ABSTRACT
In the recent past, cross-kingdom movement of miRNAs, small (20–25 bases), and endogenous regulatory RNA molecules has emerged as one of the major research areas to understand the potential implications in modulating the plant’s biotic stress response. The current review discussed the recent developments in the mechanism of cross-kingdom movement (long and short distance) and critical cross-talk between host’s miRNAs in regulating gene function in bacteria, fungi, viruses, insects, and nematodes, and vice-versa during host-pathogen interaction and their potential implications in crop protection. Moreover, cross-kingdom movement during symbiotic interaction, the emerging role of plant’s miRNAs in modulating animal’s gene function, and feasibility of spray-induced gene silencing (SIGS) in combating biotic stresses in plants are also critically evaluated. The current review article analysed the horizontal transfer of miRNAs among plants, animals, and microbes that regulates gene expression in the host or pathogenic organisms, contributing to crop protection. Further, it highlighted the challenges and opportunities to harness the full potential of this emerging approach to mitigate biotic stress efficiently.
1. Introduction
MicroRNAs (miRNAs) are endogenous, small non-coding RNA molecules with sizes ranging from 20 to 25 bases [Citation1,Citation2], which negatively regulate gene expression at the post-transcriptional level [Citation3]. They are one of the most abundant classes of gene regulatory molecules, regulating the expression of many growths and development associated protein-coding genes during the entire cycle of a multicellular organism [Citation4]. MicroRNA was first discovered as lin-4 in Caenorhabditis elegans (C. elegans) [Citation5–7]. Since then, thousands of miRNAs have been identified in plants, animals, and other eukaryotic organisms [Citation8]. In plants, miRNAs were first discovered in Arabidopsis thaliana and subsequently in other plant species [Citation9,Citation10]. The latest release of miRbase (v22) was reported to contain 38,589 hairpin precursors and 48,860 mature microRNAs sequences from 271 organisms showing a continuous increase in the miRNA pool [Citation11,Citation12]. So far, about 8433 miRNAs from 121 plant species have been archived in the plant miRNA database (miRBase) [Citation13]. Moreover, 16,422 novel miRNAs from 88 plant species were archived in the plant miRNA Encyclopaedia (PmiREN, http://www.pmiren.com/) [Citation14]. The PmiREN v.2.0 latest release contains 38,186 known miRNAs belonging to 7,838 families with a predicted 141, 327 miRNA-targets pairs in 179 plant species [Citation15]. These miRNAs can control a broad range of biological processes by modulating their corresponding target genes expression [Citation16,Citation17], involved in a vast range of plant functions, including leaf morphogenesis [Citation18], root development [Citation19,Citation20], growth transition [Citation21], reproductive stage [Citation22], disease resistance [Citation23,Citation24], etc.
The miRNAs involved in modulating diseases response regulate their target gene expression either through up or down-regulation upon fungal infection [Citation25,Citation26]. For instance, Gupta et al. [Citation26] reported a significant accumulation of miR1138 in bread wheat infected with P. graminis f.sp. tritici (62G29-1). The earlier speculation supports the idea of miRNAs targeting the pathogen’s genes in the host cell upon infection, and to counter the host defence, the pathogen’s small RNA mediates the targeting of host defence-related genes. The miRNAs targeting pathogen’s genes can be achieved by the cross-kingdom transfer of small RNAs from the host to the pathogens. The first report of cross-kingdom transfer of small RNA from host to pathogen and vice-versa in Botrytis cinerea-Arabidopsis and Lycopersicon esculentum pathosystem [Citation27] has unlocked a new area on small RNA-based plant-pathogen interaction for further exploration. This, during the last decade, enabled extensive work on cross-kingdom systemic, i.e. host (plant & animal) to the pathogen (bacteria, fungi, viruses, insects, etc.) and vice-versa, movement of small RNA [Citation28]. Moreover, with rapid advancement in molecular understanding, the research area on the potential applications of cross-kingdom movement of small RNAs in crop protection is gaining more familiarity [Citation29,Citation30]. Considering the quantum of information coming daily on the cross-kingdom movement of small RNAs, we synthesized this review to critically evaluate the existing trends, challenges and opportunities in utilizing this approach in crop protection against biotic stresses.
2. Biogenesis of miRNAs: miRNA transcription and maturation
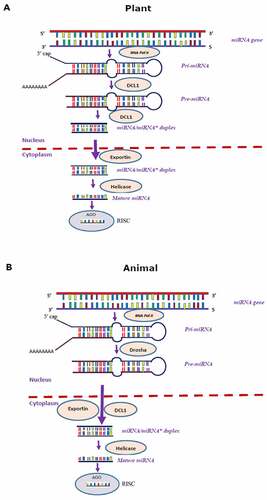
miRNA sequence specificity with its corresponding target gene is necessary for regulating their expression in both plants and animals [Citation20]. Earlier reports suggest that most animal and plant miRNAs regulate the expression of their corresponding target genes by triggering translational repression and mRNA cleavage, respectively [Citation21,Citation22]. In contrast, few reports suggest miRNA-mediated translational inhibition in plants [Citation23,Citation24]. Despite the cohesion in the mode of action of miRNAs in plants and animals, there are significant differences in their biogenesis [Citation8,Citation31]. The loci that produce miRNAs have distinct genomic arrangements in each kingdom, and miRNAs are excised from precursor transcripts by different pathways in the two kingdoms [Citation8]. The biogenesis pathway of miRNA in both plants and animals is depicted in . The miRNAs are primarily synthesized as primary transcripts (pri-miRNA) with 5ʹ capping and polyadenylation at 3ʹ end by RNA polymerase II and III in plants and animals [Citation32]. In-plant cells, the pri-miRNAs are processed using Dicer-like 1 protein (DCL 1) to remove poly-A tail generating pre-miRNAs [Citation33–36]. The looped secondary structure of pre-miRNAs are further processed by DCL 1, resulting in miRNA-miRNA* (guide-passenger strand) duplexes [Citation37], which is transported from the nucleus to cytoplasm with the help of exportin transporter [Citation38]. Finally, the duplex gets separated in the cytoplasm, and matured strand of miRNA is incorporated with an RNA-induced silencing complex (RISC) that acts as a guide for mature miRNA to recognize the complementary site of its target gene [Citation32].
