ABSTRACT
The protein domain arrangement known as the La-module, comprised of a La motif (LaM) followed by a linker and RNA recognition motif (RRM), is found in seven La-related proteins: LARP1, LARP1B, LARP3 (La protein), LARP4, LARP4B, LARP6, and LARP7 in humans. Several LARPs have been characterized for their distinct activity in a specific aspect of RNA metabolism. The La-modules vary among the LARPs in linker length and RRM subtype. The La-modules of La protein and LARP7 bind and protect nuclear RNAs with UUU-3ʹ tails from degradation by 3ʹ exonucleases. LARP4 is an mRNA poly(A) stabilization factor that binds poly(A) and the cytoplasmic poly(A)-binding protein PABPC1 (also known as PABP). LARP1 exhibits poly(A) length protection and mRNA stabilization similar to LARP4. Here, we show that these LARP1 activities are mediated by its La-module and dependent on a PAM2 motif that binds PABP. The isolated La-module of LARP1 is sufficient for PABP-dependent poly(A) length protection and mRNA stabilization in HEK293 cells. A point mutation in the PAM2 motif in the La-module impairs mRNA stabilization and PABP binding in vivo but does not impair oligo(A) RNA binding by the purified recombinant La-module in vitro. We characterize the unusual PAM2 sequence of LARP1 and show it may differentially affect stable and unstable mRNAs. The unique LARP1 La-module can function as an autonomous factor to confer poly(A) protection and stabilization to heterologous mRNAs.
INTRODUCTION
Central aspects of mRNA metabolism are determined by the length of the 3ʹ poly(A) tail (PAT) and the cytoplasmic poly(A) binding protein (PABP, PABPC1) that binds to it [Citation1,Citation2]. PABP is a translation factor that also binds eukaryotic initiation factor eIF4G, physically linking the mRNA 5ʹ initiation factors with the 3ʹ PAT [Citation3–5].
Beyond its influence on stabilizing translation initiation complexes, PABP appears to be dynamically involved in the control of mRNA deadenylation and associated mRNA turnover(reviewed in [Citation2,Citation6,Citation7]). This aspect of PABP activity requires its conserved C-terminal MLLE domain which contains a binding site for PABP-interacting motif-2 (PAM2) peptides. PAM2 motifs are found in multiple proteins that promote or attenuate deadenylation, either generally or in an mRNA or pathway specific manner [Citation2,Citation6]. PAM2-containing proteins most relevant to this topic are the PAN3 deadenylase subunit of the PAN2/3 complex, the Tob1 and Tob2 protein interaction partners of CCR4/NOT1 [Citation8,Citation9], the Ccr4 and Caf1 deadenylase complex [Citation10–12], and the poly(A) mRNA stabilization factors LARP4 [Citation13–15] and LARP4B [Citation16].
Precursor mRNAs are polyadenylated in the nucleus and, after further processing, exit to the cytoplasm with PATs of ~250 nt. Deadenylation rates vary depending on the presence or absence of mRNA-specific sequences. Thus, deadenylation and decay of total mRNA reflect the combined effects of gene-specific, multifaceted kinetics involving several deadenylase activities and modulators thereof [Citation2,Citation17–19]. Stable mRNAs, e.g., those with short UTRs that lack destabilizing elements are deadenylated and degraded by a default pathway [Citation17,Citation18]. Unstable mRNAs typically contain destabilizing sequences in their 3ʹ UTRs such as AU-rich elements (ARE) or complementary sites for miRNAs that are recognized by specific binding proteins. These in turn recruit the CCR4/NOT1 deadenylases that accelerate mRNA decay. The stable ribosomal protein (rp)-mRNAs and the GFP reporter mRNA typically have half-lives of ~12 hours compared to ~60 minutes for unstable mRNAs such as c-Fos and the β-blobin-ARE reporter [Citation20–22].
LARP4 (aka LARP4A) is a cytoplasmic mRNA stabilization protein that binds poly(A) [Citation13]. Its La-module is related to those in nuclear La protein and LARP7 both of which bind and protect the 3ʹ oligo(U) tails of RNAs transcribed by RNA polymerase III from degradation. LARP4 poly(A) length protection and associated mRNA stabilization activity requires its two PABP-interaction motifs [Citation15]. Transcriptome-wide single-molecule poly(A) tail sequencing (SM-PAT-seq) indicate that LARP4 protects mRNA from deadenylation [Citation14]. Kinetic analysis suggests that LARP4 opposes deadenylation when mRNA-PATs are relatively short and PABP is sensitive to dissociation [Citation14].
LARP1 is a cytoplasmic La-module containing protein that represses translation of mRNAs with a 5ʹTOP (terminal oligopyrimidine) motif in response to reduction in certain signalling pathways [Citation23–25]. LARP1 also stabilizes TOP mRNAs [Citation26,Citation27] and was shown to have activities for poly(A) length protection and mRNA stabilization in the assays used to characterize LARP4 [Citation15].
Using assays for stable and unstable reporter mRNAs as well as endogenous cellular rp-mRNA, we show here that the poly(A) length protection and mRNA stabilization activities of LARP1 are intrinsic to its La-module. We redefine the sequence of the LARP1 PAM2 motif by uncovering its functionally important N-terminal phenylalanine, and show that it mediates high-affinity binding to the MLLE of PABP. Mutation of the PAM2 motif decreases LARP1 La-module activity for mRNA stabilization and PABP binding. Thus, the LARP1 La-module contains a bona fide PAM2 involved in mRNA poly(A) metabolism. This is a novel architecture among La-modules that can protect mRNAs from deadenylation and consequent destabilization as an autonomous trans-acting unit. The data suggest potential for general activity in poly(A) metabolism relevant to large sets of LARP1-associated mRNAs [Citation15,Citation24,Citation26–36]. Although LARP1 and LARP4 are very divergent beyond their La-modules, they appear to share similar activities related to mRNA poly(A) 3ʹ end metabolism.
RESULTS
The poly(A) mobility shift and mRNA stabilization assays
As reviewed by ourselves [Citation7] and others [Citation20,Citation21], Tet-Off promoters were developed to characterize poly(A) length and decay/stability of β-globin mRNA reporters with and without a destabilizing ARE in their 3ʹUTR. Newly synthesized mRNA from transfected β-globin genes and partial deadenylated intermediates were assessed by their relative mobility on northern blots [Citation20,Citation21]. Such assays were fundamental to studies that led to the biphasic model of deadenylation and decay [Citation20,Citation21] (see [Citation7]).
A Tet-Off β-globin with the TNFα ARE in its 3ʹUTR (βG-ARE) had revealed that increased accumulation levels reflects increased stabilization of the mRNA [Citation15]. Using this assay, it was shown that LARP4-mediated mRNA stabilization is accompanied by the apparent lengthening of poly(A) on the mRNA, although such length effects are more robustly manifested on the stable GFP mRNA reporter [Citation15]. In transfection assays, PATs are longer in LARP4 expressing cells because the mRNAs undergo slower deadenylation than in control cells [Citation15]. Consistent with this, mRNAs in mouse cells genetically deleted of LARP4 have shorter PATs than in the wild-type cells and decay faster [Citation14]. PAT length protection and mRNA stabilization require the PABP-interaction motifs of LARP4 [Citation15].
While poly(A) lengthening is most robust and tractable on GFP mRNA, the βG-ARE mRNA is appropriate for quantification of stabilization activity. βG-ARE is a reliable reporter of stability because of its short half-life and the relationship of this to the levels of its accumulation. The steady-state level of an RNA reflects its synthesis and decay rates. At uniform synthesis, achieving steady state requires 5–6 half-lives, at which point the ratio of RNA levels at steady state should equal the ratio of the RNA half-lives [Citation37,Citation38]. With a half-life of 75–90 min [Citation15,Citation39] the βG-ARE mRNA is a reporter whose accumulation levels can reliably serve as a proxy for mRNA stabilization in 48 h transfection experiments [Citation15]. The stable GFP mRNA also responds to stabilizing effects, e.g., higher accumulation with LARP4 but not LARP4 mutated proteins [Citation15], although depending on the extent of further stabilization and half-life extension, may not achieve steady state.
While the βG-ARE mRNA is reliable for use in transfection experiments, its degree of mobility shift due to poly(A) length is not well correlated with its stabilization [Citation15]. Yet this is not unexpected for an unstable mRNA, whose lifespan is dominated by irregular and asynchronous deadenylation kinetics that are known to not correlate with decay [Citation17] (recently reviewed in [Citation7]). Specifically, the TNFα ARE in βG-ARE is recognized by TTP which recruits the CCR4-NOT1 deadenylase to the mRNA [Citation40,Citation41]. Deadenylation by CCR4-NOT1 is asynchronous and PAT shortening is very difficult to correlate with mRNA decay [Citation17] even by high-resolution methods [Citation10,Citation42], although it could be modelled[Citation43].
The biphasic deadenylation model can also explain why poly(A) protection by LARP4 is more robustly detected and tractable on stable mRNA. GFP mRNA does not have an ARE to recruit deadenylases. Accordingly long PATs protected from deadenylation endure longer in the first phase of deadenylation (see in reference 7), and shorten slower relative to unstable mRNA [Citation15].
