Citation302]. In addition, treating yeast with oxidants and alkylating agents results in stress-specific changes in tRNA modifications that correlate with increased translation of specific transcripts required to respond to the stress [Citation303]. These examples suggest that dynamic tRNA modification controls gene expression post-transcriptionally by altering tRNA decoding efficiency, resulting in enhanced translation of specific transcripts required to deal with changing environmental conditions [Citation304,Citation305]. Also supporting this model, Torrent et al. [Citation306] demonstrated that in response to stress, yeast change their tRNA pool to favour the expression of stress response transcripts with select codon bias.
The importance of tRNA modification is underscored by the association of mutations in tRNA modifying enzymes with disease [reviewed in Citation307]. MELAS (Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes) and MERRF (Myoclonic epilepsy with ragged-red fibres) were first associated with mutations in mitochondrial tRNAs that lead to hypomodification of U34 [Citation308,Citation309]. Mutations in genes encoding the enzymes responsible for these modifications, GTPBP3 and MTO1, result in similar outcomes [Citation310,Citation311]. FTSJ1 encodes a methyltransferase that modifies positions 32 and 34 on tRNALeu, tRNAPhe and tRNATrp [Citation312,Citation313]. Mutation of FTSJ1 is linked with intellectual disabilities [Citation314,Citation315]. Both mouse and fly models reveal that FTSJ1 mutations impact transcription, small RNA silencing and disease associations [Citation312,Citation316]. Other tRNA modification enzymes, NSUN2 [Citation317], ADAT3 [Citation318] and TRMT1 [Citation319], are also implicated in intellectual disability. Related to this, mutations in the gene encoding the IKAP subunit of Elongator, a multi-subunit, multifunctional complex first described for its association with elongating RNA polymerase II [Citation320], result in the neuropathy familial dysautonomia [Citation321]. The mechanism involves Elongator’s role in the U34 modification of a number of tRNAs [reviewed in Citation322]. Interestingly, overexpression of NSUN2 [Citation323], Elongator components [Citation324, reviewed in Citation325] and other tRNA modifying enzymes are associated with cancer [reviewed in Citation326]. Some caution must be taken in interpreting how mutations in these modifying enzymes are acting, as many of these enzymes modify additional RNA substrates [reviewed in Citation327]. For example, NSUN2 modifies cytoplasmic tRNA, as well as mitochondrial tRNAs [Citation328], mRNAs [Citation329] and miRNAs [Citation330].
tRNA-dependent mistranslation
Mistranslation, when an amino acid that differs from what is specified by the codon is inserted into a protein during synthesis, occurs naturally in all cells at frequencies ranging from 1 in 3000 to 1 in 106 depending on the amino acid [Citation331–334, reviewed in Citation233]. A number of factors contribute to mistranslation frequency, with many centring on tRNAs. Since the ribosome reads the codon and not the acylated amino acid, misacylation of tRNAs leads to mistranslation [Citation11]. Mistranslation also results from codon–anticodon misreading by the ribosome through nonconventional base pairing at all three positions, as discussed in the previous section, and can be influenced by tRNA modification, which affects codon-anticodon pairing [Citation271,Citation335–339]. Furthermore, mistranslation increases upon protein overexpression, likely by altering the pools of aa-tRNAs when genes with rare codons are introduced [Citation340,Citation341].
Mistranslation should not be viewed simply as an error. Like protein modification, it provides diversity and is often an adaptive response. A well-studied example occurs in response to oxidative stress. Cysteine residues are often found within enzyme active sites or have regulatory roles [Citation342]. Their oxidation often results in enzyme inhibition [for example, Citation343]. The Pan lab has shown that, in organisms including E. coli, yeast and mammals, mistranslation of methionine at non-methionine codons occurs at a frequency as high as 10% in response to oxidative stress [Citation344–348]. These surface exposed methionines create a high concentration of oxidant scavengers that protect key cysteine residues in the protein [Citation349].
Mistranslation is also a response to amino acid starvation. This was first documented in E. coli strains lacking a stringent response [Citation350,Citation351]. The stringent response requires the alarmone guanosine tetraphosphate (ppGpp, Magic spot). ppGpp dramatically increases upon nutrient stress, with one consequence being the inhibition of translation [Citation352, reviewed in Citation353]. In the absence of translational inhibition, specific amino acid substitutions appear when amino acids are depleted. Near-cognate substitutions such as leucine for phenylalanine [Citation236,Citation354] or lysine for asparagine [Citation237] are observed. Starvation-induced increases in mistranslation are also observed in mammalian cells [Citation189,Citation351,Citation355–357]. Interestingly, mistranslation makes cells more resistant to other stresses, perhaps by priming stress response pathways [Citation358–360].
Some tRNA variants significantly increase the frequency of mistranslation. Many of these were first identified as suppressor mutations that compensate for nonsense or missense mutations in E. coli, yeast [reviewed in Citation361–363], mammalian cells [Citation364] and archaea [Citation365]. Intergenic suppressor mutations that had broad cellular consequences as a result of altering information flow from DNA to protein were termed informational suppressors [Citation366]. A number of the original tRNA suppressor mutations were found based upon second-site mutations that restored tryptophan auxotrophy in alleles of trpA with nonsense [Citation367] or missense mutations [Citation368–370]. These studies identified, for example, a tRNAGly with a C36 to U base change that resulted in decoding of arginine codons with glycine. Similarly, missense and nonsense suppressor tRNAs have been identified in several eukaryotic organisms [e.g. Citation188, Citation371-374].
Mistranslation results when the anticodon and aminoacylated tRNA are not a match defined by the genetic code. For example, mutations that result in a G3:U70 base pair in a tRNA cause mistranslation to alanine (). These occur spontaneously in yeast within tRNAProUGG and cause alanine insertion at proline codons [Citation188]. The effect of the mistranslation caused by the tRNA variant is buffered by multiple copies of wild-type tRNAPro with the frequency of mistranslation being approximately 3% [Citation188]. Protein quality control mechanisms such as the heat shock response and proteasomal degradation of misfolded proteins allow cells to cope with the mistranslation with minimal phenotypic consequences [Citation375,Citation376]. As the anticodon plays no role in aminoacylation, tRNASer variants with anticodon changes similarly cause high levels of mistranslation. In fact, if they are not buffered by secondary mutations that reduce their functionality or by additional copies of the wild-type tRNA, they are lethal (; [Citation377]). We and others have demonstrated the insertion of serine at a number of different codons in yeast and mammalian cells [Citation192,Citation378,Citation379]. The same type of anticodon variation in tRNAAla or tRNALeu will result in misincorporation of alanine or leucine, respectively. The impact of each of these tRNA variants on cells depends on both the extent of mistranslation and the type of substitution (Berg et al., in preparation). The threshold frequency at which cells no longer tolerate sustained mistranslation in yeast is approximately 8%, as compared to approximately 10% in E. coli [Citation192,Citation376]. The ability of tRNA variants to effectively fix mutations in protein-encoding genes [for example, Citation188, Citation380] raises their therapeutic potential in diseases resulting from missense or nonsense mutations [reviewed in Citation381].
Figure 6. tRNA variants can increase levels of mistranslation. (A) A S. cerevisiae proline tRNA variant with a C70U mutation that creates a G3:U70 base pair in the acceptor stem is aminoacylated with alanine instead of proline, leading to alanine misincorporation [Citation188]. The toxicity of mistranslation is buffered by other wild-type copies of tRNAPro that compete for decoding of CCA codons with the mistranslating tRNA variant. (B) A serine tRNA variant with a UGG anticodon is aminoacylated with serine, but decodes proline codons, leading to serine misincorporation. A secondary mutation of G26A dampens tRNA function allowing non-lethal levels of mistranslation [Citation377]
![Figure 6. tRNA variants can increase levels of mistranslation. (A) A S. cerevisiae proline tRNA variant with a C70U mutation that creates a G3:U70 base pair in the acceptor stem is aminoacylated with alanine instead of proline, leading to alanine misincorporation [Citation188]. The toxicity of mistranslation is buffered by other wild-type copies of tRNAPro that compete for decoding of CCA codons with the mistranslating tRNA variant. (B) A serine tRNA variant with a UGG anticodon is aminoacylated with serine, but decodes proline codons, leading to serine misincorporation. A secondary mutation of G26A dampens tRNA function allowing non-lethal levels of mistranslation [Citation377]](/cms/asset/f9e67150-d205-4c0d-9583-346e245b398d/krnb_a_1809197_f0006_c.jpg)
The occurrence of other complex mechanisms that lead to mistranslation is perhaps best demonstrated in an example from E. coli. Hirsh [Citation382] identified a tRNATrp that inserts tryptophan at UGA stop codons. The tRNA contains a G to A change in the D-arm resulting in U-A rather than U-G base pair, demonstrating that tRNA properties independent of the anticodon can affect decoding. Kinetic and structural studies show that binding of the aa-tRNA with EF1A to the A site of the ribosome and interactions with the mRNA occurs in discrete steps that require flexibility in the tRNA structure [Citation383–385]. The Hirsh mutation alters tRNA flexibility enabling codon–anticodon mismatching. Further studies have highlighted the dynamics of the tRNA throughout the translation process [Citation386].
Interestingly, similar mutations within tRNA encoding genes that cause mistranslation in yeast are found in human tRNA genes [Citation381,Citation387]. Using a DNA capture array, we sequenced the tRNA genes from 84 individuals. On average each individual contained 65 variations from the reference genome with nine of these being found in less than 5% of the population. Included amongst these were 17 variants creating a G3:U70 base pair or anticodon changes that have a high potential to mistranslate. As mistranslation would increase proteotoxic stress, we have hypothesized that the tRNA variants may act as modifiers of disease, particularly diseases characterized by proteotoxic stress. Our findings in yeast reveal that mistranslating tRNA variants, even those with similar mistranslation frequency, impact cell growth, heat shock response and the cellular proteome differently and have different genetic interactions. From this we propose that individual tRNA variants may contribute to distinct diseases, depending on the type of mistranslation.