In animals, the pri-miRNAs poly-A tails are removed with the help of a microprocessor complex, minimally composed of Drosha, an RNase III enzyme, resulting in pre-miRNA hairpin [Citation39,Citation40], with a 5’ monophosphate group and a 2-nt 3’-end overhang [Citation40]. The pre-miRNAs are simultaneously processed and exported from the nuclease to the cytoplasm with the help of DCL 1 and exportin-5 (XPO5) in the presence of its Ran-GTP co-factor, forming miRNA/miRNA* duplex [Citation41–43]. Once in the cytoplasm, GTP hydrolysis resulted in the dissociation of pre-miRNA from XPO5 [Citation40]. The RNA poly III enzymes cleave the pre-miRNA hairpin loops to produce a ~ 22 bp mature miRNA duplex [Citation44,Citation45]. Then, the mature miRNA is formed by helicase. Finally, the RNA binding proteins and PACT (protein activator of PKR) associate with Dicer in vivo, facilitating the assembly of matured miRNA into RISC to perform its regulatory function [Citation46,Citation47].
A difference in the location of the binding site of miRNA within the target region between animals and plants was detected. For instance, in animals, the binding site usually occurs in multiples and always within the 3′ untranslated region (3′-UTR) of the mRNA, while plant miRNA-binding sites are found almost exclusively within the open reading frames (ORF) of the target genes [Citation48]. However, in few plants, the binding site of miRNAs is predicted to occur in 3′-UTR of mRNA [Citation49]. Hence, the number of miRNA binding sites and their location reflect a significant mechanistic difference between animals and plants [Citation48]. Even though there are several differences in miRNAs binding sites between animals and plants, in both kingdoms, miRNAs regulate target gene expression either by inhibiting translation through a slicer-independent mechanism [Citation50] or negatively controlling the protein-coding sequence via mRNA-directed cleavage mechanism at a post-transcriptional level [Citation26,Citation28,Citation51]. Moreover, in both plants and animals, miRNAs sequence specificity with their corresponding target is necessary to regulate gene expression [Citation20], determining whether the target gene is cleaved or translationally inhibited [Citation9,Citation22].
3. Disease pathogenesis and plant defence modulated by miRNAs
Plants are often more prone to different biotic and abiotic stresses owing to their sessile nature, and constant exposure to an unpredictable environment leading to extreme loss to crop productivity [Citation52]. The overexpression, up- or down-regulation, or knock-in of transcribed miRNA gene sequences has confirmed the involvement of miRNAs in biotic stress responses in different plant species [Citation53]. For instance, overexpression of miR396 in rice leads to an enhanced susceptibility to M. oryzae [Citation54], whereas overexpression of miR164 and miR396 significantly improved tolerance to cyst nematode [Citation53]. Furthermore, the overexpression of miR827 increased susceptibility to H. schachtii, whereas the expression of a miR827-resistant NLA decreased plant susceptibility [Citation55]. An induced expression of miR166 under Rhizoctonia solani infection in susceptible and resistant rice cultivars suggest basal response regulators [Citation56]. Similarly, an increase in the accumulation of miR166 and miR159 in cotton plants in response to fungal pathogen Verticillium dahliae infection was reported [Citation57]. Overexpression of miR393 represses auxin signalling, enhancing bacterial resistance, suggesting auxin signalling plays a vital role in plant-induced immune response [Citation58,Citation59]. The complementary strand miR393 has also been reported to play a role in antibacterial immunity by negatively regulating the expression of MEMB12 (SNARE), a protein involved in membrane fusion, thereby promoting the exocytosis of pathogenesis-related protein (PR1) [Citation52]. Natarajan et al. [Citation60] demonstrated that miR160 plays a crucial role in local defence and systemic acquired resistance (SAR) responses by regulating targets of auxin response factor (StARF10) and MAP kinase (StMAPK9) during the interaction between potato and P. infestans. Moreover, miR160a positively regulates PAMP-induced callose deposition, whereas miR398b and miR773 negatively regulate PAMP-induced callose deposition and disease resistance to bacteria, suggesting a complexity of the miRNA regulation in plant innate immunity [Citation61]. Hence, miRNAs have been shown to modulate plant defence responses at various levels as regulation of gene expression by miRNAs is a crucial mechanism in facilitating the response of plants against biotic stress [Citation62]. Despite this advancement, further targeted work on functional validation of the role of miRNA in regulating the expression of genes utilizing emerging reverse genetic technologies such as CRISPR/Cas 9 technology is critically required to broaden the current horizon of miRNA-target gene-mediated disease cross talk.
4. Advances in cross-kingdom movement and role of host miRNAs during host-pathogen interaction
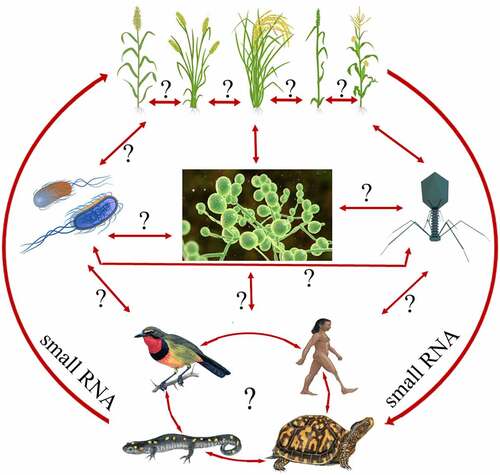
In the recent past, several reports believe that movement of sRNA, especially miRNAs have no boundary, i.e. they can move not only within cells/tissues within the individual organism but also across the kingdom in different eukaryotic species or species to species [Citation28,Citation63–67]. This type of signal transfer across the kingdom between distantly related species is termed cross-kingdom RNA interference (RNAi). represents the hypothesis of all possible interactions for the cross-kingdom movement of small RNA. Micro RNAs have been reported for their potential transfer to distantly related organisms, where they exert a regulatory role in cross-kingdom fashion [Citation68]. The conserved features of the RNA silencing machinery among eukaryotes favour cross-kingdom miRNA transfer, though taxon-specific variations exist [Citation68]. Such type of variation is mainly related to the ability of organisms to incorporate RNA molecules into other tissues/cells, silencing the target gene expression [Citation69,Citation70].
Figure 2. A hypothetical model representing the cross-kingdom movement of sRNA. Question mark (?) represent the unavailability of information in literatures.
