ABSTRACT
Subgenomic RNAs are produced by several RNA viruses through incomplete degradation of their genomic RNA by the exoribonuclease Xrn1, and have been shown to be essential for viral growth and pathogenicity. Within the flavivirus genus of the Flaviviridae family, two distinct classes of Xrn1-resistant RNA motifs have been proposed; one for mosquito-borne and insect-specific flaviviruses, and one for tick-borne flaviviruses and no-known-vector flaviviruses. We investigated tick-borne and no-known-vector flavivirus Xrn1-resistant RNA motifs through systematic in vitro mutational analysis and showed that both classes actually possess very similar structural configurations, including a double pseudoknot and a base-triple at identical, conserved locations. For the no-known-vector flavivirus Modoc virus, we show that in vivo generation of subgenomic flaviviral RNA was affected by mutations targeted at nucleotides involved in the structural features of flaviviral Xrn1-resistant RNA motifs that were defined in this work. Our results suggest that throughout the genus flavivirus Xrn1-resistant RNA motifs adopt the same topologically conserved structure.
Introduction
The family Flaviviridae comprises four genera: flaviviruses, pestiviruses and the more recently added genera hepaci- and pegiviruses [Citation1–3]. They are all enveloped, positive-stranded RNA viruses with genomes ranging between 10 to 12.5 kb. Based on their mode of transmission, the flaviviruses can be further subdivided into mosquito-borne flaviviruses (MBFV), tick-borne flaviviruses (TBFV), no-known-vector flaviviruses (NKVFV) and insect-specific flaviviruses (ISFV). Infection of a suitable host by either the mosquito- or tick-borne flavivirus strictly depends on an arthropod vector, whereas infection by the mainly bat- or small rodent-infecting NKVFVs appears to occur via direct contact with infected individuals. ISFVs replicate solely in insects and are unable to replicate in a mammalian host. The arthropod-borne flaviviruses are particularly associated with microcephaly and harmful diseases like haemorrhagic fever, encephalitis and dengue fever in humans.
The flavivirus genome is flanked by a capped 5ʹ untranslated region (UTR) that mediates translation initiation and a relatively long, highly structured, non-polyadenylated 3ʹ UTR [Citation4,Citation5]. The RNA structure of the distal part of this 3ʹ UTR is relatively well conserved within each of the four distinguishable groups of flaviviruses. It has been shown to contain conserved RNA sequences for base pairing with complementary sequences at the 5ʹ end of the genome, which is critical for replication [Citation6–8]. The flavivirus 3ʹ UTR carries several RNA stem-loop and pseudoknot structures that are generally well conserved, especially among flaviviruses belonging to the same subgroup. This region has been shown to be responsible for the accumulation of noncoding subgenomic flaviviral RNA (sfRNA) in infected cells, resulting from stalling of the host exoribonuclease Xrn1 on Xrn1-resistant RNA (xrRNA) structures after degrading roughly 95% of the flavivirus genome [Citation9–12]. These sfRNAs are implicated to interfere with cellular RNA decay, RNAi pathway, and the immune response [Citation9,Citation13–15]. Following a flavivirus infection, Xrn1 activity is decreased, which suggests that it may be sequestered by stalling on xrRNA structures, thereby altering cellular RNA homeostasis to benefit viral replication [Citation16]. Currently, the generation of sfRNA due to xrRNA in the 3ʹ UTR has only been demonstrated in vivo by members of the flavivirus genus. Analysis of intracellular RNA isolated from cells infected with the pestivirus BVDV and hepacivirus HCV previously did not show the production of sfRNA [Citation9]. However, structures in the 5ʹ UTR of BVDV and HCV are capable of stalling and sequestering Xrn1, thereby altering mRNA longevity [Citation17]. Production of sfRNA was revealed in nematodes infected with the unclassified soybean-cyst nematode virus 5 [Citation18]. Due to its homology, this virus was initially identified as a pestivirus, however it was then moved into the flavivirus genus, in part because of its production of subgenomic RNA.
The production of sfRNA requires the presence of an intricate xrRNA structure that consists of five stem elements including two pseudoknot interactions [Citation19]. These are oriented in such a way that they can halt Xrn1’s processive degradation mode. The xrRNA crystal structures of the MBFVs Zika virus (ZIKV) and Murray Valley encephalitis virus (MVEV) have been resolved and appear to form a small ring-like RNA structural fold in which the 5ʹ end of the viral RNA protrudes from the back to the front [Citation20,Citation21]. The diameter of the ring is too small for Xrn1 to protrude and continue to hydrolyse the RNA; therefore the enzyme is stalled. This structural analysis, combined with subsequent mutational studies, has revealed the requirement of additional nucleotide interactions that are critical for a functional stalling site, including a strongly conserved base triple [Citation20,Citation21]. Analysis of the xrRNA present in cell-fusing agent virus (CFAV), a typical ISFV, has indicated a highly similar structure and the absolute requirement of a homologous base triple, thus implying that MBFV and ISFV xrRNA stall Xrn1 in a similar fashion [Citation19]. In contrast, the tick-borne and no-known-vector flaviviruses have been suggested to employ a slightly different structure to resist Xrn1, which endorsed a distinction between these two classes of xrRNA structures (‘class I’ for MBFV and ISFV xrRNAs and ‘class II’ for TBFV and NKVFV xrRNAs). However, in the absence of a solved 3D structure for these class II xrRNAs, this distinction may be preliminary.
