ABSTRACT
tRNA-derived small RNA (tsRNA) is a novel class of non-coding RNA that is usually produced from tRNA following endonuclease cleavage which occurs under stress conditions. There are two types of tsRNAs: tRNA-derived fragments (tRFs) and stress-induced tRNA halves (tiRNAs), which differ in their cleavage position. Many studies have demonstrated that tsRNAs are involved in various physiological and pathological processes apart from cancer and gene expression. In this review, we briefly described the biogenesis, classification, and characteristics of tsRNAs and summarized the current research progress of tsRNAs in metabolic diseases, senescence, reproduction, stress, and organ injury, and finally put forward some problems to be solved.
Introduction
tRNAs are important non-coding RNAs in organisms that primarily drive protein synthesis. In the past, tRNAs were thought to only function in transporting the corresponding amino acids to the ribosome for peptide chain elongation, while the small RNAs derived from tRNAs, named tRNA-derived small RNAs (tsRNAs), were considered as by-products of random tRNA cleavage [Citation1]. However, further studies have proven that tsRNAs are not by-products but also played a significant role in various physiological and pathological processes, including suppressing breast cancer development via YBX1 displacement [Citation2], regulating ribosome biogenesis by binding mRNA and ribosomal proteins [Citation3], inhibiting translation by interfering with peptide bond formation in vitro [Citation4] and so on. These discoveries illustrate that tsRNAs have complex functions and suggest that tsRNAs are vital molecules in gene expression and protein synthesis. Thus, an important question was asked: what roles do tsRNAs play in the development of other complex diseases and processes outside of regulating gene expression and cancer development? In this review, we briefly describe the biogenesis, classification, and characteristics of tsRNAs, summarize the research progress of tsRNAs in metabolic diseases, senescence, reproduction, stress, and organ injury, and discuss some unknown aspects of tsRNA biology to be addressed in the future.
tsRNAs species and biogenesis
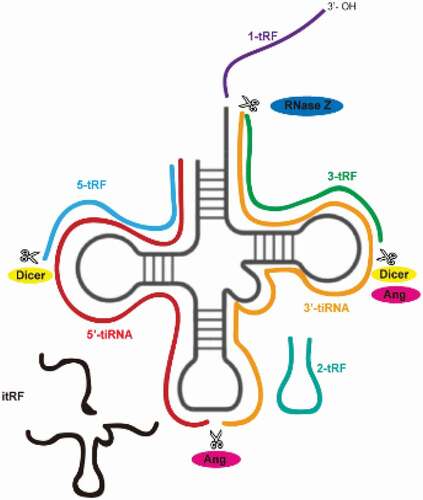
tsRNAs are derived from tRNAs. Mature tRNAs come from tRNA precursors (pre-tRNAs) through a series of processing events in the following order: RNA polymerase III transcribes the DNA sequence into RNA, ribonucleases P (RNase P) and Z (RNase Z), respectively, cut the transcript-specific sequences at the 5ʹ end and the 3ʹ end, the intron sequences are removed by tRNA endonucleases, and final post-transcriptional modifications are made, such as pseudouridylation, methylation, and the addition of the trinucleotide sequence CCA to the 3ʹ end of the transcript by tRNA nucleic acid transferases. The tertiary structure of a mature tRNA is L-shaped, including a D loop, a TψC loop, an anticodon loop, and a variable loop. When different enzymes cut the specific position of tRNA precursors or mature tRNAs, tsRNAs are produced. According to their cutting site, tsRNAs can be divided into two categories [Citation5,Citation6]. The first category is a tRNA-derived fragment (tRF), usually produced by Dicer or angiogenin, which can be divided into five subtypes described in the following table [Citation7,Citation8]. Under normal conditions, tRFs are located ubiquitously in various biofluids and their abundance is affected by diet, diseases, or external stimulation [Citation2,Citation9–11]. The second category of tsRNAs is stress-induced tRNA halves (tiRNA), which possesses a double-stranded structure and is 30–40 bases in length. Under stress conditions such as heat shock, ultraviolet radiation, starvation, hypoxia, and viral infection [Citation12–14], angiogenin cuts the mature tRNA into two halves from the anticodon loop to obtain 3ʹ-tiRNA and 5ʹ-tiRNA [Citation15] ( and ). In addition, different RNA modification has different effect on tsRNAs biogenesis. Pseudouridylation (Ψ) is an abundant and widespread nucleoside modification, which isomerize uridine to pseudouridine. Ψ is catalysed by evolutionarily conserved pseudouridine synthases (PUSs). When knocking out PUS7 in human embryonic stem cells (hESCs), tRNAs lost Ψ, leading to change in biogenesis of 5-tRFs and 3-tRFs. And a special class of 5-tRFs derived from tRNA-Ala, tRNA-Lys, tRNA-Val which contain five consecutive guanine residues at 5ʹ end decreased significantly [Citation16]. Another frequent RNA modification is methylation. Previous study has proved Dnmt2, a tRNA methyltransferase, protected tRNA from being cleaved by ribonuclease under stress [Citation17]. Dnmt2 methylate the C38 position (m5C) of tRNA-Gly whose loss promoted tRNA fragmentation [Citation18]. In Dnmt2−/- mice sperm, tsRNAGly were up-regulated [Citation19]. However, we need further investigations of how tRNA modifications influence tsRNAs biogenesis.
