ABSTRACT
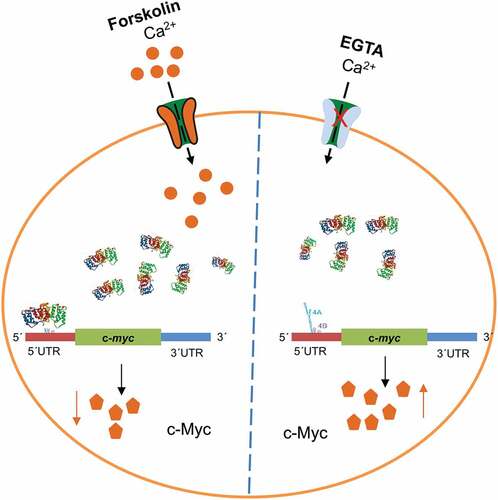
The expression and localization of the oncoprotein c-Myc is highly regulated at the level of transcription, mRNA transport, translation, as well as stability of the protein. We previously showed that Annexin A2 (AnxA2) binds to a specific localization element in the 3ʹuntranslated region (UTR) of c-myc mRNA and is involved in its localization to the perinuclear region. In the present study, we demonstrate that AnxA2 binds in a Ca2+-dependent manner to the internal ribosomal entry site (IRES) containing two pseudo-knots in the 5´UTR of the c-myc mRNA. Here, we employ an in vitro rabbit reticulocyte lysate system with chimeric c-myc reporter mRNAs to demonstrate that binding of AnxA2 to the c-myc IRES modulates the expression of c-Myc. Notably, we show that low levels of AnxA2 appear to increase, while high levels of AnxA2 inhibits translation of the chimeric mRNA. However, when both the AnxA2-binding site and the ribosomal docking site in the c-myc IRES are deleted, AnxA2 has no effect on the translation of the reporter mRNA. Forskolin-treatment of PC12 cells results in upregulation of Ser25 phosphorylated AnxA2 expression while c-Myc expression is down-regulated. The effect of forskolin on c-Myc expression and the level of Ser25 phosphorylated AnxA2 was abolished in the presence of EGTA. These findings indicate that AnxA2 regulates both the transport and subsequent translation of the c-myc mRNA, possibly by silencing the mRNA during its transport. They also suggest that AnxA2 act as a switch to turn off the c-myc IRES activity in the presence of calcium.
Abbreviations: AnxA2, Annexin A2; β2--µglob, β2-microglobulin; cpm, counts per minute; hnRNP, heterogenous nuclear ribonucleoprotein; IRES, internal ribosomal entry site; ITAF, IRES trans-acting factor; MM, multiple myeloma; PABP, poly(A)-binding protein; PCBP, poly(rC) binding protein; PSF, PTB-associated splicing factor; PTB, polypyrimidine tract binding protein; RRL, rabbit reticulocyte lysate; UTR, untranslated region; YB, Y-box binding protein.
Graphical abstract
1. Introduction
Annexin A2 (AnxA2) is a multi-functional and -compartmental protein with a variety of functions related to intercellular and intracellular communication via exosomes, cell proliferation, membrane-cytoskeleton interactions, endo- and exocytosis, as well as mRNA transport and translation [Citation1–4]. The different functions of AnxA2 are modulated by its post-translational modifications [Citation4]. Furthermore, the expression of AnxA2 is deregulated in several cancers. It is down-regulated in prostate cancer and overexpressed in a variety of cancers, including those of the pancreas, breast and brain (reviewed by [Citation5–7]). Moreover, AnxA2 is specifically upregulated in invasive and metastatic breast cancer cells, while poorly invasive cells express only low levels of the protein [Citation8]. Furthermore, the protein has been associated with increased chemotherapy resistance [Citation9].
We have previously demonstrated that AnxA2 acts as an mRNA-binding protein associated with a specific subpopulation of mRNP complexes linked to the cytoskeleton [Citation10–13]. In addition, we have mapped the mRNA-binding site of AnxA2 to specific amino acids in helices C-D of its domain IV [Citation10], as well as identified the AnxA2-binding regions of the 3´untranslated regions (UTRs) of anxA2 [Citation11] and c-myc [Citation12] mRNAs. Our initial experiments also indicated a putative AnxA2 binding-site in the c-myc 5ʹUTR. As AnxA2 binds to the c-myc mRNA [Citation11,Citation12,Citation14], it is likely to participate in the regulation of the expression of the c-Myc protein at the level of mRNA transport [Citation12] and/or translation.
The cellular proto-oncogene c-Myc encodes a basic helix-loop-helix transcription factor that acts in conjunction with its partner Max by binding to E-box DNA sequences (CACGTG). The c-Myc-Max heterodimers are activators of a critical set of target genes involved in cell proliferation, differentiation, and apoptosis [Citation15,Citation16]. Like AnxA2, deregulated expression of c-myc mRNA is associated with a wide range of cancers and their resistance to chemotherapy [Citation17]. Tumorigenesis involving c-Myc is best illustrated in the case of human Burkitt’s lymphoma, the most rapidly proliferating type of human tumour [Citation18]. The reciprocal chromosomal translocation associated with this disease recombines c-myc in juxtaposition with regulatory elements of the immunoglobulin heavy or light chain loci, leading to altered expression of c-Myc [Citation18]. Furthermore, c-Myc overexpression in transgenic mice triggers tumour development [Citation19], indicating that it affects control mechanisms that are necessary to prevent abnormal proliferation and differentiation. Increased expression of c-Myc protein is also associated with multiple myeloma (MM) and is observed in MM-derived cell-lines, which inhabit a C-U mutation in the region of c-myc mRNA thought to possess an internal ribosomal entry site (IRES) [Citation20]. The same mutation was detected in 42% of the samples derived from the bone marrow of MM patients [Citation21]. It was suggested that the mutation could result in an altered repertoire of proteins associated with the IRES, the so-called ITAFs (IRES trans-acting factors) [Citation20]. Interestingly, AnxA2 is not expressed in at least two B-cell lymphoma cell lines [Citation22].
The highly complex regulation of c-Myc expression is modulated by several signal transduction pathways at multiple levels, including transcription [Citation15,Citation16], translation [Citation23] as well as stability of the mRNA [Citation24] and protein [Citation25]. The human c-myc gene is transcribed from no less than four alternative promoters, designated as P0, P1, P2 and P3. The P1 and P2 promoters account for almost all of the cytoplasmic c-myc mRNA, with the minor promoters, P0 and P3, lying upstream and downstream of P1 and P2, respectively [Citation26]. Both human and mouse c-myc mRNAs encode two protein products of different size, c-Myc1 and c-Myc2, translated from two different initiation codons (CUG and AUG), respectively. c-Myc2 is the predominant form, which is responsible for cell growth and proliferation, whereas c-Myc1 has been reported to inhibit cell growth in the absence of c-Myc2 [Citation27]. Thus, the two isoforms are presumably involved in the regulation of different subsets of genes. The complex regulation of c-Myc expression depends on many different factors. For example, c-myc mRNA translation is regulated by the prevailing levels of the initiation factor eIF4E, which binds to the cap structure [Citation28].
The regulation of c-myc mRNA transport and translation is complicated and involves both of its UTRs and protein factors. In general, the 3ʹUTR of an mRNA is involved in its localization and stability, while the 5ʹUTR modulates gene expression by influencing the initiation of translation. However, the two UTRs interact via proteins to modulate translation. Thus, both regions are controlled via their interaction with trans-acting RNA-binding factors. Such factors recognize specific structures or sequences (cis-acting elements), present mainly in the UTRs (reviewed by [Citation29,Citation30]). In addition, small RNA molecules known as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), components of mRNP complexes, are implicated in the regulation of translation and mRNA transport. They are thought to repress translation by interacting with the 3ʹUTR of mRNA (reviewed by [Citation30,Citation31]). A stem-loop structure in the c-myc 3ʹUTR functions as a localization signal and targets c-myc mRNA to the perinuclear region for translation on cytoskeletal-bound polysomes [Citation32,Citation33]. AnxA2 binds to this localization element in the c-myc 3ʹUTR [Citation12].
The 5ʹUTR of c-myc mRNA is considerably longer than the majority of eukaryotic 5ʹUTRs and contains complex secondary structures suggesting strong regulation of the translation of the c-myc mRNA. The translation initiation of c-myc mRNA is a complex process, occurring by two different mechanisms [Citation26,Citation34]. When cap-dependent initiation is compromised, there is a shift to internal ribosomal entry site (IRES)-directed translation initiation [Citation35,Citation36]. The IRES in human c-myc mRNA is located upstream of the CUG start codon of c-Myc2. Hence, it is present in transcripts initiated from the P0, P1, and P2 promoters, and has the ability to promote cap-independent synthesis of both c-Myc isoforms (c-Myc1 and c-Myc2) [Citation26,Citation34]. However, at least in cells transformed by eIF4E, there are indications of c-Myc1 being translated by ribosomal scanning from the cap structure, while c-Myc2 is translated by internal ribosome repositioning [Citation37].
Several proteins (ITAFs) that interact with the c-myc IRES have been identified. It appears that the proteins involved in IRES-dependent translation initiation are regulated by their subcellular targeting and post-translational modifications [Citation38]. Canonical translation initiation factors are required for IRES-mediated translation, although the c-myc IRES functions in the absence of an eIF3-eIF4F interaction [Citation39]. The c-myc IRES-mediated translation is stimulated by the poly(rC) binding proteins 1 and 2 (PCBP1 and PCBP2) and heterogeneous nuclear ribonucleoprotein K (hnRNP K) [Citation40]. Interestingly, hnRNP K binds more strongly to the mutated IRES sequence found in MM, thus resulting in enhanced expression of c-Myc [Citation40]. In addition, PCBP1, PCBP2 [Citation41] and hnRNP K [Citation42] are able to shuttle between the nucleus and the cytoplasm, indicating that the proteins may interact with the c-myc IRES while still in the nucleus. Another protein, hnRNP C, binds specifically to a heptameric U sequence located between the two alternative translation initiation codons CUG and AUG in the 5ʹ-UTR of human c-myc mRNA [Citation43]. At the G2/M phase of the cell cycle, the hnRNP C is especially enriched in the cytoplasm, giving rise to an increased translation of c-myc mRNA [Citation43]. Moreover, hnRNP A1, G-rich RNA sequence binding factor 1 (GRSF-1), Y-box binding protein 1 (YB-1), polypyrimidine tract binding protein (PTB) as well as its associated splicing factor (PSF) and p54nrb (ligand of PSF) also bind to the c-myc IRES [Citation44–46]. All these proteins stimulate the translation of c-myc mRNA.
Fine-tuning of c-Myc expression is critical for key cellular functions, playing important roles in the regulation of growth, proliferation, differentiation, and apoptosis. Changes in c-Myc expression levels may shift its normal physiological role towards a pathophysiological role [Citation47]. Therefore, elucidating how the expression levels of c-Myc can be modulated is of great interest. Here, we employ an in vitro system with chimeric c-myc reporter mRNAs to demonstrate that binding of AnxA2 to the c-myc IRES modulates the expression of c-Myc.
2. Materials and methods
2.1 Recombinant Annexin A2
The bovine His-tagged AnxA2 (UniProtKB – P04272), the full-length non-RNA-binding mutant K307S-K308S-K309S-K312S-Y316S-Q320S AnxA2 [Citation10] and the His-tagged N-terminally truncated forms of bovine AnxA2 (Δ19AnxA2; starts at Pro20 and Δ33AnxA2; starts at Ala34, counting the first Ser as 1) were expressed in E. coli and subsequently purified on Ni2+-resin (Ni-NTA agarose, Qiagen) essentially as described [Citation11]. The His-maltose-binding protein (MBP)-AnxA2 (bovine), both wild type and the mutated form that does not bind RNA were also expressed and purified after tobacco etch virus (TEV) cleavage as described [Citation10,Citation11]. Note that wild type AnxA2 and mutated AnxA2 were not separated from His-MBP and His-TEV by using Q-Sepharose but were obtained by purification on a second Ni2+-affinity resin leaving the His-tagged proteins bound to the resin. All recombinant forms of AnxA2 were gel filtrated on a SuperdexTM 200 Increase 10/300 GL column (GE Healthcare Bio-Sciences AB, Sweden). The purity of recombinant AnxA2 was determined by SDS-PAGE followed by Coomassie Brilliant Blue staining (InstantBlue, Abcam, Cambridge, UK). The NanoDrop quantitation method was employed for purified AnxA2 based on its Mw, extinction coefficient (33,810 M−1cm−1) and the absorption was measured at 280 nm.