In this study, we examined the generation of sfRNA by tick-borne encephalitis virus (TBEV), Modoc virus (MODV) and Rio Bravo virus (RBV) – viruses harbouring xrRNA motifs of the proposed class II-type – using manual modelling based on phylogeny, and in vitro Xrn1-degradation assays. Through site-directed mutagenesis, important features in the three-dimensional structures responsible for stalling Xrn1 were investigated. In doing so, conserved structural elements were identified, which raises the question whether the distinction between flaviviral xrRNA classes is justified. Using MODV, the effect of these mutations was further verified by implementing them into an infectious clone in order to assess their ability to generate sfRNA in vivo.
Materials and methods
Design and production of DNA templates for in vitro RNA transcription
TBFV and NKVFV constructs used in this study were amplified through oligonucleotide templates carrying reverse complementary sequences on the 3ʹ ends. These were purchased from SigmaAldrich and Eurogentec in desalted form. Forward primers carried a T7 promoter sequence (GTAATACGACTCACTATA), followed by an AU-rich 12 nt leader sequence. A list of oligonucleotides is available on request. PCR reactions were carried out in a 50 μl volume, containing 400 nM of each oligo, 200 μM dNTPs and 2 units DreamTaq polymerase on a BioRad cycler. PCR fidelity was checked by agarose gel electrophoresis and products were purified by ethanol/NaAc precipitation at room temperature and dissolved in 25 μl Milli-Q water.
In vitro Xrn1 degradation assay
In vitro transcription reactions were carried out using T7 RiboMAX™ Large Scale RNA Production System (Promega) in 10 μl volumes, containing 5 μl PCR product (~250 ng), 5 mM of each rNTP, 1 μl Enzyme mix, in 1x Transcription Optimized buffer (40 mM Tris-HCl, 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, pH7.9 @ 25°C). After incubation at 37°C for 30 mins, 1 unit RQ1 RNase-Free DNAse was added to the reaction and incubation proceeded at 37°C for 20 mins. Reaction samples were checked on agarose gel in order to establish subsequent usage of equal amounts of RNA. Xrn1 digestion reactions were performed with ~400 ng RNA in 1x NEB3 buffer totalling 10 μl, which was equally divided over two tubes. To one of the tubes, 0.2 units of Xrn1 and 0.3 units of RppH (New England Biolabs) were added. Both tubes were incubated for 30 mins at 37°C and the reactions were terminated by adding 5 μl formamide containing trace amounts of bromophenol blue and xylene cyanol FF. After addition of 5 μl denaturing loading buffer (8 M urea, 20 mM Tris-HCl, 20 mM EDTA, trace amounts of bromophenol blue and xylene cyanol FF) samples were denatured for five minutes at 75°C. These were run on 8 M urea 10% polyacrylamide gels in TBE buffer, equilibrated at 60–65°C. Gels were stained with EtBr and each construct was subjected to this assay at least twice. Bands were quantified using the Quantity One 1-D analysis software. Xrn1 stalling positions were estimated using the y-positions of untreated RNAs and their corresponding sizes, from which an exponential function was formulated that was used for estimating the sizes of Xrn1-treated RNAs.
In vitro RNA transcription of infectious full length MODV genome RNA
Details on the construction and characterization of the MODV infectious cDNA clone that was fused at the 5ʹ end to a T7 ø2.5 promoter [Citation22] in the low copy number vector pACNR, will be published elsewhere (Jiang et al., manuscript in preparation). Plasmid DNA for in vitro run-off RNA transcription was purified using the Nucleobond Xtra Maxi DNA isolation kit (Macherey-Nagel, Düren, Germany) and linearized with AflII at a unique site directly adjacent to the MODV 3ʹ UTR. Template DNA was purified by phenol/chloroform extraction and ethanol precipitation. Approximately 1 μg of linearized DNA was used as a template for in vitro transcription using the Ampliscribe™ T7 high-yield transcription kit (Lucigen, Madison, USA). For the production of 5ʹ A-capped full-length MODV transcripts, NTP concentrations were 7.5 mM, except for ATP, which was adjusted to 2 mM. G(5ʹ)ppp(5ʹ)A (NEB, Ipswich, USA) was added as RNA cap analogue to a final concentration of 6 mM. After a 2 hr incubation at 37°C, DNase I was added and the incubation was continued for another 15 min. The RNA transcripts were subsequently purified by LiCl precipitation. The quality of the transcripts was checked by agarose gel electrophoresis and the concentration was determined by spectrophotometry.
