ABSTRACT
Recent research unveiled that LINC00857 plays a regulatory role in multiple human cancers, such as lung adenocarcinoma and gastric cancer. Nevertheless, the function of LINC00857 in pancreatic adenocarcinoma (PAAD) remains unclear. This study concentrates on LINC00857 to discuss the relevant molecular mechanism of this gene in PAAD. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) and western blot were implemented for measuring the expressions of RNAs and proteins. Wound healing and Transwell assays were used to assess cell migration and invasion, and fluorescent in situ hybridization (FISH) to locate LINC00857 in PAAD cells. Additionally, mechanism assays were conducted to validate the interaction between genes. Results indicated that LINC00857 was upregulated in PAAD cells and the knockdown of LINC00857 impeded PAAD cell migration, invasion and epithelial-mesenchymal transition (EMT). Further, it was found that LNC00857 regulates CLDN12 expression by targeting miR-150-5p. Moreover, LINC00857 was confirmed to recruit serine/arginine-rich splicing factor 1 (SRSF1) to promote the alternative splicing (AS) targeting CLDN12, affecting the phenotypes of PAAD cells. In addition, the transcription factor ZNF460 was proven to positively regulate LINC00857 expression. To sum up, LINC00857 regulated by ZNF460 upregulates CLDN12 expression by sponging miR-150-5p and recruiting SRSF1 to facilitate the progression of PAAD cells.
Introduction
Pancreatic adenocarcinoma (PAAD) has been recognized as the most frequent pancreatic cancer with the peculiarity of high local and distant invasiveness[Citation1]. Few symptoms were exhibited in early stage and most PAAD patients suffer from poor prognosis [Citation1,Citation2]. Although surgical resection is usually performed for the treatment of cancers at the early stage, the surgical intervention of PAAD remains controversial [Citation2,Citation3]. Recent years witness the development of adjuvant chemotherapy following surgical resection for PAAD. Nevertheless, the overall five-year survival rate of PAAD patients was still unsatisfactory at the probability of around 20% [Citation4,Citation5]. Considering the dismal diagnosis of PAAD [Citation6] and the trend of target treatment, it is urgent for us to explore the underlying molecular mechanisms in PAAD cells to provide novel potential therapeutic targets for PAAD.
According to previous studies, long non-coding RNAs (lncRNAs) with more than 200 nucleotides have been identified to play crucial roles in the progression of multiple cancers, including PAAD [Citation7,Citation8]. Accumulating research work unveils that long intergenic non-coding RNAs (lincRNAs), a subclass of lncRNAs, are dysregulated in various cancers, such as colorectal cancer [Citation9], hepatocellular carcinoma [Citation10] and PAAD [Citation11]. For instance, LINC01232 has been testified to act as an oncogene promoting the deterioration of PAAD [Citation11]. Moreover, LINC00857 has been reported to enhance the EMT of hepatocellular carcinoma cells [Citation12]. Aside from that, high expression of LINC00857 has been reported to be correlated with poor survival of lung cancer patients through mediating the tumour progression [Citation13]. However, referring to published reports, the association between LINC00857 and PAAD has not been well explored yet. Hence, we intended to focus on LINC00857 and probe into the relevant molecular mechanism of this gene in PAAD.
Emerging evidence supports that competing endogenous RNA (ceRNA) engages in regulating the progression of diverse cancer cells, including PAAD cells [Citation14]. Upregulated LINC00857 is unearthed to exert influences on repressing the apoptosis of lung adenocarcinoma cells and propelling the cell proliferation through miR-1179/SPAG5 axis in a ceRNA network [Citation15]. Nevertheless, the function and regulatory mechanism of LINC00857 in PAAD cells has been mentioned in limited research work. Thus, this study tried to investigate the role of LINC00857 and explore its downstream genes in the ceRNA network.
Recently, it has been reported that alternative splicing (AS) is likely to generate varieties at RNA and protein levels and this event engages in multiple pathologies including cancers [Citation16,Citation17]. Based on previous research, AS could influence cancer-related phenotypes, such as cell proliferation, apoptosis [Citation18], EMT and stemness [Citation19]. It has been unveiled that serine/arginine splicing factor 1 (SRSF1) and RNA-binding motif protein 4 (RBM4) are involved in an antagonistic mechanism which is associated with the oncogenic impacts on lung cancer cells [Citation20]. An emerging study also unveils the splicing process in PAAD which correlates with prognosis-related splicing factors (SFs) and AS, suggesting the prognostic value of AS events [Citation21]. However, the knowledge of SFs and their underlying regulatory mechanisms in PAAD are insufficient. Hence, this study was to probe into the potential mechanism of AS in regulating the progression of PAAD cells.
In brief, this research aimed to make in-depth investigation into the role of LINC00857 and found the potential ceRNA network involving LINC00857 in PAAD cells. Meanwhile, we attempted to discover other regulatory mechanism concerning AS in PAAD cells, and this study was supposed to provide more molecular information about PAAD cells.
Materials and methods
Cell culture
Normal human pancreatic ductal epithelial (HPDE) cell line and PAAD cell lines (PANC-1, Capan-1, BxPC-3) were all procured from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells used in this study were cultured in RPMI 1640 medium (Gibco, Gaithersburg, MD, USA) containing 10% foetal bovine serum (FBS; Gibco, USA). The medium was also added with 100 U/ml penicillin-streptomycin solutions and maintained in the humidified incubator with 5% CO2 at 37°C.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNAs were extracted by Trizol (Takara, Japan) following the manufacturer’s instructions. Reverse transcription kits were utilized to reversely transcribe the extracted RNAs into their corresponding cDNAs. Afterwards, qRT-PCR Kit (QR0100-1KT, Sigma-Aldrich, USA) was applied for RT-qPCR analysis. For the detection of the expression of CLDN12 isoforms, taq-man probes specifically targeting CLDN12a/b/c were used for the multiplex PCR of genotypes. β-actin acted as the internal reference for lncRNA/mRNA quantification while U6 for miRNA quantification. 2−ΔΔCt method was applied for calculation. Sequences of shRNAs were shown in .
