ABSTRACT
Antiproliferative BTG/Tob proteins interact directly with the CAF1 deadenylase subunit of the CCR4-NOT complex. This binding requires the presence of two conserved motifs, boxA and boxB, characteristic of the BTG/Tob APRO domain. Consistently, these proteins were shown to stimulate mRNA deadenylation and decay in several instances. Two members of the family, BTG1 and BTG2, were reported further to associate with the protein arginine methyltransferase PRMT1 through a motif, boxC, conserved only in this subset of proteins. We recently demonstrated that BTG1 and BTG2 also contact the first RRM domain of the cytoplasmic poly(A) binding protein PABPC1. To decipher the mode of interaction of BTG1 and BTG2 with partners, we performed nuclear magnetic resonance experiments as well as mutational and biochemical analyses. Our data demonstrate that, in the context of an APRO domain, the boxC motif is necessary and sufficient to allow interaction with PABPC1 but, unexpectedly, that it is not required for BTG2 association with PRMT1. We show further that the presence of a boxC motif in an APRO domain endows it with the ability to stimulate deadenylation in cellulo and in vitro. Overall, our results identify the molecular interface allowing BTG1 and BTG2 to activate deadenylation, a process recently shown to be necessary for maintaining T-cell quiescence.
GRAPHICAL ABSTRACT

Introduction
Members of the BTG/Tob family of proteins (BTG for B-cell translocation gene and Tob for Transducer of ErbB2) are characterized by the presence of a conserved domain at their N-terminus, the BTG domain, also called the APRO domain [Citation1]. These proteins are present in metazoans, with six members in vertebrates. The latter can be classified into three subgroups based on their overall organization and sequence similarity (): the two closely related factors with short C-terminal tails BTG1 and BTG2, the two related Tob1 and Tob2 proteins possessing the longest C-terminal moiety, and the more distantly related BTG3 and BTG4 proteins [Citation1–3]. In addition to a common organization, BTG1 and BTG2 present at the C-terminus of their BTG/APRO domains a boxC motif [Citation4], hereafter defined as the 11 amino-acid long sequence DGSICVLYEAS in BTG1 and DGSICVLYEEA in BTG2. Besides sequence similarities, the BTG/Tob proteins have been associated with anti-proliferative properties as their ectopic over-expression in cultured cells reduces cell proliferation [Citation5–12]. Consistently, their endogenous expression was often observed to correlate with cell cycle arrest [Citation13–17]. Moreover, numerous analyses have reported their connection to human cancers and suggested tumour suppressor functions (reviewed in [Citation3,Citation18,Citation19]). In particular, correlations between their level of expression, disease progression and survival outcome have been observed and, on this basis, they were proposed as candidate prognosis biomarkers [Citation20–22]. BTG/Tob proteins are also involved in cellular differentiation. In particular, BTG1 and BTG2 have been implicated in neuronal development (reviewed in [Citation23]) and haematopoiesis [Citation16,Citation17,Citation19,Citation24,Citation25].
Figure 1. Organization of BTG/Tob proteins
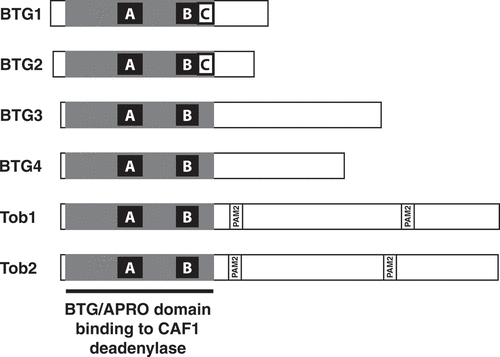
Remarkably, several BTG/Tob proteins were independently found to interact with CAF1 [Citation9,Citation26,Citation27]. The latter is a subunit of the CCR4-NOT complex, a multi-protein assembly conserved in all eukaryotes that was shown to be the predominant deadenylase catalysing the mRNA poly(A) tail shortening that initiates eukaryotic mRNA decay [Citation28–33 to cite a few]. Two subunits of the CCR4-NOT complex harbour deadenylase activities, CAF1, for which two paralogues named CNOT7 and CNOT8 exist in mammals, and CCR4, with two paralogues in mammals as well: CNOT6 and CNOT6L. BTG/Tob proteins directly bind to either the CNOT7 or the CNOT8 deadenylase (those will collectively be referred to as CAF1). This interaction has been documented at the structural level through the resolution of the crystal structure of a complex containing human CNOT7 and an N-terminal part of human Tob1 [Citation34]. This showed that the Tob1-CAF1 interaction involves residues of the strongly conserved boxA and boxB motifs found within the globular fold of the APRO domains, supporting the idea that all BTG/Tob proteins adopt a similar mode of interaction with CAF1 and that their evolutionary conserved role is to impact the functions of the CCR4-NOT complex. The resolution of the structures of free human and mouse BTG2 revealed that its 3D organization is similar to the one of the N-terminal region of Tob1 [Citation35]. In the BTG2 structure, boxC corresponds essentially to the final beta-strand of the APRO domain [Citation35]. Overall, structural analyses established that the APRO domain folds as a unique and independent globular module, which does not significantly change its conformation upon CAF1 binding.
Functionally, ectopic over-expression of BTG2 or Tob1 in cells stimulated CAF1 activity leading to deadenylation and decay of RNA reporters and endogenous transcripts [Citation36–38]. Importantly, the ability of the BTG/Tob proteins to stimulate mRNA deadenylation and decay correlated with their anti-proliferative activity [Citation11,Citation12,Citation39]. In contrast to BTG proteins, Tob factors present in their C-terminal regions two PAM2 motifs (Poly(A)-Binding-Protein-Interacting Motif 2) that mediate their association with the MLLE domain of cytoplasmic Poly(A)-Binding-Proteins (PABPC) [Citation40–42]. Mutations in PAM2 motifs that abrogate Tob1 binding to PABPC also abrogated Tob1 capacity to stimulate mRNA deadenylation [Citation36,Citation37]. These observations suggested a model by which Tob proteins stimulate RNA deadenylation by recruiting the CCR4-NOT complex to mRNA through their interaction with PABPC. This hypothesis was further supported by tethering experiments: when Tob proteins were bound directly to transcript reporters via tethering, their ability to stimulate deadenylation of the reporter did not depend anymore on intact PAM2 motifs [Citation39]. Unexpectedly, BTG1 and BTG2 were also found to interact directly with PABPC1 [Citation12]. This observation suggested that BTG1 and BTG2 use a mechanism similar to Tob factors to stimulate mRNA deadenylation, even though the motifs involved in the respective interactions of these proteins with PABPC differ. Consistently, in the absence of PAM2 motifs in BTG1 and BTG2 factors, a mutational analysis revealed that a four amino acid substitution in the boxC motif, a region conserved in the APRO domain of BTG1 and BTG2 but absent from Tob factors, BTG3 and BTG4, blocked BTG2 binding to PABPC1 and its ability to stimulate deadenylation [Citation12].
