?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
PIWI interacting RNAs (piRNAs) have emerged as important gene regulators in recent times. Since the release of our first version of piRNAQuest in 2014, lots of novel piRNAs have been annotated in different species other than human, mouse and rat. Such new developments in piRNA research have led us to develop an updated database piRNAQuest V.2. It consists of 92,77,689 piRNA entries for 25 new species of different phylum along with human, mouse and rat. Besides providing primary piRNA features which include their genomic location, with further information on piRNAs overlapping with repeat elements, pseudogenes and syntenic regions, etc., the novel features of this version includes (i) density based cluster prediction, (ii) piRNA expression profile across various healthy and disease systems and (iii) piRNA target prediction. The concept of density-based piRNA cluster identification is robust as it does not consider parametric distribution in its model. The piRNA expression profile for 21 disease systems including cancer have been hosted in addition to 32 tissue specific piRNA expression profile for various species. Further, the piRNA target prediction section includes both predicted and curated piRNA targets within eight disease systems and developmental stages of mouse testis. Further, users can visualize the piRNA-target duplex structure and the ping-pong signature pattern for all the ping-pong piRNA partners in different species. Overall, piRNAQuest V.2 is an updated user-friendly database which will serve as a useful resource to survey, search and retrieve information on piRNAs for multiple species. This freely accessible database is available at http://dibresources.jcbose.ac.in/zhumur/pirnaquest2.
Introduction
PIWI interacting RNAs (piRNAs) belong to a broad group of endogenous small non-coding RNAs(ncRNAs) [Citation1], which typically ranges in length from 25 to 33 nucleotides (nts). In mammals, these ncRNAs were first reported in mouse testes [Citation2–5]. They act as guide for PIWI proteins, which belongs to Argonaute protein family and exhibit slicer activity [Citation6–9]. Unlike other small ncRNAs, i.e. miRNAs and siRNAs, piRNAs are biogenised from both primary processing pathway as well as the amplifying ping-pong mechanism [Citation10] from single stranded precursor molecules [Citation11] via Dicer independent pathway [Citation12]. The primary piRNAs originate from individual genomic loci that are commonly known as piRNA clusters [Citation10]. In most cases, the germline clusters generate piRNAs from both strands (known as dual-strand clusters), whereas flamenco clusters of Drosophila follicle cells and murine pachytene piRNA clusters generate piRNAs from only a single DNA strand (uni-strand clusters) [Citation13]. In the ping-pong cycle, generation of sense secondary piRNAs is initiated by the antisense primary piRNAs which in turn produces secondary antisense piRNAs and the amplifying loop continues [Citation7,Citation10].
Although studies on fish, flies and mammals have shown a conserved association of piRNAs with PIWI proteins [Citation2,Citation10,Citation11], the length variation of piRNAs have been observed with evolving sequencing technologies between different species. In general, piRNAs in mammals can be categorized into two subclasses called pachytene (29–33 nts) and pre-pachytene (26–28 nts) [Citation14], whereas piRNAs in Caenorhabditis elegans are named as 21 U-RNA owing to its bias for length of 21 nts. Though the piRNAs are best seen in germ cells, several studies have shown piRNA expression in brain, kidney, lung, liver, stomach, testis and ovary [Citation15–18] as well as in different cancers [Citation19].
To maintain genome integrity in germ cell lineages, highly expressed PIWI proteins in germ and stem cells [Citation9] take part in controlling transposon activity as a defensive mechanism [Citation20]. Studies showed that, mutation in MIWI which is a PIWI homolog in mouse leads to male infertility as well as over expression of retrotransposon transcripts [Citation21]. Similar observation has been reported in case of flies [Citation12]. In association with piRNA forming piRNA-induced silencing complexes (piRISCs), PIWI-piRNA pathway silences transposons via complementary base-pair recognition between piRNA and transposon followed by endonucleolytic cleaving of the target [Citation22,Citation23].
Existing databases like piRNABank [Citation24], piRBase [Citation25], piRNAdb [https://www.pirnadb.org], piRTarBase [Citation26] and piRNA Cluster Database [Citation27] provide information on piRNAs for multiple species. Among these, piRBase is a manually curated database which hosts piRNA information on multiple species and some disease systems. ‘piRNA cluster database’ is a dedicated database for piRNA clusters where the clusters are predicted using proTRAC [Citation28]. piRDisease V1.0 [Citation29] hosts piRNA records for different diseases but is not currently accessible. Despite such extensive work on piRNAs, there still remain several unexplored areas, such as their association with long noncoding RNAs (lncRNAs) or the presence of any genomic elements within their loci which can influence their function. We published the first version of piRNAQuest to probe deep into these lesser explored domains of piRNAome. It hosted piRNA information for three species, viz. human, rat and mouse [Citation30].
Though various computational tools have characterized novel piRNAs [Citation31,Citation32] but their function remains unclear. Hence, it is important to identify potential piRNA targets and disease-related piRNAs. Further, both predicted and validated piRNA targets including mRNAs and lncRNAs are not properly curated in any of the existing databases. Moreover, identifying piRNA clusters which are hotspots of piRNA biogenesis is another big challenge in piRNA research.
In this work, we present piRNAQuest V.2, which is an extended version of piRNAQuest. This new version includes the following additional features: (i) extensive analysis on 25 new species in addition to human, mouse and rat of the previous version, (ii) density-based clustering approach [Citation33] to identify the ‘hotspots of piRNA expression’, popularly known as ‘piRNA clusters’. Since piRNA distribution varies with genomic locations in different species, identifying piRNA clusters based on their density in genome can provide new impetus to get biologically relevant clusters, (iii) tissue specific expression of piRNAs among different species, and (iv) expression of piRNAs in different disease systems with an emphasis on different types of cancers. Emerging evidences suggest that piRNAs have important roles in disease progression and diagnosis [Citation34–39]. Thus, the efficacy and potential mechanism of action of a piRNA in cancer relies on its expression in various tissues and disease systems which correlate with disease progression, (v) piRNA target prediction within both mRNAs and lncRNAs that would further help to identify the key players contributing towards disease development.