Table 1. Sequences of shRNAs
Cell transfection
Sh-RNAs (Ribobio, Nanjing, China) were utilized for the silence of LINC00857, CLDN12, SRSF1 or ZNF460, and sh-NCs were the corresponding negative controls. PcDNA3.1 was used for the overexpression of LINC00857. All constructed plasmids were transfected into cells and after 48-hour transfection, transfected cells were involved in the subsequent research. RT-qPCR was conducted to examine the efficiency of silence or overexpression.
Wound healing assay
Firstly, the cells transfected with sh-NC, sh-LINC00857 or sh-CLDN12 were planted into 6-well plates. When the cell confluence reached about 80%, the 200-μL sterile pipette tip was used for scratching the layers. Then, the scratched cells were washed away by phosphate buffer saline (PBS; Vicmed, China). 2% FBS was added into the fresh medium. The culture dishes were placed into the incubator in the same environment as the cell culture. After 24-h incubation, the scratches were captured with a microscope. MShot Image Analysis System was applied for analysing the images obtained.
Transwell assay
Transwell filter chambers (BD Biosciences, New Jersey, USA) were used in this assay according to the manufacturer’s guidance. The transfected cells were suspended in the upper chambers containing serum-free RPMI 1640 medium (Gibco, Gaithersburg, MD, USA). A matrigel-coated membrane was covered under the bottom of upper chamber for the invasion assay. The lower chamber was added with 10% FBS (Gibco, USA). After 24-hour incubation, the cells migrating or invading into the lower chambers were fixed by methanol for 10 minutes. 0.5% crystal violet was used for staining. Finally, the migrated or invaded cells were counted under the light microscope.
Western blot
Proteins were firstly isolated with protein lysis buffer (Keygen, Nanjing, China) and extracted by Total Protein Extraction Kit (PROTTOT-1KT, Sigma-Aldrich, USA). Later, the proteins extracted were treated with SDS Quick Match Gel Kit (P0670-250 ml, Beyotime Biotechnology, Shanghai, China) and then transferred onto polyvinylidene fluoride (PVDF) membranes (Vicmed, China). Afterwards, the membranes blocked by 5% skimmed milk were co-cultivated with the primary antibodies, including Anti-β-actin, Anti-CLDN12and Anti-SRSF1 antibodies, at 4°C overnight. Subsequently, secondary antibodies were added for incubation. Eventually, the expressions of target proteins were analysed by J software. β-actin served as the internal reference.
Fluorescent in situ hybridization (FISH)
After being washed by PBS (Vicmed, China), cells grown on the slides were fixed with 4% paraformaldehyde for 30 minutes. Then, cells were co-cultured with FISH probe targeting LINC00857 or control probe. The mixtures were washed by PBS and later, stained by DAPI. Confocal laser microscopy (Zeiss LSM 880) was applied for observation.
RNA binding protein immunoprecipitation (RIP) assay
Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA) was utilized in this assay. The user manual was strictly followed. Cell lysates were incubated with magnetic-bead-coated negative control anti-immunoglobulin G (IgG, GeneTex, California, USA) or human anti-Argonaute 2 (Ago2) antibody at 4°C overnight. The immunoprecipitated RNAs were purified and tested through RT-qPCR or agarose gel electrophoresis (AGE).
RNA pull-down assay
Pierce™ RNA 3ʹ End Desthiobiotinylation Kit (Thermo 20,163, USA) was used based on the supplier’s instructions. Cells were transfected for 24 hours severally with biotinylated NC (Bio-NC) for LINC00857, CLDN12 or pre-CLDN12, biotinylated wild-type LINC00857 (Bio-LINC00857-WT), biotinylated mutated LINC00857 (Bio-LINC00857-Mut), biotinylated wild-type CLDN12-3ʹuntranslated region (Bio-CLDN12-3ʹUTR-WT), biotinylated mutated CLDN12-3ʹUTR (Bio-CLDN12-3ʹUTR-Mut), biotinylated wild-type pre-CLDN12 (Bio-pre-CLDN12-WT). After obtaining cell lysates, streptavidin-coated magnetic beads were added for co-culture. The target genes or proteins bound to biotin-coupled RNA complexes were then subjected to RT-qPCR or western blot.
Luciferase reporter assay
PmirGLO and pGL3 were procured for constructing plasmids used in this assay. PmirGLO-LINC00857-WT/-LINC00857-Mut and pmirGLO-CLDN12-3ʹUTR-WT/-CLDN12-3ʹUTR-Mut plasmids with wild-type or mutant miR-150-5p binding sites were transfected into cells. These constructed plasmids were co-transfected with miR-150-5p mimics or mimics NC. PGL3-LINC00857 promoter-WT/-LINC00857 promoter-Mut plasmids with wild-type or mutant binding sites of ZNF460 were transfected into cells, and they were co-transfected with pcDNA3.1 or pcDNA3.1-ZNF460. After that, Luciferase Reporter Assay Substrate Kit (ab228530, Abcam, Shanghai, China) was utilized and the luciferase activities were detected via GloMax20/20 Luminometer (Progema).