Based on yeast 2-hybrid assay results, the protein arginine methyltransferase PRMT1 was reported to interact with BTG1 and BTG2, but not with BTG3 [Citation4,Citation43]. Deletion mapping suggested that the boxC motif was required for PRMT1 interaction with BTG1 and BTG2 [Citation4]. Following this initial analysis, expression of BTG1 or BTG2 proteins containing or lacking their boxC motif was used to interpret their biological impacts as being mediated by PRMT1, for example, during erythrocyte differentiation [Citation24], following stimulation of B lymphoma cells [Citation44] or for induction of cell-cycle arrest of pre-B cells [Citation16].
The observations that mutation or deletion of the BTG1/2 boxC motif impaired binding to PABPC1 or PRMT1, respectively, raised the need to better understand the role of this motif in mediating interaction with partners. Here, we confirm that BTG1 and BTG2 boxC is the principal site mediating interaction of their APRO domains with PABPC1. We show further that, when grafted into another APRO domain, this sequence is sufficient to promote interaction with PABPC1. In contrast, we were unable to corroborate that the boxC motif is directly involved in the BTG2–PRMT1 interaction. Thus, our results reveal that the biological effects attributed to the recruitment of PRMT1 by BTG1 and BTG2, based on their obliteration in boxC deletion mutants, should be carefully reinterpreted. Indeed, our results indicate that BTG1 and BTG2 boxC is a key determinant for their capacity to induce general deadenylation. This process was shown to contribute to various biological programmes such as the maintenance of T cells in a quiescent state [Citation17,Citation45].
Material and methods
Plasmid constructions
Plasmids and oligonucleotides used in this study are presented in Supplementary Tables 1 and 2, respectively. DNA fragments amplified with PCR were verified by sequencing.
Cell culture and transfections
HEK293 cells stably expressing CNOT7-TAP fusion protein [Citation46], and HEK293 Tet-Off cells [Citation38], were maintained in DMEM medium containing 4.5 g/l glucose, GlutaMAX, 10% foetal calf serum and 40 μg/ml gentamycine. They were transfected with TurboFect transfection reagent (Thermo Scientific) according to the manufacturer’s recommendation.
RNA extraction and RACE-PAT
Total RNA was extracted using the NucleoSpin RNA kit (Macherey-Nagel). To monitor the poly(A) tail length of the β-globin transcript reporter, a modified RACE-PAT assay was performed as described previously [Citation12,Citation46]. PCR products were fractionated on 3% agarose gels, stained with ethidium bromide. Those were digitized using Typhoon FLA 9500 (GE Healthcare) and quantification was done using the ImageJ software [Citation47].
Protein co-immunoprecipitation and western blot
Co-immunoprecipitation experiments were performed using GFP-Trap magnetic agarose beads (Chromotek). Specifically, cells grown in 10 cm dishes were lysed 24 hours after transfection in 200 μl lysis buffer (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.5% Igepal CA-630) supplemented with protease inhibitors (Complete Protease Inhibitor Cocktail EDTA-free, Roche). After centrifugation, 300 μl of wash buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.5 mM EDTA) was added to the supernatant. Cell lysates were then incubated with 10 μl GFP-Trap magnetic beads for 1 hour at 4 °C. After recovery, beads were washed three times with wash buffer and eluted for 10 min at 95°C with Laemmli sample buffer (60 mM Tris-HCl pH 6.8, 10% glycerol, 0.002% bromophenol blue, 2% sodium dodecyl sulphate and 5% dithiothreitol).
Western blotting was performed by standard procedures and visualized with the Amersham Imager 600 (GE Healthcare). CNOT7-TAP, GFP fusion proteins and PRMT1 were revealed with peroxidase anti-peroxidase soluble complex (P1291, Sigma–Aldrich) used at 1/3000 dilution, monoclonal antibody anti-GFP (JL-8, Clontech) used at 1/2000 dilution and monoclonal antibody anti-PRMT1 (PRMT1-171, Sigma–Aldrich) used at 1/1000 dilution, respectively. The Luminata Crescendo Western HRP Substrate (Millipore) was used as HRP substrate.
Yeast 2-hybrid assay
The diploid yeast strain Y187/L40 (MATα/MATa, ade2-101/ade2, his3-200/his3Δ200, leu2-3,-112/leu2-3,-112, lys2-801am::LYS2::(lexAop)4-HIS3/LYS2, MEL1/?, met-/MET, trp1-901/trp1-901, gal4Δ/gal4-542, gal80Δ/gal80-538, ura3-52::URA3::GAL1UAS-GAL1TATA-lacZ/URA3::(lexAop)8-lacZ) was transformed simultaneously with the LexA-Binding-Domain and Gal4-Activating-Domain-derived plasmids by standard LiAc procedure. Interaction between the different chimeric proteins was monitored by β-galactosidase production using the Beta-Glo Assay System (Promega) according to the manufacturer’s recommendation.
Purification of recombinant proteins
Recombinant proteins were produced in E. coli BL21-CodonPlus strain (Stratagene) grown in autoinduction media (Formedium).
GST and GST-BTG2(APRO). Bacteria were lysed by sonication in buffer (30 mM HEPES-KOH pH 7.5, 100 mM NaCl, 1 mM CHAPS and 2 mM dithiothreitol (DTT)) supplemented with protease inhibitors (Complete Protease Inhibitor Cocktail EDTA-free, Roche) and benzonase (BaseMuncher, Expedeon). GST-tagged proteins were purified with affinity chromatography on GSTrap FF column and then on Sephacryl S200 16/50 column (GE Healthcare Life Sciences) with PBS 1X and stored in PBS 1X.
6His-PABPC1(1–190) and 6His-CNOT7: Bacteria were resuspended in a lysis buffer (30 mM HEPES-KOH pH 7.5, 300 mM NaCl and 1 mM DTT) supplemented with protease inhibitors (Complete Protease Inhibitor Cocktail EDTA-free, Roche) and Benzonase (BaseMuncher, Expedeon) and lysed by sonication. Proteins were purified with affinity chromatography on HisTrap FF crude column (GE Healthcare Life Sciences) followed by a size exclusion chromatography step on Superdex 75 column (GE Healthcare Life Sciences). Proteins were stored in gel filtration buffer containing 20 mM HEPES-KOH pH 7.5, 50 mM NaCl and 0.5 mM TCEP. 6His-CNOT7 sometime appeared as a doublet, probably owing to trimming of a few N- or C-terminal unstructured and less conserved residues with this construct [Citation34].
GST-Tob1(APRO) and GST-Tob1-DGSICVLYEEA. Bacteria were lysed by sonication in buffer (50 mM Tris-HCl pH 8, 150 mM NaCl and 2 mM dithiothreitol (DTT)) supplemented with protease inhibitors (Complete Protease Inhibitor Cocktail EDTA-free, Roche). GST-tagged proteins were purified with affinity chromatography on GSTrap FF column and then on Sephacryl S200 16/50 column (GE Healthcare Life Sciences) with PBS 1X and stored in PBS 1X.