Table 1. tsRNAs species
The latest progress of tsRNAs
tsRNAs involved in the occurrence and development of metabolic diseases ()
Obesity is a risk factor for diabetes, non-alcoholic fatty acid diseases, and cardiovascular disease [Citation24,Citation25]. All the time, scientists turn to seek robust methods to reduce weight, but there is no completely reliable one up to now. Fat accumulation directly results in obesity, which is associated with pre-adipocytes proliferation and differentiation, and mature adipocyte hypertrophy. Emerging evidence indicates that tsRNAs are involved in adipocyte fate and function. Zhu Li group found that a tRF from tRNAGlu-TTC, referred to as tRFGlu-TTC, promoted the proliferation of preadipocytes while inhibiting differentiation in the prerenal adipose tissue of rats fed a high-fat diet. This tRF not only promoted 3T3-L1 preadipocyte proliferation via increasing the expression of cell cycle regulatory factors (CDK4, CyclinD1, and Cyclin E), but also targeted the key pro-adipogenic transcription factors KLF9, KLF11, and KLF12 directly, which led to decreases in aP2, PPARγ, and C/EBPα expression. tRFGlu-TTC reduced and increased the expression of fatty acid oxidation-related and synthesis-related genes, respectively, and the expression patterns of these genes were consistent with studies focused on fatty acid phenotypes in other systems [Citation26–28]. What’s more, a new study demonstrated that a lack of tsRNAs led to a significant decrease in the expression levels of PPARγ, FABP4, and C/EBPα [Citation29]. According to the importance of PPARγ and C/EBPα in the regulation of adipogenesis, we inferred that tsRNAs might facilitate adipogenesis in adipocytes to support lipid accumulation and meanwhile promote pre-adipocytes proliferation and differentiation, so that influence obesity. However, this hypothesis requires further study. Besides this, a tsRNA named tsRNA-06018, promoted adipogenic differentiation by targeting the 3ʹUTR of Stanniocalcin-2 (STC2) mRNA via the extracellular signal-regulated kinase 1/2 (ERK1/2) [Citation29].
Figure 2. tsRNAs take part in various metabolic diseases (Created with Servier Medical Art, https://smart.servier.com)
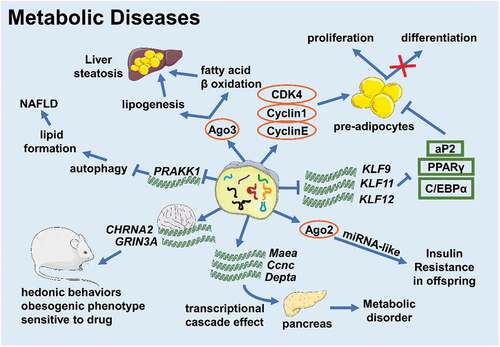
Alcoholic liver disease (ALD) and non-alcoholic fatty liver disease (NAFLD) are serious global health issues whose incidences are rising in recent decades [Citation30]. Although the pathogenesis is different, ALD and NAFLD share a similar developmental course beginning at steatosis and leading to hepatitis, cirrhosis, and hepatocellular carcinoma [Citation30]. So, there might be something common participating in disease progression. In patients with ALD, the expression level of tRFGLy increased and the expression of Sirt1 decreased, and the ALD mice models exhibited the same phenotype [Citation11]. He Songqing group further found that tRFGLy can downregulate Sirt1 expression through binding to Ago3 and acting on the 3ʹ UTR of Sirt1 mRNA to promote lipogenesis and inhibit β-oxidation of fatty acids, thus promoting the development of liver steatosis and liver damage in ALD mice [Citation11]. This outcome suggests that tsRNAs function in ALD progression. On the other hand, in mice with NAFLD, elevated tRF-300b can target and inhibit the expression of the autophagy-related gene Prkaa1 and promote lipid formation, which aggravates the development of NAFLD [Citation31]. However, the existing research on tsRNAs and liver diseases is relatively simple and we need to continue to conduct in-depth research on their mechanisms of development.
Epidemiology and basic research have shown that the health and nutritional status of parents is one of the risk factors for chronic diseases in the offspring [Citation32–34]. Studies have shown that tsRNAs are the most abundant small RNAs in mature mouse sperm [Citation35]. The role of tsRNAs in the intergenerational or transgenerational inheritance of metabolic diseases has also drawn much attention. Cropley J.E. et al. used Avy/a male mice crossed to a/a female mice to set up a congenic rodent model of obesity and pre-diabetes. Pre-diabetic obese fathers made the F1 generation potentially susceptible to hepatic insulin resistance, and the sperm of F1 males could still transmit the induced metabolic phenotype to the F2 generation without obesity, changes in diet, or any significant metabolic damage. Matching with this, it was found that approximately a quarter of the small RNAs in F1 male sperm are 5-tRFGlu-CTC and 5-tRFGly-GCC, and the content of 5-tRFGlu-CTC was reduced about 30% in the sperm of the F1 of obese paternal mice compared to the control group. It was suspected that 5-tRFGlu-CTC combined with Ago2 and might act in a miRNA-like manner in the zygote. This example suggests that tsRNAs might influence the intergenerational inheritance of metabolic phenotypes without extrinsic factors, which means that tsRNAs might act as a novel epigenetic factor [Citation36]. Chinese scientists further verified this conjecture, as they produced a study showing that the expression profile of 5ʹ-tiRNAs changed in the sperm of the parents fed on a high-fat diet. After the injection of purified tsRNAs into the zygote, multiple metabolic regulation-related genes were downregulated in the early embryonic stage. Such changes would affect the expression of metabolic genes through the transcriptional cascade effect into adulthood, and influence the reprogramming of the pancreatic islets of the F1 generation, leading to metabolic disorders [Citation19]. Studies have also demonstrated that the metabolic profile of the maternal line could be passed to offspring through sperm tsRNAs. Sarker G. et al. observed that compared to male offspring of female mice fed a chow diet, sperm tsRNA expression increased in those fed a high-fat diet. After microinjecting all or part of specific sperm tsRNAs of these F1 males into normally fertilized eggs, scientists found that the F2 generation who received 30–34 nt sperm tsRNAs had similar obese bodies and hedonistic behaviours as the father, with a preference for delicious food and alcohol with increased sensitivity to central stimulant drugs. This result indicated that sperm tsRNAs were indeed carriers of genetic information, which helped to spread maternal high-fat diet-induced hedonistic behaviour and obesogenic phenotypes across generations via targeting CHRNA2 and GRIN3A in the brain [Citation37].