DNA pull-down assay
DNA pull-down assay was performed to confirm the binding relationship between LINC00857 promoter and potential transcription factors. The biotinylated probes targeting wild-type LINC00857 promoter (Bio-LINC00857 promoter-WT) and mutated LINC00857 promoter (Bio-LINC00857 promoter-Mut) were utilized and then incubated with cell lysates. Streptavidin magnetic beads were added to help extract the DNA-protein complexes. The protein levels were further measured through western blot.
Statistical analysis
SPSS 22.0 software was applied for statistical analysis. Each experiment was performed at least three times, and the collected data were demonstrated as mean ± standard deviation (SD). Student’s t-test, one-way analysis of variance (ANOVA) or two-way ANOVA was used for difference analysis. P < 0.05 was regarded as statistically significant in differences.
Results
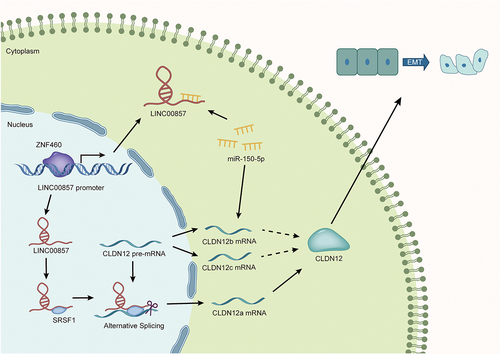
LINC00857 promotes malignant phenotypes of PAAD cells
LINC00857 expressions in normal HPDE cells and three PAAD cell lines (PANC-1, Capan-1, and BxPC-3) were measured by RT-qPCR (). The outcomes revealed that LINC00857 was upregulated in PAAD cells, especially in PANC-1 and BxPC-3, which were selected for the following study. The efficiency of LINC00857 knockdown was detected subsequently. As shown in , sh1-LINC00857 and sh2-LINC00857 were more efficient than sh3-LINC00857 and were used in the later loss-of-function assays. We performed CCK-8 assay to evaluate the proliferation of PAAD cells after the knockdown of LINC00857, finding that LINC00857 depletion led to the decrease of cell proliferation of PAAD (Fig. S1A). To understand the influence of LINC00857 on PAAD cell migration, wound healing assays were conducted. Based on the collected data, PAAD migration capability was restrained due to LINC00857 knockdown (). Then, Transwell assays were performed to test the migration and invasion ability of PAAD cells. The results demonstrated that migration and invasion capabilities of PANC-1 and BxPC-3 cells were significantly hindered after the knockdown of LINC00857 (). We also observed the shapes of PANC-1and BxPC-3 before and after the knockdown of LINC00857, and the captured images showed that LINC00857 knockdown reduced the spindle phenotype of cells which is an EMT phenotype, indicating that LINC00857 downregulation would hamper EMT of PAAD cells (). The expressions of EMT-related proteins were measured through western blot. After the knockdown of LINC00857, the levels of epithelial marker proteins (E-cadherin and ZO-1) increased, while that of mesenchymal marker proteins (N-cadherin and vimentin) decreased (). The outcomes further indicated that EMT of PAAD cells were inhibited when LINC00857 was knocked down. After that, we performed FISH assay to determine the distribution of LINC00857 (). The fluorescent signal presented that LINC00857 was mainly located in cytoplasm, which made us speculate if LINC00857 might regulate the progression of PAAD in a ceRNA-related manner. To summarize, LINC00857 promotes malignant phenotypes of PAAD cells. The underlying mechanism of LINC00857 was probed afterwards.
Figure 1. LINC00857 promotes malignant phenotypes of PAAD cells.
LINC00857 regulates the expression of CLDN12 by sponging miR-150-5p in PAAD cells
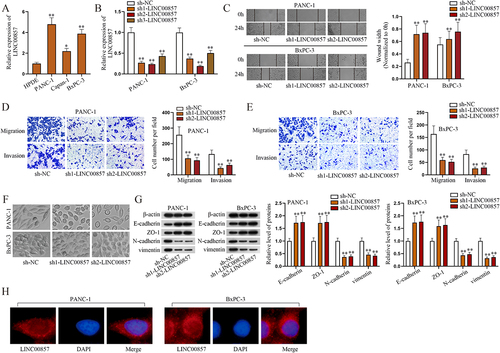
In this section, we designed and implemented a series of experiments to explore the mechanism of LINC00857 in PAAD cells. In support of the abovementioned speculation, RIP assays were conducted. Through RT-qPCR, LINC00857 could be detected in PANC-1 and BxPC-3 cells and the enrichment of LINC00857 in Anti-Ago2 group was shown in . We inferred that LINC00857 might exert influences through ceRNA network, as Ago2 is the main component of RNA-induced silencing complex (RISC). Reasonably, we used LncBase Predicted v.2 website (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted) under the condition of threshold ≥ 0.9 to predict the candidate miRNAs. As shown in , hsa-miR-6736-3p, hsa-miR-1290 and hsa-miR-150-5p were selected out. Through RNA pull-down assay in PANC-1 cells, we found that miR-150-5p was most inclined to combine with bio-LINC00857-WT (). Moreover, the results of RT-qPCR showed that miR-150-5p was notably low-expressed in PANC-1, Capan-1 and BxPC-3 cells compared with HPDE cells (Fig. S1B). Thereof, miR-6736-3p and miR-1290 were excluded. The following results of RNA pull-down assay further confirmed that the wild type of LINC00857 could bind to miR-150-5p, rather than its mutant form (). Subsequently, RIP assays and RT-qPCR were done in PANC-1 cells to detect the enrichments of miR-150-5p as well as LINC00857 (). Apart from that, we intended to complete the ceRNA network involved in this study by identifying the target mRNA. StarBase v2.0 website (http://starbase.sysu.edu.cn) was utilized to screen out candidate mRNAs likely to interact with miR-150-5p. Under the conditions of CLIP-Data ≥10 and pan-Cancer ≥18, MANBAL, VEGFA, CLDN12 and IPO5 were screened out (). According to the previous studies, MANBAL (mannosidase, beta A, lysosomal-like) has been reported to correlate with tongue squamous cell carcinoma [Citation22]; VEGFA, representing vascular endothelial growth factor A, is able to facilitate the progression of pancreatic cancer [Citation23]; CLDN12, short for claudin 12, is allowed to regulate the development of lung squamous cell carcinoma [Citation24]; and IP05, short for importin 5, has also been verified to promote the tumourigenicity of colorectal cancer cells [Citation25]. To analyse these four candidates, we firstly performed RT-qPCR to quantify their expressions, noticing that only CLDN12 apparently declined due to LINC00857 knockdown (). Since the expressions of other candidate mRNAs had almost no variation, CLDN12 was determined to be involved in the following investigation in PANC-1 cells. After biotinylating CLDN12-3ʹUTR, we performed RT-qPCR and the consequences indicated that miR-150-5p could bind to wild type of CLDN12-3ʹUTR, rather than its mutant form (). Moreover, through RIP assay and RT-qPCR analysis, the enrichments of miR-150-5p and CLDN12 were measured and presented in in Anti-Ago2 group (). Additionally, we realized that downregulating LINC00857 would lessen the expression of CLDN12 while miR-150-5p expression hardly changed, which indicated that LINC00857 was positively with CLDN12 expression but could not affect miR-150-5p expression (). The following luciferase reporter assays verified that miR-150-5p could respectively bind to wild type of LINC00857 and CLDN12-3ʹUTR (). Referring to the above results, we could reach a conclusion that LINC00857 acted as a sponge of miR-150-5p and could regulate the expression of CLDN12. To dig into the regulatory mechanism of LINC00857, the rescue experiments were done. The outcomes of RT-qPCR and western blot suggested that the effects of LINC00857 knockdown and overexpression on CLDN12 could be greatly counteracted by the altering of the expression of miR-150-5p, which implied that LINC00857 could modulate CLDN12 expression through miR-150-5p (). Taken together, LINC00857 competitively binds to miR-150-5p and further regulate the expression of CLDN12 in PAAD cells.
Figure 2. LINC00857 regulates the expression of CLDN12 by sponging miR-150-5p in PAAD cells.
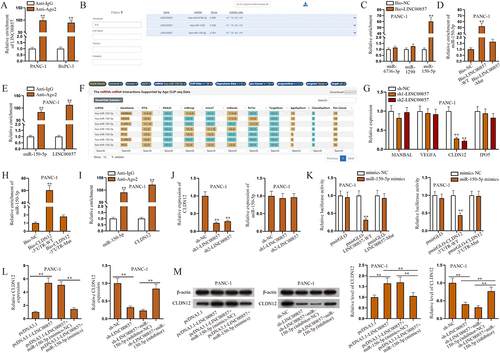
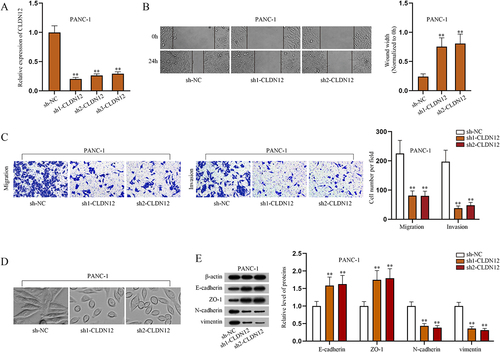
CLDN12 facilitates the malignant phenotypes of PAAD cells
In recent years, CLDN12 has been reported to promote proliferation and migration of osteosarcoma cells [Citation26]. However, the influences of CLDN12 on the phenotypes of other cancer cells including PAAD have not been well explored so far. In this study, we further probed into the effects of this gene. Firstly, RT-qPCR was applied to test the efficiency of CLDN12 knockdown in PANC-1 cells (). Sh1-CLDN12 and sh2-CLDN12 were selected for the following experiments. Based on the results of CCK-8 assay, ablated CLDN12 suppressed the proliferation of PANC-1 cells (Fig. S1C). Through wound healing and Transwell assays, it was informed that downregulating CLDN12 restricted migration and invasion of PANC-1 cells (). Moreover, we observed the shapes of PANC-1 cells before and after CLDN12 knockdown. The obtained images reflected that CLDN12 promoted EMT of PANC-1 cells (). We examined the expressions of EMT-related proteins through western blot assays, and the findings further proved that knockdown of CLDN12 inhibited EMT of PANC-1 cells (). In short, CLDN12 plays an oncogenic role in PAAD cells and facilitates the malignant phenotypes of PAAD cells.
Figure 3. CLDN12 facilitates the malignant phenotypes of PAAD cells.