2.2 Seamless cloning of the cDNAs coding for the different regions of the chimeric mRNAs
The BbsI restriction enzyme type two was used in the seamless cloning procedures [Citation48] and was introduced in the primers used for seamless cloning (). It cleaves at N + 2 (3ʹend) and N + 6 (5ʹend), counted from the last base of its recognition site. After PCR production of each of the cDNA fragments of the chimeric mRNAs and restriction enzyme digestion, the purified PCR fragments were ligated with T4 ligase. The DNA fragments were ligated in a stepwise manner (initially two fragments were ligated, then the third), and incubated 30 min each at RT. Finally, the vector was added, and the whole reaction was ligated ON at 4°C. The constructs were ligated into the EcoRI and XbaI sites of the pGEM®-3Zf(+) plasmid (Promega, Madison, USA) for storage. All constructs were sequenced in both directions to verify their orientation and sequences. The cDNAs used for the construction of the chimeric cDNAs were from the mouse c-myc (variant 2: NM_001177353.1) and the codon-optimized Renilla luciferase cDNA (a generous gift from Dr. Beate Stern, University of Bergen, Norway).
Table 1. Primers for seamless BbsI cloning (recognition site in bold) of the c-myc chimera 1A and the and β2-µglob chimera and subsequent synthesis of PCR products with T7 promoter
2.3 In vitro transcription
RNA probes were uniformly [α-32P] UTP-labelled (3000 Ci/mmol; EasyTide, Waltham, PerkinElmer, USA) by T7-driven in vitro transcription of PCR products () for 90 min, by standard procedures as described in the Promega protocol. A specific activity of 1.0–2.5 × 108 cpm/μg RNA was obtained. Primers for the different regions of the c-myc 5ʹUTR are given in . Labelled RNA probes were analysed for integrity and purity by agarose gel electrophoresis. Annealing of the NA and NB was performed by mixing the two transcripts at a 1:1 molar ratio in Binding Buffer (10 mM triethanolamine, pH 7.4 (Fluka, Thermo Fisher Scientific, Waltham, USA), 50 mM KCl (Merck Millipore, Burlington, USA), 1 mM DTT (Sigma-Aldrich, Saint-Louis, USA), 2 mM MgSO4 (Sigma-Aldrich, Saint-Louis, USA), 70 µM CaCl2 (Sigma-Aldrich, Saint-Louis, USA), 1 mg/mL yeast tRNA (Invitrogen, Thermo Fisher Scientific, Waltham, USA). Subsequently, the RNA probes were heated to 95°C for 5 min and allowed to cool slowly to RT.
Table 2. Primers for making PCR products to produce RNA probes
2.4 Capture of AnxA2 from the cytoskeletal fraction by in vitro transcribed c-myc 5´UTR with or without the IRES
The c-myc 5´UTR with or without the IRES (region 4) were in vitro transcribed from the corresponding PCR products containing a T7 promoter site using the HiScribe T7 High Yield RNA Synthesis Kit (New England BioLabs, Boston, USA) according to the manufacturer. The DNA template was removed by treatment with RQ1 RNase-free DNase (Promega, Madison, USA) for 15 min at 37°C. After phenol/chloroform/isoamyl alcohol extraction and subsequent precipitation with 3 M sodium acetate (pH 5.2; Thermo Fisher Scientific, Waltham, USA) and ethanol (Vinmonopolet, Oslo, Norway), purified RNA transcripts were resuspended in ddH2O after centrifugation at 16,000 g for 30 min and two washes with 70% (v/v) ethanol. An antisense biotin-5´DNA oligomer (5ʹ-CGTCGTGGCTGTCTGCGGGG-3ʹ) to the 3´end of the full-length c-myc 5´UTR as well as the c-myc 5´UTR lacking region 4 was bound to the PBS pre-washed streptavidin magnetic beads (50 µL) for 3 h at room temperature in PBS containing 1 M NaCl. After three washes of the coated beads in PBS, RNA transcripts (20 pmoles) were first heated to 72°C and then cooled slowly before incubation ON at 4°C with the biotin-5´DNA oligomer (Sigma-Aldrich, Saint-Louis, USA) coupled to the magnetic streptavidin beads (Dynal, Thermo Fisher Scientific, Waltham, USA) in 130 mM KCl buffer (130 mM KCl, 5 mM MgSO4, 8.6% (w/v) sucrose, 10 mM triethanolamin; pH 7.4) containing RNasin (Promega, Madison, USA). Subsequently, the beads were washed twice in 130 mM KCl buffer and incubated with 40 µg protein from the cytoskeletal fraction derived from PC12 cells in the same buffer prepared as previously described [Citation49,Citation50] for 3 h at RT in the presence of protease inhibitors (cOmplete EDTA-free; Roche, Basel, Switzerland) and RNasin (Promega, Madison, USA) in the absence or presence of EGTA or CaCl2 as indicated in . The coated beads were washed and incubated with RNase A/T1 mix (Thermo Fisher Scientific, Waltham, USA) for 40 min at 37°C to release the proteins associated with the RNA transcripts. The released proteins were concentrated by precipitation ON with 4x volume cold acetone at −20°C. The samples were centrifuged 16,000 rpm x 30 min and the pellets were dried before resuspension in H2O and denaturation buffer. The samples were heated for 15 min at 56°C before SDS-PAGE and Western blot analyses.
2.5 Annexin A2-RNA binding
The interaction of AnxA2 with RNA was assayed by UV-crosslinking experiments, performed essentially as described [Citation11]. RNA transcripts were preheated for 3 min at 72°C, and then gradually cooled for 15 min to RT to allow folding of secondary structures. AnxA2 and mRNA were incubated in Binding Buffer for 20 min at RT in solution before UV-crosslinking. After SDS-PAGE, proteins in the gels were stained by Coomassie Brilliant Blue. Subsequently, the gels were dried and exposed to screens.
2.6 In vitro coupled transcription-translation system
The TNT® T7 Quick for PCR DNA is an in vitro coupled transcription/translation system based on rabbit reticulocyte lysate (RRL) (Promega, Madison, USA). Prior to these assays, DNA templates were PCR amplified using forward primers containing a T7 promoter site. The primers were the same for all the four c-myc chimeric constructs: 5′-TAATACGACTCACTATAGG-ACCCCTGGCTGCGCTGCTCT (5´UTR c-myc forward with a T7 promoter site) and 5ʹ-GTATTTTTTCCAATTATTTT (c-myc 3´UTR reverse). PCR products were subjected to agarose gel electrophoresis; bands of correct sizes were excised from the gel and purified.
The reaction mixture was incubated for 60 min at 30°C. Two parallels of 1 μL were spotted directly onto filter to measure the total radioactivity in the mixture. Two parallels were withdrawn for scintillation counting at 0, 15 and 30 min, while triplicates were withdrawn at 60 min and each aliquot was transferred into 1 mL of 1 M NaOH with 2% H2O2, which was subsequently incubated for 10 min at 37°C. After incubation, 2 mL of ice-cold 25% trichloroacetic acid (TCA) containing 2% (w/v) casein hydrolyzate (Sigma-Aldrich, Saint-Louis, USA) was added, and the samples were placed on ice for at least 30 min. The precipitate was filtrated on glass fibre filters and washed 7 times with 5% TCA. Subsequently, the filters were washed with 96% ethanol and 4 mL of scintillation liquid (Opti-Fluor, PerkinElmer, Waltham, USA) was added to the filters in scintillation vials. Incorporated [35S]-Met into the RLuc reporter and total radioactivity in the reaction mixture were measured in the liquid scintillation counter (Packard).
2.7 Cell cultures and drug treatments
The adherent sub-clone of rat pheochromocytoma (PC12) cells were maintained as described previously [Citation51] in RPMI 1640 medium (R-8758, Sigma-Aldrich, Saint-Louis, USA), with 10% (v/v) horse serum (PAA Laboratories GmbH, GE Healthcare, Chicago, USA), 5% (v/v) foetal bovine serum (F-9665, Sigma-Aldrich, Saint-Louis, USA), 100 units/ml penicillin, and 100 μg/mL streptomycin (P-0781; Sigma-Aldrich, Saint-Louis, USA) in a humidified 5% CO2 atmosphere at 37°C . The cells were treated with 100 μM EGTA (E-4378; Sigma-Aldrich, Saint-Louis, USA) and/or 20 μM forskolin (344282; Sigma-Aldrich, Saint-Louis, USA) for 1 h or 2 h, as indicated. Cells were briefly washed with PBS and lysed in Lysis buffer (50 mM Hepes, 150 mM NaCl, 1 mM EDTA, 0.5% (w/v) NP-40, 1 mM dithiothreitol, 1 mM Na3VO4, 1 mM NaF, and 1x EDTA-free protease inhibitor cocktail from Roche (11836170001; Roche, Basel, Switzerland).
2.8 SDS-PAGE and immunoblot analysis
Samples from lysates were heated at 70°C for 10 min in Laemmli sample buffer (Bio-Rad Laboratories, Hercules, USA) and resolved in 10% or 4–20% (w/v) SDS-PAGE gels. Proteins were transferred to nitrocellulose membranes (#162-0112; Bio-Rad Laboratories, Hercules, USA), which were then blocked with 5% (w/v) BSA, probed with antibodies and developed using enhanced chemiluminescence reagents (WesternBright Sirius, Advansta; San Jose, USA). The blots were imaged using Gel DOC XRS+ (Bio-Rad Laboratories, Hercules, USA) and densitometric analyses were performed with Image J software (NIH, Bethesda, USA). Blots treated with phospho-specific antibody were stripped with restore stripping buffer (21059; Thermo Fisher Scientific, Waltham, USA) as per manual instruction, blocked and re-probed with antibody recognizing total protein. Densitometric values were expressed per unit of protein applied to the gel lane. Proteins were normalized to loading control. Antibodies used for immunoblotting were as follows: Annexin AII (1:1000; 610069; BD Biosciences, Franklin Lakes, USA), Annexin AII (pSer25) (1:1000; OAAF00618; Aviva Systems Biology, San Diego, USA), c-Fos (1:1000; PC-38; Merck Millipore, Burlington, USA), GAPDH (1:5000; 32233; Santa Cruz Biotechnology, Dallas, USA), c-Myc (1:1000; 32072; Abcam; Cambridge, UK), P-S133-CREB (1:1000; NB300-273; Novus Biologicals, Centennial, USA), p53 (1:1000; 263; Santa Cruz Biotechnology, Dallas, USA).
2.9 Statistical analysis
The arbitrary unit values are reported as mean ± SEM. Statistical comparisons were calculated with the student t-test using GraphPad Prism 9.0. Significance level was set at p < 0.05. Data correspond to representative of 3 or 4 independent experiments.
3. Results
3.1 Annexin A2 binds to the IRES of the c-myc mRNA
We previously showed that AnxA2 interacts with the localization signal in the mouse c-myc 3ʹUTR [Citation12]. Our previous UV-crosslinking experiments revealed that it may also bind to the c-myc 5ʹUTR, including both a 1–517 nt and a full-length 1–580 nt transcript of this region (data not shown). To identify the precise region of c-myc 5ʹUTR that interacts with AnxA2, we first used the Sfold platform (Wadsworth Bioinformatics Center, http://sfold.wadsworth.org/) to fold the mouse c-myc 5ʹUTR (1–517 nt) in silico into structures of predicted minimum free energy, ΔGo [Citation52]. Consequently, the use of these proposed structures allowed the division of the 5ʹUTR (1–517 nt) into fragments whose transcription would leave the originally predicted secondary structure of the c-myc 5ʹUTR (1–517 nt) as unperturbed as possible. Thus, radio-labelled region 1 (1–127 nt; red), region 2 (128–444 nt; marked by black bar on top of the structure) and region 3 (445–517 nt; yellow) of c-myc were transcribed in vitro from the corresponding PCR fragments containing a T7 promoter (, Panels A and B). When the binding of the transcripts to AnxA2 were tested in UV-crosslinking experiments, region 2 was observed to bind AnxA2, while regions 1 and 3 failed to show binding (). Subsequently, we generated radiolabelled RNA transcripts of subdomains of region 2. These included: region 4 (grey; 196–393 nt; consisting of regions 4A and 4B); the separate region 4A (196–331 nt; light blue) and region 4B (332–393 nt; purple) domains, as well as the 4Ai (238–290 nt; pink) domain. Additionally, we generated the stem region denoted NegA (NA; orange) and NegB (NB; green), corresponding to the lower part of region 2 (, Panels A and B). Interestingly, we found that AnxA2 preferentially binds to region 4A of the c-myc 5ʹUTR (). When comparing the regions of mouse c-myc 5ʹUTR () with those of human c-myc IRES ()[Citation53,Citation54] by nt alignment, it was clear that the AnxA2-binding site in region 4A of c-myc 5ʹUTR is situated within the IRES. This site (region 4A) includes one side of the two pseudo-knots (, compare Panels A and B), while the other side is in region 4B. Region 4Ai contains only 2 nt at the 3ʹend of one of the pseudo-knots. It should be noted that when the two strands of the stem consisting of NegA and NegB were tested separately with AnxA2, a faint signal was observed, indicating weak interaction. However, when the double-stranded stem was tested with AnxA2, no binding was detected ().