Genetic code evolution
Mutations in tRNA genes that cause mistranslation provide the material for genetic code evolution. The ambiguous intermediate theory for genetic code evolution predicts that tRNA mutations can lead to decoding of a single codon as two different amino acids [Citation388]. In time, codons that cannot tolerate ambiguous decoding will be recoded to other synonymous codons. An example of this is seen in Candida species, where the CUG codon has been recoded from leucine to serine due to a serine tRNA variant that decodes leucine codons [Citation389]. Early in this evolution, the CUG codon would have been decoded by both the novel tRNASer variant and the canonical tRNALeu. This evolutionary intermediate has been retained in the related species, Ascoidea asiatica [Citation390]. Over time, CUG codons that could not tolerate being decoded as serine were replaced by synonymous leucine codons and the tRNALeu that decoded CUG codons was lost. In other yeast species, CUG is decoded as alanine [Citation391,Citation392].
More commonly, deviations from the ‘standard’ genetic code are seen in mitochondria [reviewed in Citation29]. In yeast mitochondria, CUN codons are decoded as threonine instead of leucine and the UGA stop codon is decoded as tryptophan [Citation393,Citation394]. In the case where the UGA stop codon is decoded as tryptophan, post-transcriptional editing or a mutation converts the tRNATrp anticodon from CCA to UCA [Citation395, Citation396. Recent approaches combining large-scale sequencing, analysis of amino acid distributions at conserved positions and characterization of organellar tRNA complements have identified many additional mitochondrial genetic code deviations in diverse species arising from novel tRNA variants [Citation397, Citation398].
Woese [Citation399] has discussed how the genetic code has co-evolved with the translational machinery in primitive cells. Central to this is that translational accuracy could not have existed early in evolution and that ambiguous codon assignments were common. Mistranslation was thus an inherent property of the primordial cell. Woese [Citation400] classifies any protein that arose from ambiguous translation as a statistical protein, that is a heterogeneous mixture of molecules, made from the same genetic template but differing at a subset of amino acid positions. As such, statistical proteins provided cells with a wider range of functions than would be achievable from perfect translation. As stated by Woese [Citation401]: ‘Statistical proteins form the basis of a powerful strategy for searching protein phase space, finding novel proteins.’ Some degree of mistranslation has been maintained to provide rapid functional diversity in response to a changing environment, not unlike protein modification. Wang and Pan [Citation402 demonstrated that mistranslation of methionine in response to high calcium levels creates statistical Ca2+/calmodulin-dependent protein kinase II (CaMKII). In HEK293T cells, CaMKII produced under high calcium conditions has greater kinase activity than CaMKII produced under non-stress conditions. CaMKII constructs containing mutations of specific amino acids to methionine had different activation profiles and subcellular localizations. One can imagine other circumstances in development and differentiation where mistranslation could provide the required diversity. Furthermore, statistical proteins created with tRNA variants provide an approach to produce proteins in synthetic biology and biotechnology applications with a wider range of binding interactions or activities than the homogenous form. For example, mistranslation makes it possible to create an antibody pool with broadened specificity.
Genetic code expansion
From primordial times to the present, the potential of the proteome has increased by incorporating additional amino acids into proteins [Citation29,Citation403–406]. The addition of selenocysteine and pyrrolysine as the 21st and 22nd amino acids are two natural examples of the increased repertoire of chemistries that an expanded genetic code provides [Citation407]. The genetic code does restrict the degree of expansion and as a result, post-translational modification has evolved as a mechanism to enhance protein function. However, the ability to expand protein chemistry with targeted genetic code expansion has advantages and has become a significant tool in modern synthetic biology [reviewed in Citation408–410]. Genetic code expansion is able to produce proteins with novel chemistries and thus also novel structures and functions. Moreover, non-canonical amino acids allow state-of-the-art biophysical, physiological and cell biological experiments either directly or combined with chemoselective reactions to specifically label a protein [reviewed in Citation408]. Synthetically expanding the code requires an open or largely open codon (to date generally a stop codon), a non-canonical amino acid that can enter the cell, a tRNA that decodes the open codon and an aaRS that specifically ligates the non-canonical amino acid to the tRNA [Citation411,Citation412]. Each orthogonal translation system must be optimized to be compatible with the translational machinery, to enhance specificity and minimize competition with native factors [Citation409]. Combined with the ability to make totally synthetic genomes where codons can be vacated and then reassigned [e.g. Citation413], and the development of an expanded genome alphabet [Citation414,Citation415], these strategies will allow the design of proteins and even organisms with unique potential.
Other functions of tRNAs
There is vast complexity within an organism’s tRNA molecules related to the presence of isoacceptors and isodecoders as well as the numerous possible base modifications. tRNAs are also ancient molecules originating with the enzymatic synthesis of proteins, even likely predating the ribosome [see, for example, Citation416]. It is not surprising then that tRNAs have evolved functions outside their direct role in translation.
The first discovered function for tRNAs outside of translation was the enzymatically catalysed transfer of arginine from tRNAArg to N-terminal Asp or Glu residues by Arg-tRNA-protein transferase [R-transferase; Citation417, Citation418]. Arginine is a primary destabilizing residue for the N-end rule signalling ubiquitylation then proteasomal turnover of the protein. Conjugation of arginine to these proteins or peptides increases their turnover in a regulated fashion [reviewed in Citation419]. Bacteria similarly transfer leucine or phenylalanine to N-terminal arginine or lysine residues, targeting their turnover by the ClpP protease [Citation420]. In a somewhat related process, bacteria use a remarkable dual-purpose tRNA-mRNA, SsrA, to extend stalled peptides with a tag that then targets them for degradation [reviewed in Citation421]. Bacteria also use aa-tRNAs to transfer amino acids to peptides and lipids, in the synthesis of antibiotics and for the interbridge peptide synthesis of peptidoglycan [reviewed in Citation422, Citation423].
In eukaryotes, tRNAs regulate the cellular proteome in response to amino acid starvation. This has been studied in detail in yeast [reviewed in Citation424], whereupon starvation for one or more amino acids, uncharged tRNAs bind to the histidyl-tRNA synthetase (HisRS)-like domain in Gcn2 activating its kinase activity. Activated Gcn2 phosphorylates the eukaryotic translation initiation factor 2 (eIF2) derepressing the translation of GCN4 mRNA, which encodes the transcriptional activator responsible for inducing amino acid biosynthetic genes [Citation425]. Phosphorylated eIF2 also decreases global translation allowing cells to reduce amino acid consumption while increasing amino acid biosynthesis [Citation426]. The latter is a common mechanism shared with mammalian cells in response to stress.
The third function for intact tRNAs in mammalian cells is their priming of reverse transcriptase catalysed cDNA synthesis of retroviral RNA genomes for subsequent integration into the host [reviewed in Citation427]. The tRNA involved depends on the virus. HIV-1, HTLV-1 and Rous sarcoma virus use specific tRNALys, tRNAPro and tRNATrp isoacceptors, respectively [Citation428–430]. Similarly, reverse transcription of some retrotransposons is primed by specific tRNAs or tRNA fragments [see, for example, Citation431, reviewed in Citation432]. Interestingly, tRNA fragments also interfere with reverse transcription and retrotransposon mobility [Citation433].
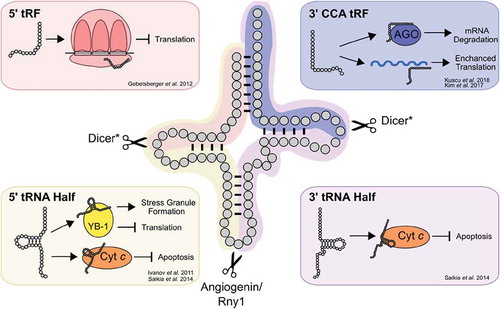
Recent studies have revealed the abundance of tRNA fragments and their roles in diverse cellular functions [Citation434, reviewed in Citation435, Citation436]. Interestingly, Torres et al. [Citation437] suggest that the differential expression of human tRNA isodecoders may relate to their roles as tRNA fragments rather than translation per se. Many tRNA fragments originate from cleavage of mature tRNAs rather than tRNA biogenesis intermediates [reviewed in Citation438]. Four types of tRNA-derived small RNAs can be produced from mature tRNAs (). tRNA halves are 31-40 nucleotides long and originate from cleavage within the anticodon loop by Rny1 in yeast [Citation439] or angiogenin in mammals [Citation440,Citation441], generating 5ʹ and 3ʹ halves. Smaller tRNA fragments (tRFs) are generated by Dicer dependent and independent mechanisms [Citation442–444] in either the D-loop, giving rise to a 5ʹ tRF, or in the T-loop, giving rise to a 3ʹ CCA tRF. Specific tRNA fragments are observed in response to numerous types of stress including starvation [Citation445], oxidative stress [Citation441,Citation446,Citation447] and hypoxia [Citation440]. As a result, many of the functions ascribed to tRNA fragments relate to stress response [reviewed in Citation448]. Both intact tRNAs and tRNA halves bind cytochrome C, blocking caspase activation and reducing stress-responsive apoptosis [Citation449, Citation450, reviewed in Citation451]. tRNA fragments also regulate translation. Stress-responsive translational inhibition occurs as a result of tRNA fragment interactions with the translational machinery [Citation441,Citation446,Citation452–454], as well as through binding of tRNA fragments to complementary sites in the mRNA [Citation444,Citation455]. Other processes, including modulating the cellular response to DNA damage, are regulated by specific tRNA fragments [Citation456].
Figure 7. tRNA-derived small RNAs regulate diverse cellular processes. Mature tRNAs can be cleaved into 5ʹ (yellow) and 3ʹ tRNA (purple) halves by Rny1 in yeast or angiogenin in mammals. Cleavage can also occur in the D- or T-loops by Dicer in mammals creating 5ʹ and 3ʹ CCA tRNA fragments (tRFs). Selected examples of the roles of tRNA fragments in regulating cellular processes are shown in the boxes. *For some tRFs, other nucleases have been implicated
The ability of tRNA fragments to regulate gene expression extends to epigenetic inheritance. tRNA fragments are abundant in mammalian sperm [Citation457]. Sharma et al. [Citation458] show that the composition of tRNA fragments in mammalian sperm is diet dependent. These tRNA fragments regulate the expression of approximately 70 genes associated with the long terminal repeat of the retroelement MERVL in the resulting embryo. Chen et al. [Citation459] observed inheritance of metabolic disorders upon injection of sperm derived tRNA fragments from mice fed a high-fat diet into normal zygotes. These effects were unrelated to changes in DNA methylation at CpG-enriched sequences.