In addition to these extensive features, we have updated another section of the database, viz. ‘Tools’, where users will be able to predict piRNA clusters using customized parameters, check ping-pong pattern overlap in their sequences and predict piRNA targets using miRanda [Citation40].
Overall, piRNAQuestV.2 is a user friendly database for multi-species piRNA survey, search and retrieval. piRNA expression within normal tissues and cancer as well as the information about piRNA targets will serve as a valuable resource for piRNA researchers. The database is freely accessible at http://dibresources.jcbose.ac.in/zhumur/pirnaquest2.
Results
piRNAQuest V.2 (an updated version of piRNAQuest) hosts information on 92,77,689 piRNAs corresponding to 28 species (consisting of 25 new species in addition to human, mouse and rat) which are from different phylum ranging from nematode to chordate (). Apart from the coverage of species, this new version has included several additional features which add to the significance of this database as compared to other piRNA database. The set of updated features of this new version compared to the old version has been put up in . We have also put up feature wise comparison of piRNAQuest V.2 with other piRNA database (Supplementary File S1).
Table 1. Comparison of features between piRNAQuest V.2 and piRNAQuest
Figure 1. Taxonomical representation of species included in piRNAQuest V.2.
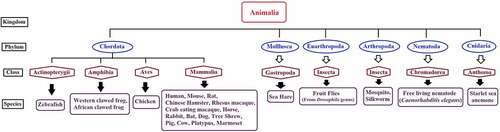
Among 28 species, 9 species (viz. Chinese hamster, Sea hare, Tree Shrew, Brown Bat, Silkworm, Mosquito, Drosophila virilis, Drosophila erecta and Starlet sea anemone) has not been annotated yet. Hence, genomic localization and related features could only be provided for the rest of the 19 species. Among rest of the 9 species, we have been able to identify repeat-associated piRNAs for 4 species (viz. Chinese hamster, Tree Shrew, Drosophila virilis and Drosophila erecta), as their repeat annotations were available from UCSC [Citation41] and this information can be visualized in graphical format from the ‘Statistics’ submenu under ‘Help Menu’ of the database.
Genomic localization based distribution of piRNAs
piRNAQuest V.2 hosts multispecies piRNA information where there is a remarkable increase in the number of piRNA entries compared to that in the previous version. Among the 28 species, distribution of piRNAs across different chromosomes has been mapped only for 19 species (as mentioned above) (Supplementary Figure SF1 and SF2). Interestingly, chromosome 15 in human contains the maximum number of piRNAs which is similar to our observation reported in the earlier version of piRNAquest [Citation30]. In this connection, it is important to note that chromosome 15 in human has been reported to harbour large number of low copy repeats popularly known as duplicons [Citation42] which facilitate nonhomologous recombination events [Citation42] that leads to genome instability [Citation43]. Presence of maximum number of piRNAs in the same chromosome might be to overcome such adverse situation of genome instability, as piRNAs are known to play significant role towards maintaining genome integrity [Citation12].
Further, chromosome 7 and 1 of mouse and rat respectively harbours the maximum number of piRNAs. Among the newly added species, chromosome IV of Caenorhabditis elegans (which has also been reported earlier [Citation44]) and Chromosome 2R of Drosophila melanogaster contains maximum number of piRNAs.
Abundance of piRNAs in intergenic regions is mostly predominant as compared to that in intronic region for most of the species (. One of the significant functions of these intergenic piRNAs is their involvement in early embryonic development [Citation45]. Further, it has been reported that intergenic regions harbour lncRNA loci [Citation46]. Hence, we have checked for the presence of lncRNA loci overlapping with piRNA clusters which consists of intergenic piRNAs (results shown later under the section ‘piRNA clusters overlapping with lncRNAs’). On the contrary, piRNA abundance in the 3’ UTR, 5’ UTR and CDS region is less, except in zebrafish (having high piRNA abundance in CDS region) and C. elegans (having high piRNA abundance in 3/ UTR and 5/ UTR regions) (.
Figure 2. Distribution of multispecies piRNAs: (a) across different genomic locations, (b) within pseudogenes and (c) within repeat family.
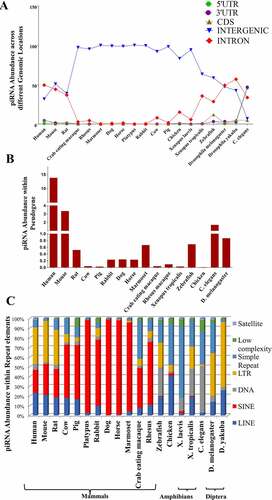
Further, it has been found that pseudogenes regulate its counter gene stability via small RNA mediated silencing [Citation47]. Recently, it has been reported that pachytene piRNAs from pseudogenes directly regulate its parent genes [Citation48]. This motivated us to check the presence of piRNAs within pseudogenes for 16 species whose pseudogene information is available [Citation49]. We obtained significant overlap between piRNAs and pseudogenes in several species (. Interestingly, maximum overlap of piRNAs with pseudogenes has been observed in human. Recently, pseudogene derived piRNAs have been found in mature human sperm cells which indicate their role in regulating expression of their parent gene in male germline cells [Citation50].
Distribution of piRNA within repeat regions
piRNAs have been reported to have originated from the repetitive regions and they silence transposons in insects and mice [Citation10,Citation51,Citation52] regulating global gene expression during embryonic development [Citation52]. The piRNA loci for all the 19 species have been mapped to the genomic locations corresponding to seven major categories of repeat elements, viz. LINE, SINE, Simple repeat, DNA, Low complexity, Satellite and LTR (. Vandewege et al. [Citation53] reported a strong piRNA response in mammals like dog and horse. These piRNAs are harboured within the SINE repeat regions which are mostly abundant within these species. In our database, we have also reported the enrichment of SINE repeat associated piRNA loci for 12 mammalian species. In addition to this, several human, mouse and rat piRNA loci overlap with LTR repeat family. On the contrary, amphibian piRNAs show a tendency to overlap with DNA and Simple repeat family. Petersen et al. [Citation54] has reported the abundance of LTR repeats within those genomic loci corresponding to the transposable elements present in Diptera (a particular order of insect class). Our study also reveals similar observation in case of the order Diptera, where piRNA enriched regions corresponding to this order overlap with LTR repeat family. Presence of such repeat regions within piRNA loci can have important implications as is shown by Halbach et al. [Citation52]. Here, it has been reported that satellite repeats modulate global gene expression via piRNA-mediated gene silencing which is important for embryonic development of Aedes.