The LARP1 La-module mediates poly(A) length protection and mRNA stabilization
Each of three LARP1 constructs () were examined using the above assays for mRNA poly(A) protection and stabilization, in their wild-type form and with an F428A mutation previously shown to decrease PABP binding [Citation27]. F428A is located in the putative PAM2 sequence, which begins in the linker downstream of the LaM and overlaps with the predicted RRM of LARP1. The constructs were examined along with LARP4 and La protein as positive and negative controls [Citation15]. The LARP1 numbering corresponds to isoform 1 (1019 a.a.); construct 310–540 represents the La-module that binds oligo(A)-25 in vitro [Citation44] whose C-terminal region is predicted to contain an extended α-helix (Discussion) whereas construct 205–509 includes the upstream conserved Tri-RG region defined as ≥3 RG (arginine-glycine) pairs separated by 0–4 residues [Citation45] which in LARP1 contains eight RGs [Citation46]. The six constructs were examined in HEK293 cells containing endogenous LARP1 and in HEK293T LARP1-KO cells in which the LARP1 gene was nullified by CRISPR/Cas9 gene editing [Citation24,Citation47]. We first describe data from HEK293 cells containing endogenous LARP1.
Figure 1. LARP1 La-module confers poly(A) length protection and mRNA stabilization. A) Schematic representation of full-length LARP1 and two La-module constructs used in this study. Amino acid (aa) numbering is according to isoform 1 (1019 aa) for the full length and the shorter constructs. The PAM2 sequence is depicted in the linker ten amino acids following the end of the LaM (see in [Citation71]) and extending into the predicted RRM. RG indicates the position of the multiple Arg-Gly repeats [Citation46] (Tri-RG region [Citation45]); CR1 and CR2 indicate the conserved regions [Citation46]. B) Northern blot analysis of total RNA isolated from HEK293 cells transfected with the constructs indicated above the lanes. A single blot was probed for βG-ARE, GFP, and histone H2A mRNAs as indicated in panels i–iii). Bottom panel shows a vertically compacted image of the EtBr stained gel prior to transfer. C) Northern blot of RNAs from HEK293T LARP1-KO cells, as in B above. D) Quantitation of βG-ARE mRNA from duplicate northern blot data from HEK293T LARP1-KO cells. Transcript levels were normalized by VA1 small ncRNA. N = 2; error bars represent the spread. E) Western blot of total protein from the same cells used for RNA shown in panel E. F) The same blot as in C showing samples from a duplicate experiment, probed for the endogenous cellular rpL35 mRNA
![Figure 1. LARP1 La-module confers poly(A) length protection and mRNA stabilization. A) Schematic representation of full-length LARP1 and two La-module constructs used in this study. Amino acid (aa) numbering is according to isoform 1 (1019 aa) for the full length and the shorter constructs. The PAM2 sequence is depicted in the linker ten amino acids following the end of the LaM (see Fig. 5 in [Citation71]) and extending into the predicted RRM. RG indicates the position of the multiple Arg-Gly repeats [Citation46] (Tri-RG region [Citation45]); CR1 and CR2 indicate the conserved regions [Citation46]. B) Northern blot analysis of total RNA isolated from HEK293 cells transfected with the constructs indicated above the lanes. A single blot was probed for βG-ARE, GFP, and histone H2A mRNAs as indicated in panels i–iii). Bottom panel shows a vertically compacted image of the EtBr stained gel prior to transfer. C) Northern blot of RNAs from HEK293T LARP1-KO cells, as in B above. D) Quantitation of βG-ARE mRNA from duplicate northern blot data from HEK293T LARP1-KO cells. Transcript levels were normalized by VA1 small ncRNA. N = 2; error bars represent the spread. E) Western blot of total protein from the same cells used for RNA shown in panel E. F) The same blot as in C showing samples from a duplicate experiment, probed for the endogenous cellular rpL35 mRNA](/cms/asset/bb110ceb-915e-4f00-8534-06292115723a/krnb_a_1860376_f0001_b.gif)
The LARP1 constructs were cotransfected into HEK293 cells along with pre-mixed plasmids for βG-ARE mRNA and GFP mRNA. A northern blot probed for βG-ARE, GFP, and endogenous histone H2A mRNA is shown in , panels i–iii. H2A is a non-polyadenylated control. The ethidium-stained gel shows no evidence of generalized degradation. βG-ARE mRNA accumulated to different levels in the presence of different LARP1 constructs reflecting differential stabilization and mobility shifts reflecting poly(A) lengths. To help define a basal unshifted state, two ‘no shift control’ and a La sample were loaded in lanes 1, 10 and 9. Supplementary Fig S1A shows quantitation of βG-ARE mRNA levels from three replicate experiments; Supp Fig S1B shows a western blot of the La-modules with expected sizes.
The representative blot in and triplicate quantitative data in Supp. Fig S1A showed that LARP1 (lane 2) increased βG-ARE mRNA levels relative to the La control (lane 9) and this was diminished by F428A. The 205–509 La-module (lane 4) also increased βG-ARE mRNA levels and with substantial reduction by the F428A mutation (lane 6). The 310–540 module exhibited no significant effect on βG-ARE mRNA levels although the F428A led to lower levels.
Certain aspects of the mobility shifts are noteworthy. Signal intensity in the bands representing the βG-ARE and GFP mRNAs are informative as their upper and lower limits reflect different fractions of poly(A) lengths, long and short [Citation15,Citation21]. LARP4 leads to upward shifts in both the upper and lower edges of βG-ARE and GFP mRNAs, as does LARP1, relative to the no shift control and La (, lanes 2, 8, 11 vs. 1, 9, 10). Comparison of lanes 4 and 5 revealed that the upper fraction of poly(A) lengths of the βG-ARE and GFP mRNAs were differentially protected more by La-module 310–540 than by the 205–509 module. From another perspective, La-module 310–540 did not protect the shorter length poly(A) fraction as well as the 205–509 module; for βG-ARE mRNA this is associated with less accumulation () which may be functionally relevant because deadenylation of short poly(A) is associated with mRNA decay. For the 310–540 module, F428A in which binding to PABP is impaired (below), the βG-ARE mRNA was more down-shifted as compared to 310–540 WT (lanes 5 vs. 7).
It is notable here as the first data in support of a recurrent theme, that while La-module 310–540 has less activity for βG-ARE mRNA relative to the 205–509 La-module, their relative activities are reversed for GFP mRNA (, compare lanes 4 and 5). This is evidence that these La-module constructs that differ at both termini exhibit differential activities for the βG-ARE mRNA and the GFP mRNA which represent unstable and stable mRNAs.
– shows data from analyses of the six constructs in the HEK293T LARP1-KO cells [Citation24,Citation47]. The LARP1-KO cells had been shown to lack mTORC1-mediated translational regulation of TOP mRNAs due to loss of LARP1 with no change in mTOR, Raptor or eIF4E levels [Citation24,Citation47]. For these cells, a plasmid with the gene for the VA1 small noncoding (nc)RNA was included with plasmids for βG-ARE mRNA and GFP mRNA; VA1 is a non-polyadenylated ncRNA transcribed by RNA polymerase III and used to gauge transfection efficiency. A northern blot probed for the different RNAs is shown in , panels i–v. La protein binds and protects VA1 RNA from decay, leading to higher levels when La is over-expressed [Citation15]. We found that the relative activities of the different LARP1 constructs for βG-ARE mRNA accumulation levels were somewhat similar in these cells to those in HEK293 cells. Most importantly, the F428A PAM2 mutation decreased βG-ARE mRNA levels for each of the three constructs in the HEK293T LARP1-KO cells ().
The differential activities exhibited by LARP1 and the La-modules for βG-ARE and GFP mRNAs observed in HEK293 was also seen in the LARP1-KO cells (, D). Again, the 205–509 La-module stabilized more βG-ARE mRNA than the 310–540 La-module (, lanes 4–7). A contrast between the two La-modules derives from and is emphasized by the F428A mutation. Although for both La-modules the amounts of mRNAs are decreased by F428A, more of what remains in the 310–540 F428A samples appears as the longer poly(A) fraction, whereas the mRNAs in the 205–509 F428A samples appear more shortened (, lanes 4–7). Again, another perspective is that the 205–509 La-module protects the shorter poly(A) mRNA and this is associated with greater mRNA abundance as compared to the 310–540 La-module which protects more of the longer poly(A) βG-ARE mRNA but not the shorter.
More striking is the contrast between the two La-module constructs on the stable vs. unstable mRNA. La-module 205–509 led to higher levels of βG-ARE mRNA than La-module 310–540, whereas this is opposite and reproducible for the GFP mRNA ( and C, compare lanes 4 and 5). Note that these data are internally controlled; there is more βG-ARE mRNA in lane 4 than in lane 5, but less GFP mRNA in lane 4 than in lane 5 on the same blot. The different La-modules preferentially stabilize a different one of the two mRNAs residing in the same cells ( and ).
Poly(A) protection of endogenous rp-mRNA by the LARP1 La-module is PAM2-dependent
The data above were obtained by following the poly(A) lengths of (transfected) reporter mRNAs. We next wanted to examine endogenous rp-mRNA, a known target of LARP1. Most rp-mRNAs are stable with long half-lives, efficiently translated and have short poly(A) tails [Citation2,Citation48]. With 5ʹ and 3ʹ UTR lengths each less than 50 nt, and a 375 nt ORF of high codon optimality, the rpL35 mRNA is typical of a short stable mRNA, bearing little if any information to signal its destabilization. Because the PATs of rp-mRNAs are short at steady state, detection of length differences among samples by gel shift can be technically challenging. It was previously found for LARP4-KO cells that examination of positive and negative samples in lanes side by side was most convincing [Citation15]. LARP4-KO cell rp-mRNAs had slightly faster gel mobility as compared to wild-type cells [Citation15] that was later shown by SM-PAT-seq of the same cells to be due to shorter poly(A) tails [Citation14].