Sharma et al. [Citation458,Citation460] demonstrate tRNA fragments in sperm arise from the fusion of small extracellular vesicles from the epididymal epithelium during spermatogenesis. tRNAs thus have a role in extracellular signalling and regulation [reviewed in Citation461, Citation462]. A related example comes from Chiou et al. [Citation463], who demonstrate that activated T cells secrete tRNA fragments that repress T cell activation. Extracellular tRNAs and tRNA fragments are also generating great interest for their diagnostic potential as they are found in biofluids [e.g. Citation464–466, reviewed in Citation467]. The exact RNA composition in extracellular vesicles is diverse and includes many classes of small noncoding RNAs [Citation468,Citation469]. Interestingly, Shurtleff et al. [Citation470] find that mature tRNAs are abundant in the vesicles released from HEK293T cells. The importance of extracellular vesicles in cellular processes such as myofiber repair [reviewed in Citation471] and in disease [Citation472] suggests that extracellular tRNAs have a broad role in mammalian physiology.
Given the importance of RNA fragments in a number of aspects of gene regulation, it is not surprising that tRNA fragments have significant roles in development and disease [Citation473–475]. In a fascinating report suggesting the therapeutic potential of tRNA fragments, Goodarzi et al. [Citation476] find that tRNA fragments displace known oncogenic transcripts from the RNA binding protein YBX1, decreasing their stability and expression, and as a result suppressing metastatic progression.
Concluding remarks
tRNAs were the solution to the problem of how to convert the four bases found in the genetic code to protein sequence composed of 20 building blocks. With differences in sequence and modifications, the potential diversity in the cellular tRNA complement is tremendous. The diversity of form affects function by altering aminoacylation, stability and base pairing. The diversity also provides distinct molecules for intra- and intercellular signalling and regulation. The number of genetic copies for each isoacceptor family has many implications. Few individual tRNA genes are essential [for an exception see Citation477] and the abundance of tRNA genes provides the raw material for genetic code evolution. Many tRNA variants occur within genomes; these include variants that are neutral or result in loss or gain of function. tRNAs play a central role in biology, it is not surprising tRNA variants contribute to disease [Citation381]. But, by restoring protein sequence, mistranslating tRNA variants have the potential to treat diseases resulting from missense mutations. The use of tRNA variants in engineering of novel proteins and biopolymers is further evidence of the central role of tRNA biology.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Julie Genereaux, Josh Isaacson and Patrick O’Donoghue for their comments on the manuscript.
Additional information
Funding
References
- Hoagland MB, Stephenson ML, Scott JF, et al. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958;231:241–257.
- Crick FH. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163.
- Crick FHC, Barnett L, Brenner S, et al. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232.
- Dounce AL. Duplicating mechanism for peptide chain and nucleic acid synthesis. Enzymologia. 1952;15:251–258.
- Nirenberg M, Leder P, Bernfield M, et al. RNA codewords and protein synthesis, VII. On the general nature of the RNA code. Proc Natl Acad Sci. 1965;53:1161–1168.
- Holley RW, Apgar J, Everett GA, et al. Structure of a ribonucleic acid. Science. 1965;147:1462–1465.
- Sharp SJ, Schaack J, Cooley L, et al. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19:107–144.
- Maglott EJ, Goodwin JT, Glick GD. Probing the structure of an RNA tertiary unfolding transition state. J Am Chem Soc. 1999;121:7461–7462.
- Nissen P, Kjeldgaard M, Thirup S, et al. The ternary complex of aminoacylated tRNA and EF-Tu-GTP. Recognition of a bond and a fold. Biochimie. 1996;78:921–933.
- Hamashima K, Tomita M, Kanai A. Expansion of noncanonical V-arm-containing tRNAs in eukaryotes. Mol Biol Evol. 2016;33:530–540.
- Chapeville F, Lipmann F, Ehrenstein GV, et al. On the role of soluble ribonucleic acid in coding for amino acids. Proc Natl Acad Sci. 1962;48:1086–1092.
- Crothers DM, Seno T, Soll DG. Is there a discriminator site in transfer RNA?. Proc Natl Acad Sci. 1972;69:3063–3067.
- Kim SH, Suddath FL, Quigley GJ, et al. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440.
- Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA. 2000;6:1091–1105.
- Nissen P, Kjeldgaard M, Thirup S, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472.
- Zhang J, Ferré-D’amaré AR. The tRNA elbow in structure, recognition and evolution. Life. 2016;6:1–11.
- Giegé R, Jühling F, Pütz J, et al. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip Rev RNA. 2012;3:37–61.
- Lorenz C, Lünse CE, Mörl M. TRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017;7. DOI:10.3390/biom7020035.
- Hubert N, Sturchler C, Westhof E, et al. The 9/4 secondary structure of eukaryotic selenocysteine tRNA: more pieces of evidence. RNA. 1998;4:1029–1033.
- Schön A, Böck A, Ott G, et al. The selenocysteine-inserting opal suppressor serine tRNA from E.coli is highly unusual in structure and modification. Nucleic Acids Res. 1989;17:7159–7165.
- Arcari P, Brownlee GG. The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 1980;8:5207–5212.
- de Bruijn MHL, Schreier PH, Eperon IC, et al. A mammalian mitochondrial serine transfer RNA lacking the “dihydrouridine” loop and stem. Nucleic Acids Res. 1980;8:5213–5222.
- Wende S, Platzer EG, Jühling F, et al. Biological evidence for the world’s smallest tRNAs. Biochimie. 2014;100:151–158.
- Wolstenholme DR, Macfarlane JL, Okimoto R, et al. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc Natl Acad Sci. 1987;84:1324–1328.
- Butcher SE, Jan E. tRNA-mimicry in IRES-mediated translation and recoding. RNA Biol. 2016;13:1068–1074.
- Giegé R, Frugier M, Rudinger J. tRNA mimics. Curr Opin Struct Biol. 1998;8:286–293.
- Nakamura Y, Ito K. tRNA mimicry in translation termination and beyond. Wiley Interdiscip Rev RNA. 2011;2:647–668.
- Kisselev L. Polypeptide release factors in prokaryotes and eukaryotes: same function, different structure. Structure. 2002;10:8–9.
- Ling J, O’Donoghue P, Söll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat Rev Microbiol. 2015;13:707–721.
- Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:D184–9.
- Bloom-Ackermann Z, Navon S, Gingold H, et al. A comprehensive tRNA deletion library unravels the genetic architecture of the tRNA pool. PLoS Genet. 2014;10:e1004084.
- Sprinzl M, Horn C, Brown M, et al. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153.
- Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237.
- Weinmann R, Roeder RG. Role of DNA-dependent RNA polymerase III in the transcription of the tRNA and 5S RNA genes. Proc Natl Acad Sci. 1974;71:1790–1794.
- Hossenlopp P, Chambon P, Wells D. Animal DNA‐dependent RNA polymerases: partial purification and properties of three classes of RNA polymerases from uninfected and adenovirus‐infected HeLa cells. Eur J Biochem. 1975;58:237–251.
- Gabrielsen OS, Sentenac A. RNA polymerase III (C) and its transcription factors. Trends Biochem Sci. 1991;16:412–416.
- Cramer P, Armache K-J, Baumli S, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352.
- Leśniewska E, Boguta M. Novel layers of RNA polymerase III control affecting tRNA gene transcription in eukaryotes. Open Biol. 2017;7:170001.
- Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975;122:855–865.
- Galli G, Hofstetter H, Birnstiel ML. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981;294:626–631.
- Hofstetter H, Kressmann A, Birnstiel ML. A split promoter for a eucaryotic tRNA gene. Cell. 1981;24:573–585.
- Sharp S, DeFranco D, Dingermann T, et al. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci. 1981;78:6657–6661.
- Segall J, Matsui T, Roeder RG. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980;255:11986–11991.
- Lassar A, Martin P, Roeder R. Transcription of class III genes: formation of preinitiation complexes. Science. 1983;222:740–748.
- Schramm L. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16(20):2593–2620.
- Bieker JJ, Martin PL, Roeder RG. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985;40:119–127.
- Setzer DR, Brown DD. Formation and stability of the 5S RNA transcription complex. J Biol Chem. 1985;260:2483–2492.
- Cormack BP, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696.
- Taggart AKP, Fisher TS, Pugh BF. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028.
- Han Y, Yan C, Fishbain S, et al. Structural visualization of RNA polymerase III transcription machineries. Cell Discov. 2018;4:40.
- Kassavetis GA, Letts GA, Geiduschek EP. The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. Embo J. 2001;20:2823–2834.
- Vorländer MK, Khatter H, Wetzel R, et al. Molecular mechanism of promoter opening by RNA polymerase III. Nature. 2018;553:295–300.
- Turowski TW, Tollervey D. Transcription by RNA polymerase III: insights into mechanism and regulation. Biochem Soc Trans. 2016;44:1367–1375.
- Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem. 2005;280:19551–19562.
- Arimbasseri AG, Maraia RJ. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol Cell. 2015;58:1124–1132.
- Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252.
- Dieci G, Bosio MC, Fermi B, et al. Transcription reinitiation by RNA polymerase III. Biochim Biophys Acta - Genet Regul Mech. 2013;1829:331–341.
- Carbon P, Krol A. Transcription of the Xenopus laevis selenocysteine tRNA(Ser)Sec gene: a system that combines an internal B box and upstream elements also found in U6 snRNA genes. Embo J. 1991;10:599–606.
- Aeby E, Ullu E, Yepiskoposyan H, et al. tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans. Nucleic Acids Res. 2010;38:5833–5843.
- Vannini A, Ringel R, Kusser AG, et al. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70.
- Towpik J, Graczyk D, Gajda A, et al. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J Biol Chem. 2008;283:17168–17174.
- Wei Y, Tsang CK, Zheng XFS. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. Embo J. 2009;28:2220–2230.
- Gerber A, Ito K, Chu C-S, et al. Gene-specific control of tRNA expression by RNA polymerase II. Mol Cell. 2020;78:765–778.e7.
- Campbell KJ, White RJ. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb Perspect Med. 2014;4:1–12.
- Cairns CA, White RJ. p53 is a general repressor of RNA polymerase III transcription. Embo J. 1998;17:3112–3123.