Figure 1. Annexin A2 binds to the c-myc IRES. Panel A: Schematic representation of the predicted secondary structure of mouse c-myc 5ʹUTR using Sfold and the different regions (transcripts) of c-myc 5ʹUTR used in the binding experiments. Panel B: Schematic representation of the locations of regions 4A, 4Ai and 4B of mouse c-myc IRES in the experimentally deduced human c-myc IRES (adopted from [Citation53]). Panel C: The binding of different radiolabelled [α32P]-rUTP c-myc 5ʹUTR transcripts to 3 μM AnxA2 in the presence of 70 µM Ca2+. 15 fmoles of regions 1–4Ai and the stem region of region 2, consisting of NegA (NA) and NegB (NB), as indicated in Panel A, were UV-crosslinked with AnxA2. AnxA2 stained with Coomassie Brilliant Blue served as a loading control. Panel D: UV-crosslinking of 3 μM of recombinant AnxA2 bound to 20 fmoles of uniformly radiolabelled full-length c-myc 5´UTR transcript alone (lanes 2 and 5), and to increasing amounts (10- or 50-fold molar excess) of the unlabelled c-myc full-length 5´UTR (lanes 3 and 4, respectively), and region 4A of the 5´UTR (lanes 6 and 7, respectively). Lane 1 contained only the radiolabelled full-length c-myc 5´UTR transcript to assess the effect of the RNAses added after UV-exposure. AnxA2 stained with Coomassie Brilliant Blue served as a loading control. Note that the protein bands in the upper gel were slightly blurred during drying before exposure to screens and phosphor imaging. Panels C and D: The transcripts were covalently bound to AnxA2 in the presence of 1 μg/μL yeast tRNA to inhibit non-specific RNA binding and then subjected to 10% SDS-PAGE. The binding was visualized using screens and phosphor imaging after ON exposure. AnxA2 is indicated to the right. Panel E: Histogram of the results of the binding competition; UV-crosslinking of 3 μM of recombinant AnxA2 bound to 20 fmoles of uniformly radiolabelled full-length c-myc 5´UTR transcript alone (columns 1 and 4), and to increasing amounts (10- or 50-fold molar excess) of the unlabelled c-myc full-length 5´UTR (columns 2 and 3, respectively), or region 4A of the 5´UTR (columns 5 and 6, respectively). The mean of two experiments is shown
![Figure 1. Annexin A2 binds to the c-myc IRES. Panel A: Schematic representation of the predicted secondary structure of mouse c-myc 5ʹUTR using Sfold and the different regions (transcripts) of c-myc 5ʹUTR used in the binding experiments. Panel B: Schematic representation of the locations of regions 4A, 4Ai and 4B of mouse c-myc IRES in the experimentally deduced human c-myc IRES (adopted from [Citation53]). Panel C: The binding of different radiolabelled [α32P]-rUTP c-myc 5ʹUTR transcripts to 3 μM AnxA2 in the presence of 70 µM Ca2+. 15 fmoles of regions 1–4Ai and the stem region of region 2, consisting of NegA (NA) and NegB (NB), as indicated in Panel A, were UV-crosslinked with AnxA2. AnxA2 stained with Coomassie Brilliant Blue served as a loading control. Panel D: UV-crosslinking of 3 μM of recombinant AnxA2 bound to 20 fmoles of uniformly radiolabelled full-length c-myc 5´UTR transcript alone (lanes 2 and 5), and to increasing amounts (10- or 50-fold molar excess) of the unlabelled c-myc full-length 5´UTR (lanes 3 and 4, respectively), and region 4A of the 5´UTR (lanes 6 and 7, respectively). Lane 1 contained only the radiolabelled full-length c-myc 5´UTR transcript to assess the effect of the RNAses added after UV-exposure. AnxA2 stained with Coomassie Brilliant Blue served as a loading control. Note that the protein bands in the upper gel were slightly blurred during drying before exposure to screens and phosphor imaging. Panels C and D: The transcripts were covalently bound to AnxA2 in the presence of 1 μg/μL yeast tRNA to inhibit non-specific RNA binding and then subjected to 10% SDS-PAGE. The binding was visualized using screens and phosphor imaging after ON exposure. AnxA2 is indicated to the right. Panel E: Histogram of the results of the binding competition; UV-crosslinking of 3 μM of recombinant AnxA2 bound to 20 fmoles of uniformly radiolabelled full-length c-myc 5´UTR transcript alone (columns 1 and 4), and to increasing amounts (10- or 50-fold molar excess) of the unlabelled c-myc full-length 5´UTR (columns 2 and 3, respectively), or region 4A of the 5´UTR (columns 5 and 6, respectively). The mean of two experiments is shown](/cms/asset/3753ecc3-79e4-41c1-9550-8e54e9f47dc7/krnb_a_1947648_f0001_c.jpg)
Subsequently, employing UV-crosslinking experiments, the binding of AnxA2 to full-length radiolabelled c-myc 5´UTR (, lanes 2 and 5) was competed with 10x and 50x molar excess concentrations of unlabelled full-length c-myc 5´UTR (, lanes 3 and 4) or region 4A of the IRES that binds AnxA2 (region Δ196-331 nt) (, lanes 6 and 7), respectively. These results (, Panels E and D) confirmed that AnxA2 binds to region 4A of the c-myc IRES.
3.2 The binding of Annexin A2 to the c-myc IRES is Ca2+-dependent
In vitro UV-crosslinking experiments showed that AnxA2 binds to the IRES of c-myc 5´UTR (). To provide evidence that this interaction also takes place in vivo, we used in vitro transcribed full-length c-myc 5´UTR with the IRES to capture AnxA2 from the cytoskeletal fraction of PC12 cells () as we have previously shown that this fraction is enriched in AnxA2 [Citation13]. In addition, equal pmoles of c-myc 5´UTR without the IRES (without region 4) was used as a control (). By hybridizing the 3´end of the c-myc 5´UTR to a biotinylated DNA oligomer, the 5´end was left free for interactions (). Using this approach, we confirmed that AnxA2 binds to the c-myc IRES only in the presence of Ca2+ (). It is also evident that there was considerably less AnxA2 left in the corresponding unbound fraction (not bound to the RNA transcript) than in the other two unbound fractions in which either no Ca2+ or EGTA was present during the ‘capture’ of AnxA2 from the cytoskeletal fraction of PC12 cells (). Control experiments were also included, showing that AnxA2 does not interact with the biotin-5´DNA oligomer either in the absence or presence of Ca2+, and that RNase treatment is necessary to release the AnxA2 bound to the c-myc 5´UTR with the IRES (). The latter property is a strong indication of its direct binding to the c-myc IRES (). Only the c-myc 5´UTR was used in these assays since c-myc mRNA also contains an AnxA2-binding site in its 3´UTR [Citation12]. Previously, AnxA2 pull-down experiments were performed by immunoprecipitating AnxA2 followed by phenol extraction and precipitation of associated mRNAs. Subsequently, RT-PCR with specific primers showed that c-myc and anxA2 mRNAs are associated with the AnxA2 protein, while the β2 – microglobulin (β2-µglob) mRNA is not [Citation11].
The binding of AnxA2 to its cognate mRNA and the c-myc 3ʹ- and 5´-UTRs is strictly Ca2+-dependent ( and , Panels B and C) [Citation10–12]. This Ca2+-dependency has also been shown by others for the binding of AnxA2 to mRNA [Citation14,Citation55]. Calcium is only supposed to induce minor changes in the conformation of AnxA2 [Citation1]. However, it renders the RNA-binding site accessible, possibly by changing the position of the first amino acids of the N-terminus of AnxA2 relative to the core structure, thus increasing the accessibility of amino acids in the RNA-binding site. To investigate the effect of AnxA2 on the translation of the chimeric c-myc reporter mRNAs, we wanted to use a lysate, the rabbit reticulocyte lysate (RRL), which has been used for numerous studies of translation. Due to the inclusion of Ca2+-dependent RNases in the commercial RRL, it was not possible to add exogenous Ca2+ to the reactions. To circumvent this problem, we used His-tagged AnxA2, which we found to bind RNA in the absence of Ca2+, most likely due to the exposure of its RNA-binding site (, lane 4).
Normally, Pro21 is the first visible amino acid in the crystal structure [Citation56], indicating that the most N-terminal part of the tail is unstructured and very flexible. Thus, we reasoned that if AnxA2 would contain a His-tag, this would also render it able to bind RNA in a Ca2+-independent manner, as the presence of a longer tail would expose the RNA binding site – presumably by allowing greater flexibility and reducing the interaction of the N-terminus with the core structure. Indeed, this appears to be the case (, lanes 4). Thus, the His-tagged AnxA2 may also be relevant in other assays, such as those related to AnxA2-actin interactions, which are Ca2+-dependent and involve domain IV of AnxA2.
Thus, taking into account that the mRNA-binding site of AnxA2 resides in its Domain IV [Citation10], we produced three recombinant versions of the protein; a full-length AnxA2 and two N-terminally truncated versions, namely, Δ19AnxA2 (devoid of the first 19 amino acids; counting the first Ser as amino acid no. 1) and Δ33AnxA2 (devoid of the first 33 amino acids). The Δ19AnxA2 form lacks the flexible part of the N-terminus, while Δ33AnxA2 lacks the entire N-terminus. The latter serves as a control, since it is folded, with the hypothesis being that part of the N-terminus of AnxA2 can render the RNA-binding site inaccessible in the absence of Ca2+. Accordingly, while full-length AnxA2 is unable to interact with region 2 of c-myc 5ʹUTR in the absence of Ca2+, both N-terminally truncated versions of AnxA2 show binding ().