Recombinant proteins for NMR studies. Recombinant 6His-PABPC1 RRM2 (amino acids 85 to 190) and 6His-GB1-fusion proteins (PABPC1 RRM1, amino acids 1 to 99, BTG2 and BTG2(APRO), amino acids 1 to 126) were over expressed in BL21(DE3) Codon plus (RIL) cells (Novagen). Cells were grown in LB rich or M9 minimal media supplemented with 15NH4Cl or 15NH4Cl and 13C-glucose at 37°C until OD600 reached 0.6–0.8, then protein expression was induced with 0.5 mM isopropyl -D-thiogalactoside (IPTG) at 20°C for 19 to 22 hours. Cells were harvested at 4°C, 20 min at 6,000 rpm and pellets were lysed with a microfluidizer at 75 PSI (0.52 MPa) in 20 mM Tris-HCl pH 8, 800 mM NaCl, 2 mM MgSO4, 5 mM CaCl2, 10 mM imidazole, 10 mM β-mercaptoethanol, 0.25 mg/mL lysozyme, 10 µg/mL DNase, one complete EDTA-free protease inhibitor tablet (Roche) in a total volume of 50 mL. Lysate cleared by centrifugation at 4°C for 30 min at 17,000 rpm was applied onto a 5 ml HiTrap Chelating HP column (GE Healthcare) charged with nickel on an AKTA prime system, washed with 20 mM Tris-HCl pH 8, 800 mM NaCl, 10 mM imidazole and 10 mM β-mercaptoethanol and eluted with 50% of the same buffer but containing 500 mM imidazole. Pooled fractions were dialysed against 3 L buffer (20 mM Na2HPO4 pH 7, 100 mM NaCl, 1 mM EDTA, 1 mM DTT) in the presence of an in-house-made TEV protease (1:50 ratio mg-TEV: mg-purified protein) at room temperature overnight to remove the 6His-GB1-tag. Additional dialysis against 2 L of the same buffer but without EDTA was performed for 4 hours. The protein was further purified by reloading the sample on the HiTrap Chelating HP column to remove the 6His-GB1-tag, overnight dialysed as above, followed by size exclusion chromatography using the Superdex75 prep grade size exclusion column (GE Healthcare) in 20 mM Na2HPO4 pH 7, 100 mM NaCl, 1 mM DTT. 6His-PABPC1-RRM2 was purified as above but without the TEV cleavage and the second HiTrap Chelating HP column step. The proteins were stored at −80°C.
NMR spectroscopy
Backbone experiments were performed in 20 mM MES pH 5.5, 50 mM NaCl, 1 mM DTT, 10% D2O, 298 K for PABPC1 RRM1 and in 20 mM Na2HPO4 pH 7, 100 mM NaCl, 1 mM DTT, 298 K for BTG2(APRO) at 600 MHz. For the 1H, 15N and 13C assignments of the protein backbone of PABPC1 RRM1 and BTG2(APRO), 2D 1H,15N-HSQC and 3D HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH and 3D 15N-NOESY-HSQC (mixing time of 100 ms) spectra were recorded.
NMR titrations were done in 20 mM Na2HPO4 pH 7, 100 mM NaCl,1 mM DTT, 10% D2O at 298 K using Bruker AVIII-500 MHz, AVIII-600 MHz and AVIII-700 MHz (equipped with cryoprobes). As the backbone experiments of RRM1 were recorded in a different buffer, an additional 3D HNCACB of PABPC1 RRM1 validated the backbone assignments also in the phosphate buffer. For titration experiments, unlabelled proteins were titrated into 15N labelled proteins at 100–400 μM. The combined chemical shift difference was calculated according to (δH2+(δN/6.51)2)1/2, were δH and δN were the differences in the 1H and 15N chemical shifts, respectively. NMR data were processed with Topspin 3.1 (Bruker) and the analysis performed using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California San Francisco). The backbone assignments of PABPC1 RRM1 and BTG2(APRO) have been deposited in the BMRB data bank under Accession Number 50526.
In vitro deadenylation assays
In vitro deadenylation assays were performed in 60 μL final reaction volume using 100 nM of a synthetic 20(A) RNA oligonucleotide labelled with Fluorescein at its 5′ end (Dharmacon), 0.7 µM of 6His-PABPC1(1–190), 1.8 µM of GST, GST-BTG2(APRO), GST-Tob1(APRO) or GST-Tob1-DGSICVLYEEA fusion proteins, and 0.4 µM of 6His-CNOT7. Reactions were performed at 30°C in buffer containing 20 mM HEPES-KOH pH 8.0; 100 mM KCl; 0.5 mM DTT; 0.1% Igepal CA-630; 5 mM MgAc. At the indicated time points, one aliquot (10 µL) of the reaction was mixed with an equal volume of RNA dye buffer (100 mM Tris-HCl pH 7.5; 1 mM EDTA; 0.001% bromophenol blue; 0.001% xylene cyanol; 0.05% sodium dodecyl sulphate; and 92% formamide) and kept on ice. Products were fractionated by electrophoresis on 15% denaturing polyacrylamide gel, detected with a Typhoon FLA 9500 (GE Healthcare), and quantified with the ImageJ software.
Results
NMR mapping of residues mediating the BTG2-PABPC1 interaction strengthens the key role of the boxC motif
We previously reported that BTG1 and BTG2 APRO domains interact with the first RRM domain of the cytoplasmic Poly(A)-Binding Protein PABPC1 [Citation12]. Furthermore, a four amino acid substitution in BTG2 boxC specifically impaired binding with PABPC1 RRM1 [Citation12]. Yet, as the structure of the BTG2(APRO) – PABPC1 RRM1 heterodimer remains unknown, one could not exclude that the boxC mutation indirectly affected association of these two factors without targeting directly the interaction interface. To identify residues involved in protein contact between BTG2 and the RRM1 domain of PABPC1, we resorted to NMR chemical shift mapping. We carried out backbone resonance assignments of BTG2(APRO) and titrations of recombinantly expressed 15N labelled BTG2(APRO) and unlabelled PABPC1 RRM1, or RRM2 as a control, monitoring resonances in 1H-15N-HSQC spectra ( and Supplementary Fig. 1). The chemical shift perturbations of 15N labelled BTG2(APRO) upon PABPC1 RRM1 addition are mostly in intermediate to fast exchange regime on the NMR time scale ( and B). In contrast, upon RRM2 addition, only a few chemical shifts show a fast exchange regime, indicating a very weak or unspecific binding of BTG2(APRO) to PABPC1 RRM2 ( blue and Supplementary Fig. 1). These data confirm the specificity of the BTG2(APRO)-PABPC1 RRM1 interaction. In these spectra, 73% of the BTG2(APRO) backbone residues are assigned, others being absent most likely because some exchanges cause line broadening and/or because of multiple conformations. Nevertheless, all central β-strands of the known human BTG2 structure (pdb code 3DJU, [Citation35], ) could be assigned. Mapping residues whose chemical shift perturbations disappear at a ratio of BTG2(APRO)-RRM1 above 1:0.25/1:0.5 and those with large chemical shift perturbations (twice above the standard deviation) on the protein sequence and on the 3D-structure identifies a region of BTG2 that is likely to mediate the interaction with PABPC1 RRM1 (). Interestingly, this region encompasses the boxC motif as well as the contiguous C-terminal region of boxB and adjacent amino acids in the folded protein (notably at the C-terminus of the first α-helix). Most importantly, it contains the residues whose substitution was previously reported to block interaction with PABPC1 RRM1 [Citation12]. Specificity was demonstrated by the fact that chemical shifts were not observed anymore when the 15N labelled mutant BTG2(APRO) domain carrying the four amino acid substitution (BTG2-kGpvkVLYEEA, residues substituted in boxC are in lowercase) was mixed with unlabelled PABPC1 RRM1 ().