Biogenesis of piRNAs – the piRNA clusters and ping-pong amplification
piRNA clusters are also known as the hotspots of piRNA biogenesis. Initially, in the first version of piRNAQuest, the method described by Lau et al. [Citation55] was followed to identify the piRNA clusters within a chromosome. Here a fixed window length of 20 kilobases (kb) was used to identify the clusters. Later in 2016, Rosenkranz reported that the piRNA clusters are not equally distributed across the chromosomes and is not even related to the length of the chromosome [Citation27]. As the piRNA read distribution varies across the genome corresponding to different chromosomes, one should not fix the window size for detecting piRNA cluster. Hence, in this new version of our database, piRNAQuest V.2, we have adopted density based clustering approach [Citation33] to identify the piRNA clusters (Supplementary Figures SF3 and SF4) which was found to be effective to recognize clusters successfully in chicken germ cell [Citation56].
We obtained maximum no. of clusters in chromosome 15 and chromosome IV for human (Supplementary Figure SF3) and C. elegans respectively (Supplementary Figure SF4). We also found the same for human previously. In C. elegans, it is reported that maximum clusters lie within chromosome IV [Citation44]. Though the function is still unknown, it has been found that among the sex determining chromosomes, ‘X’ chromosomal piRNAs mainly originate from clusters compared to the ‘Y’ chromosomal piRNAs [Citation27]. Our analyses also have revealed more piRNA clusters in ‘X’ chromosomes than that in ‘Y’ chromosome of human, mouse and rat.
piRNA clusters overlapping with lncRNAs: We have studied the distribution of piRNA clusters within the lncRNA loci obtained from LncRBase V.2 [Citation57]. As mentioned earlier, in the first point of this result section, we observed a significant overlap of piRNA clusters with the intergenic lncRNAs (Supplementary Figure SF5A) which are transcribed from in between two gene loci. This goes in line with previous reports [Citation58]. In addition, we looked at the overlap of piRNA clusters with repeat regions (Supplementary Figure SF5B) and found similar observation as that obtained from the distribution of piRNAs in repeat elements (as shown in ).
Motifs within piRNA clusters: Characteristic motifs have been identified for each of the clusters. These highly conserved motifs within the piRNA clusters may provide us information on possible common piRNA binding sites within its target gene. piRNAs from a cluster generated from coding gene regions can also regulate its ‘host’ gene expression [Citation59]. A significant % of total piRNA clusters have been found to be overlapping with coding regions in many species (Supplementary Figure SF5C).
piRNAs are also generated via secondary biogenesis or the ping-pong amplification loop. Studies on fly have shown that somatic piRNAs generally do not show ping-pong pattern, suggesting that the ping-pong loop may work mainly in germline cells [Citation60,Citation61]. The distribution of ping-pong piRNAs among the different chromosomes was determined. Chromosome 15 in human shows predominance for ping-pong piRNAs and has also been reported very recently by Ray and Pandey [Citation62]. More than 50% of the ping-pong piRNAs in human are found to overlap with protein-coding genes which indicates towards piRNA-dependent gene regulation [Citation63]. Further, less than 10% of these piRNAs are found to overlap with repeat elements among which SINE repeat family is predominant. Previously, Das et al. [Citation64] showed that ping-pong amplification does not occur in nematode, but surprisingly in our analysis, 509 ping-pong piRNAs are found in chromosome IV of C. elegans which may instigate the role of ping-pong loop in nematode as well.
Tissue-specific expression of piRNAs
Initially, piRNAs were observed to be expressed exclusively in germline cells [Citation3]. But gradually they have been identified in somatic cells and the somatic piRNA pathway have been seen to regulate germline transpositions [Citation65]. Hence, we have analysed 243 small RNA sequencing samples for 32 tissue types corresponding to 25 species (Supplementary File S2) in which 13 tissue types are from human. Supplementary Figure SF6 reveals the expression pattern of piRNAs among different tissue types corresponding to all these 25 species. For human, we have found the presence of maximum number of piRNAs in brain followed by colon, testis, spermatozoa which indicates the role of piRNAs not only in the germline cells, but also in other somatic cells. Previously, it was shown that there are piRNA complexes in mouse dendritic spines of brain and knockdown of those piRNAs resulted in lower spine density in the axons [Citation45]. Recent studies also indicate that piRNAs in brain are associated in suppressing retrotransposons. This has a significant role in brain pathology [Citation66]. It has been found that the piRNA length distribution is related to the age of the individual belonging to a particular species, e.g. in Drosophila the length of piRNAs becomes shorter with age [Citation67]. Further, loss of methyltranferase result in piRNA instability and reduction in piRNA length and volume, which ultimately leads to male sterility during spermatogenesis [Citation68]. Interestingly, in our study, we have found the presence of piRNAs, which are around 36 nts in length in human sperm samples, whereas such longer piRNAs have been seen to be expressed very less in any somatic cells.
Disease specific expression of piRNAs
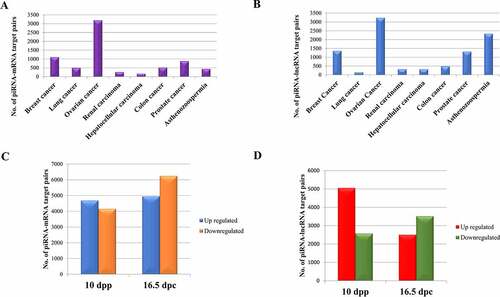
With developments in pathological research, studies have highlighted the importance of piRNAs in disease systems. piRNAs and PIWI proteins are found to be expressed abnormally in several cancer systems that increases their importance as potential novel biomarkers for therapeutic research [Citation19]. Recent evidences suggest that genomic stability of neurons may be disturbed by dysregulation of the piRNA pathways which results in various neurodegenerative disorders [Citation69]. As genes involved in the biogenesis of piRNAs have an essential role in spermatogenesis, mutation in those genes may lead to male infertility [Citation70]. Besides, piRNAs are shown to regulate Th2 cell development by downregulating IL-4, thus inhibiting allergic inflammation and asthma [Citation71] and have specific binding partners in synovial fibroblasts, suggesting its role in inflammatory processes like Rheumatoid Arthritis [Citation72]. Here, we have analysed 211 samples corresponding to 21 disease types (Supplementary File S3) in which 16 types of cancer are present. The distribution of piRNAs ( among different cancers shows the higher contribution of piRNAs in germ cell cancers like ovarian and testicular cancer. Here, our observation goes in line with the established role of piRNAs towards maintaining germ cells [Citation73].