The cross-kingdom miRNA transfer has been observed in host-pathogen relations, inhibiting invasive pathogen powers [Citation68]. Plants are attacked by a large number of pathogens such as bacteria, fungi, mycoplasma, nematodes, viruses, viroids, and parasites, and they have developed a defence strategy against these pathogens [Citation71,Citation72]. Due to evolved nature of plants, they have developed a sophisticated mechanism of resistance against pathogens through miRNA-guided transcriptional or post-transcriptional silencing of pathogenic mRNA of virulence genes. Growing reports have sufficiently demonstrated the potential implications of many plant’s miRNAs in defence response against various pathogens [Citation32,Citation72], see review [Citation73]. To enumerate a few, resistance mechanism in cotton plants against fungal pathogen has been demonstrated by miRNA-based targeting of virulence gene [Citation74]. The miR1138 was highly accumulated in wheat infected with P. graminis f.sp. tritici (62G29-1) [Citation26]. Similarly, Yin et al. [Citation75] have reported the potential role of cotton miRNAs enhancing resistance against Verticillium dahlia infection. Overexpression of miR160a and miR398b in transgenic rice displayed enhanced resistance against Magnaporthe oryzae infection, resulting in decreased fungal growth and up-regulation of defence-related genes [Citation76]. Antibacterial immunity was activated by miRNA393-AGO1 mediated suppression of auxin receptors [Citation77]. Recently, Kuntala and Niraj [Citation78] have reviewed the status of miRNAs’ role in plant-insect interactions. Moreover, the cross-kingdom plant-derived miR159a, miR166a-3p, and the novel-7703-5p were demonstrated to influence cellular and metabolic processes in P. xylostella [Citation68,Citation79]. Despite significant efforts that have been made in deciphering the role of host miRNA during host-pathogen interaction along with cross-boundary movement, further comprehensive work involving several hosts and pathogens could be useful in reorienting our current understanding. This understanding will help molecular breeders and pathologists devise a suitable strategy to mitigate pathogen infestations.
5. Promising mechanism of long and short distance cross-kingdom movement of miRNAs
Plant-derived miRNAs can be transferred to the animal via diet/plant vegetables [Citation80]. Diet/plant-derived miRNAs were reported in the serum of human/plant-feeding animals, regulating gene expression in recipients in a sequence-specific manner [Citation3]. Plant miRNAs can act as a bioactive constituent of the plant, which has the potential of travelling from plants to animals via the gastrointestinal (GI) tract to access its target, modulating gene expression in the recipients [Citation81]. It is proposed that diet/plant-derived miRNAs are absorbed by the intestinal epithelial cell and packaged into microvesicles (MVs) to shelter degradation and subsequently released into blood circulation [Citation81]. The miRNAs are then distributed to various tissues/cells, where they perform regulation of target gene expression [Citation3]. Plant-derived miRNAs can also be associated with animal AGO2 protein forming RNA-induced silencing complex (RISC) to perform their function in the animal system [Citation3]. Small RNAs can move locally between cells through plasmodesmata and over long distances through phloem [Citation82]. In addition, sRNA can also move via symplast and apoplast in the plant.
During the long-distance travel of plant miRNAs to animals, questions arise about how they can survive in the animal’s gastrointestinal tract (GI), enter the blood circulatory system, and eventually identify their potential target genes [Citation81]. For degradation resisting in the animal’s gut, the 3′-terminal nucleotide of plant’s miRNAs is 2’-O-methylated, enhancing the stability of miRNAs to ensure their regulatory function in animals [Citation81,Citation83]. Most plant miRNAs displayed modest resistance in the acidic gastric environment of animals [Citation84]. The increased stability in an animal might also be ensured by the high GC content of plant-derived miRNAs [Citation81]. For instance, a high GC content of MIR2911 may increase its digestive stability [Citation85,Citation86]. Most importantly, the carriers of plant-derived miRNAs are more likely to protect the miRNAs from enormously punitive surroundings and support their movement into mammals [Citation87]. Moreover, plant-derived miRNAs can be orally administered to animals for the treatment for therapeutic application. For instance, oral administration of miR159 mimic significantly suppressed the xenograft breast tumours in mice [Citation88].
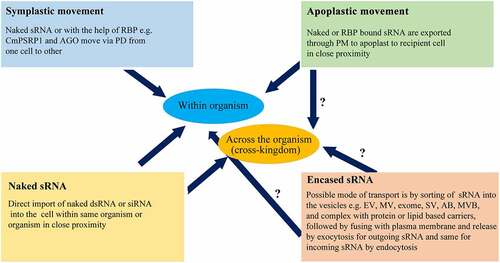
Wang et al. [Citation80] have analysed two different mechanisms by which endogenous miRNAs can be incorporated into distantly related species, i.e. the use of the systemic RNA interference deficient (SID) transmembrane channel-mediated proteins and microvesicle (MV) compartments. Moreover, evidence supported that the sRNA is transferred either as a naked molecule or mediated by vesicles encasing. Different strategies utilized for sRNA movement have been described in . For instance, in and between plants and fungi, the sRNA can be transported through naked form, combined with RNA-binding proteins, or enclosed by vesicles [Citation89]. In trans-kingdom transportation of small RNAs between plant and fungi, small RNAs inside vesicles can be transported from cell to cell through plasmodesmata (PM) which secreted through the plant plasma membrane (PPM) and then plant cell wall (PCW) to extracellular spaces, where they can also be taken up by fungal cell through fungal cell wall [Citation89]. This transportation of sRNAs can be bidirectional, i.e. the small RNAs can be transferred through the fungal plasma membrane (FPM)–fungal cell wall (FCW)–extra‐invasive hyphae matrix (EIHMx)‐extra‐invasive hyphae membrane (EIHM) and then to plant cytoplasm pathway [Citation89]. Even though different strategies for the cross-kingdom movements of miRNA were explained, the mechanism by which the exogenous miRNAs are loaded onto Argonaute proteins of distantly related species to produce a functional miRNA form has still needs to be explored in detail [Citation80]. Therefore, the fungal cell wall plays an indispensable role in controlling sRNA movement between host and fungal cells.
6. Role of host miRNA in regulating the pathogen’s gene expression
MicroRNAs play an essential role in regulating the host’s biological, biochemical and physiological pathways against pathogen (viruses, fungi, parasite, and bacterial) infection by modulating the gene expression and deviation in cellular alignments [Citation90]. The host’s RNAi silencing machinery has the potential capacity to directly target the RNA genome and related transcripts of several pathogens such as viruses, virus satellites, and viroids, to regulate the transcripts accumulation [Citation91]. This silencing is performed by exporting specific plant sRNAs, including miRNAs, to induce cross-kingdom gene silencing in pathogenic fungi, thereby conferring disease resistance [Citation30,Citation74]. For example, siRNAs enter Oomycete Phytophthora via extracellular vesicles, silencing Phytophthora virulence genes to confer resistance in Arabidopsis during infection [Citation30]. Similarly, Arabidopsis miR166 was exported to V. dahlia fungal hyphae to suppress pathogenicity [Citation92]. A comprehensive list of sRNA moving from plants to the pathogen is given in . Moreover, Zhang et al [Citation74]. have investigated the transfer of the two miRNAs (i.e. miR159 and miR166) from the cotton plant into Verticillium dahliae hyphae after infection. These two miRNAs have targeted the expression of the Verticillium genes coding for Ca2+-dependent cysteine protease (Clp-1) and isotrichodermin C-15 hydroxylase (HiC-15), respectively, associated with triggering fungal virulence [Citation74]. Tinoco and co-workers reported translocation of silencing signals across the germinated spores from transgenic tobacco into F. verticillioides cells [Citation93].