- Chu WM, Wang Z, Roeder RG, et al. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761.
- Altman S. A view of RNase P. Mol Biosyst. 2007;3:604–607.
- Lechner M, Rossmanith W, Hartmann RK, et al. Distribution of ribonucleoprotein and protein-only RNase P in Eukarya. Mol Biol Evol. 2015;32:3186–3193.
- Guerrier-Takada C, Gardiner K, Marsh T, et al. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857.
- Jarrous N, Gopalan V. Archaeal/Eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res. 2010;38:7885–7894.
- Perederina A, Berezin I, Krasilnikov AS. In vitro reconstitution and analysis of eukaryotic RNase P RNPs. Nucleic Acids Res. 2018;46:6857–6868.
- Kirsebom LA, Trobro S. RNase P RNA-mediated cleavage. IUBMB Life. 2009;61:189–200.
- Carrara G, Calandra P, Fruscoloni P, et al. Site selection by Xenopus laevis RNAase P. Cell. 1989;58:37–45.
- Nichols M, Soll D, Willis I. Yeast RNase P: catalytic activity and substrate binding are separate functions. Proc Natl Acad Sci. 1988;85:1379–1383.
- Paisley TE, Va Tuyle GC. The processing of wild type and mutant forms of rat nuclear pre-tRNA Lys by the homologous RNase P. Nucleic Acids Res. 1994;22:3347–3353.
- Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip Rev RNA. 2011;2:362–375.
- Stefano JE. Purified lupus antigen la recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154.
- Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta - Genet Regul Mech. 2010;1799:365–378.
- Schiffer S, Rösch S, Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. Embo J. 2002;21:2769–2777.
- Copela LA, Fernandez CF, Sherrer RL, et al. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA. 2008;14:1214–1227.
- Ozanick SG, Wang X, Costanzo M, et al. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:298–308.
- Deutscher MP, Hilderman RH. Isolation and partial characterization of Escherichia coli mutants with low levels of transfer ribonucleic acid nucleotidyltransferase. J Bacteriol. 1974;118:621–627.
- Zhu L, Deutscher MP. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. Embo J. 1987;6:2473–2477.
- Heinemann IU, Nakamura A, O’Donoghue P, et al. TRNA His-guanylyltransferase establishes tRNAHis identity. Nucleic Acids Res. 2012;40:333–344.
- Cooley L, Appel B, Soll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5ʹ terminus of histidine tRNA. Proc Natl Acad Sci. 1982;79:6475–6479.
- Gu W. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5ʹ end of tRNAHis. Genes Dev. 2003;17:2889–2901.
- Chen AW, Jayasinghe MI, Chung CZ, et al. The role of 3′ to 5′ reverse RNA polymerization in tRNA fidelity and repair. Genes (Basel). 2019;10:250.
- Jahn D, Pande S. Histidine tRNA guanylyltransferase from Saccharomyces cerevisiae. II. Catalytic mechanism. J Biol Chem. 1991;266:22832–22836.
- Jackman JE, Phizicky EM. tRNAHis guanylyltransferase adds G-1 to the 5ʹ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA. 2006;12:1007–1014.
- Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs?. FEBS Lett. 2010;584:265–271.
- Limbach PA, Crain PF, Mccloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196.
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860.
- Hopper AK, Banks F, Evangelidis V. A yeast mutant which accumulates precursor tRNAs. Cell. 1978;14:211–219.
- Melton DA, De Robertis EM, Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980;284:143–148.
- Nishikura K, De Robertis EM. RNA processing in microinjected Xenopus oocytes. J Mol Biol. 1981;145:405–420.
- Huh W-K, Falvo JV, Gerke LC, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691.
- Zasloff M. tRNA transport from the nucleus in a eukaryotic cell: carrier-mediated translocation process. Proc Natl Acad Sci. 1983;80:6436–6440.
- Chatterjee K, Nostramo RT, Wan Y, et al. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: location, location, location. Biochim Biophys Acta - Genet Regul Mech. 2018;1861:373–386.
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314.
- Hellmuth K, Lau DM, Bischoff FR, et al. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386.
- Kutay U, Lipowsky G, Izaurralde E, et al. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369.
- Arts GJ, Kuersten S, Romby P, et al. The role of exportin-t in selective nuclear export of mature tRNAs. Embo J. 1998;17:7430–7441.
- Cook AG, Fukuhara N, Jinek M, et al. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461:60–65.
- Huang H-Y, Hopper AK. In vivo biochemical analyses reveal distinct roles of β-importins and eEF1A in tRNA subcellular traffic. Genes Dev. 2015;29:772–783.
- Lipowsky G, Bischoff FR, Izaurralde E, et al. Coordination of tRNA nuclear export with processing of tRNA. RNA. 1999;5:S1355838299982134.
- Shibata S, Sasaki M, Miki T, et al. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34:4711–4721.
- Cook A, Bono F, Jinek M, et al. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671.
- Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085.
- Chatterjee K, Majumder S, Wan Y, et al. Sharing the load: mex67–Mtr2 cofunctions with Los1 in primary tRNA nuclear export. Genes Dev. 2017;31:2186–2198.
- Wu J, Bao A, Chatterjee K, et al. Genome-wide screen uncovers novel pathways for tRNA processing and nuclear–cytoplasmic dynamics. Genes Dev. 2015;29:2633–2644.
- Watanabe K, Shinma M, Oshima T, et al. Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem Biophys Res Commun. 1976;72:1137–1144.
- Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2005;102:11290–11295.
- Takano A, Endo T, Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142.
- Hurto RL, Tong AHY, Boone C, et al. Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae. Genetics. 2007;176:841–852.
- Whitney ML, Hurto RL, Shaheen HH, et al. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell. 2007;18:2678–2686.
- Huynh L-N, Thangavel M, Chen T, et al. Linking tRNA localization with activation of nutritional stress responses. Cell Cycle. 2010;9:3184–3190.
- Huang H-Y, Hopper A. Multiple layers of stress-induced regulation in tRNA biology. Life. 2016;6:16.
- Murthi A, Shaheen HH, Huang H-Y, et al. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol. Biol Cell. 2010;21:639–649.
- Takano A, Kajita T, Mochizuki M, et al. Cytosolic Hsp70 and co-chaperones constitute a novel system for tRNA import into the nucleus. Elife. 2015;4:1–22.
- Ohira T, Suzuki T. Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc Natl Acad Sci. 2011;108:10502–10507.
- Kessler AC, Kulkarni SS, Paulines MJ, et al. Retrograde nuclear transport from the cytoplasm is required for tRNATyr maturation in T. Brucei RNA Biol. 2018;15:528–536.
- Schwenzer H, Jühling F, Chu A, et al. Oxidative stress triggers selective tRNA retrograde transport in human cells during the integrated stress response. Cell Rep. 2019;26:3416–3428.e5.
- Hopper AK, Nostramo RT. tRNA processing and subcellular trafficking proteins multitask in pathways for other RNAs. Front Genet. 2019;10. DOI:10.3389/fgene.2019.00096.
- Grosshans H, Hurt E, Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 2000;14:830–840.
- Sarkar S, Azad AK, Hopper AK. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad Sci. 1999;96:14366–14371.
- Francklyn CS, Mullen P. Progress and challenges in aminoacyl-tRNA synthetase-based therapeutics. J Biol Chem. 2019;294:5365–5385.
- Heinemann IU, Söll D, Randau L. Transfer RNA processing in archaea: unusual pathways and enzymes. FEBS Lett. 2010;584:303–309.
- Yoshihisa T. Handling tRNA introns, archaeal way and eukaryotic way. Front Genet. 2014;5:1–16.
- Paushkin SV, Patel M, Furia BS, et al. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell. 2004;117:311–321.
- Trotta CR, Miao F, Arn EA, et al. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858.
- Greer CL, Peebles CL, Gegenheimer P, et al. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546.
- Phizicky EM, Schwartz RC, Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986;261:2978–2986.
- Greer CL, Söll D, Willis I. Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:76–84.
- Reyes VM, Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988;55:719–730.
- Robertis EM, Black DP, Nishikura K. Intranuclear location of the tRNA splicing enzymes. Cell. 1981;23:89–93.
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, et al. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell. 2003;14:3266–3279.
- Knapp G, Ogden RC, Peebles CL, et al. Splicing of yeast tRNA precursors: structure of the reaction intermediates. Cell. 1979;18:37–45.
- Peebles CL, Gegenheimer P, Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983;32:525–536.
- Mori T, Ogasawara C, Inada T, et al. Dual functions of yeast tRNA ligase in the unfolded protein response: unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol Biol Cell. 2010;21:3722–3734.
- Popow J, Englert M, Weitzer S, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764.
- Kanerva PA, Mäenpää PH. Codon-specific serine transfer ribonucleic acid synthesis in avian liver during vitellogenin induction. Acta Chem Scand B. 1978;32:561–568.
- Karnahl U, Wasternack C. Half-life of cytoplasmic rRNA and tRNA, of plastid rRNA and of uridine nucleotides in heterotrophically and photoorganotrophically grown cells of Euglena gracilis and its apoplastic mutant W3BUL. Int J Biochem. 1992;24:493–497.
- Nwagwu M, Nana M. Ribonucleic acid synthesis in embryonic chick muscle, rates of synthesis and half-lives of transfer and ribosomal RNA species. J Embryol Exp Morphol. 1980;Vol. 56:253–267.
- Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics. 2013;194:43–67.
- Anderson J, Phan L, Cuesta R, et al. The essential Gcd10p-Gcd14p nuclear complex 15 required for 1- methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662.
- Kadaba S, Krueger A, Trice T, et al. Nuclear surveillance and degradation of hypomodified initiator tRNA Met in S cerevisiae. Genes Dev. 2004;18:1227–1240.
- Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521.
- Gudipati RK, Xu Z, Lebreton A, et al. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell. 2012;48:409–421.
- Alexandrov A, Chernyakov I, Gu W, et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96.
- Chernyakov I, Whipple JM, Kotelawala L, et al. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5ʹ-3ʹ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380.
- Dewe JM, Whipple JM, Chernyakov I, et al. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA. 2012;18:1886–1896.