Figure 3. General expression profile of piRNAs in (a) different cancers and (b) other disease systems.
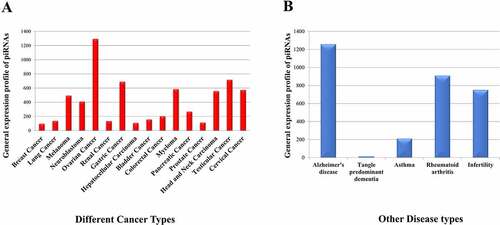
Among other diseases (, we found the presence of 1274 piRNAs among which hsa_piRNA_425 is highly abundant and hsa_piRNA_28207 is lowly abundant as compared to the abundance of other piRNAs in Alzheimer’s disease. These have been reported previously [Citation36]. The number of piRNAs expressed in asthma and rheumatoid arthritis are 278 and 910 respectively. Another interesting observation in this dataset is regarding the length of the piRNAs. In our study, we have observed the presence of longer piRNAs in sperm sample where maximum length of piRNAs is 32 nts in case of infertile samples indicating the significance of piRNA length towards spermatogenesis [Citation68].
Beside the general expression profile of piRNAs in different diseases, differential expression analysis has been also performed using DESeq [Citation74] to see the differential mode of regulation of piRNAs in seven cancer systems and asthenozoospermia (based on the availability of both test and control datasets). shows the number of differentially expressed piRNAs among which the expression of some piRNAs corroborated with that obtained from literature evidences. For example, hsa_piRNA_9871 and hsa_piRNA_27200 are found to be upregulated in breast and lung cancer respectively [Citation75]. This has also been observed in our study. The upregulated piRNAs hsa_piRNA_7806 and hsa_piRNA_31147 promote proliferation and invasiveness in colon [Citation76] and renal cancer [Citation77], respectively, and are also observed to be upregulated in our analysis.
Table 2. Differentially expressed piRNAs, genes and lncRNAs in different cancer systems, Asthenozoospermia and different developmental stages of mouse testis
piRNA-target gene interaction
Beside piRNA mediated cleavage of transposable elements, piRNAs are also known to target mRNAs and lncRNAs and subsequently regulate their expression. The involvement of piRNAs in regulating mRNAs has been studied extensively [Citation36,Citation78,Citation79]. In a way, similar to the slicing of mRNAs, PIWI-piRNA complex can target lncRNAs which has been observed in multiple organisms [Citation80]. It has been reported that a decrease in the expression levels of the target may correspond to an increase in the expression levels of the targeting piRNA, and vice versa [Citation81]. Hence, for precise target prediction, we have screened those piRNAs and mRNAs as well as piRNAs and lncRNAs whose expression are negatively correlated. This has limited our analysis to those cancer datasets where both long and small RNA seq datasets are available. Hence, we have been able to predict piRNA-mRNA and piRNA-lncRNA interaction for 7 cancer systems (viz. lung, breast, renal, hepatocellular, ovarian, prostate and colorectal). The input dataset have been shown in Supplementary File S4 and the differential analysis was performed using the ‘New Tuxedo’ protocol [Citation82]. Sequence based target prediction has been done using miRanda. Tissue and cell line data have been analysed separately. In order to highlight the role of piRNAs in different developmental stages, we have analysed the small RNA data corresponding to different developmental stages of mouse testis viz. 10dpp (days post-partum) and 16.5dpc (days postcoitum) as compared to that of six months old adult mouse testis. Further, piRNAs have been analysed corresponding to another disease system named asthenozoospermia where the sperm motility gets reduced in semen sample. The differentially expressed mRNAs, lncRNAs and piRNAs are mentioned in . The final set of piRNA-mRNA and piRNA-lncRNA target pairs for 7 cancer types is shown in , respectively. shows the number of piRNA targets within mRNAs and lncRNAs respectively in two developmental stages of mouse testis. Moreover, we have also curated experimentally validated piRNA–mRNA target pairs for human, mouse and C. elegans.
Discussion
There has been an increase in the number of piRNAs that have been identified in different species as well as in different cells since our first release of piRNAQuest in 2014. Initially, we developed piRNAQuest, with a goal to develop a non-redundant comprehensive catalogue of human, mouse and rat piRNAs so as to provide a better understanding regarding their genomic localization, overlaps with genomic elements and their association with other lncRNAs. Although, initial reports reveal the main functions of piRNAs to be transposon silencing [Citation51] and maintenance of gene integrity mainly in germline cells [Citation9], but later it has been identified in somatic cells as well in many species [Citation10,Citation12]. All these put forward, the increasing importance of its diverse functions not only in transposon silencing but also in gene expression regulation. Hence, we have come up with this new version of piRNAQuest named as piRNAQuest V.2, where we have expanded our study to 25 new species (apart from those included in previous version) covering different phylum or classes. Along with the previous features, piRNAQuest V.2 has focused on several new aspects such as directionality of piRNA cluster, piRNA expression among normal tissues and disease systems and its targets among protein coding genes and lncRNAs. These will open up novel avenues for piRNA research.
Over time, many studies have demonstrated the mechanism of primary biogenesis of piRNAs from piRNA clusters [Citation22,Citation23]. Several protocols have been developed to identify them. However, lack of uniform distribution of piRNAs among the chromosomes lead us to consider the density based clustering approach to identify piRNA clusters. It will help in understanding the distribution of piRNAs throughout the genome and the formation of clusters which are the ‘hotspots’ of piRNAs for primary biogenesis. Besides, secondary biogenesis via ping-pong amplification is also important for generation of piRNAs and its role towards silencing of its target. Emphasizing on this, we checked the ping-pong overlap among the piRNAs and have also provided options to visualize the ping-pong signature within the piRNAs. In human, we have seen the presence of maximum piRNAs in chromosome 15 where the maximum number of piRNA clusters and ping-pong piRNAs are also present.