Mutagenesis of the MODV infectious cDNA clone
The shuttle vector pBluescript-MODV9651-10,605 that contained the MODV sequences encoding the COOH-terminal part of NS5 and the complete 3ʹ UTR was used as a template to create mutations within the MODV xrRNA using the Quick Change strategy. The MODV inserts of the mutated plasmids were sequenced to verify the presence of the mutations and to exclude unintended base changes. Correct plasmids were cut with KasI and XhoI and the ~1kb MODV insert was cloned into KasI- and XhoI-cut pACNR-FLMODV61ΔSS, which contains the complete MODV infectious cDNA except for the 3ʹ terminal 93 nucleotides. Correct colonies were identified by colony PCR and used to isolate plasmid DNA.
Transfection of BHK21J cells with infectious MODV RNA transcripts
BHK-21 J cells were transfected by electroporation with 5 μg of full-length MODV RNA as described previously [Citation23] and seeded in 75 cm2 flasks. At 72 hr p.e. the medium was harvested from the transfected cells and centrifuged to remove cell debris. The supernatant was aliquoted and stored at −80°C. MODV titres in these supernatants were determined by plaque assays [Citation23]. For RNA analysis, 2.5 ml (approximately 1.5 × 106 cells) of the transfected BHK-21 J cell suspension was seeded in a 10 cm2 plate. Total RNA was isolated from the transfected cells at 30 h post electroporation (p.e) using Tripure (Roche, Mannheim, Germany) according to the manufacturer.
Analysis of sfRNA production in MODV infected cells
BHK-21 J cells were grown to 80% confluency in 9.6 cm2 plates, washed once with PBS and infected with MODV at MOI 5. After 1 hr the inoculum was replaced by 2 ml DMEM + 2% FCS. At 24 hr post infection (p.i.) the infected cells were lysed and total RNA was isolated using Tripure (Roche, Mannheim, Germany). Samples containing 7.5 μg of total RNA from infected and mock infected cells were analysed for the production of MODV sfRNA production by agarose gel electrophoresis under denaturing conditions [Citation24] and transferred to a Hybond N+ membrane (GE Healthcare, Buckinghamshire, UK) by classical capillary blotting. Membranes were subsequently treated as described previously [Citation23] and hybridized with a 5ʹ 32P-labelled oligonucleotide (NKV 4) that is complementary to positions 10,573 to 10,600 of the MODV genome.
Results
Secondary and tertiary interactions that are involved in the formation of xrRNA structures of the representative mosquito-borne and insect-specific flaviviruses ZIKV () and CFAV have previously been elucidated through mutational analyses, sequence comparison and x-ray crystallography [Citation19,Citation21]. Similar secondary interactions have been proposed for xrRNA structures found within the 3ʹ UTR of the tick-borne TBEV, and the no-known vector flaviviruses MODV and RBV [Citation19,Citation25]. Supposed differences in structural element configuration within xrRNA structures from MBFV/ISFV and TBFV/NKVFV species have warranted the division of these xrRNAs into two different classes (Supplementary Figure S1) [Citation19]. However, it is possible to map the expected interactions that occur within the ‘class II’ TBEV, MODV and RBV xrRNA and to model these to illustrate the typical three-dimensional contacts present in ‘class I’ MBFV and ISFV xrRNA structures, according to the known ZIKV and CFAV conformation ().