- Kotelawala L, Grayhack EJ, Phizicky EM. Identification of yeast tRNA Um44 2′-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNASer species. RNA. 2008;14:158–169.
- Guy MP, Young DL, Payea MJ, et al. Identification of the determinants of tRNA function and susceptibility to rapid tRNA decay by high-throughput in vivo analysis. Genes Dev. 2014;28:1721–1732.
- Payea MJ, Sloma MF, Kon Y, et al. Widespread temperature sensitivity and tRNA decay due to mutations in a yeast tRNA. RNA. 2018;24:410–422.
- Payea MJ, Hauke AC, De Zoysa T, et al. Mutations in the anticodon stem of tRNA cause accumulation and Met22-dependent decay of pre-tRNA in yeast. RNA. 2020;26:29–43.
- Whipple JM, Lane EA, Chernyakov I, et al. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011;25:1173–1184.
- Berg P, Offengand EJ. An Enzymatic mechanism for linking amino acids to RNA. Proc Natl Acad Sci. 1958;44:78–86.
- Sprinzl M. Chemistry of aminoacylation and peptide bond formation on the 3′ terminus of tRNA. J Biosci. 2006;31:489–496.
- Hoagland MB. An enzymic mechanism for amino acid activation in animal tissues. Biochim Biophys Acta. 1955;16:288–289.
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650.
- Griffin BE, Jarman M, Reese CB, et al. Some observations relating to acyl mobility in aminoacyl soluble ribonucleic acids. Biochemistry. 1966;5:3638–3649.
- Weinger JS, Strobel SA. Participation of the tRNA A76 hydroxyl groups throughout translation. Biochemistry. 2006;45:5939–5948.
- Wang C, Guo Y, Tian Q, et al. SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res. 2015;43:10534–10545.
- Wu XQ, Gross HJ. The long extra arms of human tRNA(Ser)Sec and tRNASer function as major identity elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res. 1993;21:5589–5594.
- Carlson BA, Xu X-M, Kryukov GV, et al. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci. 2004;101:12848–12853.
- Leinfelder W, Zehelein E, MandrandBerthelot M, et al. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988;331:723–725.
- Xu XM, Carlson BA, Mix H, et al. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:0096–0105.
- Yuan J, Palioura S, Salazar JC, et al. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc Natl Acad Sci. 2006;103:18923–18927.
- Gonzalez-Flores JN, Shetty SP, Dubey A, et al. The molecular biology of selenocysteine. Biomol Concepts. 2013;4:349–365.
- Woese CR, Olsen GJ, Ibba M, et al. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236.
- Cathopoulis T, Chuawong P, Hendrickson TL. Novel tRNA aminoacylation mechanisms. Mol Biosyst. 2007;3:408–418.
- Curnow AW, Hong KW, Yuan R, et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci. 1997;94:11819–11826.
- Sauerwald A, Zhu W, Major TA, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972.
- Mukai T, Crnković A, Umehara T, et al. RNA-dependent cysteine biosynthesis in bacteria and archaea. MBio. 2017;8:1–13.
- De Duve C. Transfer RNAs: the second genetic code. Nature. 1988;333:117–118.
- Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035.
- Goldgur Y, Mosyak L, Reshetnikova L, et al. The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNA(Phe). Structure. 1997;5:59–68.
- Commans S, Lazard M, Delort F, et al. tRNA anticodon recognition and specification within subclass IIb aminoacyl-tRNA synthetases. J Mol Biol. 1998;278:801–813.
- Chang KY, Varani G, Bhattacharya S, et al. Correlation of deformability at a tRNA recognition site and aminoacylation specificity. Proc Natl Acad Sci. 1999;96:11764–11769.
- Limmer S, Hofmann HP, Ott G, et al. The 3ʹ-terminal end (NCCA) of tRNA determines the structure and stability of the aminoacyl acceptor stem. Proc Natl Acad Sci. 1993;90:6199–6202.
- Puglisi EV, Puglisi JD, Williamson JR, et al. NMR analysis of tRNA acceptor stem microhelices: discriminator base change affects tRNA conformation at the 3ʹ end. Proc Natl Acad Sci. 1994;91:11467–11471.
- Francklyn C, Schimmel P. Enzymatic aminoacylation of an eight-base-pair microhelix with histidine. Proc Natl Acad Sci. 1990;87:8655–8659.
- Himeno H, Hasegawa T, Ueda T, et al. Role of the extra G-C pair at the end of the acceptor stem of tRNAHis in aminoacylation. Nucleic Acids Res. 1989;17:7855–7863.
- Francklyn C, Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989;337:478–481.
- Hou YM, Schimmel P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989;28:6800–6804.
- McClain WH, Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3ʹ acceptor end. Science. 1988;240:793–796.
- Ripmaster TL, Shiba K, Schimmel P. Wide cross-species aminoacyl-tRNA synthetase replacement in vivo: yeast cytoplasmic alanine enzyme replaced by human polymyositis serum antigen. Proc Natl Acad Sci. 1995;92:4932–4936.
- Hoffman KS, Berg MD, Shilton BH, et al. Genetic selection for mistranslation rescues a defective co-chaperone in yeast. Nucleic Acids Res. 2017;45:3407–3421.
- Lant JT, Berg MD, Sze DH, et al. Visualizing tRNA-dependent mistranslation in human cells. RNA Biol. 2017;15:567–575.
- Berg MD, Genereaux J, Zhu Y, et al. Acceptor stem differences contribute to species-specific use of yeast and human tRNASer. Genes (Basel). 2018;9:612.
- Asahara H, Himeno H, Tamura K, et al. Escherichia coli seryl-tRNA synthetase recognizes tRNASer by its characteristics tertiary structure. J Mol Biol. 1994;236:738–748.
- Berg MD, Zhu Y, Genereaux J, et al. Modulating mistranslation potential of tRNASer in Saccharomyces cerevisiae. Genetics. 2019;213:849–863.
- Biou V, Yaremchuk A, Tukalo M, et al. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science. 1994;263:1404–1410.
- Himeno H, Yoshida S, Soma A, et al. Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNA(Leu) in vitro. J Mol Biol. 1997;268:704–711.
- Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78.
- Baldwin AN, Berg P. Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J Biol Chem. 1966;241:839–845.
- Eldred EW, Schimmel PR. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J Biol Chem. 1972;247:2961–2964.
- Yarus M. Phenylalanyl-tRNA synthetase and isoleucyl-tRNAPhe: a possible verification mechanism for aminoacyl-tRNA. Proc Natl Acad Sci. 1972;69:1915–1919.
- Fersht AR. Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry. 1977;16:1025–1030.
- Nureki O, Vassylyev DG, Tateno M, et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582.
- Beebe K, Mock M, Merriman E, et al. Distinct domains of tRNA synthetase recognize the same base pair. Nature. 2008;451:90–93.
- Du X, Wang ED. Tertiary structure base pairs between D- and TΨC-loops of Escherichia coli tRNALeu play important roles in both aminoacylation and editing. Nucleic Acids Res. 2003;31:2865–2872.
- Yao P, Zhu B, Jaeger S, et al. Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps. Nucleic Acids Res. 2008;36:2728–2738.
- Farrow MA, Nordin BE, Schimmel P. Nucleotide determinants for tRNA-dependent amino acid discrimination by a class I tRNA synthetase. Biochemistry. 1999;38:16898–16903.
- Hale SP, Auld DS, Schmidt E, et al. Discrete determinants in transfer RNA for editing and aminoacylation. Science. 1997;276:1250–1252.
- Calendar R, Berg P. D-Tyrosyl RNA: formation, hydrolysis and utilization for protein synthesis. J Mol Biol. 1967;26:39–54.
- Soutourina J, Plateau P, Blanquet S. Metabolism of D-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J Biol Chem. 2000;275:32535–32542.
- Wydau S, Ferri-Fioni ML, Blanquet S, et al. GEK1, a gene product of Arabidopsis thaliana involved in ethanol tolerance, is a D-aminoacyl-tRNA deacylase. Nucleic Acids Res. 2007;35:930–938.
- An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J Biol Chem. 2004;279:42359–42362.
- Wong FC, Beuning PJ, Silvers C, et al. An isolated class II aminoacyl-tRNA synthetase insertion domain is functional in amino acid editing. J Biol Chem. 2003;278:52857–52864.
- Ahel I, Korencic D, Ibba M, et al. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci. 2003;100:15422–15427.
- Chong YE, Yang XL, Schimmel P. Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J Biol Chem. 2008;283:30073–30078.
- Guo M, Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat Chem Biol. 2013;9:145–153.
- Miller DL, Cashel M, Weissbach H. The interaction of guanosine 5′-diphosphate, 2′ (3′)-diphosphate with the bacterial elongation factor Tu. Arch Biochem Biophys. 1973;154:675–682.
- Gavrilova LP, Kostiashkina OE, Koteliansky VE, et al. Factor-free (“Non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J Mol Biol. 1976;101:537–552.
- Johnson AE, Miller DL, Cantor CR. Functional covalent complex between elongation factor Tu and an analog of lysyl-tRNA. Proc Natl Acad Sci. 1978;75:3075–3079.
- Dreher TW, Uhlenbeck OC, Browning KS. Quantitative assessment of EF-1α.GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J Biol Chem. 1999;274:666–672.
- Rodnina MV, Pape T, Fricke R, et al. Initial binding of the elongation factor Tu-GTP-aminoacyl-tRNA complex preceding codon recognition on the ribosome. J Biol Chem. 1996;271:646–652.
- Hamel E, Koka M, Nakamoto T. Requirement of an Escherichia coli 50S ribosomal protein component for effective interaction of the ribosome with T and G factors and with guanosine triphosphate. J Biol Chem. 1972;247:805–814.
- Loveland AB, Demo G, Grigorieff N, et al. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature. 2017;546:113–117.
- Ogle JM, Brodersen DE, Clemons WM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902.
- Shao S, Murray J, Brown A, et al. Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell. 2016;167:1229–1240.e15.
- Yoshizawa S, Fourmy D, Puglisi JD. Recognition of the codon-anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725.
- Schmeing TM, Voorhees RM, Kelley AC, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694.
- Ogle JM, Murphy FV IV, Tarry MJ, et al. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732.
- Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. Embo J. 1999;18:3800–3807.
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E.coli ribosome. Embo J. 1998;17:7490–7497.