However, in a sparrow study, by comparing the small RNA expression profiles of mature sperm of adult and old sparrows, there was no statistical difference in tRFs between the two, suggesting that the carrier of epigenetic information might also be related to RNA modification [Citation38]. Chen Qi group injected endogenous tsRNAs from high-fat diet-fed mice and artificially synthesized tsRNAs into fertilized eggs, respectively, and found that endogenous tsRNAs can produce offspring with impaired glucose tolerance and insulin resistance but synthetized tsRNA cannot, indicating that the modification of RNA affected the function of tsRNAs [Citation19]. The scientists further found that compared to the tsRNAs of mice fed a normal diet, the m5C and m2G modifications of tsRNAs of mice fed with a high-fat diet were significantly up-regulated [Citation19]. Although the function of m2G was unknown, early studies had shown that m5C could improve RNA stability and was related to RNA-mediated cross-generational inheritance [Citation18,Citation39]. Another study found that tsRNAsGly levels rose in Dnmt2-/- sperm. Dnmt2 deletion caused tRNA m5C hypomethylation, changed tRNA secondary structure significantly, and promoted tRNA fragmentation, thereby eliminating the ability of sperm tsRNAs to induce the metabolic changes of the offspring. In addition, three artificially synthesized tsRNAsGly with varying degrees of m5C modification induced different transcriptome responses in specific gene categories [Citation40]. Therefore, we believe that tsRNAs are possible carriers of epigenetic information that relate to RNA modification, but how and where tsRNAs obtain this epigenetic information needs further study.
tsRNAs have multiple roles in the ageing process ()
Recent studies have shown that the abundance of tsRNAs changed in ageing insects and animals. The expression of Drosophila tsRNAs increased in an age-dependent manner [Citation41]. C. elegans tsRNA levels generally rose during ageing [Citation42]. American scientists found that the level of 5ʹ-tiRNA in mice serum changed significantly with age [Citation43]. The abundance of 3-tRF in mouse brain cells increased with age, and the abundance of 3-tRF in older rats was also higher than that in younger rats [Citation44]. These data above show that tsRNAs might participate in, or are at least associated with, the ageing process.
Figure 3. tsRNAs participate in diseases related to senescence (Created with Servier Medical Art, https://smart.servier.com)
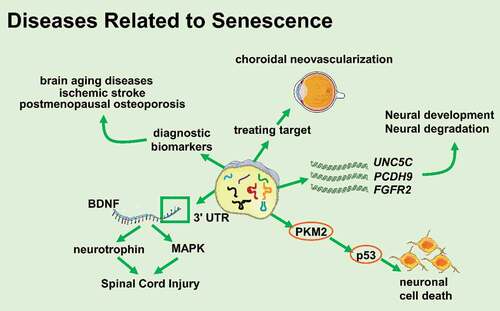
The degeneration of the nervous system is coupled with ageing. Many studies have shown that tsRNAs were involved in the development or degeneration of the nervous system and the occurrence and development of related diseases. Early studies demonstrated that compared to miRNAs, the levels of age-related mRNAs targeted by tRFs decreased more significantly. The targets of tRFs included UNC5C, PCDH9, FGFR2, etc. UNC5C mutations are associated with the susceptibility of Alzheimer’s disease and PCDH9 is related to the development and maintenance of the central nervous system structure. FGFR2 is a powerful regulator during the development of the nervous system [Citation44–47]. A study has also demonstrated that 5ʹ-tRFTyr derived from tyrosine pre-tRNA was more likely to cause p53-dependent neuronal death than other types of tRFs. 5ʹ-tRFTyr contained the 5ʹ leader sequence and 5ʹ exon of tRNATry, directly bound to PKM2, enhanced p53 activation, and led to neuronal cell death. Injecting 5ʹ-tRFTyr into zebrafish single-cell embryos caused embryonic development defects and induced zebrafish larvae microcephaly [Citation48]. All these data suggest that tsRNAs are closely related to the development and degeneration of the nervous system. A study has also proved that tiRNAGly-GCC−001 might participate in the MAPK and neurotrophic factor pathways by targeting brain-derived neurotrophic factor (BDNF) and regulating the pathophysiological processes following spinal cord injury [Citation49], prompting that tsRNAs might play an important role in injury and recovery of the nervous system.
In addition, it has also been reported that patients with Parkinson’s disease and healthy controls had a set of tRFs that could clearly distinguish the two groups in the brain prefrontal cortex, cerebrospinal fluid, and serum [Citation50]. Changes in the expression profiles of tRFs have been detected in the brains of SAMP8 mice (a model of neurodegenerative disorders) and SAMR1 mice (a control model) [Citation51]. These pieces of evidence indicate that tsRNAs are related to brain ageing-related diseases. Angiogenin is one of the key enzymes in the production of tRFs, and at the same time, angiogenin mutations have been reported in patients with amyotrophic lateral sclerosis and Parkinson’s disease [Citation52]. Angiogenin directly participated in the pathways leading to motor neuron degeneration or dopaminergic neuron degeneration. Therefore, angiogenin might be involved in the occurrence and development of neurodegenerative diseases by protecting central neurons or making changes in tRFs levels [Citation50,Citation52]. Zhang Shuai group found that the differential expression of target genes of tRFs in the brains of SAMP8 mice and SAMR1 mice were involved in various brain functions, including synapse formation and synaptic vesicle circulation pathways. They speculated that tRFs might participate in the pathogenesis of Alzheimer’s disease and Parkinson’s disease through miRNA-like patterns, so the expression pattern of tRFs could be used as a potential biomarker for the diagnosis of brain ageing and related diseases [Citation51].
In addition to brain ageing, stroke is also one of the diseases that seriously affects the quality of life of the elderly. Elkordy et al. found that various oxidative stresses induced the production of tRNA fragmentation in neuronal PC12 cells of rats and that there was tiRNA production during ischaemia-reperfusion injury. Interestingly, tiRNAs were produced before severe cell damage and death, and their production was related to the severity of the cellular injury. When the cells had recovered from stresses after a few hours, the content of tiRNAs decreased significantly, showing that tiRNAs could be a potential early biomarker of ischaemic stroke [Citation53,Citation54].