Figure 1. (A) Representations of ZIKV, TBEV, MODV and RBV xrRNA structures, illustrating two- (above) and three-dimensional (below) contacts. The proposed stem-interactions α, β, γ, δ and ε are given in blue, green, magenta, red and orange, respectively. Nucleotides involved in the base triple interactions are shown in bold. (B) Sequences of ZIKV and CFAV xrRNA, in structural alignment with a collection of various tick-borne and no-known-vector flaviviruses, revealing covariations in this region. Grey columns in the background depict which nucleotides are involved in the base triple interaction. Accession numbers used for sequences were: Zika virus isolate 15555 (ZIKV), MN025403; Cell fusing agent virus isolate Guadeloupe (LR694081); Tick-borne encephalitis virus Sofjin-HO (TBEV), AB062064; Spanish goat encephalitis virus (SGEV), NC_027709; Tick-borne encephalitis virus Neudoerfl (WEST), U27495; Langat virus strain TP21 (LANG), AF253419; Louping ill virus LI3/1 (LOUP), KP144331; Karshi virus strain LEIV 2247 (KARS), AY863002; Deer tick virus strain ctb30 (DEER), AF311056; Powassan virus strain LB (POWA), L06436; Modoc virus (MODV), NC_003635; Rio Bravo virus (RBV), JQ582840; Montana myotis leukoencephalitis virus (MMLV), NC_004119; Apoi virus (APOIV), AF452050
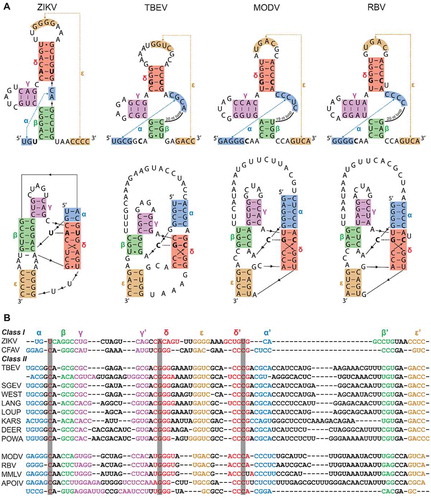
Through BLAST searches on the TBEV sequence in GenBank, we found analogous putative xrRNA sequences in a large variety of other TBFV members (). Furthermore, closer inspection of xrRNA structures in NKVFVs showed that the second Xrn1-stalling site of TBEV, MMLV and both stalling sites in APOIV can be folded according to the MBFV model as well (Supplementary Figure S2). Interestingly, these viruses – together with MODV and RBV – have already demonstrated the production of sfRNA in infected cells [Citation19]. Structural alignment of these sequences reveals that, while not very conserved in sequence and stem-lengths, a double pseudoknot with five stem elements (here termed stems α through ε) is found in all sequences. Notably, many covariations exist between the class I and class II species, and also among TBFVs and NKVFVs. Of note, one instance of such a covariation includes the strongly conserved C•G-C base triple, substituting the ZIKV U•A-U interaction. These findings strongly suggest the presence of functional xrRNA within the 3ʹ UTR of the listed TBFV members, forming a structure that is equivalent throughout the genus flavivirus.
One of the differences previously postulated between MBFV/ISFV and TBFV/NKVFV xrRNA are the Xrn1-stalling positions. In MBFV, it was mapped just 5ʹ of the actual structure, while in TBEV and MODV xrRNA Xrn1 has been found to stall within a ‘bulge’ that is putatively formed within the first stem it encounters when processing the RNA in a 5ʹ->3ʹ fashion (Supplementary Figure S1) [Citation19]. Note that in , the three-dimensional structure is modelled without the sequence 5ʹ of this supposed Xrn1-stalling site. To test whether this sequence is initially required for Xrn1-resistance, we replaced it with a 12-nt AU-rich sequence that is unlikely to form any stable interactions with the rest of the structure and additionally serves as a landing path for Xrn1. Such a ‘minimal’ construct was designed for TBEV, MODV and RBV xrRNA sequences, which were transcribed in vitro, subjected to an in vitro Xrn1 degradation assay and their resistance thereof assessed on a denaturing gel (). An equivalent ZIKV xrRNA was included in order to compare between the two putative xrRNA classes (Supplementary Figure S3). The resulting gels indicated that these constructs are capable of stalling Xrn1. By comparing relative migration distances (see Materials and methods), we estimated that Xrn1 stalls 1 nt (±1 nt) before stem α, coinciding with the Xrn1-stalling sites previously observed by MacFadden et al [Citation19].
Figure 2. In vitro Xrn1 degradation assays probing (A) the Xrn1-resistance of ‘minimal’ RNA constructs for TBEV, MODV and RBV, and (B) the Xrn1-resistance of these constructs carrying loop deletions. RNAs, incubated with or without Xrn1, were loaded on denaturing polyacrylamide gels shown here with the corresponding names of constructs given above. Data below the gels depict the average percentage (± SD) of Xrn1-resistant RNA
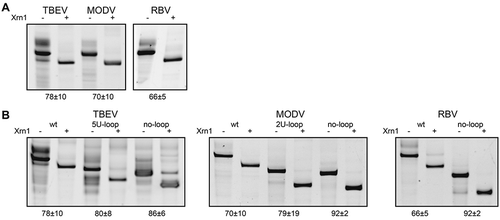
MBFV and ISFV sequences differ from their tick-borne and no-known-vector flavivirus counterparts by the absence of a long stretch of nucleotides between the α and β stems. In order to further assess the similarity between class I and class II xrRNA, we targeted this loop sequence by replacing it with five uracils in TBEV (5 U-loop), two uracils in MODV (2 U-loop) or by deleting the loop completely in TBEV, MODV or RBV (no-loop), mimicking the lack of such a loop in class I xrRNA structures (). These changes did not decrease Xrn1-stalling capacity by any of the tested constructs. This result suggests that this loop is redundant for proper function of the xrRNA.