Table 1. List of sRNAs moving from plants to pathogens
Zhu and co-workers reported that compared to royal jelly, beebread harbour more plant miRNAs that decrease ovary and body size in honeybees. This hinders the differentiation of larvae into queens leading to more worker bees [Citation70]. Plant-parasitic nematodes are responsible for considerable crop losses worldwide [Citation68]. The most scientific literature on gene silencing mechanisms comes from nematodes, specifically from Caenorabditis elegans [Citation68]. However, most of these studies emphasize on uptake of dsRNAs from the surroundings than on the cross-kingdom movement of plant miRNAs [Citation68,Citation94,Citation95]. Over the years, significant progress has been made in deciphering the role of plant miRNAs against phytonematodes infection [Citation68,Citation96–99]. Zhang and co-workers observed that miR166a-3p, miR159a, and the novel-7703-5p target BJHSP2, BJHSP1 ((basic juvenile hormone-suppressible protein 1 and 2) and PPO2 (polyphenol oxidase subunit 2) genes which affects metabolic and cellular processes in P. xylostella [Citation53]. For instance, Zhang and co-workers confirmed a modest level of plant-derived miR168 in Lepidoptera and Coleoptera species [Citation100]. Wang and co-workers predicted 13 sorghums (Sorghum bicolour) miRNAs and three barley miRNAs in Aphid targeting aphid genes playing essential roles in sucrose and starch metabolism and detoxification [Citation101]. Despite this, the precise role of exogenous plant miRNAs on herbivore gene expression still needs to be functionally elucidated.
7. Evidence and advances on the role of pathogen’s miRNA in modulating the host gene expression
The evidence-based science of cross-kingdom movement of sRNA has recently gained significant attention, with a plethora of research being performed in different hosts and pathogens. Available reports suggested that sRNAs derived from pathogens can also work as an effector molecule and modulate host gene expression as a counter defence strategy. For instance, The novel miRNA (Pst-milR1) in Puccinia striiformisf. sp.tritici takes part in cross-kingdom RNA interference (RNAi) events by binding the pathogenesis-related 2 (PR2) (b-1,3-glucanase SM638) gene in wheat [Citation102] that might suppress the host-mediated defence strategy in its counter defence. Similarly, Bc-sRNAs derived from Botrytis cinerea binds with Argonaute 1 (AGO1) and capture the host RNAi machinery leading to selective silencing of host immunity genes [Citation27], suggesting that the B. cinerea transfers virulent sRNA effector molecules into host plant cells to suppress host immunity as a counter defence strategy to achieve infection [Citation27]. Wang and co-workers functionally validated the role of Bc-siR37 as an effector molecule that is predicted to target several Arabidopsis genes associated with disease pathogenesis, such as receptor-like kinases, WRKY transcription factors, and cell wall-modifying enzymes upon B. cinerea infection [Citation103] Brilli et al. [Citation104] identified bidirectional interaction between pathogen-host, i.e. the sRNA produced by Plasmopara viticola triggered the cleavage of grapevine (Vitis vinifera) genes, while the sRNAs produced from grapevine target the P.viticola mRNAs. An updated list of sRNA moving from pathogen to plants and their regulatory roles has been given in .
Table 2. List of sRNAs that move from pathogens to plants
In addition to the pathogens’ miRNAs modulating host defence response, various molecules or effectors from pathogen reported to interfere with the host defence mechanism during pathogen interaction. Interestingly, plant viruses encode viral suppressors of RNA silencing (VSRs) molecule, interfering with host RNA silencing through multiple modes of action [Citation105,Citation106]. The plant virus-encoded VSR physically interacts with AGO1 to prevent miRNA or siRNA loading or degrading AGO1 protein [Citation14,Citation107]. For instance, the tombusvirus P19 protein (a type of VSRs) binds and sequesters plant miRNAs to suppress their activity in AGO, resulting in the increased loading of miR168 into AGO1 and subsequently reduced accumulations of cellular AGO1 [Citation108,Citation109]. Further research in the area of comprehensive characterization of pathogen’s miRNAs and their functional validation in several models and non-model plants would broaden our current understating, which will guide us in devising suitable mitigation strategies against pathogen mediated crop losses.
8. Cross-kingdom movement of miRNAs during symbiotic interaction
Small RNA-based cell-to-cell communication occurs between an organism of different species by transporting regulatory molecules across the cellular boundaries between the host and its interacting pathogens/symbionts [Citation67]. The cross-kingdom transfer of miRNAs between symbiotic or mutualistic relations impacts mutualistic relations and the performance of different agricultural crop plants [Citation68]. The miRNAs cross-transferred from the plant through symbiotic/mutualist relation reported influencing the growth and developmental stage of the receiving organisms [Citation110]. The Arbuscular Mycorrhizal Fungi (AMF) is an important component of the host plant’s root providing several benefits, including improving nutrient uptake and tolerance to various stress. Even though little is now about RNAi mechanism and sRNAs occurrence in Arbuscular Mycorrhizal Fungi (AMF), several fungal sRNAs have the potential to target transcripts, including some specific mRNA in Medicago truncatula roots upon Arbuscular Mycorrhizal Fungi (AMF colonization [Citation111]. The transfer of fungal sRNAs in symbiosis interaction modulates plant metabolic pathways and defence response [Citation111]. Hence, the fungal sRNAs positively affect the symbiotic interaction between fungi and their host plant.
Moreover, in the mutualistic relation of plant-pollinator, the dietary intake of the plant miR162a was shown to regulate caste development at the larval stage of honey [Citation68,Citation110]. Hence, silencing TOR (target of rapamycin) by plant-derived miR162a blocks queen fate and results in individuals with worker morphology. A contrary report on the uptake of plant-derived miRNAs by recipient organisms has been observed. Snow et al. [Citation112] observed negligible delivery of plant-derived miRNAs in recipient honeybees despite oral uptake of pollen containing these molecules, suggesting that the horizontal delivery of plant-derived miRNAs via dietary ingestion was neither a robust nor a frequent mechanism to maintain steady-state microRNA levels in receiving organisms. However, Masood et al. [Citation113] revealed an accumulation of plant miRNAs after pollen ingestion in adult bees’ midguts without evidencing their biological role. They supported the premise that pollen miRNAs ingested as part of a typical diet were not robustly transferred across barrier epithelia of adult honey bees under normal conditions. The reports signifying cross transfer and accumulation of miRNA involved in the symbiotic relationship of plants and other organisms are limited. Moreover, more specialized or specific delivery mechanisms for more efficient cross-transfer of miRNAs between symbiotic/mutualistic relations will be required to be explored.