- Demeshkina N, Jenner L, Westhof E, et al. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259.
- Rozov A, Westhof E, Yusupov M, et al. The ribosome prohibits the G•U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res. 2016;44:6434–6441.
- Dell VA, Johnson AE, Dell VA, et al. Effects of nucleotide- and aurodox-induced changes in elongation factor Tu conformation upon its interactions with aminoacyl transfer RNA. A fluorescence study. Biochemistry. 1990;29:1757–1763.
- Thompson RC, Stone PJ. Proofreading of the codon anticodon interaction on ribosomes. Proc Natl Acad Sci. 1977;74:198–202.
- Jenner L, Demeshkina N, Yusupova G, et al. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17:1072–1078.
- Joshi K, Cao L, Farabaugh PJ. The problem of genetic code misreading during protein synthesis. Yeast. 2019;36:35–42.
- Crick FH. Codon—anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–555.
- Parker J, Holtz G. Control of basal-level codon misreading in Escherichia coli. Biochem Biophys Res Commun. 1984;121:487–492.
- Parker J, Precup J. Mistranslation during phenylalanine starvation. Mol Gen Genet. 1986;204:70–74.
- Precup J, Parker J. Missense misreading of asparagine codons as a function of codon identity and context. J Biol Chem. 1987;262:11351–11355.
- Joshi K, Bhatt MJ, Farabaugh PJ. Codon-specific effects of tRNA anticodon loop modifications on translational misreading errors in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2018;46:10331–10339.
- Manickam N, Nag N, Abbasi A, et al. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014;20:9–15.
- Zhang Z, Shah B, Bondarenko PV. G/U and certain wobble position mismatches as possible main causes of amino acid misincorporations. Biochemistry. 2013;52:8165–8176.
- Grosjean HJ, de Henau S, Crothers DM. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci. 1978;75:610–614.
- Sugimoto N, Kierzek R, Freier SM, et al. Energetics of internal GU mismatches in ribooligonucleotide helixes. Biochemistry. 1986;25:5755–5759.
- Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96.
- Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol. 2008;15:192–198.
- Derrick WB, Horowitz J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993;21:4948–4953.
- Hall KB, Sampson JR, Uhlenbeck OC, et al. Structure of an unmodified tRNA molecule. Biochemistry. 1989;28:5794–5801.
- Maglott EJ, Deo SS, Przykorska A, et al. Conformational transitions of an unmodified tRNA: implications for RNA folding. Biochemistry. 1998;37:16349–16359.
- Perret V, Garcia A, Grosjean H, et al. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990;344:787–789.
- Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed. In Vitro Proc Natl Acad Sci. 1988;85:1033–1037.
- Serebrov V, Vassilenko K, Kholod N, et al. Mg2+ binding and structural stability of mature and in vitro synthesized unmodified Escherichia coli tRNA(Phe). Nucleic Acids Res. 1998;26:2723–2728.
- Vermeulen A, McCallum SA, Pardi A. Comparison of the global structure and dynamics of native and unmodified tRNAVal. Biochemistry. 2005;44:6024–6033.
- Yue D, Kintanar A, Horowitz J. Nucleoside modifications stabilize Mg2+ binding in Escherichia coli tRNAVal: an imino proton NMR investigation. Biochemistry. 1994;33:8905–8911.
- Agris PF, Koh H, Söll D. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch Biochem Biophys. 1973;154:277–282.
- Kowalak JA, Dalluge JJ, McCloskey JA, et al. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994;33:7869–7876.
- McCloskey JA, Graham DE, Zhou S, et al. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29:4699–4706.
- Väre VYP, Eruysal ER, Narendran A, et al.. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017;7:1–32.
- Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026.
- Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol. 1999;285:115–131.
- Yarian CS, Basti MM, Cain RJ, et al. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA’s position 39. Nucleic Acids Res. 1999;27:3543–3549.
- Dalluge JJ, Hashizume T, Sopchik AE, et al. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 1996;24:1073–1079.
- Dalluge JJ, Hamamoto T, Horikoshi K, et al. Posttranscriptional modification of tRNA in psychrophilic bacteria. J Bacteriol. 1997;179:1918–1923.
- Noon KR, Guymon R, Crain PF, et al. Influence of temperature on tRNA modification in archaea: methanococcoides burtonii (optimum growth temperature [Topt], 23°C) and Stetteria hydrogenophila (Topt, 95°C). J Bacteriol. 2003;185:5483–5490.
- Nobles KN, Yarian CS, Liu G, et al. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2002;30:4751–4760.
- Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266.
- Jackman JE, Montange RK, Malik HS, et al.. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585.
- Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118.
- Behm-Ansmant I, Urban A, Ma X, et al. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:ψ-synthase also acting on tRNAs. RNA. 2003;9:1371–1382.
- Xing F, Martzen MR, Phizicky EM. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA. 2002;8:370–381.
- Xing F, Hiley SL, Hughes TR, et al.. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J Biol Chem. 2004;279:17850–17860.
- Agris PF, Eruysal ER, Narendran A, et al. Celebrating wobble decoding: half a century and still much is new. RNA Biol. 2018;15:537–553.
- Murphy FV, Ramakrishnan V, Malkiewicz A, et al.. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol. 2004;11:1186–1191.
- Weixlbaumer A, Murphy FV IV, Dziergowska A, et al. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol. 2007;14:498–502.
- Torres AG, Piñeyro D, Filonava L, et al. A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS Lett. 2014;588:4279–4286.
- Muramatsu T, Nishikawa K, Nemoto F, et al. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181.
- Agris PF, Sierzputowska-Gracz H, Smith W, et al. Thiolation of uridine carbon-2 restricts the motional dynamics of the transfer RNA Wobble position nucleoside. J Am Chem Soc. 1992;114:2652–2656.
- Sheng J, Larsen A, Heuberger BD, et al.. Crystal structure studies of RNA duplexes containing s2U:A and s2U:U base Pairs. J Am Chem Soc. 2014;136:13916–13924.
- Sierzputowska-Gracz H, Agris PF, Sochacka E, et al. Chemistry and structure of modified uridines in the anticodon, wobble position of transfer RNA are determined by thiolation. J Am Chem Soc. 1987;109:7171–7177.
- Gupta R, Walvekar AS, Liang S, et al. A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife. 2019;8:1–33.
- Nedialkova DD, Leidel SA. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618.
- Cabello-Villegas J, Winkler ME, Nikonowicz EP. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J Mol Biol. 2002;319:1015–1034.
- Dao V, Guenther R, Malkiewicz A, et al. Ribosome binding of DNA analogs of tRNA requires base modifications and supports the “extended anticodon”. Proc Natl Acad Sci. 1994;91:2125–2129.
- Boccaletto P, MacHnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307.
- Pernod K, Schaeffer L, Chicher J, et al. The nature of the purine at position 34 in tRNAs of 4-codon boxes is correlated with nucleotides at positions 32 and 38 to maintain decoding fidelity. Nucleic Acids Res. 2020;48(11):6170–6183.
- Stuart JW, Gdaniec Z, Guenther R, et al. Functional anticodon architecture of human tRNA(Lys3) includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry. 2000;39:13396–13404.
- Sambhare SB, Kumbhar BV, Kamble AD, et al. Structural significance of modified nucleosides k2C and t6A present in the anticodon loop of tRNAIle. RSC Adv. 2014;4:14176.
- Schweizer U, Bohleber S, Fradejas-Villar N. The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol. 2017;14:1197–1208.
- Rajbhandary UL, Chang SH. Studies on polynucleotides: LXXXII. Yeast phenylalanine transfer ribonucleic acid: partial digestion with ribonuclease T1 and derivation of the total primary structure. J Mol Biol. 1965;14:221–IN10.
- Carlson BA, Kwon SY, Chamorro M, et al. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255:2–8.
- Konevega AL, Soboleva NG, Makhno VI, et al. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101.
- Stuart JW, Koshlap KM, Guenther R, et al.. Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J Mol Biol. 2003;334:901–918.
- Cantara WA, Bilbille Y, Kim J, et al. Modifications modulate anticodon loop dynamics and codon recognition of E. coli tRNAArg1,2. J Mol Biol. 2012;416:579–597.
- Tworowska I, Nikonowicz EP. Base pairing within the ψ32, ψ39-modified anticodon arm of Escherichia coli tRNAPhe. J Am Chem Soc. 2006;128:15570–15571.
- Han LU, Phizicky EM. A rationale for tRNA modification circuits in the anticodon loop. RNA. 2018;1277–1284. DOI:10.1261/rna.067736.118.RNA.
- Guy MP, Podyma BM, Preston MA, et al. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA. 2012;18:1921–1933.
- Guy MP, Phizicky EM. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA. 2015;21:61–74.
- Guy MP, Shaw M, Weiner CL, et al. Defects in tRNA anticodon loop 2′-O-Methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum Mutat. 2015;36:1176–1187.
- Senger B, Auxilien S, Englisch U, et al.. The modified wobble base inosine in yeast tRNA(Ile) is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry. 1997;36:8269–8275.
- Pütz J, Florentz C, Benseler F, et al.. A single methyl group prevents the mischarging of a tRNA. Nat Struct Biol. 1994;1:580–582.
- Åström SU, Byström AS. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell. 1994;79:535–546.
- Chan CTY, Dyavaiah M, DeMott MS, et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:1–9.
- Begley U, Dyavaiah M, Patil A, et al. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28:860–870.
- Chan CTY, Pang YLJ, Deng W, et al.. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3. DOI:10.1038/ncomms1938.
- Chan CTY, Deng W, Li F, et al. Highly predictive reprogramming of tRNA modifications is linked to selective expression of codon-biased genes. Chem Res Toxicol. 2015;28:978–988.
- Duechler M, Leszczyńska G, Sochacka E, et al.. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol Life Sci. 2016;73:3075–3095.
- Chan C, Pham P, Dedon PC, et al.. Lifestyle modifications: coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol. 2018;19:1–11.
- Torrent M, Chalancon G, de Groot NS, et al.. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci Signal. 2018;11:eaat6409.
- Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20:306–314.
- Kirino Y, Goto YI, Campos Y, et al.. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc Natl Acad Sci. 2005;102:7127–7132.