We examined the endogenous cellular transcript rpL35 mRNA in HEK293T LARP1-KO cells after transfection with the LARP1 constructs (, duplicate experiments). A dashed rectangle was placed on the image to orient upper and lower boundaries of the rpL35 mRNA band mobilities. Lanes 1, 10 and 19 show that the mRNA from cells with an empty vector display a relatively low amount of long-to-short rpL35 mRNA. By comparison, the rpL35 mRNA from LARP1 transfected cells in lanes 2 and 11 are shifted up. The mRNA in LARP1-F428A cells are shifted down compared to LARP1 (, compare lanes 2 & 3 and 11 & 12). La-module 205–509 is shifted up relative to the F428A mutant (compare lanes 4 & 6).
Consistent with data on the stable GFP mRNA in , the F428A mutation had less effect on La-module 310–540 than on La-module 205–509 on the stable rpL35 mRNA (). This can be appreciated by comparing both the upper and lower edges in lanes 4–7 and 13–16.
PAM2-dependent La-module binding to PABP
The LARP1 La-module 205–509 was previously shown to co-immunoprecipitate (co-IP) PABP [Citation32]. We wanted to know if this La-module and La-module 310–540 which lacks the RG repeat region and is extended on its C-terminus exhibit stable binding to cellular PABP as monitored by co-IP, in a PAM2-dependent manner. Extracts from transfected HEK293 cells were immunoprecipitated using anti-FLAG beads in the presence of RNase I or mock treatment followed by western blotting with anti-PABP antibody. shows the extract inputs (lanes 1–7) and corresponding anti-FLAG IPs (lanes 8–21); quantified data are in . The anti-FLAG beads did not pull down PABP from the control extract lacking transfected LARP1 demonstrating absence of background signal for the IP conditions (, lanes 8, 15).
Figure 2. PAM2-dependent stable binding to PABP by the isolated La-modules. A) Western blot showing extracts used for immunoprecipitation (IP) by anti-FLAG IgG (input, lanes 1–7) and products of IP in the absence (lanes 8–14) and presence of RNase I (lanes 15–21). The extracts were isolated from HEK293 cells transfected with the Flag-tagged constructs indicated above the lanes. The antibodies used to detect proteins on the blots are indicated to the right of the panels. The bottom panel shows an EtBr stained gel of a fraction of the extract after the IP and processed for RNA purification. B) Quantitation of the results of duplicate experiments, the levels of PABP that co-IP with La-module 205–509 were set to 100% in both mock and +RNase I treated extract. The error bars represent the spread
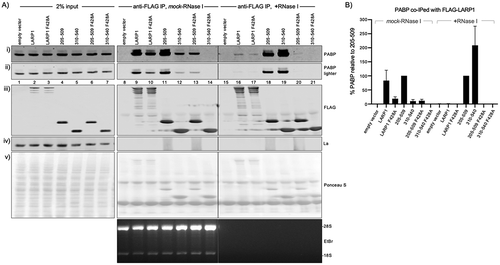
In the absence of RNase (, lanes 8–14), the amount of PABP immunoprecipitated with LARP1 was decreased by the PAM2 mutation F428A (, lane 9 vs. 10, ). PABP was co-IPed with the 205–509 La-module and the F428A mutation reduced this (, lane 11 vs. 13), both at a similar level as was found for LARP1 (). PABP was co-immunoprecipitated with the 310–540 La-module although to much less extent than with 205–509 and LARP1, yet F428A nullified the interaction (, lanes 12, 14, ).
Researchers have reported that association of some factors with LARP1 increased while other factors decreased after RNase treatment of the same extract [Citation29]. RNase I had differential effects on co-IP of PABP by the LARP1 constructs (, lanes 15–21, ). Unlike RNase A, RNase I readily cleaves poly(A) [Citation49]. The amount of PABP bound to LARP1 was decreased by extract treatment with RNase I relative to mock treatment whereas the amount bound to La-module 310–540 was much increased after RNase I treatment (, B). These data further distinguish the 310–540 La-module. This La-module may be bound by poly(A) in a way that renders it only partially accessible to binding by PABP or to IP (Discussion).
The data suggest that the La-module is largely responsible for mediating the mRNA poly(A) length protection and stabilization previously observed for LARP1 [Citation15] in a PAM2-dependent manner. F428A mutation in the PAM2 decreased this activity in LARP1, and in the La-module constructs.
The PAM2 F428A mutation does not inhibit oligo(A)-binding by the LARP1 La-module
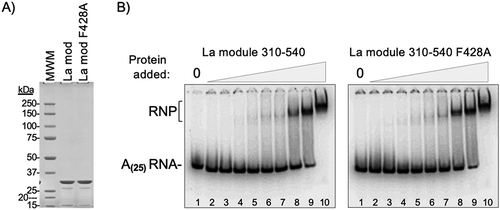
Because point mutation of consensus position 10 of the LARP4 PAM2w significantly decreases oligo(A) binding by the NTR [Citation50] we wanted to examine if the corresponding mutation F428A in LARP1 might affect oligo(A) binding by the isolated La-module. We chose to examine La-module 310–540 because it was shown to bind oligo(A) [Citation44] and lacks the RG repeats which are known to interact with RNA [Citation45]. Location of the PAM2 in the La-module of LARP1 suggests the possibility that its mutation might affect larger structural changes and/or inter-domain interactions important for RNA binding. Therefore, we compared 32P-oligo(A)(25) binding to recombinant purified La-modules 310–540 and 310–540(F428A) by EMSA [Citation44] (). The purified proteins are shown in , and EMSAs representative of highly reproducible results for the conditions used are in . These and other EMSAs using other conditions showed that both La-module proteins exhibited very similar binding avidity for 32P-oligo(A)(25). These data provide evidence that the PAM2 mutation F428A that impairs PABP binding does not interfere with the ability of the 310–540 La-module to bind poly(A).
Figure 3. The F428A PAM2 mutation does not inhibit oligo(A)-binding by the La-module. A) SDS PAGE analysis of the purified recombinant La-module 310–540 and La-module 310–540 F428A proteins used for RNA binding. The expected mass is 29.9 kDa; molecular weight markers (MWM) are indicated. B) RNA binding by Electrophoretic mobility shift analysis (EMSA). The positions of the free 32P-r(A)25 RNA and the RNA-protein (RNP) complex are indicated. The protein concentrations ranged from 0.03 μM to 7.5 μM indicated above the lanes as triangle ramps
Characterization of the LARP1 PAM2 sequence
The region in LARP1 attributed with PABP binding was identified as a PAM2-like motif localized to the La-module [Citation27]. The proposed sequence, SQL-LNCPEFVP was unusual because it contains a gap and a cysteine in place of an invariant alanine. Deletion of the proposed PAM2 sequence or mutation of phenylalanine F428A (F10 in the motif) decreased the amount of PABP that co-immunoprecipitated with LARP1 supporting the conclusion that the sequence acts as a PAM2 motif [Citation27]. In order to better understand this atypical PAM2, we used NMR spectroscopy and isothermal titration calorimetry (ITC) to study the binding of LARP1 sequences to the PABP MLLE domain.
Alignment of known PAM2s motifs showed that the sequences from eRF3 and NFX1 have an upstream phenylalanine (F1 in the motif) that could be aligned with LARP1 and 1B sequences. Including this residue allowed the LARP1 and LARP1B sequences to be aligned without gaps (). Since LARP1 PAM2 lacks a hydrophobic residue typically found at position 3, our analyses predicted that the initial phenylalanine (F1) would be critical for PABP-binding (Discussion). A comparison of the LARP1 and 1B sequences from vertebrates including fish, frogs, chicken and mammals [Citation46] confirmed the strict conservation of the initial phenylalanine and cysteine residue at position 7. Sequence LOGOs for vertebrate LARP1 and LARP1B sequences are shown in .