Figure 2. AnxA2 derived from PC12 cells and wild type recombinant AnxA2 binds the IRES of c-myc 5´UTR in the presence of calcium. Panel A: Schematic presentation of the AnxA2 ‘capture’ procedure by the c-myc 5´UTR with or without the IRES as baits. An antisense biotin-5´DNA oligomer to the 3´end of c-myc 5´UTR was bound to magnetic streptavidin beads. Subsequently, the c-myc 5´UTR with or without the IRES was bound to the immobilized antisense DNA oligomer. The now immobilized c-myc 5´UTRs were incubated with the cytoskeletal fraction of PC12 cells. After several washes, proteins were released by RNase A/T1 treatment before SDS-PAGE and Western blot analyses using AnxA2 monoclonal antibodies. Panel B: AnxA2 in the cytoskeletal fraction of PC12 cells binds to the c-myc 5´UTR containing the IRES only in the presence of 70 µM Ca2+, but not to the c-myc 5´UTR lacking the IRES. The procedure was performed in the absence (lanes 1 and 4) or presence of 70 µM added Ca2+ (lanes 2 and 5) or in the presence of 2 mM EGTA (lanes 3 and 6). Panel C: The corresponding fractions containing unbound AnxA2 (not bound to the immobilized c-myc 5´UTRs) to the fractions shown in Panel B. Panel D: Control experiments showing that AnxA2 fails to bind to the DNA linker with biotin or to streptavidin and that RNase treatment is required to release the captured AnxA2 (compare with Panel B). The first lane to the left contains 40 µg of proteins from the cytoskeletal fraction of PC12 cells. Panel E: Ca2+-dependency of the binding of full-length AnxA2, but not of its N-terminally truncated or His-tagged versions, to the c-myc IRES. 15 fmoles of region 2 of c-myc 5ʹUTR (as indicated in , Panel A) were UV-crosslinked with full-length AnxA2 (lane 1), ∆19AnxA2 (lane 2), ∆33AnxA2 (lane 3) or His-tagged AnxA2 (lane 4) in the absence or presence of 70 µM Ca2+ as indicated and subjected to 10% SDS-PAGE. The RNA transcripts containing [α32P]-rUTP were covalently bound to AnxA2 in the presence of 1 μg/μL yeast tRNA to inhibit non-specific RNA binding, subjected to 10% SDS-PAGE, whereafter the binding was visualized using screens and phosphor-imaging after a 13 h exposure (or 22 h exposure for the two lanes numbered 4). AnxA2 stained with Coomassie Brilliant Blue served as a loading control. AnxA2 is indicated to the right
![Figure 2. AnxA2 derived from PC12 cells and wild type recombinant AnxA2 binds the IRES of c-myc 5´UTR in the presence of calcium. Panel A: Schematic presentation of the AnxA2 ‘capture’ procedure by the c-myc 5´UTR with or without the IRES as baits. An antisense biotin-5´DNA oligomer to the 3´end of c-myc 5´UTR was bound to magnetic streptavidin beads. Subsequently, the c-myc 5´UTR with or without the IRES was bound to the immobilized antisense DNA oligomer. The now immobilized c-myc 5´UTRs were incubated with the cytoskeletal fraction of PC12 cells. After several washes, proteins were released by RNase A/T1 treatment before SDS-PAGE and Western blot analyses using AnxA2 monoclonal antibodies. Panel B: AnxA2 in the cytoskeletal fraction of PC12 cells binds to the c-myc 5´UTR containing the IRES only in the presence of 70 µM Ca2+, but not to the c-myc 5´UTR lacking the IRES. The procedure was performed in the absence (lanes 1 and 4) or presence of 70 µM added Ca2+ (lanes 2 and 5) or in the presence of 2 mM EGTA (lanes 3 and 6). Panel C: The corresponding fractions containing unbound AnxA2 (not bound to the immobilized c-myc 5´UTRs) to the fractions shown in Panel B. Panel D: Control experiments showing that AnxA2 fails to bind to the DNA linker with biotin or to streptavidin and that RNase treatment is required to release the captured AnxA2 (compare with Panel B). The first lane to the left contains 40 µg of proteins from the cytoskeletal fraction of PC12 cells. Panel E: Ca2+-dependency of the binding of full-length AnxA2, but not of its N-terminally truncated or His-tagged versions, to the c-myc IRES. 15 fmoles of region 2 of c-myc 5ʹUTR (as indicated in Fig. 1, Panel A) were UV-crosslinked with full-length AnxA2 (lane 1), ∆19AnxA2 (lane 2), ∆33AnxA2 (lane 3) or His-tagged AnxA2 (lane 4) in the absence or presence of 70 µM Ca2+ as indicated and subjected to 10% SDS-PAGE. The RNA transcripts containing [α32P]-rUTP were covalently bound to AnxA2 in the presence of 1 μg/μL yeast tRNA to inhibit non-specific RNA binding, subjected to 10% SDS-PAGE, whereafter the binding was visualized using screens and phosphor-imaging after a 13 h exposure (or 22 h exposure for the two lanes numbered 4). AnxA2 stained with Coomassie Brilliant Blue served as a loading control. AnxA2 is indicated to the right](/cms/asset/d608b7a4-1770-4f75-a772-59ae5c8cca67/krnb_a_1947648_f0002_c.jpg)
3.3 Effect of the c-myc UTRs on translational efficiency of a chimeric reporter mRNA
It is well accepted that the 5ʹUTR interacts with the 3ʹUTR via specific proteins for efficient translation and that these dynamic interactions are highly regulated [Citation57]. By lacking endogenous AnxA2, the RRL system for in vitro translation provides an ideal system, since it allows precise control of the concentration of exogenously added AnxA2. Therefore, we employed the coupled transcription/ translation RRL system to direct T7-driven expression of chimeric c-myc mRNAs from PCR products to investigate the effect of the c-myc UTRs on translation. To analyse the effect of c-myc UTRs on the efficiency of the translation of the reporter, Renilla luciferase (Rluc), six different chimeric mRNAs were produced containing no or different combinations of the 5ʹUTRs (1–580 nt) and 3´UTRs of c-myc and/or β2-microglobulin (β2-µglob) mRNAs as indicated in . This resulted in the following cDNAs: i) the coding region of the reporter, RLuc, alone, ii) c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 1A), iii) c-myc-5ʹUTR-RLuc (chimera 2), iv) RLuc-c-myc-3ʹUTR (chimera 3), v) β2-µglob-5ʹUTR-RLuc-β2-µglob-3ʹUTR (chimera 4) and vi) β2-µglob-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 5) (). The latter contains a short 54 nt 5ʹUTR with the nucleotide sequence: 5ʹ-GATTTTCAGTGGCTGCTACTCGGCGCTTCAGTCGCGGTCGCTTCAGTCGTCAGC. It should be noted that we have previously shown that the β2-µglob-mRNA is not associated with AnxA2 using IP of AnxA2 and RT-PCR of immunoprecipitated AnxA2 [Citation11]. The T7-driven expression of the RLuc reporter was performed in the RRL in vitro coupled transcription/translation system and measured as the amount of [35S]-Met incorporated into the reporter protein. This translational system is a very sensitive and established approach.
The c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 1A) shows the highest translational efficiency (defined as 100%); while the coding region of RLuc alone has the lowest translational efficiency (, compare chimera 1A with RLuc). Providing RLuc with the UTRs derived from the β2-µglobulin mRNA increases the translational efficiency about 2-fold (, compare RLuc with chimera 4). Exchanging the β2-µglob 3´UTR with that of c-myc 3´UTR has no effect on translational efficiency of the reporter (, compare chimera 4 with chimera 5). However, when RLuc is equipped with the c-myc UTRs, translational efficiency is changed dramatically (, compare chimera 1A with RLuc and chimera 4). Exchanging the c-myc 5ʹUTR with the β2-µglob 5ʹUTR decreases translational efficiency by more than 50% (, compare chimera 1A with chimera 5) indicating specific crosstalk between the c-myc 5ʹUTR and 3ʹUTR, presumably via protein interactions. This is corroborated by the results showing that the chimeric c-myc mRNA containing a c-myc 5ʹUTR, but no c-myc 3ʹUTR, also greatly diminishes the translation of the reporter (, compare chimera 1A with chimera 2), while the absence of a 5ʹUTR, as expected, also has an inhibitory effect on the efficiency of translation (, compare chimera 1A with chimera 3).
Figure 3. High translational efficiency of the chimeric c-myc reporter mRNA in an in vitro coupled transcription/translation system depends on the joint presence of the c-myc 5ʹUTR and 3ʹUTR. Panel A: Schematic representation of the different chimeric mRNAs used in Panel B: The T7-driven expression of chimeric mRNAs from PCR products (8 ng/μL) containing regions coding for the CDS of RLuc alone, c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 1A), c-myc-5ʹUTR-RLuc (chimera 2), RLuc-c-myc-3ʹUTR (chimera 3), β2-µglob-5ʹUTR-RLuc-β2-µglob-3ʹUTR (chimera 4) or β2-µglob-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 5) mRNAs. The transcription reaction and subsequent translation of the RLuc reporter were performed at 30°C. The incorporation of [35S]-Met is expressed as the % relative to c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 1A; 100%). It was measured as counts per minute (cpm)/mole cDNA and determined using the mean value of the triplicates withdrawn at 60 min to indicate the efficiency of each chimeric mRNA in the in vitro system. * Indicates p < 0.05 based on two-tailed Student’s t-test compared to control (RLuc alone)
![Figure 3. High translational efficiency of the chimeric c-myc reporter mRNA in an in vitro coupled transcription/translation system depends on the joint presence of the c-myc 5ʹUTR and 3ʹUTR. Panel A: Schematic representation of the different chimeric mRNAs used in Panel B: The T7-driven expression of chimeric mRNAs from PCR products (8 ng/μL) containing regions coding for the CDS of RLuc alone, c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 1A), c-myc-5ʹUTR-RLuc (chimera 2), RLuc-c-myc-3ʹUTR (chimera 3), β2-µglob-5ʹUTR-RLuc-β2-µglob-3ʹUTR (chimera 4) or β2-µglob-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 5) mRNAs. The transcription reaction and subsequent translation of the RLuc reporter were performed at 30°C. The incorporation of [35S]-Met is expressed as the % relative to c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR (chimera 1A; 100%). It was measured as counts per minute (cpm)/mole cDNA and determined using the mean value of the triplicates withdrawn at 60 min to indicate the efficiency of each chimeric mRNA in the in vitro system. * Indicates p < 0.05 based on two-tailed Student’s t-test compared to control (RLuc alone)](/cms/asset/af2d296c-2732-4c60-acb7-445ecd016d0b/krnb_a_1947648_f0003_b.gif)
3.4 High concentrations of Annexin A2 inhibit translation of the chimeric reporter mRNA
The effect of AnxA2 on the T7-driven expression of the RLuc reporter in the RRL system was studied in the absence or presence of 10 or 20 μM AnxA2 using four different chimeric mRNAs, all coding for the reporter RLuc and full-length c-myc 3ʹUTR. In addition, chimera 1A contains full-length 5ʹUTR, while chimera 1B contains the 5ʹUTR without region 4A (196–331 nt), chimera 1 C contains the 5ʹUTR without region 4B (331–393 nt) and chimera 1D contains the 5ʹUTR without region 4 (196–393 nt) of c-myc mRNA (depicted in , Panels A-D). Quantitation of the expression after 60 min incubation was carried out by scintillation counting of the incorporation of [35S]-Met into the RLuc reporter protein (, Panels E-H, M and N).
The presence of AnxA2 in the RRL system significantly reduced the expression of the reporter protein from chimeras 1A (full-lengt c-myc 5´UTR), 1B (lacking region 4A) and 1C (lacking region 4B) (, Panels E, F, and G as well as Panels I, J, and K). Apparently, the translational inhibition by AnxA2 is less in the case of chimera 1B, at least at higher concentrations of the protein (). Strikingly, AnxA2 had little or no effect on the expression of RLuc from the c-myc chimeric mRNA lacking region 4 (chimera 1C), that is, lacking the two intact pseudo-knots as well as the ribosomal docking site (, Panels H and L).
Next, we analysed the kinetics of translation of the four chimeric mRNAs (, Panels I-L). Deleting region 4A of the c-myc IRES (chimera 1B) had little effect on the kinetics of translation, as compared to the chimeric mRNA containing full-length c-myc UTRs (chimera 1A) (, compare Panels E and F). However, when both region 4A and the ribosomal docking site (region 4B) – which together form region 4 (see ) – were deleted, translation proceeded rapidly until 30 min, but never reached the same efficiency as when using the two other chimeric mRNAs (, compare Panel L with Panels I and J). Noticeably, chimera 1C lacking the ribosomal docking site follows the same kinetics of translation as chimera 1D (, compare Panels K and L) and has a much lower translational efficiency in the RRL than the other c-myc chimeras (, Panel M). Several control experiments were performed: A mutated AnxA2 that does not bind RNA [Citation11,Citation58] and bovine serum album (BSA) that has no effect on the translation of the chimeric mRNA containing full-length c-myc UTRs (chimera 1A) (). Furthermore, no binding of AnxA2 to the coding region of the reporter RLuc was observed ().