One of the crucial interactions for adopting the MBFV (class I) fold is the interaction forming stem β. While it was missed in previous predictions, the formation of this stem is supported by covariations between all assessed species (). The importance of β was demonstrated by systematic disruption and restoration of these β interactions in the TBEV, MODV and RBV minimal xrRNA constructs (). For TBEV, mutations targeting the 5ʹ or 3ʹ side of the putative β stem resulted in the loss of Xrn1-resistance, which could be rescued by combining these complementary mutations. Mutating the 5ʹ side of MODV putative β stem, leaving a single potential base pair, was sufficient to almost entirely disrupt the Xrn1 resistance of the MODV construct. Restoration of these three base pairs through complementary mutations on the 3ʹ side, completely restored Xrn1-stalling ability. Interestingly, this construct retained Xrn1 resistance when just the 3ʹ side bases were substituted, although this could be due to the fact that a single nucleotide shift in the loop between α and β could already lead to base pairing of the two substituted nucleotides. Mutating the 5ʹ side would allow for two base pairs as well, through a single nucleotide shift in the other direction, although this would bulge out a cytosine between stems β and γ. Within the RBV construct, mutating its β interaction on either the 5ʹ or the 3ʹ side leads to complete degradation, whereas Xrn1 resistance is re-established in the combined double mutant. These results indicate that the β stem plays an important structural role in the TBEV, MODV and RBV xrRNAs.
Figure 3. In vitro Xrn1 degradation assays performed a variety of constructs and loaded on denaturing polyacrylamide gels, testing the effect of mutations targeted at the (A) β stem, (B) ε stem and (C) the base triple interaction. Data below the gels depict the average percentage (± SD) of Xrn1-resistant RNA. ‘N.D.’ signifies that this value could not be determined reliably, but does not exceed 10%
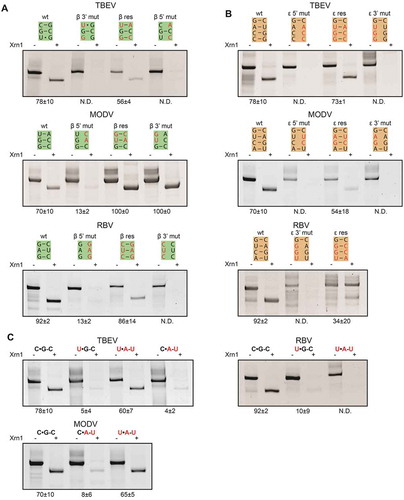
The formation and requirement of the long-range pseudoknot interaction for Xrn1-resistance has been demonstrated for a number of MBFV xrRNA species [Citation10–12]. In particular, we demonstrate that for ZIKV xrRNA, mutations targeted at this interaction disturb its resistance towards Xrn1 completely, while the combination of these mutations restored its stalling capacity (Supplementary Figure S3), in agreement with previous results [Citation21] Disrupting stem ε in our minimal TBEV construct by mutating bases on the 5ʹ or 3ʹ side of ε (), also led to complete degradation of the RNA whereas the combined mutations restored ε and prevented RNA degradation. Similar to TBEV xrRNA, its MODV counterpart makes use of a long-range pseudoknot interaction, of which the importance for Xrn1 stalling has been proven before [Citation19]. Through disrupting mutations, we show similar results within our minimal construct (), although subsequent compensating mutations of MODV stem ε yields only a very small population of Xrn1-resistant RNAs. The same procedure was carried out for RBV stem ε (), which appears to lose some inherent stability due to such mutations, as multiple bands appear on gel. These species are all degraded by action of Xrn1 however, and restoration of this stem yields partially degraded RNA again, indicating its importance for Xrn1 stalling by RBV xrRNA.
MBFV xrRNA species make use of a well-conserved U•A-U base triple, which is crucial for stalling Xrn1, and can be substituted by a C•G-C (Supplementary Figure S3) [Citation19]. In order to assess whether the C•G-C base triples modelled in TBEV, MODV and RBV are of importance as well, these nucleotides were mutated and the resulting constructs subjected to an in vitro Xrn1-degradation assay (). By changing the putative C•G-C triple in TBEV to either a U•G-C or C•A-U triple, almost all input TBEV RNA transcripts were degraded by Xrn1, while restoration of this triple to a U•A-U rescued its Xrn1-stalling capacity. MODV has a similar putative C•G-C triple; mutating it to C•A-U reduced its Xrn1 resistance, which was restored almost completely by the compensating mutation that forms a U•A-U triple. Similar mutations were tested within the RBV construct, which lost its innate Xrn1 resistance completely when changing its C•G-C triple to form a U•G-C, revealing its importance. However, subsequent restoration to a U•A-U triple did not recover Xrn1 stalling ability, indicating a more stringent requirement of the wild-type base triple forming nucleotides.