Figure 4. Characterization of a LARP1 PAM2 sequence that binds MLLE with high affinity. A) Multiple sequence alignment reveals distinctive features of the LARP1 PAM2 sequence. ClustalW alignment performed using MacVector (Methods). The PAM2 consensus numbering is indicated above the alignment. A separate alignment of the human TNRC6A/B/C sequences, offset according to Kozlov et al [Citation55] (see text) is below the main alignment; it includes the high-affinity MLLE-binding peptide of GW182/TNRC6C that was characterized in detail [Citation55]. B) Sequence LOGOs derived from the mammalian, opossum, and frog PAM2 LARP1 and LARP1B sequences compiled by Deragon [Citation46]. C) 15N-1H NMR correlation spectra (left) and ITC thermograms (right) of the wild-type PAM2 peptide (TDFSQLLNCPEFVPRQHYQKE) titrated into the PABP MLLE domain. Arrows in the NMR spectra show the shifts of two glycine signals upon PAM2 binding. The pattern of chemical shift changes in the MLLE domain is typical for high-affinity PAM2 binding. D) NMR and ITC titrations with the F428A mutant PAM2 peptide (TDFSQLLNCPEAVPRQHYQKE) show no binding. E) Titrations with a recombinant LARP1 fragment 399–540 containing both the PAM2 motif and full predicted RRM domain show slightly reduced binding affinity relative to the isolated PAM2 peptide. Nonetheless, the patterns of spectral changes are identical (arrows), which confirms that both the PAM2 peptide and PAM2-RRM domain bind to the same site on the MLLE domain
![Figure 4. Characterization of a LARP1 PAM2 sequence that binds MLLE with high affinity. A) Multiple sequence alignment reveals distinctive features of the LARP1 PAM2 sequence. ClustalW alignment performed using MacVector (Methods). The PAM2 consensus numbering is indicated above the alignment. A separate alignment of the human TNRC6A/B/C sequences, offset according to Kozlov et al [Citation55] (see text) is below the main alignment; it includes the high-affinity MLLE-binding peptide of GW182/TNRC6C that was characterized in detail [Citation55]. B) Sequence LOGOs derived from the mammalian, opossum, and frog PAM2 LARP1 and LARP1B sequences compiled by Deragon [Citation46]. C) 15N-1H NMR correlation spectra (left) and ITC thermograms (right) of the wild-type PAM2 peptide (TDFSQLLNCPEFVPRQHYQKE) titrated into the PABP MLLE domain. Arrows in the NMR spectra show the shifts of two glycine signals upon PAM2 binding. The pattern of chemical shift changes in the MLLE domain is typical for high-affinity PAM2 binding. D) NMR and ITC titrations with the F428A mutant PAM2 peptide (TDFSQLLNCPEAVPRQHYQKE) show no binding. E) Titrations with a recombinant LARP1 fragment 399–540 containing both the PAM2 motif and full predicted RRM domain show slightly reduced binding affinity relative to the isolated PAM2 peptide. Nonetheless, the patterns of spectral changes are identical (arrows), which confirms that both the PAM2 peptide and PAM2-RRM domain bind to the same site on the MLLE domain](/cms/asset/c56d6206-d762-4b8d-b3be-541bce134082/krnb_a_1860376_f0004_c.jpg)
NMR was used to measure binding of peptides containing different versions of the PAM2 sequence of LARP1. NMR is a sensitive technique for detecting even weak binding and has been used previously to characterize a wide variety of PAM2 sequences [Citation13,Citation51,Citation52]. The PABP MLLE domain was labelled with 15N and 2D 15N-1H correlation spectra acquired in the presence and absence of the LARP1 peptides. Addition of a LARP1 peptide, 417–437, TDFSQLLNCPEFVPRQHYQKE produced many large changes in MLLE spectrum indicating high-affinity binding. At the early steps of titration, a large number of MLLE signals disappeared, which indicates intermediate exchange kinetics and low micromolar affinity (Supp Fig S2). Upon reaching a 1:1 molar ratio of MLLE:peptide, the changes were essentially complete and no further peak shifts were observed with a 1:2 excess of peptide (). The saturation at the 1:1 ratio suggests the Kd is significantly below the MLLE protein concentration of 100 μM in the assay. In contrast, the substitution of phenylalanine by alanine, corresponding to LARP1 F428A, completely blocked binding with negligible changes in the spectrum even at a 1:2 molar ratio (). This is in agreement with previous studies that have shown the critical role of position-10 in the PAM2 motif for MLLE binding [Citation8,Citation53–55].
To assess the importance of the initial phenylalanine (F1) of the PAM2 motif, additional NMR experiments were carried out with shorter peptides missing the phenylalanine and two preceding residues (Supp Fig S3). When peptides corresponding to LARP1 (420–437) and LARP1B (311–328) were added to 15N-labelled MLLE domain, the spectra showed chemical shift changes but the changes did not saturate even at the highest peptide concentrations tested (0.5–0.6 mM). This allows us to estimate that the Kd of interactions is above 100 μM and conclude that the phenylalanine residue at the N-terminus of the PAM2 motif is required for optimal binding. The effects of mutating this residue (F419) on mRNA stability and the LARP1-PABP interaction in cells are presented below.
To obtain more precise measurements of the binding affinity, we turned to ITC experiments. Titration of the LARP1 PAM2 peptide (417–437) into a cell containing the MLLE domain resulted in a heat release consistent with binding (, right panel). The fit of heat changes measured a Kd of 3.8 μM, which is typical for PAM2-MLLE binding [Citation6]. An ITC titration with the F428A PAM2 peptide produced no heat of binding in agreement with the NMR result that the F428A mutation abrogates the interaction ().
Next, we addressed the question of whether the LARP1 PAM2 is functional in the context of the predicted RRM domain. This is important because PAM2 motifs typically occur in unstructured regions and the LARP1 PAM2 motif overlaps with the N-terminus of the predicted RRM domain. We expressed and purified LARP1 399–540 construct and used it in the NMR titration with the 15N-labelled MLLE domain. Addition of LARP1 399–540 produced nearly identical shifts in the MLLE NMR spectrum as observed with the isolated PAM2 peptide (). An ITC titration with the same LARP1 fragment confirmed binding albeit with a slightly weaker affinity than the free PAM2 peptide. These results substantiate that the LARP1 PAM2-motif is functional and able to bind PABPC1 in the context of the full-length proteins.
The LARP1 PAM2 position 1 is critical for PABP interaction
The F419A mutation in LARP1 (F496 by isoform-2, 1096 aa, numbering) was introduced into LARP1 and the two La-modules. These were transfected with the premixed reporters into HEK293T LARP1-KO cells and assayed as for alongside the F428A and unmutated constructs. βG-ARE mRNA and GFP mRNA analysis by northern blotting as well as stable binding to cellular PABP by co-IP are shown in . The data indicate that F419 in the N-terminal part of the PAM2 and F428 in the C-terminal part each separately contribute to stabilization and accumulation of βG-ARE mRNA and GFP mRNA by LARP1 (–), and to stable binding to cellular PABP (). The data also support that LARP1-mediated poly(A) protection may have different effects on stable and unstable mRNAs.
Figure 5. LARP1 PAM2 position-1 F419 is critical for functional and physical PABP interaction. A) Northern blot analysis of total RNA isolated from HEK293T LARP1-KO cells transfected with the constructs above the lanes. A single blot was probed for βG-ARE, GFP, and histone H2A mRNAs as well as the VA1 small ncRNA as indicated for panels i–iv). The bottom panel shows an image of the EtBr stained gel prior to transfer. B & C) Quantification of duplicate northern blot data for βG-ARE mRNA and GFP mRNA from the HEK293T LARP1-KO cells. Transcript levels were normalized by VA1 small ncRNA. N = 2; error bars represent the spread. D) Western blot of protein isolated from HEK293 cells used for immunoprecipitation (IP) of transfected Flag-tagged LARP1 constructs indicated above the lanes and developed using antibodies to proteins or epitopes indicated to the right of the panels. The bottom panel shows an image of the EtBr stained gel (see text)
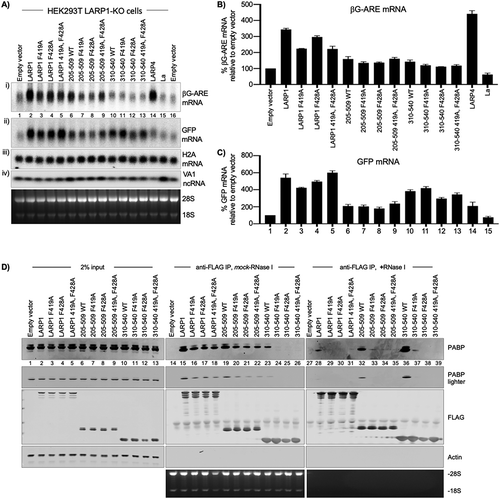
Intriguingly, the F419A mutation had differential effects in the two La-modules on the stable vs. unstable mRNAs (–). First, that a LARP can differentially affect stable vs. unstable mRNAs is most strikingly apparent by comparing effects of LARP4 on βG-ARE and GFP mRNA ( and C, and ), the detailed characterization of which will be described elsewhere (manuscript in preparation). The differential effects of the two La-modules on the unstable vs. stable mRNAs is also apparent (, compare panels i and ii, lanes 6–9 vs. 10–13).
Stabilization of βG-ARE mRNA by LARP1 was decreased by the single F419A mutation and by the single F428A mutation, whereas both together did not further decrease the stabilization (, panel i, lanes 2–5, ). Each of the single mutations also decreased βG-ARE mRNA levels in the two La-modules although at lower magnitude (, panel i, lanes 6–9 and 10–13, 5B).
The results differed for the stable GFP mRNA. While F419A alone or in combination with F248A in La-module 310–540 decreased βG-ARE mRNA levels, this same mutation did not have apparent effect or slightly increased GFP mRNA in the same cells (–, lanes 10–13). Evidence that the PAM2 mutations differentially affect stable vs unstable mRNA metabolism by full-length LARP1 was revealed by comparison of lanes 2–5 of –. One or the other PAM2 mutation decreased βG-ARE mRNA accumulation and combining both did not lead to further decrease (, B). However reprobing of the same blot for GFP mRNA (reproducibly) showed a different pattern for the PAM2 mutations than observed for βG-ARE mRNA; although each separate PAM2 mutation decreased GFP mRNA accumulation, combining both nullified the destabilization of the stable mRNA (, panels i and ii, ). We note that despite quantifications, such co-transfection experiments are not well suited for obtaining high statistical significance of mRNA levels. However, biological significance may be apparent by internal qualitative measures, e.g., two mutants that have opposite effects on different substrates. Therefore, we wish to emphasize that this type of significance is represented in lanes 4 and 5 of . Visual inspection readily reveals opposite relative amounts of the two mRNAs in the two LARP1 mutants in lanes 4 and 5 of in the same cells, supported by quantification (, C).
shows results of co-IPs ± RNase I. In the absence of RNase, F419A and F428A each separately decreased co-IP of PABP to nearly the same extent for LARP1 and for both La-modules (, lanes 14–26). Again, the amount of PABP that co-immunoprecipitated with La-module 310–540 dramatically increased with RNase treatment and this was much decreased by each of the PAM2 mutations (, lanes 36–39). The amount of PABP co-immunoprecipitated with LARP1 was decreased by RNase treatment and was further decreased by each of the PAM2 mutations (, lanes 28–31). The amount of PABP co-immunoprecipitated with La-module 205–509 was again the least affected by RNase, and was substantially reduced by each of the single PAM2 mutations (lanes 32–35). These data demonstrate that F419 of LARP1 at PAM2 position-1 is required for stable binding to PABP separate from the requirement for PAM2 position-10, F428.