Figure 2. NMR identification of the BTG2 residues involved in its interaction with PABPC1 RRM domains
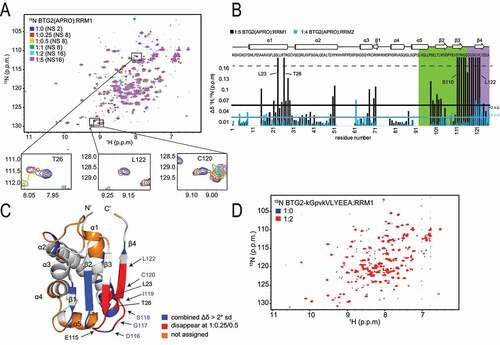
In converse experiments, using 15N labelled PABPC1 RRM1 and unlabelled BTG2, we mapped PABPC1 residues whose chemical shifts are altered upon binding. Backbone resonance assignments of RRM1 revealed the involvement of RRM1 α1, β2 ( and B) and the loop following β2. Interestingly, residues within α1, such as T23, M26 and Y28 showed the strongest shifts and multiple residues in α1 disappeared already at a 1:0.25 RRM1:BTG2 ratio. These major changes in chemical environment indicate direct interactions or conformational modifications of this element. Consistently, these shifts were not detected when the BTG2(APRO) domain carrying the four amino acid substitution (BTG2-kGpvkVLYEEA) preventing its interaction with PABPC1 RRM1 was used (). Most residues whose chemical shifts are altered upon binding of BTG2 to PABPC1 form a contiguous surface on RRM1 (, pdb code 1CVJ, [Citation48]), suggesting its direct involvement in the interaction with BTG2.
Figure 3. NMR identification of the PABPC1 RRM1 residues involved in the interaction with BTG2
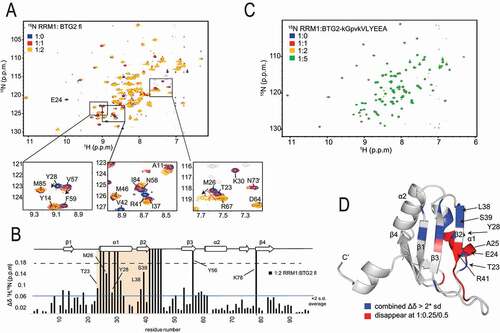
Altogether, these NMR experiments confirm the involvement of BTG2 boxC and show chemical shift changes in the C-terminal end of α1 and part of the β-sheet, notably boxC residues, upon binding of PABPC1 RRM1. Conversely, PABPC1 RRM1 recognizes BTG2 via its α1, β2 and loop following β2.
BTG2 boxC motif is sufficient to mediate interaction to PABPC1
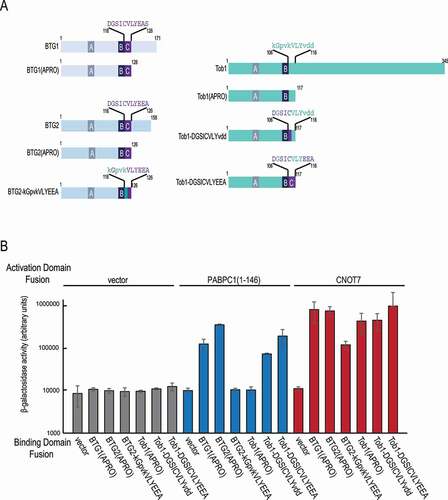
As NMR analyses confirmed a direct role of the BTG2 boxC motif in binding PABPC1, we next tested whether boxC in the context of an APRO domain, might be sufficient for this interaction. The BTG2–PABPC1 interaction was monitored through β-galactosidase production in a yeast 2-hybrid assay. We had previously used this system to show that an N-terminal region of human PABPC1 encompassing its first and part of its second RRM domains (amino acids 1 to 146) interacts with the BTG1 and BTG2 APRO domains but not with the Tob1 APRO domain [Citation12, see also BTG1(APRO], BTG2(APRO) and Tob1(APRO) in ). Using the structural similarity between the APRO domains of BTG2 and Tob1 [Citation34,Citation35] to guide site-directed mutagenesis experiments, we constructed BTG1, BTG2 and Tob1 derivatives where sequences corresponding to boxC were exchanged (). These constructs are named according to the protein backbone (BTG2 or Tob1) followed by the sequence of the boxC region with residues DGSICVLYEEA present in BTG2 (or common to BTG2 and Tob1) in the upper case and residues from Tob1 in the lower case (kGpvkVLYvdd, note the four shared residues in upper case) (). Control assays and the 1H-15N-HSQC spectra of BTG2(APRO) and BTG2(APRO) boxC mutant (BTG2-kGpvkVLYEEA) demonstrated that these mutations did not strongly affect the folding of the APRO domain in these derivatives as they were still able to interact with human CNOT7 () and the chemical shift changes were only observed in the corresponding boxC region and the neighbouring β3 strand (). Consistent with previous results [Citation12], substituting four of the five residues at the N-terminus of boxC in the BTG2 APRO domain (boxC mutant construct BTG2-kGpvkVLYEEA) abolished the interaction with PABPC1(1–146) (). Interestingly, the introduction of a partial or complete BTG2 boxC into the Tob1 APRO domain in derivatives Tob1-DGSICVLYvdd and Tob1-DGSICVLYEEA, respectively, was sufficient to create a binding site for the first RRM domains of PABPC1 as shown by a > 10-fold increase in β-galactosidase production in the 2-hybrid assay (). This gain-of-function result demonstrates the key role of boxC within the APRO domain in mediating the interaction with PABPC1. We concluded that the boxC motif in the context of an APRO fold is not only necessary but also sufficient to allow binding to the first RRM domains of PABPC1.
Figure 4. The boxC motif is sufficient and necessary to allow binding to PABPC1
Grafting a boxC motif into the Tob1 APRO domain enhances its ability to stimulate deadenylation in cellulo and in vitro
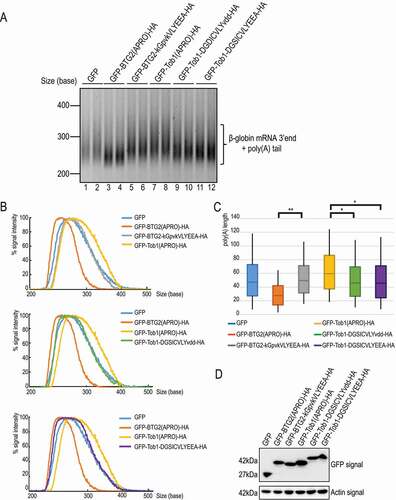
Having generated Tob1 APRO domain derivatives able to bind PABPC1, we next tested whether this property also enabled the chimeric proteins to stimulate deadenylation. To this end, constructs encoding GFP- and HA-tagged wild-type BTG2 and Tob1 APRO domains, as well as their chimeric derivatives, were transfected in HEK293 Tet-Off cells [Citation38] together with a β-globin mRNA reporter expressed under the control of a tetracycline-regulated promoter. The steady-state poly(A) tail length of the β-globin transcript was then monitored by RACE-PAT, an RT-PCR-based assay [Citation12,Citation46]. In this assay, in agreement with results published previously [Citation12], stimulation of deadenylation by the BTG2(APRO) fusion protein results in shorter poly(A) tails of the β-globin reporter as compared to control cells expressing GFP alone or the BTG2 boxC mutant (BTG2-kGpvkVLYEEA) (, note that RACE-PAT was performed in duplicate to establish reproducibility). Long poly(A) tails are observed in cells expressing the wild-type Tob1 APRO domain (). These tails appeared slightly longer than those detected in GFP transfected cells, probably as a result of ectopically expressed Tob1(APRO) acting as a competitor towards endogenous BTG/Tob factors, binding CAF1 without promoting deadenylation. When a partial or complete BTG2 boxC motif was grafted into the Tob1 APRO domain (Tob1-DGSICVLYvdd and Tob1-DGSICVLYEEA), the poly(A) tails of the β-globin reporter transcripts were shorter compared to those observed in cells expressing wild-type GFP-Tob1(APRO) (, compare lanes 9–10 and 11–12 with lanes 7–8; profiles in and box plots in ). Yet, the expression of the Tob1 derivatives was not as efficient as the expression of wild-type GFP-BTG2(APRO) in shortening the reporter poly(A) tails (, compare lanes 9–12 and lanes 3–4, respectively). Since the expression level () and binding to CNOT7 () are similar between the Tob1 chimera and the BTG2(APRO) construct, we hypothesize that the Tob1 derivatives have a less potent intrinsic ability to stimulate deadenylation possibly because of reduced binding to PABPC1 relative to BTG2(APRO) (). Nevertheless, this gain-of-function indicates that grafting the BTG2 boxC into a Tob1 backbone confers the ability to stimulate mRNA deadenylation in vivo.