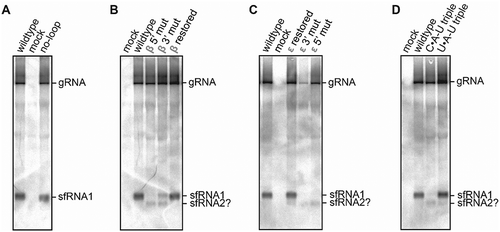
To obtain in vivo support for the proposed structural determinants in class II xrRNA, infectious MODV cDNA clones were constructed, carrying mutations corresponding to constructs that were tested for Xrn1-resistance in vitro. In vitro transcribed, full-length viral RNA was used to infect BHK-21 cells, from which RNA was isolated and analysed for the production of sfRNA by Northern blotting and hybridization. This revealed the formation of a band corresponding with a short RNA segment derived from the vector cloned with wild-type MODV xrRNA (). The same band was formed from the no-loop mutant, which agrees with its retainment of Xrn1-resistance in vitro. Disruption of the β stem on the 5ʹ or 3ʹ side led to either a complete or partial loss of a band at the same height, respectively (). This effect was rescued by combining these mutations. Mutants disrupting and restoring the MODV ε stem () and base triple () also reveal Xrn1-resistance corresponding with results in vitro. Interestingly, all mutants that demonstrate a deficiency of Xrn1-stalling at one xrRNA structure show an additional sfRNA (putatively labelled as sfRNA2 in B, C and D), corresponding with a previously unknown Xrn1-stalling site 3ʹ of the MODV xrRNA structure investigated here.
Figure 4. Northern blot analysis of total RNA isolated from BHK-21 cells transfected with RNA derived from infectious MODV cDNA clones. The clone varieties are listed above the lanes, corresponding with the no-loop (A), β stem (B), ε stem (C) and base triple (D) mutants as used in in vitro Xrn1 degradation assays
Discussion
Flaviviral structures responsible for stalling Xrn1 adopt an elaborate RNA conformation, formed by several specific interactions. Although in comparison to MBFV and ISFV xrRNA, TBFV and NKVFV xrRNAs do not appear to be very conserved on a sequence level, we were able to model these sequences into a conformation with equivalent secondary and tertiary interactions. While prior distinction between class I and class II flaviviral xrRNA motifs suggests differing modes of Xrn1 stalling, the results presented in this work imply that throughout the flavivirus genus, xrRNA motifs employ a topologically and functionally equivalent structure.
The apparent formation and necessity of the proposed β stem in all species tested here has not been described previously. The 3ʹ side of this stem is located just upstream of the 3ʹ side of the long-range pseudoknot interaction ε, on a sequence that was previously predicted to be either single-stranded (TBEV), or involved in a downstream stem-loop (MODV) that is not required for Xrn1-resistance in vitro [Citation19]. The same study shows that deleting the pentanucleotide 5ʹ–GGCCA–3ʹ, which includes the two G residues in the β stem, already inactivates the ability to stall Xrn1. Notably, through analysis of its secondary structure, this xrRNA was classified as being TBEV-like, together with several other NKVFVs. The MODV xrRNA model presented in our study shows that the guanines in this sequence are involved in the β stem, which explains why its deletion would lead to an Xrn1-stalling-deficient RNA.
All RNA transcripts used in the in vitro Xrn1 assays carried a 12-nt AU-rich sequence at their 5ʹ end that is directly fused to the most 5ʹ nucleotide involved in the formation of the putative α stem. Since all of these constructs show innate Xrn1 resistance, upstream and downstream sequences in the genomic RNA from which they originate, are less likely to be significant. This includes nucleotides implicated in an additional stem proposed by MacFadden et al [Citation19]. for class II flaviviral xrRNAs found in, for instance, TBEV. This stem employs nucleotides from the loop between stems α and γ, which has been indicated as another distinction between xrRNA structural types. It is not clear why TBFV and NKVFV possess a longer loop than MBFV and ISFV xrRNAs, but while the loop may be base-paired to a region upstream of α, it becomes single-stranded upon degradation of the 5ʹ half of the extended α stem by Xrn1. Its presence is presumably unrelated to the function of the xrRNA as the results above show that it can be deleted from TBFVs and NKVFVs without any effect on Xrn1-resistance. Therefore, by disregarding these putative interactions, as in the models proposed in this study, the xrRNA structure of NKVFVs and TBFVs becomes very similar to the structure of the class xrRNAs of the MBFVs and ISFVs. Notably, this also moves the proposed Xrn1 halt site from a position between stems, to one preceding the actual Xrn1 stalling position, just as in ZIKV and CFAV xrRNA. Our minimal constructs, including the ZIKV xrRNA, also do not contain the small hairpin that is present just downstream of class I xrRNA structures (Supplementary Fig. 1). However, previous work on the MBFV MVE xrRNA indicated that it is not required for Xrn1 resistance in vitro [Citation12]. TBFV and NKVFV species lack this hairpin and we show that they do not need sequences downstream of the ε stem. Recent work has indicated that this structure is presumably required for improving xrRNA binding affinity for Xrn1, as it has been shown to interact with the Xrn1 winged helix domain and is apparently required for MBFV sfRNA production in vivo [Citation26].