In addition to this, analysing piRNA expression profile of various normal and disease systems will help us to understand the piRNA-mediated gene regulation in those systems. In this version, we have incorporated the piRNA expression profile of 21 disease systems along with several normal tissue data corresponding to different species. As piRNAs are differentially regulated between disease and normal conditions, a decrease in the expression levels of the target should correspond to an increase in the expression levels of the targeting piRNA, and vice versa. Taking this as an opportunity, to unravel the connection between piRNA expression and disease occurrence, we have predicted probable piRNA targets which may serve as promising biomarkers for early diagnosis and act as therapeutic targets for diseases like cancer. Further, in order to show the involvement of piRNAs in different developmental stages, we have predicted piRNA targets within mRNAs and lncRNAs in different developmental stages of mouse testis.
Overall, the newly added features along with the existing ones will make piRNAQuest V.2 a user friendly, comprehensive database for piRNAs. Our future goal is to update the database regularly with newly annotated piRNAs along with its novel features in order to continue contributing to the growing piRNA knowledgebase.
Materials and method
Improved content and new features
Input dataset
piRNA entries have been extended to 25 new species in addition to human, rat and mouse. The genome builds, availability of genome annotation and repeat annotation information and the number of piRNAs corresponding to the species has been mentioned in Supplementary Table ST1. The genome builds are updated from hg19 to hg38 and rn5.0 to rn6.0 for human and rat respectively. Data are collected in different formats like fasta, gtf and bed from the respective sources. Repeat elements and Refseq annotated 5/UTR, 3/UTR, exon, intron and CDS information have been downloaded from UCSC [Citation41]. The miRNA information has been downloaded from miRBase 22 release. Annotated piRNA sequences were downloaded in.fasta format from National Centre for Biotechnology Information (NCBI) [Citation83]. Normal tissue and disease specific small and long RNA sequencing data has been obtained from NCBI Gene Expression Omnibus (GEO) [Citation84]. LncRNA information has been retrieved from LncRBase V.2 [Citation57].
Data processing and refinement
Redundancy check and ID assignment: The procedure of assigning IDs to non redundant piRNA entries is similar to that followed in piRNAQuest [Citation30]. The sequencing data were aligned to respective genome. We further filtered out those reads mapped to other ncRNAs and screened the reads predicted to be piRNAs using our in-house script. Thereafter, non-redundant reads were re-aligned with reference genome for complete alignment with no mismatches and annotated with unique piRNAQuest IDs, i.e. [three letter abbreviation of species name]_piRNA_[number]. The annotation IDs are same for human and mouse as assigned in the previous version. The only difference is in the annotation of rat from the previous one as in the last version it was not annotated as three letter abbreviation of species name. Users can find the previous IDs which are annotated in this version of the database in the ID conversion of Help menu for human, mouse and rat. To study the distribution of piRNAs within genome, we searched for the localization of piRNAs within gene, intergenic regions, intron, CDS, UTR regions, repeat elements and pseudogenes using in-house perl scripts as that followed in the first version.
Density based piRNA cluster prediction: Previously used cluster prediction protocol [Citation55] have a disadvantage of considering window size of fixed length for all species and hence does not account for the variation in read distribution among different species. To overcome this discrepancy, we have adopted density based clustering algorithm DBSCAN [Citation33] to develop a python based in-house protocol for identifying piRNA clusters which is based on the read distribution of piRNAs across the genome.
Clustering parameters: There are two parameters ‘Eps (or epsilon)’ and ‘MinReads’ which allow us to find candidate clusters. ‘Eps’ is defined as the distance of a read from a neighbourhood point and ‘MinReads’ are the minimum number of reads within ‘eps’ distance. To determine the clustering parameters, inter distance between the annotated piRNAs are calculated by performing k-dist analysis [Citation33]. We calculated the distance between each mapped read and its kth nearest neighbouring read which is referred to as k-dist which is plotted with respect to the its counts. Eventually, a sharp valley has been observed in this ‘count versus distance’ plot until the k-dist follows a uniform distribution. The distance, for a given value of k, after which the graphs follows an asymptotic decrease is termed as the eps i.e. eps represents the distance which repeats itself for maximum number of times and hence has the highest probability of defining the boundaries of a cluster containing at least, the ‘MinReads’ which represents the number of reads within the cluster. After the ‘Eps’ and ‘MinReads’ parameters are set for each chromosome corresponding to each species, clusters are detected from the coordinate file of the annotated piRNAs.
Cluster score: In order to calculate piRNA enrichment within each cluster, we have calculated cluster score for each piRNA clusters. This has been calculated as follows:
We also checked for the strand specificity of the clusters based on the directionality of the constituent piRNAs within that cluster. If a cluster contains both sense and antisense piRNAs, it is considered as ‘dual strand cluster’ and if it contains only sense piRNAs or antisense piRNAs, it is considered as ‘uni-strand cluster’.
Localization of piRNA clusters and characteristic motifs within them: (i) In-house perl scripts have been used to check for any overlap of piRNA clusters with coding genes or lncRNAs or the repeat regions. (ii) piRNAs show a strong tendency to form clusters in the syntenic regions of genome [Citation3]. We have downloaded the syntenic regions from UCSC [Citation41] and searched for piRNA clusters in the corresponding syntenic regions among different species. (iii) MEME have been used to find the presence of any significant motifs within the piRNA clusters [Citation85].