DISCUSSION
A major conclusion of this work is that the poly(A) length protection and mRNA stabilization activities of LARP1 previously demonstrated [Citation15] have been localized to its La-module and to be impaired by point mutation of its intrinsic PAM2 (, , &, & B). Moreover, these activities can be conferred, albeit to variable extent by the isolated La-module in its two forms examined here, also in a PAM2-sensitive manner. The results fit with work from the Berman lab that showed that the La-module can bind oligo(A) with high affinity [Citation44]. The data also suggest the possibility that LARP1 may affect mRNA stability in a PAM2/PABP-independent manner (below).
Here, we employed the same β-globin-ARE mRNA and GFP mRNA reporter assays previously used to characterize LARP4 activity [Citation14,Citation15]. The data strengthen the idea that the mRNA poly(A) length protection and stabilization activities are shared to some extent by LARP1 and LARP415. Analysis by SM-PAT-seq showed that LARP4 protects, blocks or inhibits long and short poly(A) mRNAs from deadenylation, the latter in conjunction with PABP to account for its stabilization activity [Citation14]. We emphasize that while the activities may appear similar, LARPs 1 and 4 have diverged very considerably in their La-modules and more so in other regions. A challenge is to understand the mechanisms by which their activities may modulate deadenylation and the relevance in cells and animals in health and disease.
A second advance that resulted from this work is definition and functional characterization of the PAM2 sequence in LARP1. Previous analysis had overlooked the importance of phenylalanine-419, which we show is essential for high-affinity binding. Addition of F419 produced a proper alignment with other PAM2s (). The new peptide containing F419 exhibits high-affinity binding to the MLLE domain of PABP (3.8 μM) similar to other PAM2 peptides (, D), whereas the peptide lacking F419 exhibited markedly weaker affinity (Supp Fig S3). Importantly, single point mutation of F419 reduced poly(A) protection and βG-ARE mRNA stabilization activity by LARP1 as well as its stable association with cellular PABP ().
A limitation of the type of approach employed here is that it mostly used transfected reporters to represent stable and unstable mRNAs, and overexpressed protein constructs. Our reproducible data not shown indicate that full-length FLAG-LARP1 is expressed at levels very similar to endogenous LARP1 in the HEK293 cells, although the La-module fragments accumulate at higher levels. We also examined endogenous rpL35 mRNA in which case full-length FLAG-LARP1 led to poly(A) length protection in a PAM2-dependent manner as did the La-module constructs (). The results provide insight into the role of LARP1 in mRNA poly(A) metabolism, its La-module and nearby regions that had previously received relatively little attention.
Distinctive features of the LARP1 PAM2 sequence
The PAM2 sequence in LARP1 is unusual in several aspects (). The presence of cysteine at position-7 is unique to LARP1 and LARP1B and not found in any of the other 150 known and predicted PAM2 sequences [Citation56]. Instead, 90% of PAM2 motifs have an alanine residue at the position. Crystal structures of the several PAM2-MLLE complexes show that the alanine acts as a bridge between two contacts sites: an N-terminal site centred around the hydrophobic PAM2 residue at position-3 (typically leucine, but sometime phenylalanine or proline), and a C-terminal site centred around phenylalanine-10. A systematic survey of binding affinities of mutant peptides showed that the hydrophobic amino acids at these positions are the most critical for binding [Citation52] (reviewed in [Citation6]). The PAM2 motifs from LARP1 and LARP1B are the only examples where position-3 is occupied by a hydrophilic amino acid, glutamine (). The requirement for phenylalanine at position-1 in LARP1/1B may act to compensate for lack of hydrophobicity at position-3, perhaps similar to the C-terminal PAM2 motif from eRF3 [Citation8]. The PAM2 sequence from eRF3 is unique in that it consists of two overlapping PAM2 motifs both of which bind with low micromolar affinity. In the crystal structure of the eRF3-C-MLLE complex, the N-terminal phenylalanine projects forward to occupy the hydrophobic binding site on the MLLE domain that is normally occupied by the leucine at position-3 [Citation8]. The C-terminal motif eRF3-C, and motif from NFX1 are the only known sequences with a phenylalanine at position-1. Unlike eRF3, no consensus PAM2 sequence overlaps with the PAM2 motif we have characterized in LARP1. Substitution of phenylalanine-10 causes complete loss of binding () consistent with essential nature of the vertebrate-invariant phenylalanine at position-1.
The bipartite nature of the PAM2 motif can be seen in the sequence conservation of LARP1/1B sequences (). The N- and C-terminal residues are highly conserved with the greatest variability at positions-3 to 5. The highest homology with other PAM2s occurs at the C-terminal end; the sequence EFV/IP is also found in Paip2, eRF3-C, PAN3 and NFX1 (). LARP1/1B also appear to differ most from most other PAM2s in their N-terminal regions (). The most divergent PAM2 sequences occur in TNRC6 (GW182) proteins [Citation57]. Binding studies and crystal structures revealed that the human TNRC6C PAM2 peptides use only the C-terminal part of the PAM2 alignment to interact with the MLLE domain [Citation55,Citation58]. The absence of the N-terminal region is compensated by additional contacts mediated by residues C-terminal to the classical alignment of PAM2 sequences (). Nonetheless, as observed for the LARP1 PAM2, point mutation of the TNRC6 phenylalanine at position-10 abolished PABP binding and activity [Citation57].
With regard to potential regulation, it may be useful to categorize the Kd values of PAM2 peptides for the MLLE domain of PABPC1 into two groups. The high-affinity group includes PAM2s of translation factors eRF3 (1 μM), Paip1 (1.4 μM), and Paip2 (0.2 μM). The lower affinity group includes the Tob1/2 subunits of the CCR4-NOT1 deadenylase complex (16 μM), the PAN3 deadenylase (40 μM), LARP4 (22 μM), and USP10 (26 μM), all involved in mRNA stability, with Kd values of 16 to 40 μM6. By this categorization, the LARP1 PAM2 forms an intermediate class – roughly four-fold weaker than the translation factors and four-fold higher affinity than LARP4, Tob1, Tob2 and the PAN3 deadenylase.
The disparate activities of La-module 310–540 that were observed for the unstable βG-ARE vs. the stable GFP mRNA hint at bipartite functionality for the LARP1 PAM2 motif (–). It is tempting to suspect that the central unique cysteine may be involved in this hypothesized bipartite functionality. More tempting is the possibility that serine-S440 (517 by isoform-2 numbering) downstream of the PAM2 that becomes phosphorylated after infection by SARS-CoV-2 [Citation59] may be involved in poly(A) metabolism. Ser-440 occupies a position relative to LARP1 PAM2 that would be adjacent to the C-terminal region of the GW182/TNRC6C peptide that interacts with MLLE.
La-modules 205-509 and 310-540 exhibit differential properties and activities
Our investigation of poly(A) length protection by subregions of LARP1 localized this activity to La-module 205–509 which contains the multiple RG repeats. Because the Berman Lab showed that the isolated La-module 310–540, which excludes the RG repeat region, could bind poly(A) [Citation44], we chose to also examine it so that potential effects of the RG region may be distinguished. We discovered that the two La-module proteins exhibited more distinctive activities than expected. These La-modules differ as follows. 205–509 contains the RG repeats which are generally known as able to interact with RNA in other proteins [Citation45]. The C-terminal extension on 310–540 but absent from 205 to 509 is predicted to form an extended α-helix by the Rosetta protein structure prediction program [Citation60].
The data observed with the 310–540 La-module in conjunction with the PAM2 F419 (position-1) mutation reveals a distinguishing activity that also suggest a PABP-independent mode of mRNA stabilization that extends to LARP1. The 310–540 La-module was associated with a longer poly(A) fraction of mRNA as compared to the 205–509 La-module (, C, ) and was also less precipitable with PABP in the absence of RNase (, ). Moreover, the large change in co-IP with PABP before and after RNase treatment observed only for the 310–540 La-module may be related to the mRNA long poly(A) mobility data and that both may reflect its relative inaccessibility to deadenylases and PABP. These considerations may help interpret the otherwise puzzling effects of the PAM2 mutations on the mRNAs in . Examination of lanes 10–13 of the GFP blot in reveals that mutation of F428 in La-module 310–540 leads to a downshift of this stable mRNA whereas the double mutation does not. A similar pattern but with regard to GFP mRNA abundance is observed for full length LARP1 in lanes 2–5. These data raise the possibility that weakening PABP binding as with one PAM2 mutation renders an associated stable mRNA more accessible to recruitment of deadenylases whereas severing PABP association as with two PAM2 mutations may isolate the mRNA from destabilizing effects. We also note that the patterns observed are consistent with the possibility that the LARP1 PAM2 motif may have capacity to function in a polar or bipartite manner as discussed above.
As noted, some mRNAs may be stabilized with long poly(A) and their decay suspended, and TOP mRNAs may be preserved with long poly(A) [Citation42,Citation61]. Also, LARPs 1, 4 and 4B [Citation13,Citation62–64] as well as other PAM2 proteins involved in mRNA metabolism localize to stress granules (SGs) of which PABP is a well-established component [Citation65]. As TOP mRNAs have been localized to SGs [Citation66,Citation67], some of the poly(A)-PABP associations observed here for the isolated La-module may apply to the high local concentration environment characteristic of SGs [Citation68] and fit with multivalent RNA-binding by LARP1 as proposed [Citation44] and possibly with stabilization during translational repression [Citation25–27,Citation61].