Figure 5. Presence of a boxC motif enhances Tob1 APRO ability to stimulate deadenylation in cellulo
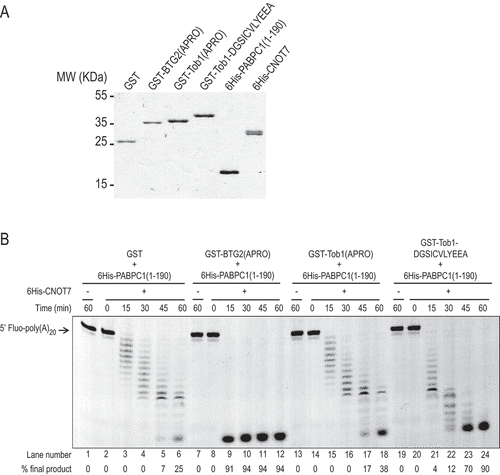
To ascertain that the introduction of a boxC motif in Tob1 directly enhances its ability to stimulate deadenylation, we conducted in vitro deadenylation assays [Citation12] with purified CNOT7, PAPBC1 and BTG/Tob derivatives. Recombinant GST-BTG2 (APRO), GST-Tob1(APRO), a Tob1(APRO) derivative carrying the BTG2 boxC (GST-Tob1-DGSICVLYEEA) and GST, as well as 6His-tagged CNOT7 and 6His-tagged PABPC1(1–190) encoding its two first RRMs were produced in E. coli (). Purified proteins were mixed with a 5ʹ-fluorescein-labelled 20-oligoadenosine long RNA substrate and samples were collected at various time points of incubation. Monitoring the resulting deadenylation by denaturing gel electrophoresis demonstrated a progressive shortening of the oligo(A) substrate by 6His-CNOT7 in presence of 6His-PABPC1(1–190) and GST over the 60 min of the reaction (, lanes 2–6). Consistent with previous results [Citation12,Citation49], GST-BTG2(APRO) strongly accelerated deadenylation with 91% of the substrate completely degraded by 15 minutes (lanes 8–12). At the same time point, no completely deadenylated product was observed in the presence of GST-Tob1(APRO) (lanes 14–18). The Tob1-DGSICVLYEEA chimera accelerated deadenylation (lanes 20–24) relative to controls, with 90% of the substrate completely deadenylated after 60 minutes (lanes 20–24). For comparison, at the same time point, only 38% of final deadenylated product was present in reactions containing wild-type Tob1(APRO) (, lane 18). The enhanced ability of BTG2(APRO) to stimulate in vitro deadenylation relative to the recombinant Tob1 chimera parallels our in cellulo results (). Importantly, no substrate degradation was detected when the RNA was incubated with the recombinant proteins in the absence of 6His-CNOT7 (, lanes 1, 7, 13 and 19) indicating that CNOT7 was the sole activity responsible for the deadenylation observed. This result confirms that the introduction of a boxC motif in the Tob1 APRO domain confers to the chimeric construct the ability to stimulate CNOT7 deadenylase activity in vitro.
Figure 6. A chimeric Tob1 APRO domain with a boxC motif stimulates CNOT7 deadenylase activity in vitro
Altogether, our results demonstrate that replacing a few residues of the Tob1 APRO domain by those of BTG2 boxC is sufficient to mediate PABPC1 binding and thereby stimulate mRNA deadenylation in cellulo and in vitro.
Association with PRMT1 is not a conserved feature of BTG/Tob proteins
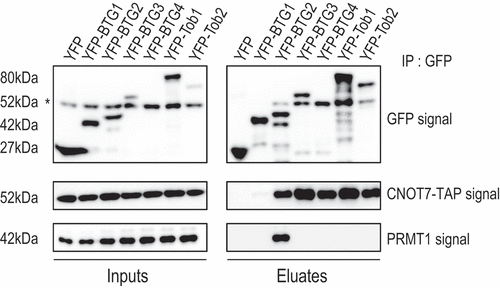
It has been shown previously that BTG1 and BTG2, but not BTG3, interacted with PRMT1 in yeast 2-hybrid assays and pull-downs of in vitro translated proteins [Citation4,Citation43]. To the best of our knowledge, there are no data documenting an interaction of Tob proteins with PRMT1. To determine which members of the human BTG/Tob protein family associate with PRMT1, we performed co-immunoprecipitation (IP) experiments with YFP-tagged full-length human BTG/Tob proteins. The proteins were expressed in HEK293 cells that stably express a TAP-tagged version of the CAF1 paralogue CNOT7 [Citation46]. All BTG/Tob proteins co-precipitated CNOT7-TAP () even though the CNOT7-TAP signal observed in the co-IP with YFP-BTG1 was reproducibly low. We have routinely observed that BTG1 co-precipitates CNOT7 less efficiently than the other BTG/Tob members even in the context of endogenous CNOT7 (Supplemental ). Nevertheless, YFP-BTG1 stimulated the decay of a co-expressed β-globin reporter mRNA as efficiently as BTG2 (Supplemental ), indicating that the YFP-BTG1 construct is functional. Assaying PRMT1 revealed efficient co-IP only with YFP-BTG2 (). As BTG1 has been reported to interact with PRMT1 in 2-hybrid and in vitro binding assays [Citation4,Citation43], we were surprised to observe that PRMT1 was not detected in co-precipitation with YFP-BTG1. However, given that YFP-BTG1 does not co-precipitate efficiently CNOT7, it is possible that the same applies to PRMT1. Overall, we observed that PRMT1 interacts with BTG2 but not BTG3 in co-IP experiments, consistent with the results of previous 2-hybrid and in vitro binding assays [Citation4,Citation43]. Our analysis shows further that BTG4, Tob1 and Tob2 do not co-IP PRMT1, indicating that its interaction with BTG2, and possibly BTG1, is not a general feature of all human BTG/Tob family members.