The WHO survey showed that osteoporosis was an age-related disease, with a very high prevalence among the elderly [Citation55]. Zhang Yan et al. found that tRFs played an important role in osteoporosis. They collected and tested 40 plasma samples of osteoporosis patients and healthy controls, and found that tRF-25, tRF-38, and tRF-18 from plasma exosomes can be used as a diagnostic biomarker of osteoporosis to prognosticate osteoporosis [Citation56]. Aside from osteoporosis, tsRNAs were found to be associated with choroidal neovascularization, which is one of the major clinical characteristics of neovascular age-related macular degeneration and a major cause of elderly blindness. Zhang Liwei group found that 72 tsRNAs were differentially expressed in choroidal neovascularization mice models and the target genes of these tsRNAs were most enriched in the NOD-like receptor signalling pathway, which plays a crucial role in angiogenesis, suggesting that tsRNAs might be a novel potential target in treating choroidal neovascularization [Citation57,Citation58].
In a study on longevity, it was found that inhibiting RNA polymerase III in the gut of adult worms or flies could prolong lifespan by reducing protein synthesis and lowering the level of pre-tRNAs. Pre-tRNAs can be processed into tsRNAs by RNase Z. This study further supported that changes in tsRNAs levels might contribute to lifespan and health [Citation59]. In summary, tsRNAs are differentially expressed in young and ageing individuals in multiple species and have special targets, which suggests that tsRNAs might not only be potential biomarkers but might also play other pivotal roles in ageing and ageing-related diseases.
Sperm gets the payload of tsRNAs in the epididymis
As mentioned above, the mature sperm cells have an abundance of tsRNAs, which promotes the intergenerational transmission of specific metabolic traits in mice. Godoy, P. M. et al. used RNA-sequencing technology to compare small RNAs systematically in 12 types of human biological fluids and found that tsRNAs are mainly found in bile, urine, seminal plasma, amniotic fluid, follicular fluid, and semen [Citation9]. Many researches regarded that extracellular tsRNAs work as potential biomarkers of diseases for tsRNAs were easy to obtain without damage to body and their abundance showed obvious differences between normal physiological body and pathological body [Citation50,Citation53,Citation56,Citation60,Citation61].
tsRNAs expression varied across different parts of the male reproductive system. The level of tsRNAs increased in the distal epididymis, and the specific tRFs expression profile was diverse in the testis, epididymal head (proximal), and epididymal tail (distal). Besides, scientists have also detected differences in the expression of tsRNAs within a cell. The sperm head contained moderate levels of tRFs. A few specific tRFs (especially tRFGly-TCC and tRFVal-Val) were relatively abundant in the sperm tail [Citation62–64]. These observations strengthened the opinion that tsRNAs are enriched in mature germ cells. Also, low levels of tRFs were detected in immature sperm cells purified from mouse testes, but studies have found that tRNAs had a large amount of cleavage in the epididymis – the place where sperm mature. Conine C.C. et al. found that epididymal epithelial cells could secrete epididymosomes containing rich 5-tRF. While sperm moving from the epididymal head to the epididymal tail, the sperm fused with 5-tRF-rich epididymosomes, suggesting that sperm might obtain tRNA fragments when passing through the epididymis [Citation63–66]. This shows that tRF pools can be transferred between different types of cells to achieve the transfer of biological information from somatic cells to germ cells and tRFs may work as information carriers and exchangers as previously described.
But where does this biological information come from? Many researchers demonstrated that diet might affect tsRNAs profiles in germaria. A human study showed that the levels of i-tRFs and 3-tRFs in sperm cells from healthy donors were upregulated and positively correlated with an increase in sperm motility, while donors accepted a two-step diet intervention-healthy diet for the first week and a high-sugar diet for the second week[Citation10]. And in mice fed by a low-protein diet, tRFGly-GCC in sperm was upregulated and it could inhibit the active endogenous reverse transcription element-related genes in the preimplantation embryo, thereby affecting the biogenesis in mammalian reproduction [Citation65]. But a newer study had different results. Colin et al. separated caput sperm and cauda sperm. The embryos produced with caput sperm showed significant overexpression of various regulatory factors and even caused embryonic lethality. Microinjection of purified small RNAs from cauda sperm into such embryos could rescue molecular defects and lethality. However, these small RNAs were miRNAs rather than tRFs, which indicated that the role of tsRNAs in embryonic development needs further studies [Citation64].
In conclusion, in reproductive and metabolic diseases mouse models, sperm tsRNAs might be a driving factor of sperm epigenetics or at least play an indispensable role [Citation19,Citation67]. Sperm tsRNAs might also interfere with mRNA synthesis of proximal gene regulation region directly, and inhibit genes driven by transposable elements [Citation19,Citation65], and this process is required for early embryonic development ().
tsRNAs participate in stress in different forms ()
Previous studies at the cellular level have shown that tsRNAs can be upregulated under various stresses (physical, chemical, and viral) [Citation68,Citation69]. Furthermore, there are abundant tsRNAs in haematopoietic and lymphatic tissues, as well as in the blood circulatory system [Citation43] . All of these observations have inspired researchers to explore whether tsRNAs are related to pathological conditions.
The level of tsRNAs existing in the circulatory system increases rapidly during inflammation, indicating that tsRNAs might play a vital role in the immune response [Citation70]. On the one hand, during the maturation of monocytes to dendritic cells, 5-tRFGlu formed a complex with the Ago-like proteins piwil4 and piwil1 and then recruited SETDB1, SUV39H1, and HP1β to the CD1A promoter to methylate histone H3K9, thereby inhibiting the expression of CD1A [Citation71]. On the other hand, tsRNAs from tRNAAla-UGC with a CCACCA sequence could directly interact with Toll-like receptors to activate the immune response of Th1 and toxic T lymphocytes. Research by Chiou et al. showed that immune activation signals promoted the secretion of vesicles containing specific tRFs that inhibited T cell activation and cytokine production [Citation72]. These results indicated that tsRNAs might work as new immune signalling molecules to participate in an immune response.