Differential mRNA poly(A) length and accumulation of βG-ARE vs. GFP mRNAs raise the possibility that the RG repeats may modulate such activity of LARP1. Arginine-glycine (RG) repeats occur in various combinatorial complexities in a large number of RNA-binding proteins and have been well documented to serve as RNA-binding motifs [Citation45]. Flexibility of glycine is believed to contribute to the intrinsic disorder of these regions and to conformational adaptation to their target RNAs [Citation69]. Whether the RG repeat region of LARP1 is involved in RNA binding and/or the poly(A) length protection by the La-module remains to be determined. We note that the presence of a polypeptide tract with potential for RNA-binding and intrinsic disorder upstream of the LARP1 La-module is reminiscent of the poly(A)-binding NTR upstream of the LARP4 La-module [Citation50].
Other considerations regarding the LARP1 La-module
The β-strands and α-helices in the predicted RRMs in multiple sequences of LARP1 and LARP1B were reported more than 10 years ago [Citation70]. A multiple sequence alignment of La-motif C-termini-linker-RRM N-terminal regions of individual LARPs, based in part on three La-module high-resolution structures, was reported [Citation71]. According to this alignment the LARP1 PAM2 sequence begins at position 10 of the interdomain linker, consistent with the original predicted topology [Citation70]. The LARP1 PAM2 would extend through the predicted β1 of the predicted RRM [Citation70] although whether such structures exist remains unknown. It is nonetheless noteworthy that the LARP1 PAM2 would be positioned very similarly to the LARP6 Crm1-type nuclear export sequence LLVYDLYL [Citation72,Citation73] that partially resides on/overlaps with the β1 strand (KMLLV) of the LARP6 RRM structure [Citation74]. A nuclear export sequence in the La-module RRM1 of La protein was functionally mapped to the α1 helix connecting β1 and β2 [Citation75]. A nuclear retention element was mapped to α3 of La RRM2a [Citation75,Citation76].
Binding analysis of the PABP MLLE domain with the LARP1 PAM2 motif as a free peptide or in the context of the LARP1 fragment 399–540 which contains the entire predicted RRM were performed by NMR and ITC (, Supp Figs S2, S3). Because the results indicated that no more binding affinity was contributed by the additional predicted RRM, it would appear that the PAM2 may constitute the protein–protein interactions of LARP1 and PABP. Each binding separately to poly(A) in addition to their protein–protein interactions would create a higher apparent binding affinity. PABP exhibits 5–10 fold higher affinity for poly(A) 25ʹmer as compared to the LARP1 La-module [Citation44]. Nonetheless, affinity enhancement via poly(A) binding may explain the PABP that co-IPs from extracts with LARP1 F419A/F428A double mutations to the PAM2 motif ().
In conclusion, this work revealed a La-module containing a newly defined and unique PAM2, that builds upon an already known versatile module of the LARP family [Citation44]. The data have shown that the La-module with upstream RG repeats, a unique PAM2 and possibly followed by an (predicted) α-helix can confer poly(A) length protection and associated mRNA stabilization in a PABP-dependent manner. This La-module may have biologically relevant characteristics and regulatory potential.
MATERIALS AND METHODS
DNA constructs
The full-length LARP1 (isoform 1) and F428A (previously designated F505A [Citation27]) constructs were described in Fonseca et al [Citation27]. The 205–509 La-module described in Fonseca et al [Citation32], and its F428A-mutated form were ordered from Genewiz and subcloned into the HindIII and BamHI sites of the pFLAG-CMV2 vector (Sigma-Aldrich). The 310–540 La-module corresponding to that described by Al-Ashtal et al [Citation44] was amplified by PCR using full-length LARP1 and LARP1-F428A as templates and subcloned into the HindIII and BamHI sites of the pFLAG-CMV2 vector (Sigma-Aldrich). The F419A mutations were introduced by site-directed mutagenesis using the Q5 kit from New England Biolabs. All constructs were verified by Sanger sequencing. The pcDNA3.1-β-globin-TNFα-ARE construct was described [Citation15] as was pcDNA-TPGFP [Citation77].
La-module for EMSA. The 310–540 WT and F428A constructs in pFLAG-CMV2 were used as templates for PCR with primers GACAACTTACTCGAGAAACTTTTTGACAATGTCA-GCAGCACC (forward) and GTGTAGAATAAGCTTTTATGATCCTTAATCCATCTGCTC-CATC (reverse) containing XhoI and HindIII restriction sites. These were cloned into the XhoI and HindIII sites of bacterial expression vector pTEV-AV2 (derivative of pET28a) and verified by bidirectional sequencing. By this approach, the310–540 La-module is preceded by MGSSHHHHHHSSGENLYFQGHMLEKL and has a calculated molecular mass of 29,888 Da.
Transfection
HEK293 cells (5.5 × 105 per 6-well) or HEK293T LARP1KO cells (1x106 per 6-well) were seeded one day prior to transfection with Lipofectamine 2000 (Invitrogen) according to manufacturers’ instructions. Per 6-well, 5 μg of LARP plasmid was transfected and a pre-mixed batch was used to transfect each sample with 100 ng of pcDNA3.1-β-globin-TNFα-ARE, 100 ng of pcDNA-TPGFP and 100 ng of pVA1 as transfection controls where indicated. Since the shorter LARP1 fragments and La accumulate to much higher levels than LARP4 and full-length LARP1, less plasmid was added of those and the difference was made up to 5 μg with empty pFLAG-CMV2 plasmid. For La, 1 μg was transfected plus 4 μg pFLAG-CMV2. For the LARP1(205–509) and LARP1(310–540) fragments, 165- and 500 ng was transfected, respectively, plus 4.835- and 4.5 μg of pFLAG-CMV2, respectively. 24 hours post-transfection, cells were split 1:5 over multiple wells and harvested 24 hours after that for protein and RNA isolation. For HEK293T LARP1 KO cells [Citation24,Citation47] the only difference was that 1 × 106 cells per 6-well were seeded one day prior to transfection.
Northern blotting
Total RNA was isolated using Tripure (Roche), separated on a 1.8% agarose-formaldehyde gel and transferred to a GeneScreen-Plus membrane (PerkinElmer). After crosslinking with UV and vacuum-baking for 2 hours at 80°C, the membrane was prehybridized in hybridization solution (6 x SSC, 2 x Denhardt’s, 0.5% SDS and 100 μg/ml yeast RNA) for 1 hour at hybridization incubation temperature (Ti). 32P-labelled oligo probes were added, and hybridization was overnight at Ti. Oligo probes used and their Ti can be found in [Citation15].
Western blotting
To isolate protein samples, cells were washed twice with PBS containing protease inhibitors (Roche) and cell lysis was performed directly in RIPA buffer (Thermo Scientific) containing protease inhibitors (Roche). The lysate was sonicated 3 times for 30 seconds on high setting in a Bioruptor sonicator (Diagenode). Proteins were size separated using SDS-PAGE and transferred to a nitrocellulose membrane. Primary antibodies used were anti-FLAG (Sigma, F1804), anti-actin (Thermo Scientific, PA1-16,890), anti-LARP1 (Abcam 86,359), anti-PABP (Abcam, ab21060), anti-La (Go), and anti-GFP (Santa-Cruz, sc-9996). Primary antibodies were detected by secondary antibodies from LI-COR Biosciences, which were conjugated to either IRDye 800CW or 680RD and western blots were scanned using the Odyssey CLx imaging system (LI-COR Biosciences).
For quantification, the signals for PABP in the IP lanes were quantified using Image Studio Light (LiCOR) and for each set, mock-treatment and +RNase I treatment, the 205–509 IP sample was set to 100%. Two biological replicates were performed; the error bars represent the spread.
Preparation of total cell extract for immunoprecipitation
HEK293 cells were co-transfected with 5 μg FLAG-LARP1 construct and 100 ng pcDNA-TPGFP construct as described above per 6-well (2 wells for each condition). 24-hour post-transfection the cells from the 2 wells were combined in a 10 cm culture dish. 48 hours post-transfection the cells were washed with 20 ml warm PBS after which they were scraped in 10 ml ice-cold PBS and transferred to a conical tube on ice. Another 5 ml ice-cold PBS was used to rinse the culture dish, and this was added to the conical tube. Cells were spun down in a pre-chilled centrifuge for 3 mins at 1400 rpm. The supernatant was discarded, and the cells were lysed in 200 μl lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.25% NP-40, 0.5 mM EDTA, 2x protease inhibitors (Roche), 2 mM PMSF added fresh). Cells were incubated for 5 mins on ice with occasional flicking of the tubes. After lysis was confirmed by inspection under the microscope, 600 μl ice-cold wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA) was added. Lysates were then spun for 15 mins at 13,000 rpm at 4°C. The supernatants were transferred to a new tube and their total protein concentration was determined by BCA (Pierce). A fraction of the clarified lysate from each sample was used as input and the remainder was used for IP (2% = 10 μg).