- Yasukawa T, Suzuki T, Ishii N, et al. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNA Lys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 2000;467(2–3):175–178.
- Bykhovskaya Y, Mengesha E, Wang D, et al. Phenotype of non-syndromic deafness associated with the mitochondrial A1555G mutation is modulated by mitochondrial RNA modifying enzymes MTO1 and GTPBP3. Mol Genet Metab. 2004;83:199–206.
- Kopajtich R, Nicholls TJ, Rorbach J, et al. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am J Hum Genet. 2014;95:708–720.
- Angelova MT, Dimitrova DG, Da Silva B, et al. TRNA 2′-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila. Nucleic Acids Res. 2020;48:2050–2072.
- Feder M, Pas J, Wyrwicz LS, et al.. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302:129–138.
- Freude K, Hoffmann K, Jensen LR, et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:305–309.
- Takano K, Nakagawa E, Inoue K, et al. A loss-of-function mutation in the FTSJ1 gene causes nonsyndromic x-linked mental retardation in a Japanese family. Am J Med Genet Part B Neuropsychiatr Genet. 2008;147:479–484.
- Jensen LR, Garrett L, Hölter SM, et al. A mouse model for intellectual disability caused by mutations in the X-linked 2′‑O‑methyltransferase Ftsj1 gene. Biochim Biophys Acta - Mol Basis Dis. 2019;1865:2083–2093.
- Khan MA, Rafiq MA, Noor A, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856–863.
- Thomas E, Lewis AM, Yang Y, et al. Novel missense variants in ADAT3 as a cause of syndromic intellectual disability. J Pediatr Genet. 2019;08:244–251.
- Zhang K, Lentini JM, Prevost CT, et al. An intellectual disability-associated missense variant in TRMT1 impairs tRNA modification and reconstitution of enzymatic activity. Hum Mutat. 2020a;41:600–607.
- Otero G, Fellows J, Yang L, et al. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118.
- Karlsborn T, Tükenmez H, Chen C, et al.. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem Biophys Res Commun. 2014;454:441–445.
- Kojic M, Wainwright B. The many faces of elongator in neurodevelopment and disease. Front Mol Neurosci. 2016;9:1–10.
- Yi J, Gao R, Chen Y, et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget. 2017;8:20751–20765.
- Delaunay S, Rapino F, Tharun L, et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med. 2016;213:2503–2523.
- Hawer H, Hammermeister A, Ravichandran KE, et al. Roles of elongator dependent tRNA modification pathways in neurodegeneration and cancer. Genes (Basel). 2019;10:1–23.
- Dong Z, Cui H. The emerging roles of RNA modifications in glioblastoma. Cancers (Basel). 2020;12:1–27.
- Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020. DOI:10.1038/s41568-020-0253-2
- Van Haute L, Lee SY, McCann BJ, et al. NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2019;47:8720–8733.
- Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m 5 C reader. Cell Res. 2017;27:606–625.
- Hussain S, Sajini AA, Blanco S, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261.
- Loftfield R. The frequency of errors in protein biosynthesis. Biochem J. 1963;89:82–92.
- Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128:1353–1356.
- Edelmann P, Gallant J. Mistranslation in E. coli. Cell. 1977;10:131–137.
- Stansfield I, Jones KM, Herbert P, et al. Missense translation errors in Saccharomyces cerevisiae. J Mol Biol. 1998;282:13–24.
- Bjork G, Wikstrom P, Bystrom A. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989.
- Maehigashi T, Dunkle JA, Miles SJ, et al.. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc Natl Acad Sci. 2014;111:12740–12745.
- Manickam N, Joshi K, Bhatt MJ, et al.. Effects of tRNA modification on translational accuracy depend on intrinsic codon–anticodon strength. Nucleic Acids Res. 2016;44:1871–1881.
- Rezgui VAN, Tyagi K, Ranjan N, et al. TRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci. 2013;110:12289–12294.
- Tükenmez H, Xu H, Esberg A, et al.. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015;43:9489–9499.
- Brinkmann U, Mattes RE, Buckel P. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene. 1989;85:109–114.
- Seetharam R, Heeren RA, Wong EY, et al. Mistranslation in IGF-1 during over-expression of the protein in Escherichia coli using a synthetic gene containing low frequency codons. Biochem Biophys Res Commun. 1988;155:518–523.
- Weerapana E, Wang C, Simon GM, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–797.
- Innes BT, Sowole MA, Gyenis L, et al. Peroxide-mediated oxidation and inhibition of the peptidyl-prolyl isomerase Pin1. Biochim Biophys Acta - Mol Basis Dis. 2015;1852:905–912.
- Gomes AC, Kordala AJ, Strack R, et al. A dual fluorescent reporter for the investigation of methionine mistranslation in live cells. RNA. 2016;22:467–476.
- Jones TE, Alexander RW, Pan T. Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc Natl Acad Sci. 2011;108:6933–6938.
- Lee JY, Kim DG, Kim B-G, et al. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J Cell Sci. 2014;127:4234–4245.
- Netzer N, Goodenbour JM, David A, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526.
- Wiltrout E, Goodenbour JM, Fréchin M, et al.. Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:10494–10506.
- Luo S, Levine RL. Methionine in proteins defends against oxidative stress. Faseb J. 2009;23:464–472.
- O’Farrell PH. The suppression of defective translation by ppGpp and its role in the stringent response. Cell. 1978;14:545–557.
- Parker J, Pollard JW, Friesen JD, et al.. Stuttering: high-level mistranslation in animal and bacterial cells. Proc Natl Acad Sci. 1978;75:1091–1095.
- Cashel M, Gallant J. Two compounds implicated in the function of RC gene. Nature. 1969;221:838–841.
- Ronneau S, Hallez R. Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol Rev. 2019;43:389–400.
- Parker J, Precup J, Fu C. Misreading of the argI message in Escherichia coli. FEMS Microbiol Lett. 1992;100:141–145.
- Harley CB, Pollard JW, Stanners CP, et al.. Model for messenger RNA translation during amino acid starvation applied to the calculation of protein synthetic error rates. J Biol Chem. 1981;256:10786–10794.
- Pollard JW, Harley CB, Chamberlain JW, et al.. Is transformation associated with an increased error frequency in mammalian cells?. J Biol Chem. 1982;257:5977–5979.
- Wen D, Vecchi MM, Gu S, et al. Discovery and investigation of misincorporation of serine at asparagine positions in recombinant proteins expressed in Chinese hamster ovary cells. J Biol Chem. 2009;284:32686–32694.
- Evans CR, Fan Y, Ling J. Increased mistranslation protects E. coli from protein misfolding stress due to activation of a RpoS-dependent heat shock response. FEBS Lett. 2019;593:3220–3227.
- Samhita L, Raval PK, Agashe D. Global mistranslation increases cell survival under stress in Escherichia coli. PLoS Genet. 2020;16:1–21.
- Zhang X, Kuang X, Cao F, et al. Effect of cadmium on mRNA mistranslation in Saccharomyces cerevisiae. J Basic Microbiol. 2020b;60:372–379.
- Celis JE, Piper PW. Nonsense suppressors in eukaryotes. Trends Biochem Sci. 1981;6:177–179.
- Hill CW. Informational suppression of missense mutations. Cell. 1975;6:419–427.
- Murgola EJ. Suppression and the code: beyond codons and anticodons. Experientia. 1990;46:1134–1141.
- Hatfield DL, Smith DWE, Lee BJ, et al.. Structure and function of suppressor tRNAs in higher eukaryote. Crit Rev Biochem Mol Biol. 1990;25:71–96.
- Bhattacharya A, Köhrer C, Mandal D, et al.. Nonsense suppression in archaea. Proc Natl Acad Sci. 2015;112:6015–6020.
- Gorini L, Beckwith JR. Suppression. Annu Rev Microbiol. 1966;20:401–422.
- Soll L, Berg P. Recessive lethal nonsense suppressor in Escherichia coli which inserts glutamine. Nature. 1969;223:1340–1342.
- Carbon J, Berg P, Yanofsky C. Studies of missense suppression of the tryptophan synthetase A-protein mutant A36. Proc Natl Acad Sci. 1966;56:764–771.
- Gupta NK, Khorana HG. Missense suppression of the tryptophan synthetase A-protein mutant A78. Proc Natl Acad Sci. 1966;56:772–779.
- Roberts JW, Carbon J. Molecular mechanism for missense suppression in E. Coli Nature. 1974;250:412–414.
- Capone JP, Sharp PA, RajBhandary UL. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. Embo J. 1985;4:213–221.
- Goodman HM, Olson MV, Hall BD. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci. 1977;74:5453–5457.
- Liebman SW, Stewart JW, Sherman F. Serine substitutions caused by an ochre suppressor in yeast. J Mol Biol. 1975;94:595–610.
- Waterston RH. A second informational suppressor, SUP-7 X, in Caenorhabditis elegans. Genetics. 1981;97:307–325.
- Kalapis D, Bezerra AR, Farkas Z, et al. Evolution of robustness to protein mistranslation by accelerated protein turnover. PLoS Biol. 2015;13:1–28.
- Ruan B, Palioura S, Sabina J, et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci. 2008;105:16502–16507.
- Berg MD, Hoffman KS, Genereaux J, et al. Evolving mistranslating tRNAs through a phenotypically ambivalent intermediate in Saccharomyces cerevisiae. Genetics. 2017;206:1865–1879.
- Geslain R, Cubells L, Bori-Sanz T, et al. Chimeric tRNAs as tools to induce proteome damage and identify components of stress responses. Nucleic Acids Res. 2010;38:e30–e30.
- Zimmerman SM, Kon Y, Hauke AC, et al. Conditional accumulation of toxic tRNAs to cause amino acid misincorporation. Nucleic Acids Res. 2018;46:7831–7843.
- Zhu Y, Berg MD, Yang P, et al.. Mistranslating tRNA identifies a deleterious S213P mutation in the Saccharomyces cerevisiae eco1-1 allele. Biochem Cell Biol. 2020. bcb-2020-0151. DOI:10.1139/bcb-2020-0151.
- Lant JT, Berg MD, Heinemann IU, et al.. Pathways to disease from natural variations in human cytoplasmic tRNAs. J Biol Chem. 2019;294:5294–5308.
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol. 1971;58:439–458.
- Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180.
- Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597.