Figure 7. Co-precipitation of PRMT1 with BTG2
BTG2 boxC motif does not mediate the interaction of BTG2 with PRMT1
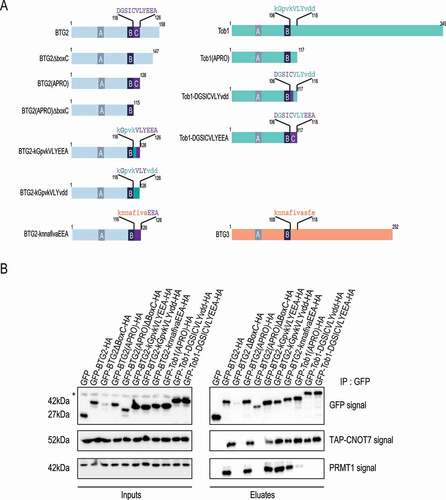
Gradual deletions from the C-terminus of BTG1 identified the boxC motif as required for its interaction with PRMT1 in a yeast 2-hybrid assay [Citation4]. An internal boxC deletion also abolished BTG1-PRMT1 heterodimer formation. Further, in a pull-down assay, in vitro translated PRMT1 precipitated with GST-BTG2 but not with a boxC-deleted GST-BTG2 protein [Citation4]. These observations led the authors to propose that the boxC motif, conserved only in the BTG1 and BTG2 APRO domains, was responsible for their association with PRMT1. As we uncovered that boxC is required for the interaction with PABPC1, we aimed to better characterize its role in mediating the association of BTG2 with PRMT1. To this end, we expressed a battery of derivatives of the BTG2 and Tob1 proteins fused to GFP and HA tags () in the HEK293 cells that stably express CNOT7-TAP and tested their ability to co-precipitate CNOT7-TAP and endogenous PRMT1 (). Deletion of the boxC motif from either the full-length BTG2 protein or its APRO domain led to their inability to co-precipitate PRMT1 (). However, these constructs did not co-precipitate CNOT7-TAP either, and were furthermore poorly expressed. Moreover, GFP aggregates could be visualized in the transfected cells (Supplemental Fig. 3). We attribute these properties to improper folding of the APRO domain lacking the boxC sequence, rendering the resulting polypeptides completely inactive. Indeed, the boxC motif contributes to the globular fold of the BTG2 APRO domain as determined by X-ray crystallography [Citation35], which likely explains the inability of boxC deletion mutants to co-precipitate CNOT7 and PRMT1. Substituting boxC residues of BTG2 by those found at equivalent positions in the structure of Tob1 (GFP-BTG2-kGpvkVLYEEA-HA and GFP-BTG2-kGpvkVLYvdd-HA) bypasses the folding problems of the mutant constructs as shown by their ability to bind CNOT7 (). These derivatives retained the ability to bind PRMT1 () revealing that the BTG2 boxC residues are not required for binding PRMT1. As some residues are conserved between the sequences of Tob1 and BTG2 (), we also introduced in the BTG2 APRO domain residues found at the equivalent location in BTG3 (construct GFP-BTG2-knnafivaEEA). Notably, BTG2 boxC and its BTG3 equivalent do not share common residues. This derivative co-precipitated CNOT7, indicating that it was at least partially folded. Moreover, it also precipitated PRMT1. Taken together, these results indicate that the boxC motif is not required for the association of BTG2 with PRMT1, supporting that earlier observations were likely the result of improper folding of deletion constructs. In parallel, we also introduced the BTG2 boxC sequence in Tob1 (GFP-Tob1-DGSCIVLYvdd-HA and GFP-Tob1-DGSCIVLYEEA-HA constructs). The resulting proteins were correctly expressed, associated with CNOT7 but failed to co-precipitate PRMT1 () despite the presence of a BTG2 boxC motif. This indicates that the boxC motif is not sufficient to mediate association with PRMT1. In summary, we confirmed that the BTG2 APRO domain co-precipitates specifically with PRMT1, yet our detailed mutational analysis failed to identify a role for boxC in this association.
Figure 8. The boxC motif is not mediating the interaction of BTG2 with PRMT1
Discussion
This study aimed to characterize the basis for binding of BTG1 and BTG2 with their PRMT1 and PABPC1 partners and the biological roles of these interactions. Our results confirm that BTG2 interacts with these two partners. While we demonstrate that its boxC motif is necessary and sufficient in the context of an APRO domain for PABPC1 binding and ensuing biological consequences, our observations indicate that, in contrast to earlier interpretations, the boxC motif is not essential for the interaction of BTG2 with PRMT1.
Evidence for the BTG1/2-PRMT1 interaction stemmed from 2-hybrid assays and pull-down of in vitro translated protein with recombinant factors [Citation4,Citation43]. Here, we confirm the association of BTG2 with PRMT1 by co-precipitation experiments consistent with previous reports [Citation50,Citation51]. We did not observe co-IP of PRMT1 with YFP-tagged BTG1 but, at this stage, cannot rule-out that this results from less efficient co-precipitation. Our data indicate further that, with the possible exception of BTG1, other BTG/Tob family members do not show interaction with PRMT1 in co-IP experiments. Original evidence for the boxC motif to be involved in the interaction of BTG1 and BTG2 with PRMT1 were based on the analysis of deletion mutants [Citation4]. This observation is not confirmed by substitution analysis, and the discrepancy is likely due to a folding problem. Indeed, we now know that the boxC motif is an integral element of the APRO domain globular fold [Citation34,Citation35]. In its absence, the entire APRO domain is likely unfolded as indicated by its inability to bind CNOT7 (), and thus also incapable to bind other partners. The specific BTG2 elements needed for its association with PRMT1 remain unknown. An increase in specific protein methylation has been observed following BTG1 or BTG2 expression, and was linked to growth arrest of haematopoietic cells [Citation16,Citation24,Citation44], inhibition of apoptosis during DNA double-strand break repair [Citation51] and regulation of transcription factor activities [Citation50,Citation52,Citation53]. Enhanced methylation was proposed to result from the direct interaction of BTG1 and BTG2 with PRMT1, and in some instances, these effects were not observed upon deletion of boxC from BTG1 or BTG2 [Citation16,Citation24,Citation44]. However, in light of our observations of the impact of this mutation on the interaction with CNOT7, PRMT1 and possibly other partners, no direct mechanistic conclusions can be drawn. Identification of the elements mediating direct or indirect association of BTG2 (or BTG1) with PRMT1 will be important to decipher the mechanisms involved in these processes and in particular to verify whether increased protein methylation is a direct consequence of BTG1 or BTG2 interaction with PRMT1.
NMR structural analyses, interaction assays as well as functional analyses demonstrate that the conserved boxC motif present in BTG1 and BTG2 mediates their association with PABPC1 RRM domains. The NMR data clearly identify boxC in BTG2 and RRM1 in PABPC1 as the two regions making direct contacts (). These observations agree with our interaction mapping and the fact that substitutions in boxC prevent BTG2-PABPC1 binding (, [Citation12]). It is noteworthy that our results rest largely on the analyses of mutants in which residues at equivalent position to boxC were swapped between different members of the BTG/Tob family. This strategy ensured that the resulting proteins are properly folded, a conclusion supported by monitoring CNOT7 binding in addition to NMR analyses ().