Contrary to the transferred role of sRNAs between plants and symbiotic/mutualistic organisms, the cross transfer of miRNAs from the plant to pathogen/parasitic or vice versa has a negative impact on the host or pathogen. For instance, the novel miRNA like RNA from Puccinia striiformis f. sp. tritici (Pst) to wheat suppressed its innate immunity [Citation102]. This part was more discussed in section 7 above.
9. Application of cross-kingdom miRNA movement in crop protection
The movement of miRNAs across different species has various applications in crop protection in an environment-friendly manner. For instance, the miRNA159 and miRNA166 constitute an example of plant miRNA transfer to pathogenic fungi from cotton (Gossypium hirsutum), which confer resistance to Verticillium dahlia [Citation74]. Hence, horizontal transfer of miRNA among plants, animals, and microbes regulates gene expression in the host or pathogenic organisms, contributing to crop protection that could efficiently be utilized in the breeding programme. The transfer of miRNAs from pathogens to hosts primarily involves suppressing plant defence mechanisms as a counter defence mechanism. Wang and co-workers showed that expressing sRNAs targeting Bc-DCL1 and Bc-DCL2 in Arabidopsis and tomato silences Bc-DCL genes and attenuates fungal pathogenicity and growth, exemplifying bidirectional cross-kingdom RNAi and sRNA trafficking between plants and fungi [Citation70]. This indicates that the cross-kingdom transfer of miRNAs suppresses the plant pathogen’s virulence and protects the crop plant. Furthermore, exogenous uptake from the environment was discovered in particular fungal pathogens, suppressing the virulence capability of the related pathogen [Citation114]. Botrytis cinerea, causing grey mould disease, has been taken external sRNAs and dsRNA through spraying on the surface of the fruit, vegetables, and flowers and targeting the fungal pathogen gene against plant infection [Citation114]. Moreover, the plant also transfers ds-siRNAs into coleopteran insects, silencing their transcription and suppressing their growth [Citation63].
10. Potential application of spray induced gene silencing (SIGS) for combating insect pests in plants
At present, crop breeders depend almost entirely on fungicides to control disease, resulting in pesticide residues that often endanger human health and the environment [Citation115]. Different resistant strains of fungi have been identified against every primary fungicide used in the agricultural production system [Citation116]. Therefore, there is an urgent need to develop an eco-friendly and effective mechanism of agricultural crop protection from pathogen invasion. Modern agriculture is now on the verge of the third green revolution; the knowledge generated by reverse genetics in the functional characterization of genes could be harnessed in agricultural pest management [Citation117]. RNA-based technologies, especially RNAi, have tremendous potential to be a practical approach for plant protection. RNAi has been explored as a strategy for pest control by expressing insect-targeted dsRNA in host plants to specifically block the expression of essential genes, resulting in insect mortality [Citation118]. Among RNAi methods, SIGS has emerged as an innovative strategy for crop protection [Citation119]. RNA sprays that result in target gene silencing have been observed with viruses [Citation120] and fungi [Citation121–123]. SIGS significantly simulates HIGS (Host-Induced Gene Silencing) without the need to develop stably transformed plants and has been demonstrated to be effective in the control of both F. graminearum and Botrytis cinerea [Citation121]. The dsRNA/siRNA-based SIGS has attracted attention due to its feasibility and low cost compared to transgenic plants, and the technology demonstrates a potential paradigm shift in crop protection [Citation117,Citation119]. The dsRNA sprayed onto plant surface enters fungal cells by two possible pathways, i.e. RNA can be taken first by the plant cell and transferred into pathogenic fungi and/or directly taken by fungal cells [Citation121]. These RNAs subsequently work in two ways: the RNAs taken up by plant cells induce the plant RNAi machinery, and then the RNAs taken up by the fungal cells induce the fungal RNAi machinery directly [Citation119]. Koch and his co-worker demonstrated that barley SIGS conferred resistance against F. graminearum by silencing CYP51 genes [Citation119]. They also demonstrated that spraying the RNA fragments of jellyfish green fluorescent protein (GFP) on barley leaves effectively silenced GFP expression in a GFP-expressing F. graminearum strain, potentially targeting any essential genes in various interacting pathogens [Citation121]. Moreover, Werner and co-workers also found that targeting ARGONAUTE and DICER genes of F. graminearum (Fg), the fungal RNAi machinery via SIGS could protect barley leaves from Fg infection [Citation124]. Additionally, the dsRNA sprays can inhibit Botrytis cinerea and Sclerotinia sclerotiorum growth on Brassica napus [Citation123]. The effectiveness of SIGS to protect pathogen invasion is dependent on the pathogen type to take up the naked miRNAs/sRNAs/RNAi. The pathogen’s RNA uptake efficiency can largely determine the success of SIGS for plant disease management, and therefore, establishing the effectiveness of SIGS across a wide range of pathogens is a critical next step in developing this technology.
11. Conclusion and prospects
Plant pathogens are continually affecting crop production throughout the world. Here, we analysed the existing cross-kingdom transfer of miRNAs during plant-animal and plant-pathogen interaction. However, there are also contradictory scenarios; plant miRNAs would not have passed through ingestion but could be mixed due to contamination during the sequencing of miRNAs. Recently, the role of miRNAs in regulating gene expression in host and pathogen have given a big concern for controlling pathogen in crop plants. Further investigation of the miRNA-mediated process in plant-pathogen interactions is needed to devise novel strategies for controlling pathogen infection in crop plants and improving crop productivity. MicroRNA-mediated gene silencing has vital significance in plant immunity. miRNAs-based SIGS techniques can be used as a mechanism of crop plant protection from pathogen invention. Moreover, miRNAs could be used to be very useful as biomarkers for disease resistance characteristics in breeding programme. Further exploration of cross-kingdom transfer of miRNAs would facilitate a more in-depth understanding of miRNAs in gene silencing in the host organism and trans regulation of a gene in host pathogens.
Author contributions
OPG conceived the program, designed the outline; TR and OPG compiled the information and wrote the first draft; OPG prepared figures; OPG, TR and VC edited the manuscript. All authors read and approved the final manuscript.
Declarations
Ethics approval and consent to participate
This research did not involve the use of any animal or human data or tissue.