Figure 4. The effect of AnxA2 on the expression of the RLuc reporter in an in vitro coupled transcription/translation system. The T7-driven transcription of chimeric mRNAs from PCR fragments (22 ng/μL) containing the following coding regions: Panels E and I: chimera 1A [c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR, as presented in Panel A]; Panels F and J: chimera 1B [presented in Panel B], Panels G and K: chimera 1 C [presented in Panel C] and Panels H and L [presented in Panel D]. Panels E, F, G and H: Translation for 60 min. The results are presented as percentage incorporated [35S]-Met, as determined by scintillation counting, relative to the reactions in the absence of His-AnxA2. Panels I, J, K and L: The equivalent time-course of the expression of the RLuc reporter in the in vitro coupled transcription/translation system to Panels E-H. Panel M: comparison of the relative translational efficiency of chimeras 1A, 1B, 1C and 1D [chimera 1A was set to 100%]. Panel N: Controls addressing the effects of 10 (blue columns) or 20 (orange columns) μM of exogenously added mutant AnxA2 that does not bind RNA [Citation10,Citation58] (dark blue and dark orange columns, respectively), or BSA (light blue and light orange columns, respectively), on the expression of chimera 1A were studied. The translation of the constructs was performed for 60 min at 30°C in the absence (green) or presence of 10 (blue) or 20 µM (orange) of His-AnxA2 as indicated. Aliquots in duplicates (0, 15, and 30 min) or triplicates (60 min) were withdrawn and the incorporation of [35S]-Met, expressed as cpm/22 ng cDNA, is given as the mean value determined for the duplicates or triplicates. The standard deviations are also indicated. The results from three independent experiments (n = 3) are shown in % relative to control with no additions (100%) and the columns also indicate the standard deviations. Statistical significance was determined by the two-tailed Student’s t-test (*p < 0.05; n = 3). Panel O: The binding of radiolabelled [α32P]-rUTP transcripts to AnxA2. 15 fmoles of full-length 5ʹUTR (lane 1), 3ʹUTR (lane 2) of c-myc mRNA or CDS of hRLuc mRNA (lane 3) were UV-crosslinked with 3 μM of purified wt AnxA2 and subjected to 10% SDS-PAGE. The RNA containing [α32P]-rUTP and covalently bound to AnxA2 was visualized using screens and phosphor-imaging following an ON exposure
![Figure 4. The effect of AnxA2 on the expression of the RLuc reporter in an in vitro coupled transcription/translation system. The T7-driven transcription of chimeric mRNAs from PCR fragments (22 ng/μL) containing the following coding regions: Panels E and I: chimera 1A [c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR, as presented in Panel A]; Panels F and J: chimera 1B [presented in Panel B], Panels G and K: chimera 1 C [presented in Panel C] and Panels H and L [presented in Panel D]. Panels E, F, G and H: Translation for 60 min. The results are presented as percentage incorporated [35S]-Met, as determined by scintillation counting, relative to the reactions in the absence of His-AnxA2. Panels I, J, K and L: The equivalent time-course of the expression of the RLuc reporter in the in vitro coupled transcription/translation system to Panels E-H. Panel M: comparison of the relative translational efficiency of chimeras 1A, 1B, 1C and 1D [chimera 1A was set to 100%]. Panel N: Controls addressing the effects of 10 (blue columns) or 20 (orange columns) μM of exogenously added mutant AnxA2 that does not bind RNA [Citation10,Citation58] (dark blue and dark orange columns, respectively), or BSA (light blue and light orange columns, respectively), on the expression of chimera 1A were studied. The translation of the constructs was performed for 60 min at 30°C in the absence (green) or presence of 10 (blue) or 20 µM (orange) of His-AnxA2 as indicated. Aliquots in duplicates (0, 15, and 30 min) or triplicates (60 min) were withdrawn and the incorporation of [35S]-Met, expressed as cpm/22 ng cDNA, is given as the mean value determined for the duplicates or triplicates. The standard deviations are also indicated. The results from three independent experiments (n = 3) are shown in % relative to control with no additions (100%) and the columns also indicate the standard deviations. Statistical significance was determined by the two-tailed Student’s t-test (*p < 0.05; n = 3). Panel O: The binding of radiolabelled [α32P]-rUTP transcripts to AnxA2. 15 fmoles of full-length 5ʹUTR (lane 1), 3ʹUTR (lane 2) of c-myc mRNA or CDS of hRLuc mRNA (lane 3) were UV-crosslinked with 3 μM of purified wt AnxA2 and subjected to 10% SDS-PAGE. The RNA containing [α32P]-rUTP and covalently bound to AnxA2 was visualized using screens and phosphor-imaging following an ON exposure](/cms/asset/1afa49c4-0213-4306-905d-959c5d5b8e91/krnb_a_1947648_f0004_c.jpg)
3.5 Low concentrations of Annexin A2 appear to stimulate the translation of c-myc IRES-containing chimeric mRNA
AnxA2 is an abundant protein in many cell types. Serum levels of AnxA2 vary between 12–148 µg/mL (0.3–4 µM) in healthy humans and between 12 and 547 µg/mL (0.3–14 µM) in pancreatic cancer patients [Citation59]. However, a literature search did not yield information on the actual molar concentration of AnxA2 in cells. To investigate whether the tested concentrations of AnxA2 used in the in vitro system () would be relevant in a physiological setting, we estimated the approximate concentration of AnxA2 in PC12 cells. Thus, using Western blot analysis we compared 30 µg and 60 µg of a whole cell lysate (612 µg total protein derived from 4 × 106 PC12 cells as determined by IR spectroscopy) with 0.1, 0.2, 0.5, 0.75, 1.0, 2.0 µg recombinant AnxA2. We found that 4 × 106 PC12 cells contain approximately 0.350 µg of AnxA2, corresponding to about 90 fg AnxA2/cell. The typical volume of mammalian cells is roughly 2000 µm3 (http://book.bionumbers.org/how-big-is-a-human-cell/). However, the PC12 cells with a diameter of 10–12 µm (https://bionumbers.hms.harvard.edu/bionumber.aspx?s=n&v=2&id=112695) are much smaller. They can be regarded as half spheres in culture (1/2 x (v = 4/3 πr3), giving an estimated volume of about 260–450 µm3. Therefore, the average concentration of AnxA2 in these cells is about 90 fg/39,000 Dalton = 0.0023 fmoles as the molecular weight of AnxA2 is about 39 kDa, which sets its average concentration in PC12 cells at about 6–7 µM. Stimulation of PC12 cells with NGF induces a 3- to 14-fold increase in the level of AnxA2 protein [Citation60,Citation61]. Thus, in response to NGF stimulation the level of AnxA2 in PC12 cells would reach the concentration range that exerts an inhibitory effect on IRES-containing c-myc mRNA. To investigate if lower concentrations of AnxA2 inhibit the translation of c-myc mRNA in a dose-dependent manner, we investigated the effect of 0.1 µM and 1 µM AnxA2 on the expression of the reporter protein translated from the full-length c-myc chimeric mRNA (). Much to our surprise, we observed that AnxA2 at low concentrations appears to exert a low stimulatory effect on the translation of the reporter mRNA containing the c-myc UTRs. Apparently, the transition between the stimulatory and inhibitory translational effects of AnxA2 lies between 1 µM and 10 µM. According to our rough calculations on the average concentration of AnxA2 in the PC12 cells, these concentrations lie within the physiologically relevant AnxA2 concentrations. In addition, local differences in subcellular AnxA2 levels may be relevant, as well as local concentrations of specific AnxA2-binding mRNAs. Most likely, cellular feed-back mechanisms are also in action.
Figure 5. Low concentrations of AnxA2 (0.1–1 µM) has a stimulatory effect on the expression of the c-myc RLuc reporter in an in vitro coupled transcription/translation system. The T7-driven transcription of chimera 1A [c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR] from a PCR fragment (22 ng/μl). Subsequent translation of the construct was performed for 60 min at 30°C in the absence or presence of 0.1, 1, 10 or 20 µM of His-AnxA2 as indicated. Aliquots in triplicates (60 min) were withdrawn. The results are presented as percentage incorporated [35S]-Met, as determined by scintillation counting, relative to the reactions in the absence (set at 100%) of His-AnxA2. The results from three independent experiments (n = 3) are shown. The standard deviations are also indicated. Statistical significance was determined by the two-tailed Student’s t-test (*p < 0.05)
![Figure 5. Low concentrations of AnxA2 (0.1–1 µM) has a stimulatory effect on the expression of the c-myc RLuc reporter in an in vitro coupled transcription/translation system. The T7-driven transcription of chimera 1A [c-myc-5ʹUTR-RLuc-c-myc-3ʹUTR] from a PCR fragment (22 ng/μl). Subsequent translation of the construct was performed for 60 min at 30°C in the absence or presence of 0.1, 1, 10 or 20 µM of His-AnxA2 as indicated. Aliquots in triplicates (60 min) were withdrawn. The results are presented as percentage incorporated [35S]-Met, as determined by scintillation counting, relative to the reactions in the absence (set at 100%) of His-AnxA2. The results from three independent experiments (n = 3) are shown. The standard deviations are also indicated. Statistical significance was determined by the two-tailed Student’s t-test (*p < 0.05)](/cms/asset/6735f7c5-c352-4cba-afe3-4051e07481cc/krnb_a_1947648_f0005_b.gif)
3.6 Forskolin treatment of PC12 cells decreases the expression of c-Myc
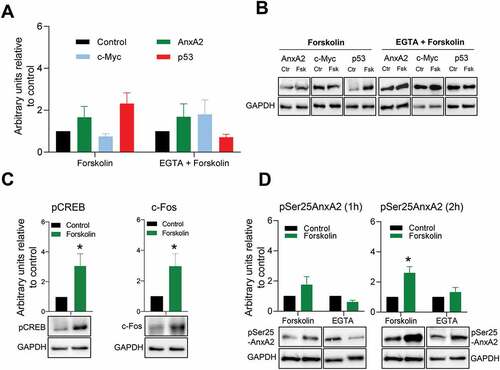
Having established that both recombinant and endogenous AnxA2 bind to the IRES of c-myc mRNA in a Ca2+-dependent manner and that AnxA2 modulates the expression of c-Myc in in vitro experiments, we next wanted to investigate if this is a physiologically relevant event in cells. Thus, AnxA2 would be expected to inhibit the translation of the c-myc mRNA at higher levels of Ca2+. Forskolin (Fsk) increases intracellular Ca2+ via regulating L-type calcium channel activities and the concentration of cyclic adenosine monophosphate (cAMP) [Citation62]. Short-term (1 hr) treatment with Fsk was therefore used to increase the intracellular Ca2+ concentration to study the effect of Ca2+ on the expression of AnxA2 and c-Myc (). Furthermore, prior to stimulation with Fsk, pre-treatment with EGTA was performed to chelate Ca2+ and thus abolish the effects of this compound (). While such a short treatment appears to increase slightly the expression of AnxA2, the expression of c-Myc was reduced and importantly this effect was abolished when Ca2+ was chelated (, Panels A and B). As it has previously been shown that the Ca2+-dependent binding of AnxA2 to the IRES of p53 mRNA increases the expression of the corresponding protein [Citation55], we also investigated the effect of Fsk on the expression of p53 in PC12 cells. As expected, Fsk resulted in an increase in the expression of p53, which was abolished by EGTA (, Panels A and B). We also verified that the pre-treatment alone with EGTA did not change the expression levels of AnxA2, c-Myc or p53 (data not shown). To confirm the specificity of the effect of Fsk, we examined the responsiveness of known Fsk targets in our experimental conditions (). The phosphorylation of CREB showed robust enhancement after Fsk stimulation [Citation63]. Furthermore, there was an increased expression of c-Fos [Citation64]. In addition, Fsk treatment caused an increase in the level of Ser25 phosphorylated AnxA2, which was even more prominent after 2 hr exposure (). This effect could be abolished by chelating Ca2+, indicating Ca2+-dependency of the phosphorylation.