- Schmeing TM, Voorhees RM, Kelley AC, et al.. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol. 2011;18:432–437.
- Agirrezabala X, Valle M. Structural insights into tRNA dynamics on the ribosome. Int J Mol Sci. 2015;16:9866–9895.
- Berg MD, Giguere DJ, Dron JS, et al. Targeted sequencing reveals expanded genetic diversity of human transfer RNAs. RNA Biol. 2019;16:1574–1585.
- Schultz DW, Yarus M. Transfer RNA mutation and the malleability of the genetic code. J Mol Biol. 1994;235:1377–1380.
- Massey SE, Moura G, Beltrão P, et al. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 2003;13:544–557.
- Mühlhausen S, Schmitt HD, Pan KT, et al. Endogenous stochastic decoding of the CUG codon by competing Ser- and Leu-tRNAs in Ascoidea asiatica. Curr Biol. 2018;28:2046–2057.e5.
- Krassowski T, Coughlan AY, Shen XX, et al. Evolutionary instability of CUG-Leu in the genetic code of budding yeasts. Nat Commun. 2018;9:1–8.
- Mühlhausen S, Findeisen P, Plessmann U, et al.. A novel nuclear genetic code alteration in yeasts and the evolution of codon reassignment in eukaryotes. Genome Res. 2016;26:945–955.
- Li M, Tzagoloff A. Assembly of the mitochondrial membrane system: sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell. 1979;18:47–53.
- Macino G, Coruzzi G, Nobrega FG, et al.. Use of the UGA terminator as a tryptophan codon in yeast mitochondria. Proc Natl Acad Sci. 1979;76:3784–3785.
- Alfonzo JD. C to U editing of the anticodon of imported mitochondrial tRNATrp allows decoding of the UGA stop codon in Leishmania tarentolae. Embo J. 1999;18:7056–7062.
- Sengupta S, Yang X, Higgs PG. The mechanisms of codon reassignments in mitochondrial genetic codes. J Mol Evol. 2007;64:662–688.
- Noutahi E, Calderon V, Blanchette M, et al.. Rapid genetic code evolution in green algal mitochondrial genomes. Mol Biol Evol. 2019;36:766–783.
- Zihala D, Eliáš M, Katz LA. Evolution and unprecedented variants of the mitochondrial genetic code in a lineage of green algae. Genome Biol Evol. 2019;11:2992–3007.
- Woese CR. The problem of evolving a genetic code. Bioscience. 1970;20:471–485.
- Woese CR. On the evolution of the genetic code. Proc Natl Acad Sci. 1965;54:1546–1552.
- Woese CR. A new biology for a new century. Microbiol Mol Biol Rev. 2004;68:173–186.
- Wang X, Pan T. Methionine mistranslation bypasses the restraint of the genetic code to generate mutant proteins with distinct activities. PLOS Genet. 2015;11:e1005745.
- Müller MM, Allison JR, Hongdilokkul N, et al. Directed evolution of a model primordial enzyme provides insights into the development of the genetic code. PLoS Genet. 2013;9:e1003187.
- O’Donoghue P, Prat L, Kucklick M, et al. Reducing the genetic code induces massive rearrangement of the proteome. Proc Natl Acad Sci. 2014;111:17206–17211.
- Prat L, Heinemann IU, Aerni HR, et al. Carbon source-dependent expansion of the genetic code in bacteria. Proc Natl Acad Sci. 2012;109:21070–21075.
- Wong JTF. Coevolution theory of genetic code at age thirty. BioEssays. 2005;27:416–425.
- Zhang Y, Baranov PV, Atkins JF, et al.. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem. 2005;280:20740–20751.
- Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408.
- O’Donoghue P, Ling J, Wang YS, et al.. Upgrading protein synthesis for synthetic biology. Nat Chem Biol. 2013;9:594–598.
- Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249.
- Cervettini D, Tang S, Fried SD, et al.. Rapid discovery and evolution of orthogonal aminoacyl-tRNA synthetase–tRNA pairs. Nat Biotechnol. 2020. DOI:10.1038/s41587-020-0479-2.
- Link AJ, Vink MKS, Agard NJ, et al. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc Natl Acad Sci. 2006;103:10180–10185.
- Iwane Y, Hitomi A, Murakami H, et al. Expanding the amino acid repertoire of ribosomal polypeptide synthesis via the artificial division of codon boxes. Nat Chem. 2016;8:317–325.
- Hoshika S, Leal NA, Kim MJ, et al. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science. 2019;363:884–887.
- Zhang Y, Lamb BM, Feldman AW, et al. A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proc Natl Acad Sci. 2017;114:1317–1322.
- Rodin AS, Szathmáry E, Rodin SN. On origin of genetic code and tRNA before translation. Biol Direct. 2011;6:1–24.
- Balzi E, Choder M, Chen W, et al.. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem. 1990;265:7464–7471.
- Kaji H, Novelli GD, Kaji A. A soluble amino acid-incorporating system from rat live. Biochim Biophys Acta. 1963;76:474–477.
- Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345.
- Tobias J, Shrader T, Rocap G, et al.. The N-end rule in bacteria. Science. 1991;254:1374–1377.
- Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–455.
- Francklyn CS, Minajigi A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2010;584:366–375.
- Katz A, Elgamal S, Rajkovic A, et al.. Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Mol Microbiol. 2016;101:545–558.
- Hinnebusch AG. Translational regulation of Gcn4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450.
- Ellenberger TE, Brandl CJ, Struhl K, et al.. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α Helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237.
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745.
- Jin D, Musier-Forsyth K. Role of host tRNAs and aminoacyl-tRNA synthetases in retroviral replication. J Biol Chem. 2019;294:5352–5364.
- Sawyer RC, Dahlberg JE. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J Virol. 1973;12:1226–1237.
- Seiki M, Hattori S, Hirayama Y, et al.. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci. 1983;80:3618–3622.
- Wain-Hobson S, Sonigo P, Danos O, et al.. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17.
- Chapman KB, Byström AS, Boeke JD. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc Natl Acad Sci. 1992;89:3236–3240.
- Martinez G. tRNAs as primers and inhibitors of retrotransposons. Mob Genet Elements. 2017;7:1–6.
- Schorn AJ, Gutbrod MJ, LeBlanc C, et al.. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71.e11.
- Lee YS, Shibata Y, Malhotra A, et al.. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23:2639–2649.
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806.
- Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:1–14.
- Torres AG, Reina O, Stephan-Otto Attolini C, et al.. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc Natl Acad Sci. 2019;201821120. DOI:10.1073/pnas.1821120116
- Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci. 2016;41:679–689.
- Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43–50.
- Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442.
- Yamasaki S, Ivanov P, Hu GF, et al.. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42.
- Cole C, Sobala A, Lu C, et al.. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160.
- Haussecker D, Huang Y, Lau A, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695.
- Kuscu C, Kumar P, Kiran M, et al. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093–1105.
- Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280:42744–42749.
- Ivanov P, Emara MM, Villen J, et al.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623.
- Thompson DM, Lu C, Green PJ, et al.. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103.
- Oberbauer V, Schaefer MR. tRNA-derived small RNAs: biogenesis, modification, function and potential impact on human disease development. Genes (Basel). 2018;9. DOI:10.3390/genes9120607.
- Mei Y, Yong J, Liu H, et al. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37:668–678.
- Saikia M, Jobava R, Parisien M, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol. 2014;34:2450–2463.
- Rashad S, Niizuma K, Tominaga T. TRNA cleavage: a new insight. Neural Regen Res. 2020;15:47–52.
- Cho H, Lee W, Kim GW, et al. Regulation of La/SSB-dependent viral gene expression by pre-tRNA 3ʹ trailer-derived tRNA fragments. Nucleic Acids Res. 2019;47:9888–9901.
- Gebetsberger J, Zywicki M, Künzi A, et al.. TRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:10–12.
- Krishna S, Yim DG, Lakshmanan V, et al. Dynamic expression of tRNA‐derived small RNAs define cellular states. EMBO Rep. 2019;20:1–18.
- Kim HK, Fuchs G, Wang S, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62.
- Maute RL, Schneider C, Sumazin P, et al. TRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci. 2013;110:1404–1409.
- Peng H, Shi J, Zhang Y, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612.
- Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396.
- Chen Q, Yan M, Cao Z, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400.
- Sharma U, Sun F, Conine CC, et al. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev Cell. 2018;46:481–494.e6.
- Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–1232.
- Tosar JP, Cayota A. Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 2020;00:1–19.
- Chiou NT, Kageyama R, Ansel KM. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 2018;25:3356–3370.e4.
- Guzman N, Agarwal K, Asthagiri D, et al. Breast cancer-specific miR signature unique to extracellular vesicles includes microRNA-like tRNA fragments. Mol Cancer Res. 2015;13:891–901.
- Hogg MC, Raoof R, El Naggar H, et al. Elevation of plasma tRNA fragments precedes seizures in human epilepsy. J Clin Invest. 2019;129:2946–2951.
- Zhao F, Cheng L, Shao Q, et al. Characterization of serum small extracellular vesicles and their small RNA contents across humans, rats, and mice. Sci Rep. 2020;10:1–16.
- Sadik N, Cruz L, Gurtner A, et al. Extracellular RNAs: a new awareness of old perspectives. Methods Mol Biol. 2018;1740:1–15.
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383.
- Tosar JP, Gámbaro F, Sanguinetti J, et al. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015;43:5601–5616.
- Shurtleff MJ, Yao J, Qin Y, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci. 2017;114:E8987–E8995.
- Bittel DC, Jaiswal JK. Contribution of extracellular vesicles in rebuilding injured muscles. Front Physiol. 2019;10:828.
- Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Bio. 2015;17:816–826.
- Guzzi N, Bellodi C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol. 2020;00:1–9.
- Qin C, Xu PP, Zhang X, et al. Pathological significance of tRNA-derived small RNAs in neurological disorders. Neural Regen Res. 2020;15:212–221.
- Zhu L, Ge J, Li T, et al.. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019;452:31–37.
- Goodarzi H, Liu X, Nguyen HCB, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802.
- Murray LE, Rowley N, Dawes IW, et al.. A yeast glutamine tRNA signals nitrogen status for regulation of dimorphic growth and sporulation. Proc Natl Acad Sci. 1998;95:8619–8624.