Acknowledgments
TR is highly thankful to the Ministry of Science and Higher Education, Ethiopia (former Ministry of Education) for sponsoring through the Fellowship Program and Department Bio and Nanotechnology, Guru Jambheshwar University of Science and Technology, Hisar, India for providing all necessary laboratory facilities. OPG is thankful to the Indian Council of Agricultural Research, Department of Agricultural Research and Education, Govt. of India for providing financial help in the form of salary.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38(3):257–268.
- O’Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402.
- Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1):107–126.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.
- Lee RC, Feinbaum RL, The AV. C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854.
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans Cell. 1993;75(5):855–862.
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901.
- Axtell MJ, Westholm JO, Lai EC. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12(4):221.
- Reinhart B, Weinstein E, Rhoades M, et al. MicroRNAs in plants. Gene Dev. 2002;16(13):1616–1626.
- Llave C, Kasschau K, Rector M, et al. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14(7):1605–1619.
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162.
- Kozomara A, Griffiths-J S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(D1):D68–D73.
- Zhang Z, Yu J, Li D, et al. PMRD: plant microRNA database. Nucleic Acids Res. 2010;38(suppl_1):806–813.
- Guo Z, Kuang Z, Wang Y, et al. PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020;48(1):1114–1121.
- Guo Z, Kuang Z, Zhao Y, et al. PmiREN2.0: from data annotation to functional exploration of plant microRNAs. In: Nucleic acids research. 2021;D1475–D1482.
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function, and decay. Nature Reviews Genetics. 2010;11(9):597–610.
- Nazarov PV, Reinsbach SE, Muller A, et al. Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res. 2013;41(5):2817–2831.
- Kim J, Jung JH, Reyes JL, et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J Cell Mol Boil. 2005;42(1):84–94.
- Guo HS, Xie Q, Fei JF, et al. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17(5):1376–1386.
- Gautam V, Singh A, Verma S, et al. Role of miRNAs in root development of model plant Arabidopsis thaliana. Indian Journal of Plant Physiology. 2017;22(4):382–392.
- Lauter N, Kampani A, Carlson S, et al. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA. 2005;102(26):9412–9417.
- Chen XA. microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–2025.
- Chen X, Liang H, Zhang J, et al. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132.
- Djami-Tchatchou AT, Sanan-Mishra N, Ntushelo K, et al. Functional roles of microRNAs in agronomically important plants-potential as targets for crop improvement and protection. Front Plant Sci. 2017;8:378.
- Zhao J-P, Jiang X-L, Zhang B-Y, et al. Involvement of microRNA-mediated gene expression regulation in the pathological development of stem canker disease in populus trichocarpa. PLoS ONE. 2012;7(9):e44968.
- Gupta OP, Permar V, Koundal V, et al. MicroRNA regulated defense responses in Triticum aestivum L. during Puccinia graminis f.sp. tritici infection. Mol Biol Rep. 2012;39(2):817–824.
- Weiberg A, Wang M, Lin FM, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342(6154):118–123.
- Zeng J, Gupta VK, Jiang Y, et al. Cross-Kingdom small RNAs among animals, plants and microbes. Cells. 2019;8(4):371.
- LaMonte G, Philip N, Reardon J, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe. 2012;12(2):187–199.
- Hua C, Zhao JH, Guo HS. Trans-Kingdom RNA silencing in plant-fungal pathogen interactions. Mol Plant. 2018;11(2):235–244.
- Pierre-Jerome E, Drapek C, Benfey PN. Regulation of division and differentiation of plant stem cells. Annual Review of Cell and Developmental Biology. 2018;34(1):289–310.
- Sanchita TR, Asif MH, Trivedi PK. Dietary plant miRNAs as an augmented therapy: cross-kingdom gene regulation. RNA Biol. 2018;15(12):1433–1439.
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci U S A. 2004;101(34):12753–12758.
- Wahid F, Shehzad A, Khan T, et al. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803(11):1231–1243.
- Wang J, Mei J, Ren G. Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci. 2019;10:360.
- Li M, Yu B. Recent advances in the regulation of plant miRNA biogenesis. RNA Biol. 2021;18(12):2087–2096.
- Tiwari M, Sharma D, Trivedi PK. Artificial microRNA mediated gene silencing in plants: progress and perspectives. Plant Mol Biol. 2014;86(1–2):1–18.
- Wang ZH, Xu CJ. Research progress of microRNA in early detection of ovarian cancer. Chin Med J (Engl). 2015;128(24):3363–3370.
- Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419.
- Xie M, Steitz JA. Versatile microRNA biogenesis in animals and their viruses. RNA Biol. 2014;11(6):673–681.
- Yi R, Qin Y, Macara IG, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016.
- Lund E, Güttinger S, Calado A, et al. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98.
- Bohnsack MT, Czaplinski K, Exportin GD. 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191.
- Grishok A, Pasquinelli AE, Conte D, et al. Genes and MeC.anisms related to RNA InterferenC. regulate expression of the small temporal RNAs that C.ntrol C. elegans developmental Timing. Elegans Developmental Timing. Cell 2001;106(1):23–34.
- Knight SW, Bass BL. A role for the RNase III Enzyme DCR-1 in RNA interference and Germ line development in caenorhabditis elegans. Science. 2001;293(5538):2269–2271.
- Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–744.
- Lee Y, Hur I, Park SY, et al. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25(3):522–532.
- Millar AA, Waterhouse PM, Millar A A, and Waterhouse PM. Plant and animal microRNAs: similarities and differences. Funct Integr Genomics. 2005;5(3):129–135.
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16(8):2001–2019.
- Lanet E, Delannoy E, Sormani R, et al. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell. 2009;21(6):1762–1768.
- Hwang DG, Park JH, Lim JY, et al. The hot pepper (C. annuum) microRNA transcriptome reveals novel and conserved targets: a foundation for understanding MicroRNA functional roles in hot pepper. PloS one. 2013;8(5):e64238.
- Rajwanshi R, Devi KJ, Sharma GR, et al. Role of miRNAs. In: Interaction P-M, Kumar M, Muthusamy A, et al., editors. In vitro plant breeding towards novel agronomic traits. Singapore: Springer; 2019. 167–195.
- Basso MF, Ferreira PCG, Kobayashi AK, et al. MicroRNAs and new biotechnological tools for its modulation and improving stress tolerance in plants. Plant Biotechnol J. 2019;17(8):1482–1500.
- Chandran V, Wang H, Gao F, et al. miR396-OsGRFs module balances growth and rice blast disease-resistance. Frontiers in Plant Science. 2019;9:1999.
- Jaubert-Possamai S, Noureddine Y, MicroRNAs FB. New players in the plant–nematode interaction. Frontiers in Plant Science. 2019;10:1180.