Ping-pong pattern within piRNAs: The secondary mode of piRNA biogenesis, i.e ping pong amplification shows a distinct sequence based feature within the piRNAs, i.e. a 10 nt overlap is found between the antisense and sense piRNAs. An in-house python script has been developed to identify these ping-pong piRNAs and visualize this ping-pong signature pattern.
piRNA profile in Normal and Disease systems: We have downloaded small RNA sequencing data for different tissue types from GEO (https://www.ncbi.nlm.nih.gov/geo). A total of 243 samples were analysed for 32 types of normal tissue samples for different species. Along with the normal dataset, we have analysed 211 samples corresponding to 21 types of disease data, which includes 16 different types of cancer data sets. To analyse the expression profile of piRNAs among the normal tissue and disease systems, BLAST [Citation86] and in-house perl scripts were used. Further the expression level of each piRNA found in a sample was normalized by counts per million (CPM) and were further screened based on the z-score [Citation87] lying between −3 and +3. Users will be able to view the expression of 200 most abundant piRNAs in each set. Additionally, we have checked the distribution of each piRNAs among all the normal tissues or disease systems which has been represented graphically to provide better understanding regarding their expression within different systems.
piRNA Target prediction: piRNA target pairs have been predicted between up regulated piRNAs and downregulated lncRNAs and mRNAs and vice versa. miRanda [Citation40] has been used for predicting piRNA targets within lncRNAs (sequences obtained from LncRBase V.2 [Citation57]) and 3/ UTR region of mRNAs (sequences downloaded from UCSC [Citation41]). The target score and energy threshold are 170 and −20 kcal/mol respectively [Citation88]. In the database, we have linked The Human Protein Atlas [Citation89] and Pathway Commons [Citation90] databases to the targeted genes for further pathway and pathology based analysis. Further, our database hosts several experimentally validated piRNA targets for human, mouse and C. elegans which have been manually curated from published reports.
The overall workflow has been outlined in (Supplementary Figure SF7).
Database execution
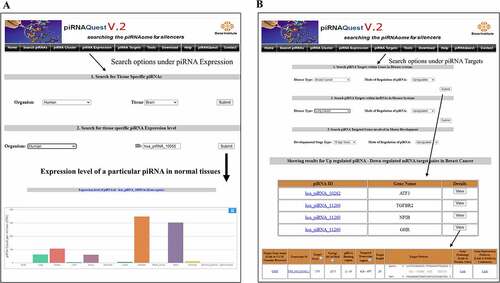
In piRNAQuest V.2, a query is basically processed via simple searching options using user’s desired selection criterion and information are presented on the web interface after retrieving related details from the database. The general information page displays basic information related to the piRNAs and provides options to probe into its further genomic details which is shown in .
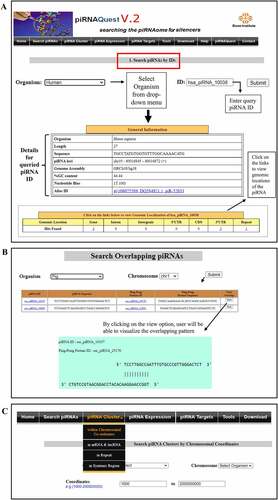
Figure 5. Web interfaces for easy access of piRNAQuest V.2 showing: (a) search options through a piRNA ID and the corresponding result page; (b) search options for pingpong piRNAs and visualizing its pattern; and (c) search options to browse piRNA clusters.
Search and output options
(a) The following options are under the ‘Search piRNAs’ menu
Search by Species Name: Users can browse all piRNAs by selecting a particular species name with the help of previous or next buttons.
Search by piRNA Accession ID/Chromosomal Co-ordinates: Users can search by piRNA accession ID for detailed information (piRNA sequence, its length, its NCBI ID (if any), %GC content, piRNA position corresponding to the genome build along with its genomic localization within genes, introns, CDS, 3/UTR, 5/UTR, intergenic regions, and repetitive elements) of selected species. A piRNA ID has already been provided as an example for each of the species. Using desired chromosomal co-ordinates, users can also get above mentioned information about piRNAs.
Search piRNAs by Sequence: Users can retrieve piRNA information by providing piRNA sequences. The sequence length should be greater than at least 20 nucleotides.
Search piRNA within Genes:User can search piRNAs present within Genes by providing a Gene Name corresponding to the selected species. The result page will show the piRNAs whose loci overlappes with this particular gene. User can also search for piRNAs within Genes of a particular species by providing chromosomal coordinates corresponding to that species.
Search piRNA within repeats: Users can search for piRNAs whose loci get mapped within repeats corresponding to genomic locations (viz. 3/UTR, 5/UTR, introns, CDS, intergenic regions) for a particular Repeat Family. Users can also search for repeat-associated piRNAs selecting their desired chromosomal location.
Search piRNAs with Ping-Pong features: User can search for 10nt overlapping piRNAs within a particular chromosome of a particular species by selecting chromosome number corresponding to that species .
(b) Search ‘piRNA clusters’
Search clusters by chromosomal co-ordinates: Users can obtain piRNA clusters by submitting a particular chromosomal location . This will fetch cluster loci, cluster score, total number of piRNAs within the cluster, cluster strandedness, prevalence of these piRNAs in minus/plus strand, and the corresponding characteristic motif of the cluster in that location. The link on the motif navigates to the website (https://meme-suite.org/meme) where users can perform further study on the motif.
Search mRNAs/lncRNAs/Repeats within piRNA Clusters: Users can check if piRNA clusters are overlapped with mRNA/lncRNA loci or with the repeat elements.
Search piRNA Clusters in Syntenic Regions: Users can search for piRNA clusters overlapping with syntenic regions by selecting a particular chromosome for both target and query organisms.
(c) Browse ‘piRNA Expression’
Search Tissue specific expression: User can view piRNA expression pattern by selecting tissue type and will be able to see the top 200 most abundantly expressed piRNAs by submitting the view option corresponding to the dataset.
Search Disease specific expression: User can also retrieve same information as above for different disease systems.
We have provided an additional option of downloading the entire set of tissue/disease wise piRNA expression information for all the samples from the download section.
An additional search option is there under both the above mentioned menus to retrieve expression level of a particular piRNA in normal tissues of selected species or that in human disease systems .
(d) Search ‘piRNA Targets’
Search Predicted Targets: Users can search piRNA targets in mRNA/lncRNA for negatively correlated dataset by selecting the disease type/different developmental stages of mouse and the mode of regulation of the piRNA. After clicking the details option, user will be able to get the detailed prediction result and visualize the piRNA-target duplex structure .
Search Curated Targets: In this section, users can find literature curated piRNA-gene target pairs.