Importantly, introduction of a boxC motif in the Tob1 APRO domain was sufficient to allow its interaction with PABPC1 and to enable stimulation of deadenylation in cellulo and in vitro. The Tob1 chimera was less efficient than BTG2(APRO) to induce poly(A) shortening in these assays, likely as a result of a lower affinity to PABPC1 () stemming from the contribution of other residues that fine-tune this interaction (). Yet, this gain-of-function result strongly argues for a mechanism by which the BTG1 and BTG2 APRO domains recruit the CAF1 deadenylase to the 3ʹ poly(A) extremity of mRNAs through their capacity to interact with CAF1 and PABPC1 simultaneously, thereby triggering deadenylation. In contrast, Tob proteins interact with CAF1 through their APRO domains and with PABPC1 through PAM2 motifs in their C-terminal region downstream of the APRO domain [Citation36,Citation37,Citation39,Citation41]. As PAM2 motifs are also found in many other proteins involved in translational regulation and/or mRNA decay [Citation40,Citation54], Tob proteins have to compete with these factors, unlike BTG1 and BTG2. The relative interactions of PAM2-containing proteins with PABPC1 depend on their differential binding affinities [Citation55] and may be regulated by post-translational modifications [Citation56]. Interestingly, BTG3 and BTG4 were also proposed to interact with poly(A)-binding proteins. Indeed, BTG4 has been shown to regulate, together with CAF1, maternal mRNA decay during late oocyte maturation and early embryogenesis in mice [Citation57–59]. This agrees with the high expression of BTG4 in oocytes [Citation57,Citation58]. The importance of BTG4 in these processes is emphasized by the identification of mutations in humans responsible for female infertility resulting from defects in embryonic development [Citation60]. Yet, the mechanism by which BTG4 targets maternal mRNAs for degradation is still under debate, and may involve the association of BTG4 with the ubiquitous nuclear poly(A) binding protein PABPN1 [Citation59], the maternally expressed PABPN1L protein [Citation61], or the embryonic cytoplasmic poly(A) binding protein PABPC1L [Citation58]. The latter study also reported an association between BTG3 and PABPC by co-IP experiments with ectopically over-expressed fusion proteins [Citation58], but the consequences of these interactions are unknown. Given that BTG3 and BTG4 lack PAM2 or boxC motifs, further analyses are required to delineate their mode of interaction with poly(A)-binding proteins. Nevertheless, these observations suggest a general mode of action by which all BTG/Tob family members regulate mRNA deadenylation via a dual interaction. They serve as adaptors that recruit CAF1 deadenylases as effector enzymes, through the conserved boxA and boxB motifs of the APRO domain, to polyadenylated mRNAs via their association with poly(A)-binding proteins through a more variable mode of interaction.
As poly(A)-binding proteins cover the poly(A) tails of all cellular mRNAs, this raises the question whether BTG/Tob factors induce the deadenylation of all mRNAs or whether some specific mRNAs are targeted. In the latter case, additional mechanisms would be required to ensure transcript specificity. Ectopic over-expression of BTG2 or Tob1 in cells has been found to target all reporters and endogenous transcripts tested [Citation36,Citation38]. However, Tob1 was also shown to interact directly with the RNA-binding protein cytoplasmic polyadenylation element-binding protein 3 (CPEB3) [Citation62], identifying a possible mechanism for preferential action. Indeed, Tob1-mediated recruitment of CAF1 to mRNAs containing CPE elements in their 3ʹ UTR triggered their enhanced deadenylation and decay [Citation62]. The region of Tob1 involved in the binding to CPEB3 is present downstream of its APRO domain and is conserved only in Tob1 and Tob2. Accordingly, Tob1 and Tob2 were shown to regulate the stability of the CPE-containing c-myc mRNA to control cell growth [Citation63]. Interestingly, a recent report showed that endogenous BTG1 and BTG2 expression in naïve T cells triggers global mRNA decay [Citation17]. This study showed further that this function is important to control naïve T cell quiescence by maintaining the activation signal below a threshold. Indeed, conditional depletion of the Btg1 and Btg2 genes in murine T cells led to their enhanced activation and proliferation even in the absence of stimulation [Citation17]. These recent results confirm the important function BTG1 and BTG2 play in controlling cellular activation states by regulating global mRNA decay. The capacity of the CCR4-NOT complex to adjust the global rate of mRNA degradation [Citation45] indicates that this is a fundamental process by which cells control gene expression patterns in organismal development and cellular differentiation programmes.
Author contributions
F.M. and B.S. designed the project; H.A., N.R., S.S. and F.M. performed experiments; F.M., B.S., F.H.-T.A., G.S., H.A. and N.R. analysed the data. F.H.-T.A. supervised the NMR work. F.M., B.S., H.A., G.S., N.R., and F.H.-T.A. wrote the paper.
Supplemental Material
Download PDF (2.9 MB)Acknowledgments
We acknowledge our team members for their support and suggestions and thank N. Karcher, N. Crespo and S. Bahit for their assistance with plasmid construction. We also thank G. Stier and A. Geerlof [European Molecular Biology Laboratory (EMBL)] for providing the petGB1-1a plasmid.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Matsuda S, Rouault J, Magaud J, et al. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001;497:67–72.
- Tirone F. The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol. 2001;187:155–165.
- Winkler GS. The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol. 2010;222:66–72.
- Berthet C, Guehenneux F, Revol V, et al. Interaction of PRMT1 with BTG/TOB proteins in cell signalling: molecular analysis and functional aspects. Genes Cells. 2002;7:29–39.
- Rouault JP, Rimokh R, Tessa C, et al. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–1670.
- Matsuda S, Kawamura-Tsuzuku J, Ohsugi M, et al. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene. 1996;12:705–713.
- Montagnoli A, Guardavaccaro D, Starace G, et al. Overexpression of the nerve growth factor-inducible PC3 immediate early gene is associated with growth inhibition. Cell Growth Differ. 1996;7:1327–1336.
- Yoshida Y, Matsuda S, Ikematsu N, et al. ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene. 1998;16:2687–2693.
- Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J, et al. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene. 1999;18:7432–7441.
- Buanne P, Corrente G, Micheli L, et al. Cloning of PC3B, a novel member of the PC3/BTG/TOB family of growth inhibitory genes, highly expressed in the olfactory epithelium. Genomics. 2000;68:253–263.
- Doidge R, Mittal S, Aslam A, et al. The anti-proliferative activity of BTG/TOB proteins is mediated via the caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the Ccr4-Not complex. PLoS One. 2012;7:e51331.
- Stupfler B, Birck C, Seraphin B, et al. BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat Commun. 2016;7:10811.
- Rouault JP, Falette N, Guehenneux F, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486.
- Iacopetti P, Michelini M, Stuckmann I, et al. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc Natl Acad Sci U S A. 1999;96:4639–4644.
- Rodier A, Marchal-Victorion S, Rochard P, et al. BTG1: a triiodothyronine target involved in the myogenic influence of the hormone. Exp Cell Res. 1999;249:337–348.
- Dolezal E, Infantino S, Drepper F, et al. The BTG2-PRMT1 module limits pre-B cell expansion by regulating the CDK4-Cyclin-D3 complex. Nat Immunol. 2017;18:911–920.