Figure 4. tsRNAs participate in stress in different forms (Created with Servier Medical Art, https://smart.servier.com)
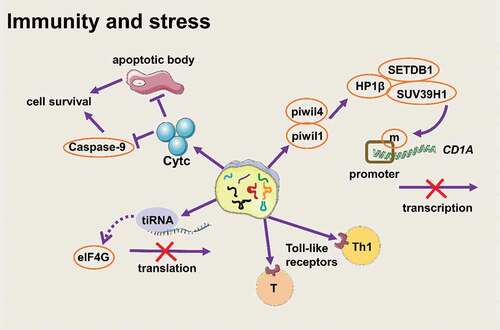
Apart from participating in stress as immune signalling molecules, tsRNAs also participate in stress response by inhibiting translation [Citation73]. When cells are exposed to unfavourable environmental conditions, they will automatically trigger the stress response to conserve energy resources for surviving. Generally, eIF2α is phosphorylated and eIF2α phosphorylation inhibits translation initiation. Then, some proteins containing domains that have low sequence complexity and a high content of glycines bind to messenger ribonucleoprotein (mRNP) rapidly and form stress granules [Citation74–76]. But tiRNAs inhibit translation in another way. Pavel Ivanov et al. found that tiRNA (mainly 5ʹ-tiRNA) was a component of stress response after stimulating stress conditions, and tiRNA could promote translation stagnation and reduce the global translation speed by 10%-15% in a phospho-eIF2α–independent way [Citation13]. Stress conditions induced the production of 5ʹ-tiRNAAla and 5ʹ-tiRNACys. The two tiRNAs contained terminal oligoguanine motif (TOG) and had ability to assemble G-quadruplexes structure (a stable guanine-rich nucleic acid structure [Citation77]), then disrupted the scanning step of translation initiation by directly binding the HEAT1 domain of eIF4G (a large scaffold protein in eIF4F) on the 5ʹ end of mRNA [Citation74,Citation78]. What’s more, a study on Trypanosoma brucei showed that a large amount of 3ʹ-tiRNAThr was produced in malnourished trypanosomes. This tiRNA was related to ribosomes and polymers. Once starvation stopped, 3ʹ-tiRNAThr promoted mRNA translation during stress recovery [Citation79]. This evidence showed that tsRNAs, especially tiRNAs, are important translation regulators during stress.
Early studies have shown that mature tRNA molecules can bind to cytochrome C and inhibit the formation of apoptotic bodies and inhibit the activity of caspase-9, thus promoting cell survival [Citation80]. Mridusmita Saikia. et al. found that when cells were exposed to a high osmotic pressure environment, angiogenin cut tRNAs to produce tiRNAs rapidly, and tiRNAs accumulated in the cytoplasm and preferentially formed a complex with cytochrome C released from mitochondria, thereby blocking autophagy [Citation81]. Thus, we can consider that tiRNAs ‘inherit’ part of the characteristics of tRNA and they play a role in stabilizing the cell state under stress.
tsRNAs are associated with multiple organ injuries ()
Several lines of research showed that tsRNAs were involved in regulating cardiomyocyte activity [Citation82]. Cardiomyocytes are prone to ischaemia. Studies have shown that tRFs were involved in tissue ischaemia [Citation83,Citation84]. In ischaemic rat brains, a mouse hindlimb ischaemia model, and a cellular hypoxia model, tRFVal and tRFGly levels increased, and they could inhibit the proliferation, migration, and tubular formation of endothelial cells, resulting in ischaemia[Citation84]. It seems like tRFs participate in myocardial ischaemia, but the relationship is not fully understood. Besides, tsRNAs were also involved in regulating cardiac hypertrophy. Zhu Li et al. used isoproterenol to induce cardiac hypertrophy in mice. In this model, tRFGly-GCC and tRFGlu-TTC were overexpressed in the heart, which both increased the surface area of H9c2 cells (rat cardiomyocyte cell line) and increased the expression of the cardiomyocyte hypertrophy markers ANF, BNP, and β-MHC. Among them, tRFGly-GCC inhibited the 3ʹ UTR of Timp3 mRNA directly, proving that tRFs were involved in the regulation of cardiac hypertrophy. [Citation85]This study also found that a variety of tRFs were highly expressed in the sperm of F0 generation mice with cardiac hypertrophy and in the heart of F1 generation mice. Compared to the control, the expression of β-MHC and ANP in the F1 generation of cardiac hypertrophic mice was higher, and fibrosis and autophagy in the heart increased. This indicated that tRFs might act as a new type of epigenetic factor to participate in the intergenerational inheritance of myocardial hypertrophy [Citation85]
Figure 5. tsRNAs are associated with multiple organs injuries (Created with Servier Medical Art, https://smart.servier.com)
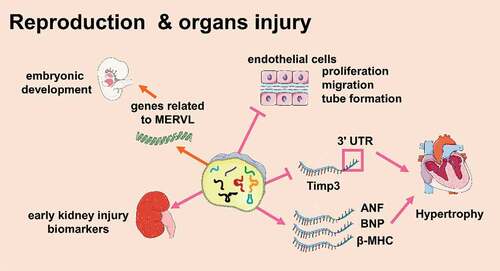
Many studies have shown that tsRNAs are associated with kidney damage. For example, the concentration of tiRNAs increased in the blood of patients with chronic kidney disease [Citation86]. The plasma level of tiRNAs increased in animals with kidney injury and renal ischaemia-reperfusion [Citation60]. Studies have shown the relationship between tsRNAs and kidney damage, when the kidney was damaged, the expression of angiogenin in renal epithelial cells significantly increased. While angiogenin participated in cell protection, it also interfered with the initiation of protein translation by producing tiRNAs. Then, tiRNAs reduced protein synthesis and promoted the adaptability of cells upon acute kidney injury [Citation87]. The levels of plasma tiRNAs and angiogenin in patients with acute or chronic kidney injury could reflect the severity of the prognosis [Citation60]. In addition, the two could also be detected in the urine, suggesting that tiRNAs and angiogenin are potential biomarkers that reflect the degree of kidney injury [Citation60].