Tools
Dynamic piRNA cluster detection: This tool can detect piRNA clusters where user can set parameters of their own, like the chromosomal coordinates, Eps distance and MinReads.
piRNA Target prediction: Users can provide the piRNA and target sequences of their own choice along with their desired energy parameters and threshold target score to predict piRNA targets.
Ping-pong signature detection: Users need to provide piRNA sequences in.fasta format to visualize ping-pong signature pattern within them.
Availability
piRNAQuest V.2 is available at http://dibresources.jcbose.ac.in/zhumur/pirnaquest2. Files can be freely downloaded and used in accordance with the GNU Public License.
Supplemental Material
Download Zip (10.8 MB)Acknowledgments
We are grateful to the Department of Science and Technology (DST) for financial support. We thank and acknowledge Samarpita Sen and S. Shanmugapriya (summer trainees) for their contribution towards building the database.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Han LC, Chen Y. Small and long non-coding RNAs: novel targets in perspective cancer therapy. Curr Genomics. 2015 Oct;16(5):319–326.
- Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006 Jul 13;442(7099):203–207.
- Girard A, Sachidanandam R, Hannon GJ, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006 Jul 13;442(7099):199–202.
- Grivna ST, Beyret E, Wang Z, et al. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006 Jul 1;20(13):1709–1714.
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006 Sep 5;103(36):13415–13420.
- Saito K, Nishida KM, Mori T, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006 Aug 15;20(16):2214–2222.
- Gunawardane LS, Saito K, Nishida KM, et al. A slicer-mediated mechanism for repeat-associated siRNA 5ʹ end formation in Drosophila. Science. 2007 Mar 16;315(5818):1587–1590.
- Nishida KM, Saito K, Mori T, et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007 Nov;13(11):1911–1922.
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25(1):355–376.
- Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007 Mar 23;128(6):1089–1103.
- Houwing S, Kamminga LM, Berezikov E, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007 Apr 6;129(1):69–82.
- Siomi MC, Sato K, Pezic D, et al. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011 Apr;12(4):246–258.
- Czech B, Hannon GJ. one loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci. 2016 Apr;41(4):324–337.
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007 Nov 2;318(5851):761–764.
- Fu A, Jacobs DI, Hoffman AE, et al. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015 Oct;36(10):1094–1102.
- Ortogero N, Schuster AS, Oliver DK, et al. A novel class of somatic small RNAs similar to germ cell pachytene PIWI-interacting small RNAs. J Biol Chem. 2014 Nov 21;289(47):32824–32834.
- Williams Z, Morozov P, Mihailovic A, et al. Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep. 2015 Oct 27;13(4):854–863.
- Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013 May 10;14(1):319.
- Liu Y, Dou M, Song X, et al. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019 Aug 9;18(1):123.
- Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33(6):2052–2059.
- Reuter M, Berninger P, Chuma S, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011 Nov 27;480(7376):264–267.
- Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012 Nov 1;26(21):2361–2373.
- Weick EM, Miska EA. piRNAs: from biogenesis to function. Development. 2014 Sep;141(18):3458–3471.
- Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008 Jan;36( Database issue):D173–7.
- Wang J, Zhang P, Lu Y, et al. piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res. 2019 Jan 8;47(D1):D175–D180.
- Wu WS, Brown JS, Chen TT, et al. piRTarBase: a database of piRNA targeting sites and their roles in gene regulation. Nucleic Acids Res. 2019 Jan 8;47(D1):D181–D187.
- Rosenkranz D. piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res. 2016 Jan 4;44(D1):D223–30.
- Rosenkranz D, Zischler H. proTRAC–a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinformatics. 2012 Jan 10;13(1):5.
- Muhammad A, Waheed R, Khan NA, et al. piRDisease v1.0: a manually curated database for piRNA associated diseases. Database. 2019 Jan 1;2019. DOI:10.1093/database/baz052
- Sarkar A, Maji RK, Saha S, et al. piRNAQuest: searching the piRNAome for silencers. BMC Genomics. 2014 Jul 4;15(1):555.
- Monga I, Banerjee I. Computational identification of piRNAs using features based on RNA sequence, structure, thermodynamic and physicochemical properties. Curr Genomics. 2019 Nov;20(7):508–518.
- Wang K, Hoeksema J, Liang C. piRNN: deep learning algorithm for piRNA prediction. PeerJ. 2018;6:e5429.
- Ester M, Kriegel HP, Sander J, et al. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD. 1996;96:226–231.
- Quek C, Bellingham SA, Jung CH, et al. Defining the purity of exosomes required for diagnostic profiling of small RNA suitable for biomarker discovery. RNA Biol. 2017 Feb;14(2):245–258.
- Bachmayr-Heyda A, Auer K, Sukhbaatar N, et al. Small RNAs and the competing endogenous RNA network in high grade serous ovarian cancer tumor spread. Oncotarget. 2016 Jun 28;7(26):39640–39653.
- Roy J, Sarkar A, Parida S, et al. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer’s disease and their probable role in pathogenesis. Mol Biosyst. 2017 Feb 28;13(3):565–576.
- Li Y, Wu X, Gao H, et al. Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol Med. 2015 May 13;21(1):381–388.
- Zhang W, Yao G, Wang J, et al. ncRPheno: a comprehensive database platform for identification and validation of disease related noncoding RNAs. RNA Biol. 2020 Jul;17(7):943–955.
- Zhang W, Zeng B, Yang M, et al. ncRNAVar: a manually curated database for identification of noncoding RNA variants associated with human diseases. J Mol Biol. 2021 May 28;433(11):166727.
- John B, Enright AJ, Aravin A, et al. Human microRNA targets. PLoS Biol. 2004 Nov;2(11):e363.
- Karolchik D, Hinrichs AS, Furey TS, et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004 Jan 1;32( Database issue):D493–6.
- Pujana MA, Nadal M, Gratacos M, et al. Additional complexity on human chromosome 15q: identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26. Genome Res. 2001 Jan;11(1):98–111.
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007 Apr;8(4):272–285.
- Ruby JG, Jan C, Player C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006 Dec 15;127(6):1193–1207.
- Lee EJ, Banerjee S, Zhou H, et al. Identification of piRNAs in the central nervous system. RNA. 2011 Jun;17(6):1090–1099.