Immunoprecipitation by anti-FLAG
50 μl of anti-FLAG M2 magnetic bead slurry (Sigma) was used per IP and washed 3 times with 1 ml of wash buffer. (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA). 500 μg total protein was added to the beads in 340 μl total volume (adjusted by wash buffer + 0.05% NP-40). To the samples that were treated with RNase I, 60 μl of RNase I (1500 U, ThermoFisher Scientific) was added. To the samples that were mock treated, 60 μl of RNase I buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.01% Triton X-100 and 50% (v/v) glycerol) was added. Samples were incubated at room temperature for 15 mins and then rotated end-over-end at 4°C for 90 minutes. To assess efficacy of RNase treatment, 100 μl of supe was taken for total RNA isolation by Maxwell (Promega) and the rest of the supe was discarded prior to washing of the beads. To the 100 μl of supe was added 200 μl of homogenization buffer containing thioglycerol (Maxwell 16 LEV simplyRNA purification kit, Promega) and the samples were vortexed. Then, the Maxwell 16 LEV simplyRNA protocol was followed using 200 μl of their lysis buffer and one sample per cartridge. RNA was eluted in 35 μl H2O and analysed on a denaturing formaldehyde-agarose gel.
The beads were washed 3 times with 500 μl wash buffer containing 0.05% NP-40. The supernatant was removed, and the beads were suspended in 12.5 μl SDS-PAGE sample buffer containing 8% β-mercaptoethanol, heated to 80°C for 3 mins, loaded and run on an SDS-PAGE gel, blotted and processed for western blotting as described above.
Purification of LARP1 La-module for EMSA
A 5 ml overnight culture of Rosetta™ (DE3) cells (Novagen) transformed by La-module 310–540 WT and F428A in pTEV-AV2-28a were inoculated into 500 ml LB (Luria broth) incubated with shaking at 225 rpm for 3 hours at 37°C then induced by addition of IPTG (isopropyl β-d-1-thiogalactopyranoside) to 0.5 mM. At this point the cultures were transferred into shaking incubator preset to 18°C and incubated for 20 hours cells after which the cells were collected by centrifugation, quick frozen in dry ice and stored at −80°C.
Protein was purified by HisTALON™ Gravity Column Purification kit 635,654 (TaKaRa Bio) according to the manufacturer’s provided protocol. The first two elution fractions containing high concentration protein were combined, an equal volume of saturated ammonium sulphate was added, and left overnight at 4°C. The precipitate was collected the next day by centrifugation for 30 minutes at 700 rpm. The soft pellet collected was diluted in protein storage buffer (0.25 mM Tris-HCl pH 8.0, 100 mM KCl, 1 mM DTT, 30% glycerol) and stored at −20°C.
RNA EMSA
RNA was obtained from Integrated DNA Technologies (IDT, Coralville, Iowa), labelled to specific activity of ~109 cpm/µg with 32Pγ-ATP (Amersham) by T4 polynucleotide kinase (3ʹ phosphatase minus, New England Biolabs) and PAGE purified prior to EMSA.
RNA binding. 2x EMSA binding buffer: 40 mM Tris, pH 8.0, 200 mM KCl, 10 mM 2-mercaptoethanol, 10% glycerol, containing 0.02% xylene cyanole and bromophenol blue gel tracking dyes. Final concentration of RNA in each reaction was ~1 nM (103 cpm), protein concentrations ranged from 0.03 μM to 7.5 μM. Binding reactions were incubated on ice for 10 min then loaded on pre-cooled 8% polyacrylamide nondenaturing tris-borate gels, and electrophoresed in a pre-cooled Mini gel system with pre-cooled tank buffer at 200 V for 30 min. The gel was transferred to Whatman filter paper, dried using a vacuum gel dryer and exposed to a PhosphorImager screen overnight and scanned on a Typhoon PhosphorImager. Quantification was performed using Image Quant software.
Proteins and peptides for NMR
The MLLE domain (residues 544–626) of human PABPC1 was expressed and purified as described previously [Citation54]. For NMR experiments, the recombinant protein was labelled by expressing in E. coli BL21 strain grown in M9 minimal medium with 15N-ammonium chloride as the sole source of nitrogen.
The LARP1 fragment (residues 399–540) was generated using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) and verified by bidirectional sequencing. The 310–540 WT construct in pTEV-AV2 was used as a template for PCR with the primer 5´-CATCATCATCATCACAGCAGGAGGGAGGAACCAG-3´. The LARP1 (399–540) was expressed in Escherichia coli BL21 (DE3) in rich (LB) medium. Upon reaching OD600 of 0.8, expression was induced by adding 1 mM IPTG and the culture was kept at 30°C for 4 hours with shaking. After harvesting, bacterial pellets were resuspended in binding buffer (50 mM HEPES, 500 mM NaCl, 5% glycerol, 5 mM imidazole, pH 7.6) and frozen at −20°C. Upon thawing, the protein was purified by affinity chromatography Ni2+-charged Sepharose beads followed by size-exclusion chromatography using a Superdex-75 column (GE Healthcare) into 10 mM MES pH 6.3, 100 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine. The protein concentration was calculated from UV absorbance at 280 nm using estimated extinction coefficient of 13,980 M−1cm−1. The 399–540 LARP1 construct is preceded by MGSSHHHHHHS and has a calculated molecular mass of 17,705 Da.
PAM2-like peptides TDFSQLLNCPEFVPRQHYQKE and TDFSQLLNCPEAVPRQHYQKE corresponding to LARP1 (417–437) and F428A mutant, respectively, were synthesized by Fmoc solid-phase peptide synthesis (GenScript, Piscataway, NJ). Peptides SQLLNCPEFVPRQHYQKE and SQLIDCPEFVPGQAFCSH corresponding to LARP1 (420–437) and LARP1B (311–328), respectively, were produced by Fmoc solid-phase peptide synthesis (Bio Basic, Markham, ON). All peptides were purified by reverse phase chromatography and verified by mass spectroscopy.
NMR spectroscopy
All NMR experiments were performed at 25°C using Bruker 600 MHz spectrometer with a TCI cryoprobe. NMR samples were in 10 mM MES pH 6.3, 100 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine. For NMR titrations, PAM2 peptides or LARP1 (399–540) were added to 0.1 to 0.2 mM 15N-labelled PABPC1 (544–626) to final molar ratios of 1:2 to 1:4 depending on the experiment. NMR spectra were processed using NMRPipe [Citation78] and analysed with SPARKY [Citation79].
Isothermal titration calorimetry. ITC experiments were performed on a MicroCal VP-ITC titration calorimeter (Malvern Instruments Ltd). The syringe was loaded with 500 μM PAM2 peptide or 300 μM LARP1 399–540, while the sample cell contained 30–35 μM MLLE domain. All experiments were carried out at 20°C with 19 injections of 15 μl with stirring at 310 rpm. Results were analysed using ORIGIN software (MicroCal) and fitted to a binding model with a single set of identical sites.
Multiple sequence alignment was performed by ClustalW using MacVector version 17.0.7. Default parameters for pairwise and multiple are matrix, Gonnet, open gap penalties of 10, with extend gap penalties of 0.1 and 0.2, respectively, and alignment speed, slow.
Abbreviations:
5ʹTOP: 5ʹ terminal oligopyrimidine, ARE: AU-rich element, IP: immunoprecipitation, KO: knockout, LaM: La motif, LARP: La-related protein, LARP1: La-related protein 1, MEFs: mouse embryonic fibroblasts, MLLE: Mademoiselle, NTR: N-terminal region, PABP: poly(A)-binding protein, PAM2: PABP-interacting motif 2, PAT: poly(A) tail, PIC: pre-initiation complex, RRM: RNA recognition motif, SG: stress granule, TE: translation efficiency, TOP: terminal oligopyrimidine, WT: wild-type.
Supplemental Material
Download PDF (2.2 MB)Acknowledgments
R.M. would like to thank Andrea Berman for helpful discussion regarding La-module 205-509. R. M. and S. M. thank Carson Thoreen for kindly providing HEK293T LARP1-KO cells, and members of The LARP Society for inspiration. This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Thompson MK, Gilbert WV. mRNA length-sensing in eukaryotic translation: reconsidering the “closed loop” and its implications for translational control. Curr Genet. 2017;63(4):613–620.
- Nicholson AL, Pasquinelli AE. Tales of detailed poly(A) Tails. Trends Cell Biol. 2019;29:191–200.
- Craig AW, Haghighat A, Yu AT, et al. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523.
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. Embo J. 1998;17:7480–7489.
- Uchida N, Hoshino S, Imataka H, et al. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J Biol Chem. 2002;277:50286–50292.
- Xie J, Kozlov G, Gehring K. The “tale” of poly(A) binding protein: the MLLE domain and PAM2-containing proteins. Biochim Biophys Acta. 2014;1839:1062–1068.
- Mattijssen S, Kozlov G, Fonseca BD, et al. LARP1 and LARP4: up close for mRNA 3ʹ poly(A) protection and stabilization. RNA Biol. 2020. In Press. g
- Kozlov G, Gehring K. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS One. 2010;5:e10169.
- Funakoshi Y, Doi Y, Hosoda N, et al. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148.
- Yi H, Park J, Ha M, et al. PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol Cell. 2018;70:1081–1088. e5
- Webster MW, Chen YH, Stowell JAW, et al. mRNA deadenylation is coupled to translation rates by differential activities of Ccr4-not nucleases. Mol Cell. 2018;70:1089–1100. e8
- Raisch T, Chang CT, Levdansky Y, et al. Reconstitution of recombinant human CCR4-NOT reveals molecular insights into regulated deadenylation. Nat Commun. 2019;10:3173.
- Yang R, Gaidamakov SA, Xie J, et al. LARP4 binds poly(A), interacts with poly(A)-binding protein MLLE domain via a variant PAM2w motif and can promote mRNA stability. Mol Cell Biol. 2011;31:542–556.