Figure 6. Short-term (1 hr) forskolin treatment increases the expression of p53 while decreasing the expression of c-Myc. Panel A: 1 hr stimulation of PC12 cells with 20 µM Fsk alone or in combination with a 1 hr pre-treatment with 100 µM EGTA and the effect on the expression of AnxA2, c-Myc and p53. The controls are indicated with black columns and are untreated cells with vehicle (DMSO) for Fsk treatment alone. For Fsk treatment with pre-incubation with EGTA, the control cells were treated with vehicle and EGTA. Panel B: Representative Western blots of the results shown in Panel A. GAPDH was used as a loading control. Panel C: The effect of 1 hr Fsk treatment on the phosphorylation of CREB and the expression of c-Fos. Untreated controls are shown as black columns and Fsk-treated cells with green columns. Representative Western blots are shown below the columns. Panel D: The effect of 1 and 2 hr Fsk treatment on the Ser25 phosphorylation of AnxA2. Proteins (12 μg) derived from total lysate from PC12 cells were subjected to 4–20% SDS-PAGE and western blot analysis. The results from three independent experiments (n = 3) are shown and were normalized to GAPDH. The standard deviations are also indicated. Statistical significance compared to control was determined by the two-tailed Student’s t-test (*p < 0.05)
4. Discussion
4.1 Annexin A2 interacts with the IRES region of c-myc 5ʹUTR containing two pseudo-knots in a Ca2+-dependent manner
AnxA2 is a multifunctional protein involved in membrane-related events as well as RNA-related events and most of its functions are regulated by calcium and multiple post-translational modifications [Citation1–4,Citation65]. Thus, AnxA2 could be regarded as a master regulator, which in response to external signalling co-ordinates vesicle and RNA events as shown by its family member, AnxA11 [Citation66]. These events are likely to be regulated by post-translational modifications such as phosphorylation. Indeed, angiotensin II activation of the AT1a receptor stimulates the AnxA2 RNA-binding activity, not by upregulating the expression of AnxA2, but by increasing its phosphorylation [Citation67]. The phosphorylation site(s) were not identified, but we have previously shown that Ser25 phosphorylated and ubiquitinated/sumoylated AnxA2 are enriched in mRNP complexes that are not active in translation [Citation50]
AnxA2 is part of mRNP complexes containing c-myc mRNA [Citation11,Citation12,Citation14] and other specific mRNAs translated on cytoskeleton-bound polysomes [Citation2,Citation13,Citation49]. Other RNA-binding proteins, such as Y-box-binding protein (YB), hnRNP K, nucleolin and poly(A)-binding protein (PABP), also interact with AnxA2 in mRNP complexes, as shown by immunoprecipitation experiments combined with mass spectrometry [Citation14,Citation68]. The poly(A)-tail with its bound PABP is important for cap-dependent translation by establishing an interaction with eIF4E via eIF4G [Citation69]. This ‘bridge’ is dispensable for the interaction of the poly(A) tail with the c-myc IRES to stimulate translation [Citation70,Citation71]. However, it is also known that PABP1 stimulates translation of at least certain viral IRES-containing RNAs [Citation72]. Thus, although AnxA2 interacts with PABP [Citation14], the role that this interaction may play in discriminating between cap-dependent and IRES-dependent initiation of translation remains unclear.
We have previously identified an AnxA2-binding site in the 3´UTR of the c-myc [Citation12] and anxA2 [Citation11] mRNAs. Furthermore, alignment studies and UV-crosslinking experiments lead us to suggest that a five nucleotide consensus sequence, 5ʹ-AA(C/G)(A/U)G, together with higher order structures of the 3ʹUTRs, are involved in the interaction with AnxA2 [Citation11]. The AnxA2-binding site in the c-myc 3´UTR involves the c-myc localization element responsible for its transport to the perinuclear region for translation [Citation12,Citation33].
Here we also identified AnxA2 as a c-myc IRES-binding protein () and determined the mRNA binding site as the c-myc 5´UTR region encompassing nt 196–331 and containing one side (strand) of the two pseudo-knots (, region 4A). This region also contains the AAGAG consensus sequence found in the c-myc 3ʹUTR. The consensus sequence is outside the two pseudoknots in the c-myc 5ʹUTR. AnxA2 can thus be regarded as a member of the c-myc ITAF proteins. In addition, we and others have shown that the interaction of AnxA2 with c-myc mRNA is Ca2+-dependent () [Citation12,Citation14].
Interestingly, AnxA2, together with PSF and PTB proteins, has previously been shown to interact with the IRES of p53, and to be required for the p53 IRES activity [Citation55]. However, while the binding of AnxA2 to the c-myc IRES leads to decreased translation of the mRNA in a dose-dependent manner (), the binding to the p53 IRES leads to increased translation of the corresponding mRNA, including both isoforms of p53 [Citation55]. It has been shown in the case of p53 mRNA that specific proteins, other than the eIF4E-eIF4G-PABP complex necessary for cap-dependent initiation of translation, can interact with both UTRs and thus regulate p53 mRNA translation [Citation73].
Not only proteins, but also miRNAs and lncRNAs regulate the translatability of an mRNA [Citation74]. For example, it has been shown that p53 inhibits the expression of c-Myc via miR-145 [Citation75]. The interaction between the 5´- and 3´UTRs via proteins that bind to both regulatory regions adds another layer to the complexity. Interestingly, during hypoxia, nucleolin binds the 3ʹUTR of the collagen prolyl 4-hydroxylase-α(I) mRNA via the association with AnxA2, while it binds directly to the 5ʹUTR of the same mRNA [Citation68], suggesting that the specific protein interactions within the mRNP complexes are highly dynamic.
Several ITAFs, such as hnRNP K and YB, bind the c-myc IRES and stimulate the expression of c-Myc [Citation40,Citation45]. Post-translational modifications of AnxA2 could alter both its interaction with other ITAFs and change the affinity for specific RNA sequences. This could change the subset of its protein interaction partners and thereby change the interaction with the c-myc IRES. It was found that the phosphor-mimicking AnxA2 mutant, AnxA2-S25E, binds RNA with higher affinity than wt AnxA2 [Citation76]. Several ITAFs have been found to possess both activator and inhibitor properties in regulating cellular IRESs [Citation77]. Therefore, the outcome of such regulation depends on the composition of the ITAF complex bound to the IRES. Thus, it is possible that by binding to the c-myc IRES, AnxA2 competes with protein factors that enhance the translation of c-myc mRNA via the IRES and/or prevent the unfolding of RNA secondary structure necessary to initiate translation under circumstances involving an increase in Ca2+ levels. For example, it has been shown that PSF and AnxA2 share overlapping binding sites on the p53 IRES and that the interplay between AnxA2, PSF and PTB regulates p53 IRES activity. Yet, of these proteins, only the binding of AnxA2 to the p53 IRES is Ca2+-dependent [Citation55], as observed here for the binding of AnxA2 to the c-myc IRES ().
However, AnxA2 binds to the pseudo-knot of infectious bronchitis virus (IBV) RNA in a Ca2+-independent manner and reduces the frameshifting efficiency from IBV pseudoknot RNA [Citation78]. At present it is not clear why the binding of AnxA2 to some mRNAs depends on Ca2+, while its binding to other mRNAs does not show this requirement. Post-translational modifications of AnxA2 could play a role in its RNA binding, since it has been shown that viral IRES activity is not only dependent on the recruitment of proteins, but also relies on the post-translational modifications of the recruited proteins [Citation79]. Ser25 phosphorylation of AnxA2 changes its conformation to the open state, which does not need Ca2+-to bind RNA [Citation50,Citation80]. It also leads to increased mRNA binding [Citation50,Citation76], as shown by the results obtained with the phosphor-mimicking S25E-AnxA2 (discussed in [Citation4]). Thus, in the absence of sufficiently high concentrations of Ca2+, Ser25 phosphorylation, possibly combined with other post-translational modifications, may regulate the binding of AnxA2 to RNA. It has been shown that the S25E mutation also increases the Ca2+-independent binding of AnxA2 to G-actin [Citation76].
4.2 Annexin A2 modulates the translation of the c-myc reporter in the RRL
The conventional nuclease-treated RRL cell-free system is ideal for our purposes, since it lacks endogenous AnxA2 [Citation81] and has low protease activity [Citation82]. Furthermore, it displays high translational efficiency, which is an important property when studying the effect of inhibitors on translation. Consequently, the RRL system has been used in numerous studies. For example, by using chimeric c-myc reporter mRNAs, this system was employed to show that hnRNP C enhances the translation of c-myc mRNA [Citation43]. Nanbru and co-workers used the RRL to translate a reporter protein from both the CUG and AUG codons in the 5ʹUTR of human c-myc mRNAs transcribed from the P0, P1, and P2 promoters [Citation26].
Our first aim was to identify the AnxA2-binding site in the c-myc mRNA. That explains why we used the P1 transcript, since our preliminary results indicated that the AnxA2-binding site is present in a c-myc exon 1 transcript. P2 is the predominant transcription start site, giving rise to 75–90% of c-myc mRNAs, while 10–25% of c-myc transcripts are generated from the P1 start site. As the AnxA2-binding site turned out to lie within the IRES, we decided to use monocistronic chimeric c-myc reporter mRNAs. It has been speculated that overexpression of bicistronic mRNAs using viral promoters may compromise the apparent efficiency of the c-myc IRES [Citation35] since ITAFs may become limiting under these conditions. Moreover, it has been shown that specific subsets of RNA-binding proteins often operate in concert, forming distinct regulatory complexes depending on intracellular conditions. Although our chimeric constructs are transcribed from the T7 promoter, the use of equal molar amounts of monocistronic chimeric mRNAs with and without the AnxA2-binding or ribosomal docking site regions of the c-myc IRES, and subsequent comparison of their properties, partly overcame this problem. However, a point of concern could be that AnxA2 acts in concert with other ITAFs and these factors may become limiting.
Since AnxA2 binds to region 4A of the IRES of the chimeric c-myc mRNA (), we initially deleted only this region in the mRNA (). To our surprise, the inhibitory effect of AnxA2 on the translation of this chimeric mRNA (chimera 1B) was only slightly reduced as compared to the chimeric c-myc mRNA containing full-length 5ʹUTR (, chimera 1A). Apparently, since AnxA2 bound to both regions 2 and 4 of c-myc 5ʹUTR with higher affinity than to region 4A alone, a new version of the chimeric c-myc mRNA (chimera 1D) was produced by seamless cloning, this time lacking both regions 4A and region 4B, the latter harbouring the ribosomal docking site and the two intact pseudo-knots. This deletion totally abolished the effect of AnxA2 on the translation of the chimeric c-myc mRNA (), indicating that the protein may interact with protein factors associating with the ribosomal docking site on the IRES or interfere with the formation of the initiation complex. For example, it has been shown that AnxA2 interacts with receptor for activated C kinase 1 (RACK1) [Citation83], a ribosome-associated protein implicated in the regulation of viral IRES activity [Citation84]. Furthermore, several ribosomal proteins of both the small and the large ribosomal subunits were found to interact with AnxA2 [Citation14], offering the possibility that AnxA2 interacts directly with the ribosome.
Subsequently, to investigate the role of the ribosomal docking site, only region 4B was deleted from the 5´UTR of the chimeric c-myc mRNA (chimera 1C) (). The translation of this chimeric mRNA was inhibited by AnxA2 in the same manner as chimeras 1A and 1B (), although not to the same extent, indicating that our assumption was correct in that region 4A is involved in the binding of AnxA2 leading to its subsequent inhibitory effect on translation. It should be noted that when only region 4B of the c-myc IRES is deleted, it has a great impact on the translational efficiency of the reporter by lowering it by about 50% () indicating its function as a ribosomal docking site.
Possible explanations for these results, could be that when only region 4A is deleted, AnxA2 may still interact with the ribosome, perhaps due to the AnxA2-binding site possibly extends slightly into region 4B. When region 4B is deleted, the main consequence would be that the translational efficiency of the mRNA is reduced due to the lack of the ribosomal docking site, but AnxA2 may still provide a sterically hindrance for a ribosome scanning from the 5´end. It is also possible that the binding of AnxA2 to region 4A combined with the deletion may result in the formation of inhibitory sequences in the c-myc 5´UTR. When both regions 4A and 4B are deleted, AnxA2 has lost its binding site and cannot interact with the ribosome or proteins factors on the ribosomal docking site. It cannot be ruled out entirely that binding of AnxA2 to the 3´UTR of c-myc affects the events taking place at the c-myc 5´UTR. However, preliminary data indicate that domain IV of AnxA2 inhibits in a dose-dependent manner the translation of a c-myc chimera only containing the c-myc 5´UTR in front of the reporter and no 3´UTR (data not shown).
Interestingly, the time-course curve of translation for the chimeric c-myc mRNA lacking region 4 of the c-myc 5ʹ-UTR indicates that translation reaches a steady state after 30 min. Furthermore, translation of this construct has a faster initial phase, while at 60 min it only amounts to about 80% of that of full-length chimeric c-myc mRNA (). This could be due to exhaustion of energy supply. Another more likely possibility is that the deletion of the ribosomal docking site hampers the binding of ribosomes to internal regions of the 5´UTR. This possibility is corroborated by the finding that deletion of the ribosomal docking site in the IRES lowers the translational efficiency of the chimeric c-myc mRNA by about 50% compared to the chimera containing the full-length 5´UTR (). In the present study, full translational activity of the chimeric c-myc reporter mRNA was only observed when the two c-myc UTRs were present, but not when the c-myc 5´UTR was exchanged with that of the β2-µglobulin mRNA. This also decreased the translatability of the mRNA by about 50% (). Considering the effect of deleting the ribosomal docking site (region 4B) from the IRES (), this suggests that this could be mainly due to the absence of an IRES in the β2-µglobulin mRNA.