Summary
As a new class of non-coding RNAs, tsRNAs encompass various types of tRNA-derived small molecules and have received extensive attention for their unique and diverse physiological and pathological functions. Recent studies on the role of tsRNAs have emphasized their important role in a variety of molecular processes, including mRNA stabilization [Citation2], miRNA-mediated silencing [Citation88,Citation89], regulation of cap-dependent and cap-independent translation [Citation79]. Also, tsRNAs are usually dysregulated in cancers,; thus,many current researches focused on the role of tsRNAs in cancer [Citation90–94].
In this review, we summarized the progression of tsRNAs in disease processes other than cancer. For metabolic diseases, tsRNAs are not only involved in development but also work as paternal or maternal intergenerational and transgenerational genetic factors. In senescence, although there is no clear evidence of tsRNAs influencing the ageing process, tsRNAs seem to have a role in senescence-related diseases and might serve as potential biomarkers. In addition, tsRNAs are regulated quickly by diet and can be transferred from somatic cells to sperm during sperm passage through the epididymal head to the epididymal tail, working as a driving factor of sperm epigenetics. For immunity and stress, tsRNAs mostly act as a regulator to maintain cellular homoeostasis. What’s more, tsRNAs play a significant role in suppressing or promoting organ injury to some extent. However, the extent of knowledge on tsRNAs in the fields of metabolic diseases, ageing, reproduction, and stress is still paltry. There are even some fields that are not yet discovered to be influenced by tsRNAs. More time is needed to explore the role and mechanism of tsRNAs during disease and physiology processes.
Although researches about tsRNAs have achieved some excellent results [Citation73,Citation77,Citation95], there are still many questions to be answered. First, there is no consensus on the classification and nomenclature of tsRNAs. For this type of small RNA derived from tRNA, some people name tRFs and tiRNAs together as tRNA-derived fragments (tDFs), and some refer to tsRNAs as tRNA-derived microRNA [Citation88]. Detailed naming is also more confusing given these numerous classifications and ppoorly definedconventions. Earlier, when we understood even less about tsRNAs, some studies mistook tsRNAs for micro RNAs [Citation96], and some were named separately [Citation88,Citation97,Citation98]. However, the current work on the classification of tsRNAs is also only to distinguish between tRFs and tiRNAs mostly, only a few with obvious characteristics such as 3-tRF are marked, which is not beneficial to the communication between researchers and the generalization of specific tsRNAs functions. So Jinghao Sheng proposed a feasible naming method -X-tsRNAsAA-NNN in 2018. Under these guidelines, tsRNAs are described with specific tRFs and tiRNAs, where X represents subclass, AA represents the amino acid corresponding to the tRNA, and NNN is the codon [Citation83]. Thus, 3ʹ-tiRNA and 5a-tRF are both derived from tRNAGly-GGG, so it can be written as 3ʹ-tiRNAGly-GGG and 5a-tRFGly-GGG.
Second, the general biogenesis process of tsRNAs is still largely undefined. Dicer, Ang, RNase, and ELAC2 are the key enzymes to produce tsRNAs, and most of tsRNAs are classified according to the cutting site and cutting enzyme. However, the length of various tsRNAs is not a certainty value, indicating that the recognition site of these endonucleases to tRNA is probably not unique and that the mechanism of these endonucleases to recognize and cleave tRNA still needs to be explored. In addition, there are more low-content tsRNAs such as 2-tRF, and it is not yet clear how they are produced. Are there other endonucleases? Does the three-dimensional structure of tRNA affect the production of tsRNAs? Except for pseudouridylation [Citation16] and methylation [Citation40], do other RNA modifications affect the production of tsRNAs? All these questions require further study and are necessary to understand the scope of tsRNA biological activities.
Thirdly, although in some studies, artificially synthesized tsRNAs can show similar activities to endogenous tsRNAs, more studies have confirmed that modifications on tsRNAs have a crucial impact on the production and activity of tsRNAs. The current research on tsRNAs is still limited by the development of technology. First of all, current sequencing methods cannot detect small RNAs with modifications. Therefore, it is often necessary to remove the modifications with a pretreatment kit before detecting the expression level of sample tsRNAs. Such processing may cause degradation of tsRNAs and affect the final test results. Secondly, compared with endogenous tsRNAs, artificially synthesized tsRNAs lose part of activity and are not as stable as endogenous tsRNAs. This is probably related to lack of RNA modification and changes in spatial structure. On the one hand, it is difficult to obtain sufficient and high-purity single specific tsRNA for mass spectrometry to verify the analysis. On the other hand, because the specific location and types of modifications on tsRNAs have not yet been fully clarified, it is difficult to modify tsRNAs precisely, leading to the endogenous tsRNAs unable to be fully reproduced. In order to obtain endogenous tsRNAs, scientists have made unremitting efforts. In 2016, Chen Qi et al. used polyacrylamide gel electrophoresis to separate tsRNAs. After gel cutting and recovery, tsRNAs with endogenous activity were obtained, but they could only be classified simply based on length, and specific individual tsRNAs could not be separated [Citation19]. In 2020, Pavel Ivanov group captured and purified single endogenous tiRNAs successfully by using DNA oligo probes complementary to target tiRNAs [Citation99]. This method is relatively efficient and can obtain high purity tsRNAs, which provides favourable conditions for the in-depth study of single tsRNAs. However, how to extract high-quality tsRNAs quickly and accurately from biological samples in large quantities and how to accurately modify artificially synthesized tsRNAs to obtain the correct spatial structure and activity requires more technological breakthroughs.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24(17):1832–1860.
- Goodarzi H, Liu X, Nguyen HCB, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802.
- Kim HK, Fuchs G, Wang S, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552(7683):57–62.
- Gebetsberger J, Zywicki M, Künzi A, et al. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea (Vancouver, BC). 2012;2012:260909.
- Shen Y, Yu X, Zhu L, et al. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med (Berl). 2018;96:1167–1176.
- Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem Sci. 2016;41:679–689.