LINC00857 recruits SRSF1 for alternative splicing to regulate CLDN12 expression in PAAD cells
Considering the results shown in , miR-150-5p could not completely counteract the effects of regulating LINC00857. Thereby, we conjectured that there might exist other ways to modulate the expression of CLDN12. Previous research work has revealed that lncRNAs are allowed to regulate expressions of mRNAs by recruiting RNA-binding proteins (RBPs) [Citation27]. Hence, we tried to find a RBP which was able to be recruited by LINC00857 and to regulate CLDN12 at the same time in PANC-1 cells. According to the searching results of starBase, we obtained a list of potential candidate RBPs which might interact with LINC00857 under the condition of CLIP-Data ≥ 1, and three candidate RBPs that are probably combined with CLDN12 under the condition of CLIP-Data ≥ 10 were screened out (). Via comparison the two results, U2AF2, ELAVL1 and SRSF1 were determined to be investigated in the following experiments. Reviewing recent studies, we found that U2AF2, short for U2 small nuclear RNA auxiliary factor 2, could propel the progression of non-small cell lung cancer [Citation28]; ELAVL1, ELAV like RNA binding protein 1, is verified to regulate the development of colorectal cancer [Citation29]; SRSF1, representing serine/arginine-rich splicing factor 1, has been reported to induce AS events in multiple cancers, such as breast cancer [Citation30], lung cancer [Citation31] and glioma [Citation32]. Since a typical splicing factor was in the list of candidate RBPs, pre-CLDN12 mRNA was involved in the subsequent research in PANC-1 cells. After RNA pull-down assay, western blot was done and the collected data suggested that only SRSF1 could bind to LINC00857 and pre-CLDN12 (). The RT-qPCR outcomes of the following RIP assay demonstrated that LINC00857 and pre-CLDN12 rather than CLDN12 could be enriched by SRSF1 (). According to these abovementioned results, we considered that SRSF1 might engage in the alternative splicing of CLDN12. Accordingly, we searched on National Center for Biotechnology Information database (NCBI; https://www.ncbi.nlm.nih.gov), finding three transcript variants of CLDN12 mRNA, CLDN12a, CLDN12b and CLDN12c. Merely one protein was translated. By utilizing SNAPGENE software, we analysed the sequences of these transcript variants and noticed the differences among them (). It was reasonable to infer that AS events might occur during the process of expressing CLDN12. Subsequently, we tested the efficiency of SRSF1 silence in PANC-1 cells via RT-qPCR. Sh1-SRSF1 and sh2-SRSF1 were more efficient than sh3-SRSF1 (). The data shown in indicated that CLDN12 expression was cut down as SRSF1 was silenced in PANC-1 cells. Furthermore, we measured the expressions of these three transcript variants via AGE and multiplex qPCR analysis before and after SRSF1 silence. The sequence-specific taq-man probes labelled with two fluorescent dyes were used in multiplex qPCR analysis. As shown in , CLDN12a expression was declined and CLDN12b&c expressions was climbed up due to SRSF1 silence, which indicated that the expression of CLDN12 was transferred from CLDN12a to CLDN12b&c. After that, the protein level of CLDN12 was detected via western blot, and the results displayed an obvious decrease in CLDN12 protein level due to SRSF1 knockdown (). As shown in , CLDN12a expression was similar to CLDN12b&c expression in terms of RNA level while the former was overtly more than the latter in terms of proteins level, which indicated that the translation efficiency of CLDN12a is the highest among all transcript variants of CLDN12. In conclusion, SRSF1 recruited by LINC00857 regulates the expression of CLDN12 via AS of pre-CLDN12.
Figure 4. LINC00857 recruits SRSF1 for alternative splicing to regulate CLDN12 expression in PAAD cells.
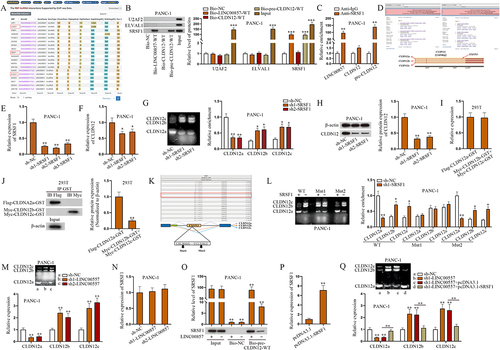
Next, we explored the specific binding site of SRSF1 on pre-CLDN12. Recent research work unveils that SRSF1 plays a main role in identifying the binding sites of exonic splicing enhancers (ESE) and activating splicing on the splice sites [Citation33]. As the former experiments confirmed that SRSF1 participated in AS on CLDN12, we then utilized ESEfinder database (http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home) to predict potential binding sites for mutations (). The outcomes of AGE and multiplex qPCR analysis indicated that the expression of CLDN12 was transferred from CLDN12a to CLDN12b&c, and the expressions of these transcript variants were not affected after the mutation of Position 1. As for the mutation of Position 2 (Mut 2), the expression of CLDN12a/b/c was still influenced. The above results indicated that SRSF1 interacts with pre-CLDN12 via Position 1 (). Further, we investigated the effect of LINC00857 on the expression of CLDN12a/b/c and SRSF1. The results showed that CLDN12a expression was reduced by LINC00857 knockdown while CLDN12b/c expression was elevated and SRSF1 had no marked change (). Additionally, the western blot results of RNA pull-down assay showed that SRSF1 was overtly abundant in pre-CLDN12 pulldown complexes but decreased due to LINC00857 knockdown (). The above results indicated that LINC00857 has no regulatory effect on SRSF1 expression but promotes the binding between SRSF1 and pre-CLDN12. Prior to rescue experiments, we overexpressed SRSF1 (). As shown in , SRSF1 overexpression reversed the inhibitory effect of LINC00857 knockdown on CLDN12a expression and its promoting effect on CLDN12b/c expression, suggesting that LINC00857 regulates AS of CLDN12 via sequestering SRSF1. To sum up, LINC00857 recruits SRSF1 for AS to regulate CLDN12 expression.