To explore whether the effects of AnxA2 on the translation of the c-myc chimeric mRNAs are specific, it was necessary to include various negative control proteins that do not bind to mRNA and therefore should have no effect on translation. We have constructed a mutant form of the full-length AnxA2, which does not bind RNA [Citation10,Citation58] and thus could be used as a negative control in the RRL. The helices C and D of domain IV of AnxA2 harbour two well-exposed regions, i.e. 307KKK309 in helix C and 315YYYIQQD321 in helix D, which together with Gly311 and Lys312 in the loop between these helices, provide the sites for interaction with a bulky RNA target. Thus, the non-RNA-binding mutant full-length AnxA2 harbours the following mutations: K307S-K308S-K309S-K312S-Y316S-Q320S [Citation10]. In addition, BSA was included as another non-mRNA-binding control protein. Indeed, the control assays confirmed that the effects exerted by AnxA2 are specific, since neither of these proteins inhibited the translation of the chimeric full-length c-myc mRNA (1A) containing the IRES (). These results argue against a possible unspecific inhibitory effect of AnxA2 on the translation of the RLuc reporter.
At present, we cannot explain the stimulatory effects on translation of chimera 1A at low concentrations of AnxA2 on the translation (). However, we have observed the same effect of AnxA2 on its cognate mRNA (results not shown). It is possible that this is related to the interaction with other proteins and as the concentration of AnxA2 increases, the stoichiometry of complex formation with specific ligands changes. This is certainly interestingly to pursue in future studies. Post-translational modifications could also be involved.
4.3 The cellular correlation between the expressions of Annexin A2 and c-Myc
Cellular control and regulation of protein expression is important and can be exerted at several levels. RNA-binding proteins play an important role at the level of mRNA transport and translation [Citation85]. Here we show that lower levels of AnxA2 apparently increase the translation of the c-myc reporter while higher concentrations of AnxA2 inhibit translation (), suggesting an inverse relationship between the cellular levels of the two proteins. We observed that AnxA2 expression is very high in MCF10A, relatively high in MDA231 and very low in MCF7 cell lines (Supplementary Fig. S1). Others have reported that c-Myc levels are high in MCF7, lower in MDA231 and very low in MCF10A cell lines [Citation86], corroborating this assumption. Furthermore, c-Myc is overexpressed in the earliest phases of prostate cancer [Citation87] when the expression of AnxA2 is very low [Citation88]. HepG2 is a human cell-line derived from hepatoblastoma. These cells express c-myc mRNA constitutively, and do not contain detectable levels of AnxA2 [Citation89,Citation90]. Thus, these findings could provide additional support for the present results, addressing the role of AnxA2 in the regulation of c-Myc expression.
It is unlikely that the correlation between the c-Myc and AnxA2 protein levels in vivo is as simple as the given examples suggest, since i) the level of c-Myc expression can be regulated at initiation of translation both by a cap-dependent and an IRES-dependent mechanism [Citation35], ii) AnxA2 binds the c-myc mRNA only in the presence of high Ca2+ concentrations, perhaps locally (if not phosphorylated on Ser25), iii) AnxA2 binds both to the c-myc 3´UTR and the 5´UTR, iv) the effect may be related to the AnxA2/mRNA (specific mRNAs binding to AnxA2) ratio and v) post-translational modifications are likely to be involved. As AnxA2 appears to harbour only one RNA-binding site [Citation10], it may bind as a homodimer or as a monomer in complex with different ligands if bound simultaneously to both UTRs. Another possibility is that AnxA2 does not bind to both UTRs simultaneously.
It is difficult to achieve a detailed understanding regarding the molecular mechanisms of cellular IRESs, since no in vitro system totally recapitulates the function of these elements [Citation91]. Since AnxA2 is a multifunctional protein and RNA-binding is only one of its many cellular functions, the use of in vitro translation systems provides a very important approach for gaining insight into its role in RNA related processes before starting with cell lines with many cellular processes taking place simultaneously.
Fsk stimulation of HUVEC cells leads to a redistribution of AnxA2 from the cytosol to the plasma membrane. Furthermore, this stimulation resulted in decreased Ser11 phosphorylation of AnxA2, leading to its subsequent binding to S100A10 and participation in secretion [Citation92], while we found that Ser25 phosphorylation of the protein was increased by Fsk treatment (). Ser25 phosphorylation of AnxA2 was highly elevated, in particular after 2 h treatment with Fsk. By contrast, Fsk stimulation of PC12 cells apparently gave rise to a modest increase in the expression of AnxA2 while slightly decreasing the expression of c-Myc (). Fsk treatment increased the expression of p53, which could be abolished by EGTA pre-treatment (). The EGTA treatment chelates extracellular Ca2+, since the stimulatory effect of Fsk is dependent on an influx of extracellular Ca2+ [Citation93]. Thus, on one hand Fsk may trigger the release of AnxA2 in secretory granules together with its partner S100A10 [Citation92]. On the other hand, increased Ser25 phosphorylation together with other post-translational modifications could lead to participation of AnxA2 in the translational silencing of specific mRNAs as pSer25AnxA2 partially colocalises with the P-body marker GW182 [Citation50]. The increased binding of AnxA2 to RNA after angiotensin II activation of the AT1a receptor was due to increased phosphorylation of the protein and not to an increase in the expression of AnxA2 [Citation67]. Thus, it is likely that Ser25 phosphorylation is important for the effects we observe in the cell system. However, due to the multifunctionality of AnxA2 it is not easy to discern between the individual functions of the protein.
The regulation of the binding of AnxA2 to RNA is not straightforward. The wild-type AnxA2 binds to RNA in the presence of Ca2+ [Citation10]. However, when AnxA2 is Ser25 phosphorylated, the protein can bind RNA in the absence of Ca2+ [Citation76]. Then the protein is in its open state [Citation80] and presumably in a state where the RNA-binding site in domain IV becomes more accessible. Thus, the effects of the Fsk treatment of PC12 cells were not clear-cut but indicated tendencies. Most importantly, the reduction in c-Myc expression and the increase in AnxA2 expression by Fsk could be reversed by EGTA treatment ().
As mentioned above, several ITAFs bind the c-myc IRES and normally promote IRES-dependent translation, while the Ca2+-dependent binding of AnxA2 to the c-myc IRES inhibits translation. Ca2+-signalling is remodelled in some cancers exerting effects on cell proliferation, invasion, propensity to apoptosis, multidrug resistance and the tumour microenvironment [Citation94]. Ca2+-fluxes also operate during mitosis [Citation95] and in the synapse [Citation96]. It is possible that under these circumstances a Ca2+-dependent switch may operate between cap-dependent and IRES-dependent mode of translation initiation. AnxA2 could mediate this switch that triggers the response to elevated Ca2+ levels, most likely in concert with other ITAFs. Future investigations should address in detail the multifunctional nature of AnxA2, whose distinct functions are regulated by its post-translational modifications and ligand interactions [Citation2] and also how this affects the regulation of the c-myc IRES. Since AnxA2 binds to both the 5ʹUTR and the 3ʹUTR, it is possible that inhibition of translation is coupled to mRNA transport. Furthermore, since AnxA2 binds to endosomes and trafficking of membranes and mRNAs are interconnected processes [Citation97], it is tempting to speculate that AnxA2 may co-ordinate transport of vesicles and specific mRNAs in response to extracellular signals.
5. Conclusions
AnxA2 binds to the IRES of c-myc 5´UTR and modulates the translation of the mRNA in the presence of Ca2+. This opens the interesting possibility that AnxA2 acts as a Ca2+-dependent switch between cap-dependent and IRES-dependent translation of c-myc mRNA.
Author Contributions
Conceptualization, A.V, E.S. and S.S.P; Data curation, H.H., S.A.S., E.S., S.S.P. and A.V.; Formal analysis, E.S., M.E.V.S., S.R., A.K.G., S.S.P. and A.V.; Funding acquisition, A.V.; Investigation, E.S., H.H, S.A.S., S.R., M.E.V.S., A.K.G., S.S.P. and A.V.; Methodology, E.S., H.H., S.A.S., S.S.P. and S.R.; Project administration, A.V.; Resources, A.V.; Supervision, A.K.G., S.S.P. and A.V.; Validation, S.S.P. and A.V.; Visualization, S.A.S., S.S.P. and M.E.V.S.; Writing – original draft, E.S., S.S.P. and A.V.; Writing – review & editing, E.S., S.A.S., S.S.R., M.E.V.S., A.K.G., S.S.P. and A.V.
Supplemental Material
Download PDF (507.7 KB)Acknowledgments
We are grateful to Prof. Jaakko Saraste (University of Bergen, Norway) for critically reading of the manuscript. We also gratefully acknowledge the generous gift from Dr. Beate Stern, University of Bergen, Norway of the codon optimised Renilla luciferase cDNA. We also acknowledge reviewers for important suggestions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82:331–371.
- Vedeler A, Hollas H, Grindheim AK, et al. Multiple roles of annexin A2 in post-transcriptional regulation of gene expression. Curr Protein Pept Sci. 2012;13:401–412.
- Bharadwaj A, Bydoun M, Holloway R, et al. A2 heterotetramer: structure and function. Int J Mol Sci. 2013;14:6259–6305.
- Grindheim AK, Saraste J, Vedeler A. Protein phosphorylation and its role in the regulation of Annexin A2 function.Biochimica Et Biophysica Acta. General Subjects. 2017;1861:2515–2529.
- Christensen MV, Hogdall CK, Jochumsen KM, et al. Annexin A2 and cancer: a systematic review. Int J Oncol. 2018;52:5–18.
- Sharma MC. Annexin A2 (ANX A2): an emerging biomarker and potential therapeutic target for aggressive cancers. Int J Cancer. 2019;144:2074–2081.
- Xu XH, Pan W, Kang LH, et al. Association of annexin A2 with cancer development (Review). Oncol Rep. 2015;33:2121–2128.
- Sharma MR, Koltowski L, Ownbey RT, et al. Angiogenesis-associated protein annexin II in breast cancer: selective expression in invasive breast cancer and contribution to tumor invasion and progression. Exp Mol Pathol. 2006;81:146–156.
- Chuthapisith S, Bean BE, Cowley G, et al. Annexins in human breast cancer: possible predictors of pathological response to neoadjuvant chemotherapy. Eur J Cancer (Oxford, England 1990). 2009;45:1274–1281. .
- Aukrust I, Hollas H, Strand E, et al. The mRNA-binding site of annexin A2 resides in helices C-D of its domain IV. J Mol Biol. 2007;368:1367–1378.
- Hollas H, Aukrust I, Grimmer S, et al. A2 recognises a specific region in the 3ʹ-UTR of its cognate messenger RNA. Biochim Biophys Acta. 2006;1763:1325–1334.
- Mickleburgh I, Burtle B, Hollas H, et al. A2 binds to the localization signal in the 3ʹ untranslated region of c-myc mRNA. FEBS J. 2005;272:413–421.
- Vedeler A, Hollas H. Annexin II is associated with mRNAs which may constitute a distinct subpopulation. Biochem J. 2000;348(Pt 3):565–572.
- Filipenko NR, MacLeod TJ, Yoon CS, et al. Annexin A2 is a novel RNA-binding protein. J Biol Chem. 2004;279:8723–8731.
- Caforio M, Sorino C, Iacovelli S, et al. Recent advances in searching c-Myc transcriptional cofactors during tumorigenesis. J Exp Clin Cancer Res. 2018;37:239.
- Carroll PA, Freie BW, Mathsyaraja H, et al. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front Med. 2018;12:412–425.
- Zhang HL, Wang P, Lu MZ, et al. c-Myc maintains the self-renewal and chemoresistance properties of colon cancer stem cells. Oncol Lett. 2019;17:4487–4493.
- Nguyen L, Papenhausen P, Shao H. The role of c-MYC in B-Cell Lymphomas: diagnostic and molecular aspects. Genes (Basel). 2017;8. DOI:10.3390/genes8040116.
- Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637.
- Paulin FE, Chappell SA, Willis AE. A single nucleotide change in the c-myc internal ribosome entry segment leads to enhanced binding of a group of protein factors. Nucleic Acids Res. 1998;26:3097–3103.
- Chappell SA, LeQuesne JP, Paulin FE, et al. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440.