- Nelson CE, Hersh BM, Carroll SB. The regulatory content of intergenic DNA shapes genome architecture. Genome Biol. 2004;5(4):R25.
- Pink RC, Wicks K, Caley DP, et al. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011 May;17(5):792–798.
- Hirano T, Iwasaki YW, Lin ZY, et al. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA. 2014 Aug;20(8):1223–1237.
- Flicek P, Amode MR, Barrell D, et al. Ensembl 2012. Nucleic Acids Res. 2012 Jan;40( Database issue):D84–90.
- Pantano L, Jodar M, Bak M, et al. The small RNA content of human sperm reveals pseudogene-derived piRNAs complementary to protein-coding genes. RNA. 2015 Jun;21(6):1085–1095.
- Vagin VV, Sigova A, Li C, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006 Jul 21;313(5785):320–324.
- Halbach R, Miesen P, Joosten J, et al. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature. 2020 Apr;580(7802):274–277.
- Vandewege MW, Platt RN 2nd, Ray DA, et al. Transposable element targeting by piRNAs in Laurasiatherians with distinct transposable element histories. Genome Biol Evol. 2016 May 9;8(5):1327–1337.
- Petersen M, Armisen D, Gibbs RA, et al. Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects. BMC Evol Biol. 2019 Jan 9;19(1):11.
- Lau NC, Seto AG, Kim J, et al. Characterization of the piRNA complex from rat testes. Science. 2006 Jul 21;313(5785):363–367.
- Jung I, Park JC, Kim S. piClust: a density based piRNA clustering algorithm. Comput Biol Chem. 2014 Jun;50:60–67.
- Das T, Deb A, Parida S, et al. LncRBase V.2: an updated resource for multispecies lncRNAs and ClinicLSNP hosting genetic variants in lncRNAs for cancer patients. RNA Biol. 2020;18(8):1–16.
- Han BW, Zamore PD. piRNAs. Curr Biol. 2014 Aug 18;24(16):R730–3.
- Barberan-Soler S, Fontrodona L, Ribo A, et al. Co-option of the piRNA pathway for germline-specific alternative splicing of C. elegans TOR. Cell Rep. 2014 Sep 25;8(6):1609–1616.
- Saito K, Ishizu H, Komai M, et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010 Nov 15;24(22):2493–2498.
- Lau NC, Robine N, Martin R, et al. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009 Oct;19(10):1776–1785.
- Ray R, Pandey P. piRNA analysis framework from small RNA-Seq data by a novel cluster prediction tool - PILFER. Genomics. 2018 Nov;110(6):355–365.
- Jehn J, Gebert D, Pipilescu F, et al. PIWI genes and piRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Commun Biol. 2018;1(1):137.
- Das PP, Bagijn MP, Goldstein LD, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008 Jul 11;31(1):79–90.
- Barckmann B, El-Barouk M, Pelisson A, et al. The somatic piRNA pathway controls germline transposition over generations. Nucleic Acids Res. 2018 Oct 12;46(18):9524–9536.
- Nandi S, Chandramohan D, Fioriti L, et al. Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proc Natl Acad Sci U S A. 2016 Nov 8;113(45):12697–12702.
- Wang H, Ma Z, Niu K, et al. Antagonistic roles of Nibbler and Hen1 in modulating piRNA 3ʹ ends in Drosophila. Development. 2016 Feb 1;143(3):530–539.
- Lim SL, Qu ZP, Kortschak RD, et al. HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLoS Genet. 2015 Oct;11(10):e1005620.
- Kim KW. PIWI proteins and piRNAs in the nervous system. Mol Cells. 2019 Dec 31;42(12):828–835.
- Kamaliyan Z, Pouriamanesh S, Soosanabadi M, et al. Investigation of piwi-interacting RNA pathway genes role in idiopathic non-obstructive azoospermia. Sci Rep. 2018 Jan 9;8(1):142.
- Zhong F, Zhou N, Wu K, et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015 Dec 2;43(21):10474–10491.
- Plestilova L, Neidhart M, Russo G, et al. Expression and regulation of PIWIL-Proteins and PIWI-interacting RNAs in rheumatoid arthritis. PloS One. 2016;11(11):e0166920.
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45(1):447–469.
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106.
- Reeves ME, Firek M, Jliedi A, et al. Identification and characterization of RASSF1C piRNA target genes in lung cancer cells. Oncotarget. 2017 May 23;8(21):34268–34282.
- Mai D, Ding P, Tan L, et al. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018;8(19):5213–5230.
- Busch J, Ralla B, Jung M, et al. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015 Jun 14;34(1):61.
- Zuo Y, Liang Y, Zhang J, et al. Transcriptome analysis identifies Piwi-interacting RNAs as prognostic markers for recurrence of prostate cancer. Front Genet. 2019;10:1018.
- Weng W, Liu N, Toiyama Y, et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. 2018 Jan 30;17(1):16.
- Wang C, Lin H. Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs. Genome Biol. 2021 Jan 8;22(1):27.
- Krishnan P, Ghosh S, Wang B, et al. Profiling of small nucleolar RNAs by next generation sequencing: potential new players for breast cancer prognosis. PloS One. 2016;11(9):e0162622.
- Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, stringtie and Ballgown. Nat Protoc. 2016 Sep;11(9):1650–1667.
- Geer LY, Marchler-Bauer A, Geer RC, et al. The NCBI bioSystems database. Nucleic Acids Res. 2010 Jan;38( Database issue):D492–6.
- Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–369.
- Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009 Jul;37( Web Server issue):W202–8.
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410.
- Hazra A, Gogtay N. Biostatistics series module 1: basics of biostatistics. Indian J Dermatol. 2016 Jan-Feb;61(1):10–20.
- Hashim A, Rizzo F, Marchese G, et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget. 2014 Oct 30;5(20):9901–9910.
- Ponten F, Jirstrom K, Uhlen M. The human protein atlas–a tool for pathology. J Pathol. 2008 Dec;216(4):387–393.
- Cerami EG, Gross BE, Demir E, et al. Pathway commons, a web resource for biological pathway data. Nucleic Acids Res. 2011 Jan;39( Database issue):D685–90.