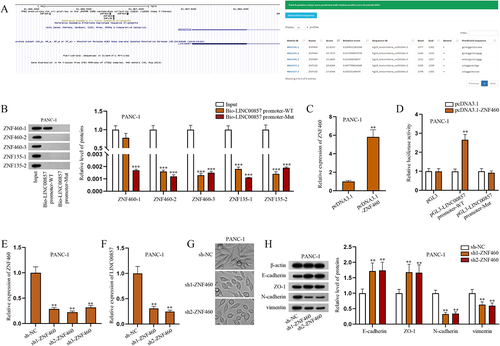
ZNF460 as the transcription factor of LINC00857 promoter regulates LINC00857 expression in PAAD cells
Given that transcription factors play a key role in regulating cancer development [Citation34], we next explored the upstream mechanism of LINC00857 in PAAD cells. The bioinformatics tools, UCSC (http://genome.ucsc.edu) and JASPAR (http://jaspar.genereg.net), were utilized to predict the possible transcription factors of LINC00857, including ZNF460 and ZNF135; three putative ZNF460 binding sites of LINC00857 and two ZNF135 ones were obtained as well (). Five putative binding sites were marked as ZNF460-1/2/3 and ZNF135-1/2 correspondingly. Referring to previous work, these two transcription factors have not been discussed yet, apart from the predictions of bioinformatics tools [Citation35,Citation36]. DNA pull-down assay followed by western blot analysis was done in PANC-1 cells to determine the most relevant transcription factor and its binding site. The results of western blot showed that only ZNF460-1 was overtly enriched in Bio-LINC00857 promoter-WT group, which indicated that transcription factor ZNF460 might bind to LINC00857 promoter via binding motif 1 in PAAD cells (). The overexpression efficiency of ZNF460 was verified via RT-qPCR (). The following luciferase reporter assay showed that ZNF460 overexpression promoted the luciferase activity of pGL3-LINC00857 promoter-WT but had no marked change on that of pGL3-LINC00857 promoter-Mut, which further verified the binding between ZNF460 and LINC00857 promoter in PAAD cells (). Thereby, we chose ZNF460 for the follow-up investigation. Subsequently, we explored the effects of ZNF460 on LINC00857 expression and EMT of PAAD cells. Before that, the efficiency of ZNF460 knockdown in PANC-1 cells was verified via RT-qPCR (). According to RT-qPCR, ZNF460 knockdown diminished the expression of LINC00857 (), indicating that ZNF460 positively regulates the expression of LINC00857. The spindle phenotype in PANC-1 cells was obviously reduced after downregulating ZNF460, which indicated that ZNF460 downregulation suppresses the EMT of PAAD cells (). In addition, the expressions of key EMT-related proteins were measured by western blot. The results showed that the protein levels of E-cadherin and ZO-1 were increased whereas those of N-cadherin and vimentin were decreased by ZNF460 knockdown, which further elucidated that ZNF460 knockdown inhibits the EMT of PAAD cells (). Accordingly, ZNF460 could promote the EMT of PAAD cells. In summary, as the transcription factor of LINC00857 promoter, ZNF460 can upregulate LINC00857 expression to enhance the EMT of PAAD cells.
Figure 5. ZNF460 as the transcription factor of LINC00857 promoter regulates LINC00857 expression in PAAD cells.
Discussion
PAAD, one of the most lethal cancers, is characterized by high aggressiveness and metastasis. Patients with PAAD are often subjected to poor survival [Citation37,Citation38]. Recently, increasing studies have exposed that lncRNAs are highly associated with the initiation and progression of multiple cancers, including PAAD [Citation39,Citation40]. For example, LINC01232 has been identified to act as an oncogene and knockdown of LINC01232 hinders the proliferation, invasion and EMT of PAAD cells [Citation11]. In this study, we aimed to investigate a novel molecular mechanism of a certain lncRNA to offer a novel sight into the comprehensive understanding of PAAD. Based on previous studies, we found that the roles and regulatory mechanism of LINC00857 in PAAD have not been studied to date. Thereby, the profiles and roles of LINC00857 were explored at the very beginning in this research. The expressions of LINC00857 in normal HPDE cell line and three PAAD cell lines (PANC-1, Capan-1, and BxPC-3) were examined by RT-qPCR. We found that LINC00857 expression was remarkably upregulated in these PAAD cell lines relative to that in HPDE cell line. After testing the efficiency of sh-LINC00857, a series of loss-of-function assays were performed and the results indicated that ablation of LINC00857 hampers the proliferation, migration, invasion and EMT of PAAD cells.
Recently, a published report has demonstrated that lncRNA-related ceRNA networks were closely associated with the pathogenesis of PAAD [Citation38]. To determine whether LINC00857-associated ceRNA network functions in PAAD cells, we firstly performed FISH assay to determine the subcellular distribution of LINC00857. The fluorescent signal presented that LINC00857 was mainly located in cytoplasm. Given that, we inferred that LINC00857 might serve as a ceRNA in PAAD cells. RIP assay further confirmed the potential of LINC00857 to act as a ceRNA. With the combination of bioinformatics, RNA pull-down assay and RT-qPCR analysis, we concluded that LINC00857 could target miR-150-5p in PAAD cells. Subsequently, we investigated the downstream target of miR-150-5p in PAAD cells. By searching on the starBase website, four putative mRNAs were sorted out, including MANBAL, VEGFA, CLDN12, and IPO5. Then, by RT-qPCR analysis, RNA pull-down, RIP and luciferase reporter assays, we found that CLDN12 was the downstream gene of miR-150-5p and positively regulated by LINC00857 in PAAD cells. The following rescue experiments testified that LINC00857 could upregulate CLDN12 expression by targeting miR-150-5p in a ceRNA-related manner.