Previous structural characterizations have shown that in class I MBFV and ISFV xrRNAs, a base triple interaction is formed, which is crucial for robust stalling of Xrn1. Our models, in vitro Xrn1 degradation assays and in vivo infection assay underpin the importance of such a triple for class II TBFV and NKVFV xrRNAs. While disruption of these triples usually allowed for a small portion of digested constructs to stall Xrn1, attempts to restore disrupted interactions through the formation of isomorphic base triples resulted in Xrn1-resistant RNA structures for TBEV and MODV. The RBV construct did not allow for exchanging of its C•G-C with a U•A-U base triple, which may be due to the fact that it uniquely flanks a bulging uracil within the δ stem, already compromising its structural integrity. It should be noted that the formation of a C•G-C triple requires protonation of the Hoogsteen-paired cytosine, which would theoretically require more acidic conditions than employed in our experiments [Citation27]. However, previous studies regarding triple helices in, for instance, MALAT1 and telomerase RNA indicate that U•A-U triples can be replaced with isosteric C•G-C triples and vice versa[Citation28,Citation29,Citation30]. This suggests that in a three-dimensional structural context, the effective pKa of the Hoogsteen-paired cytosine becomes higher than in isolation. This phenomenon has been demonstrated to occur within DNA triple helices before [Citation31].
The in vitro Xrn1 degradation assays performed in this study shed light unto the main principles behind the structures that may be formed by xrRNA sequences of TBFVs and NKVFVs. From that information alone we cannot say that the effect of the tested mutations would also convey similar effects in vivo. However, the Northern blot analysis of MODV mutants does completely agree with the in vitro results. This analysis further indicated a second band, possibly corresponding with a second putative xrRNA site in the 3ʹ UTR of MODV. Unlike MBFV or TBFV viral species, NKVFVs are not known to carry as many, if any, structural duplications in their 3ʹ UTR [Citation4], although we were able to structurally align and model two stalling sites for APOIV (Supplementary Figure S2). It is however likely that structures of another type are capable of stalling Xrn1 as well, as is evident for instance from Northern blot analyses on TBEV and West Nile virus 3ʹ UTRs, revealing at least three Xrn1 stalling sites for both species, while they carry only two motifs consistent with the one studied here [Citation12,Citation19,Citation32]. Since the region downstream of MODV xrRNA1 contains other dumbbell and stem-loop structures of which equivalents can be found in MBFV and TBFV 3ʹ UTRs [Citation25], one of these is likely responsible for the second stalling site.
The structural models and mutants investigated in this study highlight several parameters that seem to be conserved within flaviviral xrRNA. For instance, while the number of base pairs forming stems α and δ varies, added together they always total 8–10 base pairs in a stacked manner. Furthermore, our structural models show that bulges or mismatches are never present between the β and γ stems. The MODV β stem mutants tested for Xrn1-stalling capacity indeed indicate that these stems have to be coaxially stacked to confer structural integrity. In contrast, the junction formed between of the β and ε stems is characterized by mismatches or non-Watson-Crick base pairs. A G-A pair is almost invariantly present at the top of the ε stem throughout the flavivirus genus. Interestingly, changing this pair into a conventional Watson-Crick base pair completely disturbs Xrn1 resistance in TBEV (Supplementary Figure S4). Furthermore, optimizing base pairing of the nucleotides flanking the β stem yields the same effect. These, and other structural parameters that we have obtained in this study contributed to the development of an algorithm [Citation33] that may aid the identification of novel, flavivirus-like xrRNA sites in other viruses belonging to the Flaviviridae family or even beyond.
Supplemental Material
Download MS Word (2.7 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- Calisher CH, Gould EA. Taxonomy of the virus family Flaviviridae. Adv virus res. 2003:59:1–19.
- Smith DB, Becher P, Bukh J, et al. Proposed update to the taxonomy of the genera hepacivirus and pegivirus within the Flaviviridae family. J Gen Virol. 2016;97:2894–2907.