- Chiang Y, Davis RG, Vishwanatha JK. Altered expression of annexin II in human B-cell lymphoma cell lines. Biochim Biophys Acta. 1996;1313:295–301.
- Shi Y, Sharma A, Wu H, et al. D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J Biol Chem. 2005;280:10964–10973.
- Misquitta CM, Chen T, Grover AK. Control of protein expression through mRNA stability in calcium signalling. Cell Calcium. 2006;40:329–346.
- Farrell AS, Sears RC. MYC degradation. Cold Spring Harb Perspect Med. 2014;4. DOI:10.1101/cshperspect.a014365.
- Nanbru C, Lafon I, Audigier S, et al. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066.
- Hann SR, Dixit M, Sears RC, et al. The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev. 1994;8:2441–2452.
- Zimmer SG, DeBenedetti A, Graff JR. Translational control of malignancy: the mRNA cap-binding protein, eIF-4E, as a central regulator of tumor formation, growth, invasion and metastasis. Anticancer Res. 2000;20:1343–1351.
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5ʹ-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416.
- Mayya VK, Duchaine TF. Ciphers and executioners: how 3ʹ-untranslated regions determine the fate of messenger RNAs. Front Genet. 2019;10:6.
- Swier LJYM, Dzikiewicz-Krawczyk A, Winkle M, et al. Intricate crosstalk between MYC and non-coding RNAs regulates hallmarks of cancer. Mol Oncol. 2019;13:26–45.
- Veyrune JL, Campbell GP, Wiseman J, et al. A localisation signal in the 3ʹ untranslated region of c-myc mRNA targets c-myc mRNA and beta-globin reporter sequences to the perinuclear cytoplasm and cytoskeletal-bound polysomes. J Cell Sci. 1996;109(Pt 6):1185–1194.
- Chabanon H, Mickleburgh I, Burtle B, et al. AU-rich stem-loop structure is a critical feature of the perinuclear localization signal of c-myc mRNA. Biochem J. 2005;392:475–483.
- Stoneley M, Paulin FE, Le Quesne JP, et al. Myc 5ʹ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428.
- Stoneley M, Chappell SA, Jopling CL, et al. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. 2000;20:1162–1169.
- Creancier L, Mercier P, Prats AC, et al. c-myc Internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol Cell Biol. 1833-1840;2001(21). DOI:10.1128/mcb.21.5.1833-1840.2001.
- Carter PS, Jarquin-Pardo M, De Benedetti A. Differential expression of Myc1 and Myc2 isoforms in cells transformed by eIF4E: evidence for internal ribosome repositioning in the human c-myc 5ʹUTR. Oncogene. 1999;18:4326–4335.
- Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035.
- Spriggs KA, Cobbold LC, Jopling CL, et al. Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Mol Cell Biol. 2009;29:1565–1574.
- Evans JR, Mitchell SA, Spriggs KA, et al. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020.
- Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA (New York, N.Y.). 2002;8:265–278.
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598.
- Kim JH, Paek KY, Choi K, et al. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol Cell Biol. 2003;23:708–720.
- Shi Y, Frost PJ, Hoang BQ, et al. 6-induced stimulation of c-myc translation in multiple myeloma cells is mediated by myc internal ribosome entry site function and the RNA-binding protein, hnRNP A1. Cancer Res. 2008;68:10215–10222.
- Cobbold LC, Spriggs KA, Haines SJ, et al. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol. 2008;28:40–49.
- Cobbold LC, Wilson LA, Sawicka K, et al. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene. 2010;29:2884–2891.
- Meristoudis C, Trangas T, Lambrianidou A, et al. Systematic analysis of the contribution of c-myc mRNA constituents upon cap and IRES mediated translation. Biol Chem. 2015;396:1301–1313.
- Zanier K, Nomine Y, Charbonnier S, et al. Formation of well-defined soluble aggregates upon fusion to MBP is a generic property of E6 proteins from various human papillomavirus species. Protein Expr Purif. 2007;51:59–70.
- Vedeler A, Pryme IF, Hesketh JE. The characterization of free, cytoskeletal and membrane-bound polysomes in Krebs II ascites and 3T3 cells. Mol Cell Biochem. 1991;100:183–193.
- Aukrust I, Rosenberg LA, Ankerud MM, et al. Post-translational modifications of Annexin A2 are linked to its association with perinuclear nonpolysomal mRNP complexes. FEBS Open Bio. 2017;7:160–173.
- Grindheim AK, Hollas H, Raddum AM, et al. Reactive oxygen species exert opposite effects on Tyr23 phosphorylation of the nuclear and cortical pools of annexin A2. J Cell Sci. 2016;129:314–328.
- Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA (New York, N.Y. 2005;11:1157–1166.
- Le Quesne JP, Stoneley M, Fraser GA, et al. Derivation of a structural model for the c-myc IRES. J Mol Biol. 2001;310:111–126.
- Bernard O, Cory S, Gerondakis S, et al. Sequence of the murine and human cellular myc oncogenes and two modes of myc transcription resulting from chromosome translocation in B lymphoid tumours. EMBO J. 1983;2:2375–2383.
- Sharathchandra A, Lal R, Khan D, et al. Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol. 2012;9:1429–1439.
- Raddum AM, Hollas H, Shumilin IA, et al. The native structure of annexin A2 peptides in hydrophilic environment determines their anti-angiogenic effects. Biochem Pharmacol. 2015;95:1–15.
- Handshakes DE. Fights: the regulatory interplay of RNA-binding proteins. Front Mol Biosci. 2017;4:67.
- Solbak SMO, Abdurakhmanov E, Vedeler A, et al. Characterization of interactions between hepatitis C virus NS5B polymerase, annexin A2 and RNA - effects on NS5B catalysis and allosteric inhibition. Virol J. 2017;14:236.
- Karabulut M, Afsar CU, Serilmez M, et al. Circulating annexin A2 as a biomarker in patients with pancreatic cancer. J Cancer Res Ther. 2020;16:S110–s115.
- Fox MT, Prentice DA, Hughes JP. Increases in p11 and annexin II proteins correlate with differentiation in the PC12 pheochromocytoma. Biochem Biophys Res Commun. 1991;177:1188–1193.
- Jacovina AT, Zhong F, Khazanova E, et al. Neuritogenesis and the nerve growth factor-induced differentiation of PC-12 cells requires annexin II-mediated plasmin generation. J Biol Chem. 2001;276,:49350–49358.
- Zhang X, Li F, Guo L, et al. Forskolin regulates L-Type calcium channel through interaction between Actinin 4 and β3 subunit in osteoblasts. PloS One. 2015;10:e0124274.
- Michael LF, Asahara H, Shulman AI, et al. The phosphorylation status of a cyclic AMP-responsive activator is modulated via a chromatin-dependent mechanism. Mol Cell Biol. 2000;20:1596–1603.
- Simpson JN, McGinty JF. Forskolin increases phosphorylated-CREB and Fos immunoreactivity in rat striatum. NeuroReport. 1994;5:10.
- Functional MK. Association between Regulatory RNAs and the Annexins. Int J Mol Sci. 2018;19:591.
- Liao YC, Fernandopulle MS, Wang G, et al. RNA granules hitchhike on lysosomes for long-distance transport, using Annexin A11 as a molecular tether. Cell. 2019;179:147–164.e120.
- Fähling M, Paliege A, Jönsson S, et al. NFAT5 regulates renal gene expression in response to angiotensin II through Annexin-A2-mediated posttranscriptional regulation in hypertensive rats. Am J Physiol Renal Physiol. 2019;316:F101–F112.
- Fahling M, Mrowka R, Steege A, et al. Translational control of collagen prolyl 4-hydroxylase-alpha(I) gene expression under hypoxia. J Biol Chem. 2006;281:26089–26101.
- Harvey RF, Smith TS, Mulroney T, et al. Trans-acting translational regulatory RNA binding proteins. Wiley Interdiscip Rev RNA. 2018;9:e1465.
- Thoma C, Bergamini G, Galy B, et al. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol Cell. 2004;15:925–935.
- Thoma C, Fraterman S, Gentzel M, et al. Translation initiation by the c-myc mRNA internal ribosome entry sequence and the poly(A) tail. RNA (New York, N.Y. 2008;14:1579–1589.
- Svitkin YV, Imataka H, Khaleghpour K, et al. A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA (New York, N.Y. 2001;7:1743–1752.
- Chen J, Kastan MB. 5ʹ-3ʹ-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev. 2010;24:2146–2156.
- Li N, Ponnusamy M, Li M-P, et al. The Role of MicroRNA and LncRNA–MicroRNA Interactions in Regulating Ischemic Heart Disease. J Cardiovasc Pharmacol Ther. 2016;22:105–111.
- Sachdeva M, Mo -Y-Y. p53 and c-myc: how does the cell balance “yin” and “yang”? Cell Cycle. 2009;8:1303.
- Grindheim AK, Hollas H, Ramirez J, et al. Effect of serine phosphorylation and Ser25 phospho-mimicking mutations on nuclear localisation and ligand interactions of annexin A2. J Mol Biol. 2014;426:2486–2499.
- Godet A-C, David F, Hantelys F, et al. IRES Trans-Acting Factors, Key Actors of the Stress Response. Int J Mol Sci. 2019;20:924.
- Kwak H, Park MW, Jeong S. Annexin A2 binds RNA and reduces the frameshifting efficiency of infectious bronchitis virus. PloS One. 2011;6:e24067.
- Barrera A, Ramos H, Vera-Otarola J, et al. Post-translational modifications of hnRNP A1 differentially modulate retroviral IRES-mediated translation initiation. Nucleic Acids Res. 2020;48:10479–10499.
- Ecsedi P, Kiss B, Gogl G, et al. Regulation of the equilibrium between closed and open conformations of Annexin A2 by N-Terminal phosphorylation and S100A4-binding. Structure. 2017;25:1195–1207.e1195.
- Aareskjold E, Grindheim AK, Hollas H, et al. Two tales of Annexin A2 knock-down: one of compensatory effects by antisense RNA and another of a highly active hairpin ribozyme. Biochem Pharmacol. 2019;166:253–263.
- Jackson RJ, Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74.
- Nilsson J, Sengupta J, Frank J, et al. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141.
- Yu Y, Ji H, Doudna JA, et al. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci. 2005;14:1438–1446.
- Müller-McNicoll M, Rossbach O, Hui J, et al. Auto-regulatory feedback by RNA-binding proteins. J Mol Cell Biol. 2019;11:930–939.
- Zhang X, Farrell AS, Daniel CJ, et al. Mechanistic insight into Myc stabilization in breast cancer involving aberrant Axin1 expression. Proc Natl Acad Sci U S A. 2012;109:2790–2795.
- Koh CM, Bieberich CJ, Dang CV, et al. MYC and Prostate Cancer. Genes Cancer. 2010;1:617–628.
- Liu J-W, Shen -J-J, Tanzillo-Swarts A, et al. Annexin II expression is reduced or lost in prostate cancer cells and its re-expression inhibits prostate cancer cell migration. Oncogene. 2003;22:1475–1485.
- Huber BE, Thorgeirsson SS. Analysis of c-myc expression in a human hepatoma cell line. Cancer Res. 1987;47:3414–3420.
- Puisieux A, Ji J, Ozturk M. Annexin II up-regulates cellular levels of p11 protein by a post-translational mechanisms. Biochem J. 1996;313(Pt 1):51–55.
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745.
- Brandherm I, Disse J, Zeuschner D, et al. cAMP-induced secretion of endothelial von Willebrand factor is regulated by a phosphorylation/dephosphorylation switch in annexin A2. Blood. 2013;122:1042–1051.
- Kojima I, Kojima K, Rasmussen H. Role of calcium and cAMP in the action of adrenocorticotropin on aldosterone secretion. J Biol Chem. 1985;260:4248–4256.
- Monteith GR, Prevarskaya N, Roberts-Thomson SJ. The calcium–cancer signalling nexus. Nat Rev Cancer. 2017;17:367.
- Capiod T. Cell proliferation, calcium influx and calcium channels. Biochimie. 2011;93:2075–2079.
- Südhof TC. Calcium control of neurotransmitter release. Cold Spring Harb Perspect Biol. 2012;4:a011353–a011353.
- Jansen RP, Niessing D, Baumann S, et al. mRNA transport meets membrane traffic. Trends Genet. 2014;30:408–417.