Previous studies have identified that CLDN12 stimulates the proliferation and migration of osteosarcoma cells [Citation26]. Hence, we further investigated the influences of CLDN12 on the phenotypes of PAAD cells. Prior to a line of loss-of-function assays, we tested the efficiency of CLDN12 silencing via RT-qPCR. Afterwards, the results of wound healing and Transwell assays confirmed that CLDN12 functions as an oncogene to promote cell migration and invasion in PAAD. By observing the changes of PAAD cell shapes and detecting the expressions of EMT-associated proteins, we also reached a conclusion that CLDN12 overexpression facilitates the EMT of PAAD cells.
Based on the results of rescue experiments, miR-150-5p inhibitor could not completely offset the inhibitory effect of LINC00857 knockdown on CLDN12 expression. We speculated that CLDN12 might be regulated in other potential approaches. Notably, lncRNAs have been reported to modulate mRNA expression by recruiting RBPs [Citation27]. Given that, we utilized the bioinformatics tool starBase to predict the potential RBPs of LINC00857 and CLDN12. Thereby, we obtained three mutual RBPs of LINC00857 and CLDN12. Furthermore, SRSF1, one of candidate RBPs, has been newly reported to act as a splicing factor in non-small cell lung cancer [Citation41]. Plus, the expression of several splicing factors has been reported to be modulated during EMT and each is capable of regulating hundreds of pre-mRNA (precursor mRNA) targets [Citation19]. Accordingly, pre-CLDN12 mRNA was taken into consideration in the following study. Afterwards, we found that SRSF1 binds to LINC00857 and pre-CLDN12 though RNA pull-down and RIP assays. Based on the results of RT-qPCR, western blot and bioinformatics, SRSF1 knockdown causes the expression of CLDN12a transferring to CLDN12b/c and meanwhile reduces the protein level of CLDN12. Therefore, we concluded that SRSF1 promotes the protein level of CLDN12 via AS of pre-CLDN12. Furthermore, we detected the mRNA and protein levels of CLDN12 in 293 T cells transfected with Flag-CLDN12a-GST, Myc-CLDN12c-GST and Myc-CLDN12c-GST. We found that under the same gene expression level, CLDN12a displayed a higher protein level compared to CLDN12b/c, indicating that the translation efficiency of different isoforms varies. After predicting the binding sites of SRSF1 on CLDN12, AGE and RT-qPCR were utilized to determine that binding site 1 was the exact position. Then we found that LINC00857 could not influence SRSF1 expression but to facilitate the binding between SFSF1 and pre-CLDN12. Also we verified that LINC00857 propelled the binding between SRSF1 and CLDN12 pre-mRNA and regulated the alternative splicing of CLDN12.
In addition, we further explored the transcription factor of LINC00857 expression. By using UCSC and JASPAR databases, ZNF460 and ZNF135 were projected to be the candidate transcription factor of LINC00857 promoter. Dependent on the results of western blot, DNA pull-down and luciferase reporter assays, ZNF460 was validated to serve as the transcription factor of LINC00857 promoter. Moreover, we found that knockdown of ZNF460 would inhibit the EMT of PAAD cells.
To summarize, this study for the first time confirmed that LINC00857 could regulate CLDN12 expression by sponging miR-150-5p and recruiting SRSF1 for AS to propel the malignant phenotypes of PAAD cells. Additionally, ZNF460 was firstly proven to be the transcription factor of LINC00857 promoter to modulate LINC00857 expression in PAAD cells. However, in vivo experiments were not involved in this research, which need to be complemented in the future investigation.
Supplemental Material
Download Zip (660.1 KB)Acknowledgments
Members involved are appreciated.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Capasso M, Franceschi M, Rodriguez-Castro KI, et al. Epidemiology and risk factors of pancreatic cancer. Acta Biomed. 2018;89:141–146.
- Tan E, Song J, Lam S, et al. Postoperative outcomes in elderly patients undergoing pancreatic resection for pancreatic adenocarcinoma: a systematic review and meta-analysis. Int J Surg. 2019;72:59–68.
- Henrikson NB, Aiello Bowles EJ, Blasi PR, et al. Screening for pancreatic cancer: updated evidence report and systematic review for the US preventive services task force. Jama. 2019;322(5):445–454.
- Sánchez Acedo P, Herrera Cabezón J, Zazpe Ripa C, et al. Survival, morbidity and mortality of pancreatic adenocarcinoma after pancreaticoduodenectomy with a total mesopancreas excision. Revista espanola de enfermedades digestivas: organo oficial de la Sociedad Espanola de Patologia Digestiva. 2019;111:609–614.
- Parmar A, Chaves-Porras J, Saluja R, et al. Adjuvant treatment for resected pancreatic adenocarcinoma: a systematic review and network meta-analysis. Crit Rev Oncol Hematol. 2020;145:102817.
- Motoi F, Unno M. Adjuvant and neoadjuvant treatment for pancreatic adenocarcinoma. Jpn J Clin Oncol. 2020;50(5):483–489.
- Song Y, Zou L, Li J, et al. LncRNA SNHG8 promotes the development and chemo-resistance of pancreatic adenocarcinoma. Eur Rev Med Pharmacol Sci. 2018;22:8161–8168.
- Jin X, Ye L, Lin M, et al. lncRNA-CCHE1 is involved in migration and invasion but not in proliferation of pancreatic adenocarcinoma cells possibly by interacting with ROCK1. Oncol Lett. 2019;18:1218–1224.