- Lee YS, Shibata Y, Malhotra A, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23:2639–2649.
- Zhu L, Ge J, Li T, et al. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019;452:31–37.
- Godoy PM, Bhakta NR, Barczak AJ, et al. Large differences in small RNA composition between human biofluids. Cell Rep. 2018;25:1346–1358.
- Nätt D, Kugelberg U, Casas E, et al. Human sperm displays rapid responses to diet. PLoS Biol. 2019;17(12):e3000559.
- Zhong F, Hu Z, Jiang K, et al. Complement C3 activation regulates the production of tRNA-derived fragments Gly-tRFs and promotes alcohol-induced liver injury and steatosis. Cell Res. 2019;29(7):548–561.
- Hu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442.
- Yamasaki S, Ivanov P, Hu G-F, et al. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42.
- Wang Q, Lee I, Ren J, et al. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21:368–379.
- Emara MM, Pavel I, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968.
- Guzzi N, Cieśla M, Ngoc PCT, et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173:1204–16.e26.
- Schaefer M, Pollex T, Hanna K, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595.
- Tuorto F, Liebers R, Musch T, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905.
- Chen Q, Yan M, Cao Z, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science (New York, NY). 2016;351:397–400.
- Balatti V, Nigita G, Veneziano D, et al. tsRNA signatures in cancer. Proc Natl Acad Sci U S A. 2017;114:8071–8076.
- Kumar P, Anaya J, Mudunuri SB, et al. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78.
- Telonis AG, Loher P, Honda S, et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015;6:24797–24822.
- Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43–50.
- Vecchié A, Dallegri F, Carbone F, et al. Obesity phenotypes and their paradoxical association with cardiovascular diseases. Eur J Intern Med. 2018;48:6–17.
- World Health Organization. Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ Tech Rep Ser. 2000;894:i–xii, 1–253.
- Shen L, Li Q, Wang J, et al. miR-144-3p promotes adipogenesis through releasing C/EBPα from Klf3 and CtBP2. Front Genet. 2018;9:677.
- Du J, Shen L, Tan Z, et al. Betaine supplementation enhances lipid metabolism and improves insulin resistance in mice fed a high-fat diet. Nutrients. 2018;10. DOI:https://doi.org/10.3390/nu10020131
- Shen L, Tan Z, Gan M, et al. tRNA-derived small non-coding RNAs as novel epigenetic molecules regulating adipogenesis. Biomolecules. 2019;9:274.
- Wang T, Cao L, He S, et al. Small RNA sequencing reveals a novel tsRNA-06018 playing an important role during adipogenic differentiation of hMSCs. J Cell Mol Med. 2020;24(21):12736–12749.
- Singh S, Osna NA, Kharbanda KK. Treatment options for alcoholic and non-alcoholic fatty liver disease: a review. World J Gastroenterol. 2017;23(36):6549–6570.
- Zhu J, Cheng M, Zhao X. A tRNA-derived fragment (tRF-3001b) aggravates the development of nonalcoholic fatty liver disease by inhibiting autophagy. Life Sci. 2020;257:118125.
- Watkins AJ, Sinclair KD. Paternal low protein diet affects adult offspring cardiovascular and metabolic function in mice. Am J Physiol Heart Circ Physiol. 2014;306(10):H1444–52.
- Hur SSJ, Cropley JR, Suter CM. Paternal epigenetic programming: evolving metabolic disease risk. J Mol Endocrinol. 2017;58:R159–r68.
- Slyvka Y, Zhang Y, Nowak FV. Epigenetic effects of paternal diet on offspring: emphasis on obesity. Endocrine. 2015;48:36–46.
- Peng H, Shi J, Zhang Y, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612.
- Cropley JE, Eaton SA, Aiken A, et al. Male-lineage transmission of an acquired metabolic phenotype induced by grand-paternal obesity. Mol Metab. 2016;5:699–708.
- Sarker G, Sun W, Rosenkranz D, et al. Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proc Natl Acad Sci U S A. 2019;116:10547–10556.
- Matsushima W, Brink K, Schroeder J, et al. Mature sperm small-RNA profile in the sparrow: implications for transgenerational effects of age on fitness. Environ Epigenet. 2019;5:dvz007.
- Kiani J, Grandjean V, Liebers R, et al. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498.
- Zhang Y, Zhang X, Shi J, et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol. 2018;20:535–540.
- Karaiskos S, Naqvi AS, Swanson KE, et al. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol Direct. 2015;10:51.
- Kim SS, Lee SV. Non-Coding RNAs in Caenorhabditis elegans Aging. Mol Cells. 2019;42:379–385.
- Dhahbi JM, Spindler SR, Atamna H, et al. 5ʹ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298.
- Karaiskos S, Grigoriev A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Res. 2016;5:2758.
- Wetzel-Smith MK, Hunkapiller J, Bhangale TR, et al. A rare mutation in UNC5C predisposes to late-onset Alzheimer’s disease and increases neuronal cell death. Nat Med. 2014;20:1452–1457.
- Redies C. Cadherins in the central nervous system. Prog Neurobiol. 2000;61:611–648.
- Ford-Perriss M, Abud H, Murphy M. Fibroblast growth factors in the developing central nervous system. Clin Exp Pharmacol Physiol. 2001;28:493–503.
- Inoue M, Hada K, Shiraishi H, et al. Tyrosine pre-transfer RNA fragments are linked to p53-dependent neuronal cell death via PKM2. Biochem Biophys Res Commun. 2020;525:726–732.
- Qin C, Feng H, Zhang C, et al. Differential expression profiles and functional prediction of tRNA-derived small RNAs in rats after traumatic spinal cord injury. Front Mol Neurosci. 2019;12:326.
- Magee R, Londin E, Rigoutsos I. TRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease. Parkinsonism Relat Disord. 2019;65:203–209.
- Zhang S, Li H, Zheng L, et al. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging (Albany NY). 2019;11:10485–10498.
- Prehn JHM, Jirström E. Angiogenin and tRNA fragments in Parkinson’s disease and neurodegeneration. Acta Pharmacol Sin. 2020;41:442–446.