- Smith DB, Meyers G, Bukh J, et al. Proposed revision to the taxonomy of the genus pestivirus, family Flaviviridae. J Gen Virol. 2017;98:2106–2112.
- Villordo SM, Carballeda JM, Filomatori CV, et al. RNA structure duplications and flavivirus host adaptation. Trends Microbiol. 2016;24:270–283.
- Ng W, Soto-Acosta R, Bradrick S, et al. 5′ and 3′ untranslated regions of the flaviviral genome. Viruses. 2017;9:137.
- Khromykh AA, Meka H, Guyatt KJ, et al. Essential role of cyclization sequences in flavivirus RNA replication. J Virol. 2001;75:6719–6728.
- Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239.
- Basu M, Brinton MA. West Nile virus (WNV) genome RNAs with up to three adjacent mutations that disrupt long distance 5′-3′ cyclization sequence basepairs are viable. Virology. 2011;412:220–232.
- Pijlman GP, Funk A, Kondratieva N, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591.
- Funk A, Truong K, Nagasaki T, et al. RNA structures required for production of subgenomic flavivirus RNA. J Virol. 2010;84:11407–11417.
- Silva PAGC, Pereira CF, Dalebout TJ, et al. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol. 2010;84:11395–11406.
- Chapman EG, Moon SL, Wilusz J, et al. RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. Elife. 2014;2014:1–25.
- Bidet K, Garcia-Blanco MA. Flaviviral RNAs: weapons and targets in the war between virus and host. Biochem J. 2014;462:215–230.
- Clarke BD, Roby JA, Slonchak A, et al. Functional non-coding RNAs derived from the flavivirus 3ʹ untranslated region. Virus Res. 2015;206:53–61.
- Moon SL, Dodd BJT, Brackney DE, et al. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology. 2015;485:322–329.
- Moon SL, Anderson JR, Kumagai Y, et al. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. Rna. 2012;18:2029–2040.
- Moon SL, Blackinton JG, Anderson JR, et al. XRN1 stalling in the 5ʹ UTR of hepatitis C virus and bovine viral diarrhea virus is associated with dysregulated host mRNA stability. PLOS Pathog. 2015;11:e1004708.
- Bekal S, Domier LL, Gonfa B, et al. A novel flavivirus in the soybean cyst nematode. J Gen Virol. 2014;95:1272–1280.
- MacFadden A, O’Donoghue Z, Silva PAGC, et al. Mechanism and structural diversity of exoribonuclease-resistant RNA structures in flaviviral RNAs. Nat Commun. 2018;9:1–11.
- Chapman EG, Costantino DA, Rabe JL, et al. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344:307–310.
- Akiyama BM, Laurence HM, Massey AR, et al. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science. 2016;354:1148–1152.
- Coleman TM, Wang G, Huang F. Superior 5ʹ homogeneity of RNA from ATP-initiated transcription under the T7 φ2.5 promoter. Nucleic Acids Res. 2004;32:e14.
- Silva PAGC, Molenkamp R, Dalebout TJ, et al. Conservation of the pentanucleotide motif at the top of the yellow fever virus 17D 3′ stem-loop structure is not required for replication. J Gen Virol. 2007;88:1738–1747.
- Meinkoth J, Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984;138:267–284.
- Leyssen P, Charlier N, Lemey P, et al. Complete genome sequence, taxonomic assignment, and comparative analysis of the untranslated regions of the Modoc virus, a flavivirus with no known vector. Virology. 2002;293:125–140.
- Zhang Q-Y, Li X-F, Niu X, et al. Short direct repeats in the 3ʹ untranslated region are involved in subgenomic flaviviral RNA production. J Virol. 2020;94:e01175–19.
- Xodo LE, Manzini G, Quadrifoglio F, et al. Effect of 5-methylcytosine on the stability of triple-stranded DNA - a thermodynamic study. Nucleic Acids Res. 1991;19:5625–5631.
- Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15:634–640.
- Wilusz JE, JnBaptiste CK, Lu LY, et al. A triple helix stabilizes the 3ʹ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407.
- Brown JA, Valenstein ML, Yario TA, et al. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–19207.
- Asensio JL, Lane AN, Dhesi J, et al. The contribution of cytosine protonation to the stability of parallel DNA triple helices. J Mol Biol. 1998;275:811–822.
- Göertz GP, Fros JJ, Miesen P, et al. Noncoding subgenomic flavivirus RNA is processed by the mosquito RNA interference machinery and determines West Nile virus transmission by culex pipiens mosquitoes. J Virol. 2016;90:10145–1059.
- Zammit A, Helwerda L, Olsthoorn RCL, et al. A database of flavivirus RNA structures with a search algorithm for pseudoknots and triple base interactions. Bioinformatics. 2020. DOI:10.1093/bioinformatics/btaa759