- Wang G, Li Z, Zhao Q, et al. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/β-catenin signaling pathway. Oncol Rep. 2014;31(4):1839–1845.
- Jia M, Jiang L, Wang YD, et al. lincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through Notch signaling-induced epithelial-mesenchymal transition. Hepatol Res. 2016;46(11):1137–1144.
- Li Q, Lei C, Lu C, et al. LINC01232 exerts oncogenic activities in pancreatic adenocarcinoma via regulation of TM9SF2. Cell Death Dis. 2019;10(10):698.
- Xia C, Zhang XY, Liu W, et al. LINC00857 contributes to hepatocellular carcinoma malignancy via enhancing epithelial-mesenchymal transition. J Cell Biochem. 2018.
- Wang L, He Y, Liu W, et al. Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget. 2016;7(10):11487–11499.
- Zhao R, Ni J, Lu S, et al. CircUBAP2-mediated competing endogenous RNA network modulates tumorigenesis in pancreatic adenocarcinoma. Aging (Albany NY). 2019;11(19):8484–8501.
- Wang L, Cao L, Wen C, et al. LINC00857 regulates lung adenocarcinoma progression, apoptosis and glycolysis by targeting miR-1179/SPAG5 axis. Hum Cell. 2020;33(1):195–204.
- Yang X, Coulombe-Huntington J, Kang S, et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell. 2016;164(4):805–817.
- Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212(1):13–27.
- Climente-González H, Porta-Pardo E, Godzik A, et al. The functional impact of alternative splicing in cancer. Cell Rep. 2017;20(9):2215–2226.
- Pradella D, Naro C, Sette C, et al. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017;16:8.
- Chang HL, Lin JC. SRSF1 and RBM4 differentially modulate the oncogenic effect of HIF-1α in lung cancer cells through alternative splicing mechanism. Biochim Biophys Acta Mol Cell Res. 2019;1866(12):118550.
- Rong MH, Zhu ZH, Guan Y, et al. Identification of prognostic splicing factors and exploration of their potential regulatory mechanisms in pancreatic adenocarcinoma. PeerJ. 2020;8:e8380.
- Yin K, Zhang Y, Zhang S, et al. Using weighted gene co-expression network analysis to identify key modules and hub genes in tongue squamous cell carcinoma. Medicine (Baltimore). 2019;98(37):e17100.
- Liu A, Huang C, Cai X, et al. Twist promotes angiogenesis in pancreatic cancer by targeting miR-497/VEGFA axis. Oncotarget. 2016;7(18):25801–25814.
- Sun L, Feng L, Cui J. Increased expression of claudin-12 promotes the metastatic phenotype of human bronchial epithelial cells and is associated with poor prognosis in lung squamous cell carcinoma. Exp Ther Med. 2019;17:165–174.
- Zhang W, Lu Y, Li X, et al. IPO5 promotes the proliferation and tumourigenicity of colorectal cancer cells by mediating RASAL2 nuclear transportation. J Exp Clin Cancer Res. 2019;38(1):296.
- Tian X, He Y, Han Z, et al. The cytoplasmic expression of CLDN12 predicts an unfavorable prognosis and promotes proliferation and migration of osteosarcoma. Cancer Manag Res. 2019;11:9339–9351.
- Luo X, He S, Hu Y, et al., Sp1-induced LncRNA CTBP1-AS2 is a novel regulator in cardiomyocyte hypertrophy by interacting with FUS to stabilize TLR4. Cardiovascular pathology: the official journal of the Society for Cardiovascular Pathology. 2019;42:21–29.
- Li J, Cheng D, Zhu M, et al. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9(1):179–195.
- Li K, Huang F, Li Y, et al. Stabilization of oncogenic transcripts by the IGF2BP3/ELAVL1 complex promotes tumorigenicity in colorectal cancer. Am J Cancer Res. 2020;10:2480–2494.
- Anczuków O, Akerman M, Cléry A, et al. SRSF1-regulated alternative splicing in breast cancer. Mol Cell. 2015;60(1):105–117.
- Sheng J, Zhao Q, Zhao J, et al. SRSF1 modulates PTPMT1 alternative splicing to regulate lung cancer cell radioresistance. EBioMedicine. 2018;38:113–126.
- Zhou X, Wang R, Li X, et al. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J Clin Invest. 2019;129(2):676–693.
- Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84(1):291–323.
- Vizcaíno C, Mansilla S, Portugal J. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharmacol Ther. 2015;152:111–124.
- Raghuram V, Weber S, Raber J, et al. Assessment of mutations in KCNN2 and ZNF135 to patient neurological symptoms. Neuroreport. 2017;28(7):375–379.
- Dai J, Li Y, Ji C, et al. Characterization of two novel KRAB-domain-containing zinc finger genes, ZNF460 and ZNF461, on human chromosome 19q13.1–>q13.4. Cytogenet Genome Res. 2003;103(1–2):74–78.
- Zhu W, Gao W, Deng Y, et al. Identification and development of long non-coding RNA associated regulatory network in pancreatic adenocarcinoma. Onco Targets Ther. 2020;13:12083–12096.
- Shi X, Zhao Y, He R, et al. Three-lncRNA signature is a potential prognostic biomarker for pancreatic adenocarcinoma. Oncotarget. 2018;9(36):24248–24259.
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914.
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407.
- Wang J, Liu T, Wang M, et al. SRSF1-dependent alternative splicing attenuates BIN1 expression in non-small cell lung cancer. J Cell Biochem. 2020;121(2):946–953.