- Elkordy A, Rashad S, Shehabeldeen H, et al. tiRNAs as a novel biomarker for cell damage assessment in in vitro ischemia-reperfusion model in rat neuronal PC12 cells. Brain Res. 2019;1714:8–17.
- Elkordy A, Mishima E, Niizuma K, et al. Stress-induced tRNA cleavage and tiRNA generation in rat neuronal PC12 cells. J Neurochem. 2018;146:560–569.
- Kanis JA. Assessment of fracture risk and its application to screening for postmenopausal osteoporosis: synopsis of a WHO report. WHO Study Group. Osteoporos Int. 1994;4:368–381.
- Zhang Y, Cai F, Liu J, et al. Transfer RNA-derived fragments as potential exosome tRNA-derived fragment biomarkers for osteoporosis. Int J Rheum Dis. 2018;21:1659–1669.
- Zhang L, Liu S, Wang J-H, et al. Differential expressions of microRNAs and transfer RNA-derived small RNAs: potential targets of choroidal neovascularization. Curr Eye Res. 2019;44:1226–1235.
- Deng Y, Han X, Yao Z, et al. PPARα agonist stimulated angiogenesis by improving endothelial precursor cell function via a NLRP3 inflammasome pathway. Cell Physiol Biochem. 2017;42:2255–2266.
- Filer D, Thompson MA, Takhaveev V, et al. RNA polymerase III limits longevity downstream of TORC1. Nature. 2017;552:263–267.
- Mami I, Pallet N. tRNA fragmentation and protein translation dynamics in the course of kidney injury. RNA Biol. 2018;15:1147–1156.
- Zong T, Yang Y, Zhao H, et al. tsRNAs: novel small molecules from cell function and regulatory mechanism to therapeutic targets. Cell Prolif. 2021;54:e12977.
- Schuster A, Tang C, Xie Y, et al. SpermBase: a database for sperm-borne RNA contents. Biol Reprod. 2016;95:99.
- Sharma U, Sun F, Conine CC, et al. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev Cell. 2018;46:481–94.e6.
- Conine CC, Sun F, Song L, et al. Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev Cell. 2018;46:470–80 e3.
- Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396.
- Leslie M. EPIGENETICS. Sperm RNA fragments modify offspring metabolism. Science. 2016;351:13.
- Guzzi N, Bellodi C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol.2020;17:1214–1222.
- Zhou J, Liu S, Chen Y, et al. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J Gen Virol. 2017;98:1600–1610.
- Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219.
- Zhang Y, Zhang Y, Shi J, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol. 2014;6:172–174.
- Zhang X, He X, Liu C, et al. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-Interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol. 2016;196:1591–1603.
- Chiou N-T, Kageyama R, Ansel KM. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 2018;25:3356–70.e4.
- Ivanov P, O’Day E, Emara MM, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111:18201–18206.
- Lyons SM, Kharel P, Akiyama Y, et al. eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function. Nucleic Acids Res. 2020;48:6223–6233.
- Aulas A, Fay MM, Lyons SM, et al. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci. 2017;130:927–937.
- Wolozin B. Physiological protein aggregation run amuck: stress granules and the genesis of neurodegenerative disease. Discov Med. 2014;17:47–52.
- Lyons SM, Gudanis D, Coyne SM, et al. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun. 2017;8:1127.
- Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623.
- Fricker R, Brogli R, Luidalepp H, et al. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun. 2019;10:118.
- Mei Y, Yong J, Liu H, et al. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37:668–678.
- Saikia M, Jobava R, Parisien M, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol. 2014;34:2450–2463.
- Liapi E, Bilsen M, Verjans R, et al. tRNAs and tRNA fragments as modulators of cardiac and skeletal muscle function. Biochim Biophys Acta Mol Cell Res. 2020;1867:118465.
- Li S, Xu Z, Sheng J. tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes (Basel). 2018;9:246.
- Li Q, Hu B, Chen C-Y HG-W, et al. tRNA-derived small non-coding RNAs in response to ischemia inhibit angiogenesis. Sci Rep. 2016;6:20850.
- Shen L, Gan M, Tan Z, et al. A novel class of tRNA-derived small non-coding RNAs respond to myocardial hypertrophy and contribute to intergenerational inheritance. Biomolecules. 2018;8(3):54.
- Mishima E, Inoue C, Saigusa D, et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. JASN. 2014;25:2316–2326.
- Mami I, Bouvier N, Karoui KE, et al. Angiogenin mediates cell-autonomous translational control under endoplasmic reticulum stress and attenuates kidney injury. J Am Soc Nephrol. 2016;27:863–876.
- Maute RL, Schneider C, Sumazin P, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409.
- Martinez G, Choudury SG, Slotkin RK. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017;45:5142–5152.
- Zhu P, Yu J, Zhou P. Role of tRNA-derived fragments in cancer: novel diagnostic and therapeutic targets tRFs in cancer. Am J Cancer Res. 2020;10:393–402.
- Londin E, Magee R, Shields CL, et al. IsomiRs and tRNA-derived fragments are associated with metastasis and patient survival in uveal melanoma. Pigment Cell Melanoma Res. 2020;33:52–62.
- Zhang M, Li F, Wang J, et al. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. Onco Targets Ther. 2019;12:6371–6383.
- Braicu C, Zimta -A-A, Harangus A, et al. The function of non-coding RNAs in lung cancer tumorigenesis. Cancers (Basel). 2019;11:605.
- Shao Y, Sun Q, Liu X, et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90:730–738.
- Schorn AJ, Gutbrod MJ, LeBlanc C, et al. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71.e11.
- Schopman NCT, Heynen S, Haasnoot J, et al. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 2010;7:573–576.
- Haussecker D, Huang Y, Lau A, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695.
- Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816–25.
- Akiyama Y, Kharel P, Abe T, et al. Isolation and initial structure-functional characterization of endogenous tRNA-derived stress-induced RNAs. RNA Biol. 2020;17:1116–1124.