- Chopperla R, Mangrauthia SK, Bhaskar RT, et al. Comprehensive analysis of MicroRNAs expressed in susceptible and resistant rice cultivars during rhizoctonia solani AG1-IA infection causing sheath blight disease. Int J Mol Sci. 2020;21(21):7974.
- Jin Y, Guo HS. Plant small RNAs responsive to fungal pathogen infection. Methods Mol Biol. 2018;1848:67–80.
- Navarro L, Dunoyer P, Jay F, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signalling. Science. 2006;312(5772):436–439.
- Peláez P, Small SF. RNAs in plant defense responses during viral and bacterial interactions: similarities and differences. Front Plant Sci. 2013;4:343.
- Natarajan B, Kalsi HS, Godbole P. MiRNA160 is associated with local defense and systemic acquired resistance against Phytophthora infestans infection in potato. J Exp Bot. 2018;69(8):2023–2036.
- Li Y, Zhang Q, Zhang J, et al. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiology. 2010;152(4):2222–2231.
- Yang X, Zhang L, Yang Y, et al. miRNA mediated regulation and interaction between plants and pathogens. Int J Mol Sci. 2021;22(6):2913.
- Zhang B, Li W, Zhang J, et al. Roles of small RNAs in virus-plant interactions. Viruses. 2019;11(9):827.
- Tomilov A, Tomilova NB, Wroblewski T, et al. Yoder JI Trans-specific gene silencing between host and parasitic plants. Plant J. 2008;56(3):389–397.
- Melnik BC, John SM, Schmitz G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr J. 2013;12(1):103.
- Knip M, Constantin ME, Thordal-Christensen H. Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 2014;10(9):e1004602.
- Weiberg A, Bellinger M, Jin H. Conversations between kingdoms: small RNAs. Curr Opin Biotechnol. 2015;32:207–215.
- Gualtieri C, Leonetti P, Macovei A. Plant miRNA cross-kingdom transfer targeting parasitic and mutualistic organisms as a tool to advance modern agriculture. Frontiers in Plant Science. 2020;11:930.
- Wang M, Weiberg A, Jin H. Pathogen small RNAs: a new class of effectors for pathogen attacks. Molecular Plant Pathology. 2015;16(3):219–223.
- Wang M, Weiberg A, Lin FM, et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants. 2016;2(10):16151.
- Islam W, Noman A, Qasim M, et al. Plant responses to pathogen attack: small RNAs in focus. International Journal of Molecular Sciences. 2018;19(2):515.
- Islam W, Qasim M, Noman A, et al. Plant microRNAs: front line players against invading pathogens. Microb Pathog. 2018;118:9–17.
- Gupta OP, Sharma P, Kumar GR, et al. Current status on role of miRNAs during plant-fungus interaction. Physiological and Molecular Plant Pathology. 2014;85:1–7.
- Zhang T, Zhao YL, Zhao JH, et al. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants. 2016;2(10):16153.
- Yin Z, Li Y, Han X, et al. Genome-wide profiling of miRNAs and other small 799 non-coding RNAs in the verticillium dahliae–Inoculated cotton roots. PLoS One. 2012;7:800 e35765.
- Li Y, Lu YG, Shi Y, et al. Multiple rice microRNAs are involved in immunity against the blast fungus magnaporthe oryzae. Plant Physiol. 2014;164(2):1077–1092.
- Zvereva AS, Pooggin MM. Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses. 2012;4(11):2578–2597.
- Bordoloi KS, Agarwala N. MicroRNAs in plant-insect interaction and insect pest control. Plant Genet. 2021;26:100271. 2352-4073.
- Zhang LL, Jing XD, Chen W, et al. Host plant-derived miRNAs potentially modulate the development of a cosmopolitan insect pest Plutella xylostella. Biomolecules. 2019a;9(10):602.
- Wang W, Liu D, Zhang X, et al. Plant micrornas in cross-kingdom regulation of gene expression. Int J Mol Sci. 2018;19(7):1–12.
- Li Z, Xu R, Li N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr Metab (Lond). 2018;15(1):68.
- Liu L, Chen X. Intercellular and systemic trafficking of RNAs in plants. Nat Plants. 2018;4(11):869–878.
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687.
- Winter N, Kragler F. Conceptual and methodological considerations on mRNA and proteins as intercellular and long-distance signals. Plant Cell Physiol. 2018;59(9):1700–1713.
- Zhou Z, Li X, Liu J, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A virus. Cell Res. 2015;25(1):39–49.
- Yang J, Hotz T, Broadnax L, et al. Anomalous uptake and circulatory characteristics of the plant-based small RNA MIR2911. Sci Rep. 2016;6(1):26834.
- Xie W, Weng A, Melzig MF. MicroRNAs as new bioactive components in medicinal plants. Planta Med. 2016;82(13):1153–1162.
- Chin AR, Fong MY, Somlo G, et al. Cross-kingdom inhibition of breast cancer growth by plant MIR159. Cell Res. 2016;26(2):217–228.
- Wang M, Dean RA. Movement of small RNAs in and between plants and fungi. Mol Plant Pathol. 2020;21(4):589–601.
- Behrouzi A, Alimohammadi M, Nafari AH, et al. The role of host miRNAs on mycobacterium tuberculosis. ExRNA. 2019;1(1):40.
- Huang J, Yang M, Lu L, et al. Diverse functions of small RNAs in different plant-pathogen communications. Front microb. 2016;7:1552.
- Cai Q, Qiao L, Wang M, et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360(6393):1126–1129.
- Tinoco MLP, Dias BB, Dall’Astta RC, et al. In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 2010;8(27). DOI:10.1186/1741-7007-8-27.
- Huang G, Allen R, Davis EL, et al. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proceedings of the National Academy of Sciences. 2006;103(39):14302–14306.
- Tian B, Li J, Vodkin L, et al. Host derived gene silencing of parasite fitness genes improves resistance to soybean cyst nematodes in stable transgenic soybean. Theoretical and Applied Genetics. 2019;132(9):2651–2662.
- Hewezi T, Howe P, Maier TR, et al. Arabidopsis small RNAs and their targets during cyst nematode parasitism. Molecular Plant-Microbe Interactions®. 2008;21(12):1622–1634.
- Li X, Wang X, Zhang S, et al. Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PloS One. 2012;7(6):e39650.
- Lei P, Han B, Wang Y, et al. Identification of microRNAs that respond to soybean cyst nematode infection in early stages in resistant and susceptible soybean cultivars. International Journal of Molecular Sciences. 2019;20(22):5634.