- Hwang SS, Lim J, Yu Z, et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science. 2020;367:1255–1260.
- Lim IK. TIS21 (/BTG2/PC3) as a link between ageing and cancer: cell cycle regulator and endogenous cell death molecule. J Cancer Res Clin Oncol. 2006;132:417–426.
- Yuniati L, Scheijen B, Van Der Meer LT, et al. Tumor suppressors BTG1 and BTG2: beyond growth control. J Cell Physiol. 2019;234:5379–5389.
- Mollerstrom E, Kovacs A, Lovgren K, et al. Up-regulation of cell cycle arrest protein BTG2 correlates with increased overall survival in breast cancer, as detected by immunohistochemistry using tissue microarray. BMC Cancer. 2010;10:296.
- Bai Y, Qiao L, Xie N, et al. Expression and prognosis analyses of the Tob/BTG antiproliferative (APRO) protein family in human cancers. PLoS One. 2017;12:e0184902.
- Shen S, Zhang R, Guo Y, et al. A multi-omic study reveals BTG2 as a reliable prognostic marker for early-stage non-small cell lung cancer. Mol Oncol. 2018;12:913–924.
- Micheli L, Ceccarelli M, Farioli-Vecchioli S, et al. Control of the normal and pathological development of neural stem and progenitor cells by the PC3/Tis21/Btg2 and Btg1 genes. J Cell Physiol. 2015;230:2881–2890.
- Bakker WJ, Blazquez-Domingo M, Kolbus A, et al. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J Cell Biol. 2004;164:175–184.
- Tijchon E, Van Emst L, Yuniati L, et al. Tumor suppressors BTG1 and BTG2 regulate early mouse B-cell development. Haematologica. 2016;101:e272–6.
- Bogdan JA, Adams-Burton C, Pedicord DL, et al. Human carbon catabolite repressor protein (CCR4)-associative factor 1: cloning, expression and characterization of its interaction with the B-cell translocation protein BTG1. Biochem J. 1998;336(Pt 2):471–481.
- Rouault JP, Prevot D, Berthet C, et al. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J Biol Chem. 1998;273:22563–22569.
- Tucker M, Valencia-Sanchez MA, Staples RR, et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in saccharomyces cerevisiae. Cell. 2001;104:377–386.
- Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455.
- Temme C, Zaessinger S, Meyer S, et al. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871.
- Wiederhold K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem Soc Trans. 2010;38:1531–1536.
- Yi H, Park J, Ha M, et al. PABP Cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol Cell. 2018;70:1081–8 e5.
- Raisch T, Chang CT, Levdansky Y, et al. Reconstitution of recombinant human CCR4-NOT reveals molecular insights into regulated deadenylation. Nat Commun. 2019;10:3173.
- Horiuchi M, Takeuchi K, Noda N, et al. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J Biol Chem. 2009;284:13244–13255.
- Yang X, Morita M, Wang H, et al. Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic Acids Res. 2008;36:6872–6881.
- Ezzeddine N, Chang TC, Zhu W, et al. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol. 2007;27:7791–7801.
- Funakoshi Y, Doi Y, Hosoda N, et al. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148.
- Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048.
- Ezzeddine N, Chen CY, Shyu AB. Evidence providing new insights into TOB-promoted deadenylation and supporting a link between TOB’s deadenylation-enhancing and antiproliferative activities. Mol Cell Biol. 2012;32:1089–1098.
- Albrecht M, Lengauer T. Survey on the PABC recognition motif PAM2. Biochem Biophys Res Commun. 2004;316:129–138.
- Okochi K, Suzuki T, Inoue J, et al. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells. 2005;10:151–163.
- Kozlov G, Menade M, Rosenauer A, et al. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J Mol Biol. 2010;397:397–407.
- Lin WJ, Gary JD, Yang MC, et al. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044.
- Hata K, Nishijima K, Mizuguchi J. Role for Btg1 and Btg2 in growth arrest of WEHI-231 cells through arginine methylation following membrane immunoglobulin engagement. Exp Cell Res. 2007;313:2356–2366.
- Sharma S, Poetz F, Bruer M, et al. Acetylation-dependent control of global poly(A) RNA degradation by CBP/p300 and HDAC1/2. Mol Cell. 2016;63:927–938.
- Mauxion F, Preve B, Seraphin B. C2ORF29/CNOT11 and CNOT10 form a new module of the CCR4-NOT complex. RNA Biol. 2013;10:267–276.
- Schneider CA, Rasband WR, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675.
- Deo RC, Bonanno JB, Sonenberg N, et al. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845.
- Pavanello L, Hall B, Airhihen B, et al. The central region of CNOT1 and CNOT9 stimulates deadenylation by the Ccr4–Not nuclease module. Biochem J. 2018;475:3437–3450.
- Passeri D, Marcucci A, Rizzo G, et al. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol Cell Biol. 2006;26:5023–5032.
- Choi KS, Kim JY, Lim SK, et al. TIS21(/BTG2/PC3) accelerates the repair of DNA double strand breaks by enhancing Mre11 methylation and blocking damage signal transfer to the Chk2(T68)-p53(S20) pathway. DNA Repair (Amst). 2012;11:965–975.
- Van Galen JC, Kuiper RP, Van Emst L, et al. BTG1 regulates glucocorticoid receptor autoinduction in acute lymphoblastic leukemia. Blood. 2010;115:4810–4819.
- Yuniati L, Van Der Meer LT, Tijchon E, et al. Tumor suppressor BTG1 promotes PRMT1-mediated ATF4 function in response to cellular stress. Oncotarget. 2015;7:3128–3143.
- Tritschler F, Huntzinger E, Izaurralde E. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nat Rev Mol Cell Biol. 2010;11:379–384.
- Ruan L, Osawa M, Hosoda N, et al. Quantitative characterization of Tob interactions provides the thermodynamic basis for translation termination-coupled deadenylase regulation. J Biol Chem. 2010;285:27624–27631.
- Huang KL, Chadee AB, Chen CY, et al. Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. RNA. 2013;19:295–305.
- Yu C, Ji SY, Sha QQ, et al. BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat Struct Mol Biol. 2016;23:387–394.
- Liu Y, Lu X, Shi J, et al. BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis. J Mol Cell Biol. 2016;8:366–368.
- Pasternak M, Pfender S, Santhanam B, et al. The BTG4 and CAF1 complex prevents the spontaneous activation of eggs by deadenylating maternal mRNAs. Open Biol. 2016;66:160184.
- Zheng W, Zhou Z, Sha Q, et al. Homozygous mutations in BTG4 cause zygotic cleavage failure and female infertility. Am J Hum Genet. 2020;107:24–33.
- Zhao LW, Zhu YZ, Chen H, et al. PABPN1L mediates cytoplasmic mRNA decay as a placeholder during the maternal-to-zygotic transition. EMBO Rep. 2020;21:e49956.
- Hosoda N, Funakoshi Y, Hirasawa M, et al. Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. EMBO J. 2011;30:1311–1323.
- Ogami K, Hosoda N, Funakoshi Y, et al. Antiproliferative protein Tob directly regulates c-myc proto-oncogene expression through cytoplasmic polyadenylation element-binding protein CPEB. Oncogene. 2014;33:55–64.