- Mattijssen S, Iben JR, Li T, et al. Single molecule poly(A) tail-seq shows LARP4 opposes deadenylation throughout mRNA lifespan with most impact on short tails. Elife. 2020;9:e59186
- Mattijssen S, Arimbasseri AG, Iben JR, et al. LARP4 mRNA codon-tRNA match contributes to LARP4 activity for ribosomal protein mRNA poly(A) tail length protection. Elife. 2017;6. DOI:10.7554/eLife.28889.
- Grimm C, Pelz JP, Schneider C, et al. Crystal structure of a variant PAM2 motif of LARP4B bound to the MLLE domain of PABPC1. Biomolecules. 2020;10. DOI:10.3390/biom10060872
- Yamashita A, Chang TC, Yamashita Y, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063.
- Chen CA, Shyu AB. Emerging themes in regulation of global mRNA turnover in cis. Trends Biochem Sci. 2017;42:16–27.
- Jalkanen AL, Coleman SJ, Wilusz J. Determinants and implications of mRNA poly(A) tail size–does this protein make my tail look big? Semin Cell Dev Biol. 2014;34:24–32.
- Xu N, Loflin P, Chen CY, et al. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 1998;26:558–565.
- Chen CY, Yamashita Y, Chang TC, et al. Versatile applications of transcriptional pulsing to study mRNA turnover in mammalian cells. RNA. 2007;13:1775–1786.
- Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA. 2011;2:167–183.
- Fonseca BD, Lahr RM, Damgaard CK, et al. LARP1 on TOP of ribosome production. Wiley Interdiscip Rev RNA. 2018;e1480. DOI:10.1002/wrna.1480
- Philippe L, van den Elzen AMG, Watson MJ, et al. Global analysis of LARP1 translation targets reveals tunable and dynamic features of 5ʹ TOP motifs. Proc Natl Acad Sci U S A. 2020;117:5319–5328.
- Haneke K, Schott J, Lindner D, et al. CDK1 couples proliferation with protein synthesis. J Cell Biol. 2020;219. DOI:10.1083/jcb.201906147.
- Aoki K, Adachi S, Homoto M, et al. LARP1 specifically recognizes the 3ʹ terminus of poly(A) mRNA. FEBS Lett. 2013;587:2173–2178.
- Fonseca BD, Zakaria C, Jia JJ, et al. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J Biol Chem. 2015;290:15996–16020.
- Merret R, Descombin J, Juan YT, et al. XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep. 2013;5:1279–1293.
- Hong S, Freeberg MA, Han T, et al. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. Elife. 2017;6:e25237
- Mura M, Hopkins TG, Michael T, et al. LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene. 2015;34:5025–5036.
- Berman AJ, Thoreen CC, Dedeic Z, et al. Controversies around the function of LARP1. RNA Biol. 2020;1–11. DOI:10.1080/15476286.2020.1733787
- Fonseca BD, Hollensen AK, Pointet R, et al. LARP1 is a major phosphorylation substrate of mTORC1. BioRxiv (Not Peer Reviewed). 2020.
- Zhang Y, Wang ZH, Liu Y, et al. PINK1 inhibits local protein synthesis to limit transmission of deleterious mitochondrial DNA mutations. Mol Cell. 2019;73:1127–1137.
- Zhang Y, Chen Y, Gucek M, et al. The mitochondrial outer membrane protein MDI promotes local protein synthesis and mtDNA replication. Embo J. 2016;35:1045–1057.
- Plissonnier ML, Cottarel J, Piver E, et al. LARP1 binding to hepatitis C virus particles is correlated with intracellular retention of viral infectivity. Virus Res. 2019;271:197679.
- To TL, Cuadros AM, Shah H, et al. A compendium of genetic modifiers of mitochondrial dysfunction reveals intra-organelle buffering. Cell. 2019;179:1222–1238. . e17
- Wada T, Becskei A. Impact of methods on the measurement of mRNA turnover. Int J Mol Sci. 2017;18. DOI:10.3390/ijms18122723
- Perez-Ortin JE. Genomics of mRNA turnover. Brief Funct Genomic Proteomic. 2007;6:282–291.
- Fialcowitz EJ, Brewer BY, Keenan BP, et al. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J Biol Chem. 2005;280:22406–22417.
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005.
- Fabian MR, Frank F, Rouya C, et al. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol. 2013;20:735–739.
- Eisen TJ, Eichhorn SW, Subtelny AO, et al. The dynamics of cytoplasmic mRNA metabolism. Mol Cell. 2020;77:786–799. e10
- Eisen TJ, Eichhorn SW, Subtelny AO, et al. MicroRNAs cause accelerated decay of short-tailed target mRNAs. Mol Cell. 2020;77:775–785. e8
- Al-Ashtal HA, Rubottom CM, Leeper TC, et al. The LARP1 La-module recognizes both ends of TOP mRNAs. RNA Biol. 2019;1–11. DOI:10.1080/15476286.2019.1669404
- Thandapani P, O’Connor TR, Bailey T, et al. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623.
- Deragon JM. Distribution, organization an evolutionary history of La and LARPs in eukaryotes. RNA Biol. 2020; 1–9. DOI:10.1080/15476286.2020.1739930.
- Philippe L, Vasseur JJ, Debart F, et al. La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 2018;46:1457–1469.
- Lima SA, Chipman LB, Nicholson AL, et al. Short poly(A) tails are a conserved feature of highly expressed genes. Nat Struct Mol Biol. 2017;24:1057–1063.
- Aryani A, Denecke B. In vitro application of ribonucleases: comparison of the effects on mRNA and miRNA stability. BMC Res Notes. 2015;8:164.
- Cruz-Gallardo I, Martino L, Kelly G, et al. LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA. Nucleic Acids Res. 2019;47:4272–4291.
- Kozlov G, Trempe JF, Khaleghpour K, et al. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci U S A. 2001;98:4409–4413.
- Kozlov G, De Crescenzo G, Lim NS, et al. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. Embo J. 2004;23:272–281.
- Lim NS, Kozlov G, Chang TC, et al. Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein. Implications for HYD function. J Biol Chem. 2006;281:14376–14382.
- Kozlov G, Menade M, Rosenauer A, et al. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J Mol Biol. 2010;397:397–407.
- Kozlov G, Safaee N, Rosenauer A, et al. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J Biol Chem. 2010;285:13599–13606.
- Albrecht M, Lengauer T. Survey on the PABC recognition motif PAM2. Biochem Biophys Res Commun. 2004;316:129–138.
- Huntzinger E, Braun JE, Heimstadt S, et al. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. Embo J. 2010;29:4146–4160.
- Jinek M, Fabian MR, Coyle SM, et al. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol. 2010;17:238–240.
- Bouhaddou M, Memon D, Meyer B, et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712. . e19
- Yang J, Anishchenko I, Park H, et al. Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci U S A. 2020;117:1496–1503.
- Ogami K, Oishi Y. TN, K Sakamoto, S Hoshino LARP1 facilitates translational recovery after amino acid refeeding by preserving long poly(A)-tailed TOP mRNAs. bioRxiv Preprint Not Peer-reviewed. 2020.
- Schaffler K, Schulz K, Hirmer A, et al. A stimulatory role for the La-related protein 4B in translation. RNA. 2010;16:1488–1499.
- Hopkins TG, Mura M, Al-Ashtal HA, et al. The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res. 2016;44:1227–1246.
- Wilbertz JH, Voigt F, Horvathova I, et al. Single-molecule imaging of mRNA localization and regulation during the integrated stress response. Mol Cell. 2019;73:946–958. e7
- Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol. 2019;11. DOI:10.1101/cshperspect.a032813
- Damgaard CK, Lykke-Andersen J. Translational coregulation of 5ʹTOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25:2057–2068.
- Hopkins KC, Tartell MA, Herrmann C, et al. Virus-induced translational arrest through 4EBP1/2-dependent decay of 5ʹ-TOP mRNAs restricts viral infection. Proc Natl Acad Sci U S A. 2015;112:E2920–9.
- Van Treeck B, Parker R. Emerging roles for intermolecular RNA-RNA interactions in RNP Assemblies. Cell. 2018;174:791–802.
- Hentze MW, Castello A, Schwarzl T, et al. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–341.
- Bousquet-Antonelli C, Deragon JM. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009;15:750–764.
- Maraia RJ, Mattijssen S, Cruz-Gallardo I, et al. The LARPs, La and related RNA-binding proteins: structures, functions and evolving perspectives. WIREs RNA. 2017;e1430. DOI:10.1002/wrna.1430
- Weng H, Kim C, Valavanis C, et al. Acheron, an novel LA antigen family member, binds to CASK and forms a complex with Id transcription factors. Cell Mol Biol Lett. 2009;14:273–287.
- Valavanis C, Wang ZH, Sun H, et al. Acheron, a novel member of the Lupus Antigen family, is induced during the programmed cell death of skeletal muscles in the moth Manduca sexta. Gene. 2007;393:101–109.
- Martino L, Pennell S, Kelly G, et al. Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res. 2015;43:645–660.
- Bayfield MA, Kaiser TE, Intine RV, et al. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol Cell Biol. 2007;27:3303–3312.
- Jacks A, Babon J, Kelly G, et al. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure (Camb). 2003;11:833–843.
- Hogg JR, Goff SP. Upf1 senses 3ʹUTR length to potentiate mRNA decay. Cell. 2010;143:379–389.
- Delaglio F, Grzesiek S, Vuister GW, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293.
- Goddard T, D. K. Sparky—NMR assignment and integration software. USA: Sparky 3 University of California, San Francisco; 2008.