- Pan X, Nichols RL, Li C, et al. MicroRNA-target gene responses to root knot nematode (Meloidogyne incognita) infection in cotton (Gossypium hirsutum L.). Genomics. 2019;111(3):383–390.
- Zhang Y, Wiggins BE, Lawrence C, et al. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics. 2012;13(1):381.
- Wang H, Zhang C, Dou Y, et al. Insect and plant-derived miRNAs in greenbug (Schizaphis graminum) and yellow sugarcane aphid (Sipha flava) revealed by deep sequencing. Gene. 2017b;599:68–77.
- Wang B, Sun Y, Song N, et al. Puccinia striiformis f. sp. tritici mi croRNA -like RNA 1 (Pst -milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017a;215(1):338–350.
- Wang M, Weiberg A, E D Jr, et al. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017;14(4):421–428.
- Brilli M, Asquini E, Moser M, et al. A multi-omics study of the grapevine-downy mildew (Plasmopara viticola) pathosystem unveils a complex protein coding- and noncoding-based arms race during infection. Sci Rep. 2018;8(1):757.
- Burgyan J, Havelda Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011;16(5):265–272.
- Wang MB, Masuta C, Smith NA, et al. RNA silencing and plant viral diseases. Molecular Plant-Microbe Interactions®. 2012;25(10):1275–1285.
- Moon JY, Park JM. Cross-talk in viral defense signaling in plants. Frontiers in Microbiology. 2016;7:2068.
- Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nature Reviews Microbiology. 2013;11(11):745–760.
- Liu SR, Zhou JJ, Hu CG, et al. MicroRNA-mediated gene silencing in plant defense and viral counter-defense. Front Microbiol. 2017;8:1801.
- Zhu K, Liu M, Fu Z, et al. Plant microRNAs in larval food regulate honeybee caste development. PLOS Genetics. 2017;13(8):e1006946.
- Silvestri A, Fiorilli V, Miozzi L, et al. In silico analysis of fungal small RNA accumulation reveals putative plant mRNA targets in the symbiosis between an arbuscular mycorrhizal fungus and its host plant. BMC Genomics. 2019;20(1):169.
- Snow JW, Hale AE, Isaacs SK, et al. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 2013;10(7):1107–1116.
- Masood M, Everett CP, Chan SY, et al. Negligible uptake and transfer of diet-derived pollen microRNAs in adult honey bees. RNA Biol. 2016;13(1):109–118.
- Wang M, Thomas N, Jin H. Cross-kingdom RNA trafficking and environmental RNAi for powerful innovative pre- and post-harvest plant protection. Current Opinion in Plant Biology. 2017;38:133–141.
- Qiao L, Lan C, Capriotti L, et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol J. 2021;19(9):1756–1768.
- Fisher MC, Hawkins NJ, Sanglard D, et al. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 2018;360(6390):739–742.
- Cagliari D, Dias NP, Galdeano DM, et al. Management of pest insects and plant diseases by non-transformative RNAi. Frontiers in Plant Science. 2019;10:1319.
- Van EE, Powell CA, Shatters RG, et al. Control of larval and egg development in Aedes aegypti with RNA interference against juvenile hormone acid methyl transferase. J Insect Physiol. 2014;70:143–150.
- Song XS, Gu KX, Duan XX, et al. Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol Plant Pathol. 2018;19(12):2543–2560.
- Niehl A, Soininen M, Poranen MM, et al. Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol J. 2018;16(9):1679–1687.
- Koch A, Biedenkopf D, Furch A, et al. An RNAi-based control of fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016;12(10):e1005901.
- Wang M, Jin H. Spray-Induced gene silencing: a powerful innovative strategy for crop protection. Trends in Microbiology. 2017;25(1):4–6.
- McLoughlin AG, Wytinck N, Walker PL, et al. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and botrytis cinerea. Scientific Reports. 2018;8(1):7320.
- Werner BT, Gaffar FY, Schuemann J, et al. RNA-spray-mediated silencing of fusarium graminearum AGO and DCL genes improve barley disease resistance. Front Plant Sci. 2020;11:476.
- Chen W, Kastner C, Nowara D, et al. Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J Exp Bot. 2016;67(17):4979–4991.
- Govindarajulu M, Epstein L, Wroblewski T, et al. Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol J. 2015;13(7):875–883.
- Song Y, Thomma B. Host-induced gene silencing compromises verticillium wilt in tomato and Arabidopsis. Molecular Plant Pathology. 2018;19(1):77–89.
- Andrade C, Tinoco M, Rieth A, et al. Host-induced gene silencing in the necrotrophic fungal pathogen sclerotinia sclerotiorum. Plant Pathol. 2015;65(4):626–632.
- Jahan SN, Åsman AK, Corcoran P, et al. Plant-mediated gene silencing restricts growth of the potato late blight pathogen phytophthora infestans. J Exp Bot. 2015;66(9):2785–2794.
- Zhou B, Bailey A, Niblett CL, et al. Control of brown patch (Rhizoctonia solani) in tall fescue (Festuca arundinacea Schreb.) by host induced gene silencing. Plant Cell Rep. 2016;35(4):791–802.
- Nowara D, Gay A, Lacomme C, et al. HIGS: host-Induced gene silencing in the obligate biotrophic fungal pathogen blumeria graminis. Plant Cell. 2010;22(9):3130–3141.
- Zhang H, Guo J, Voegele RT, et al. Functional characterization of calcineurin homologs PsCNA1/PsCNB1 in puccinia striiformis f. sp. tritici using a host-induced RNAi system. PLoS ONE. 2012;7(11):e49262.
- Zhu X, Qi T, Yang Q, et al. Host-induced gene silencing of the MAPKK gene PsFUZ7 confers stable resistance to wheat stripe rust. Plant Physiol. 2017;175(4):1853–1863.
- Qi T, Zhu X, Tan C, et al. Host-induced gene silencing of an important pathogenicity factor PsCPK1 in puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol J. 2018;16(3):797–807.
- Thakur A, Sanju S, Siddappa S, et al. Artificial MicroRNA mediated gene silencing of phytophthora infestans single effector Avr3a gene imparts moderate type of late blight resistance in potato. Plant Pathology Journal. 2015;14(1):1–12.
- Hou Y, Zhai Y, Feng L, et al. A phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe. 2019;25(1):153–165.e5.
- Guo H, Song X, Wang G, et al. Plant-generated artificial small RNAs mediated aphid resistance. PLoS One. 2014;9(5):e97410.
- Saini RP, Raman V, Dhandapani G, et al. Silencing of HaAce1 gene by host-delivered artificial microRNA disrupts growth and development of Helicoverpa armigera. PloS One. 2018;13(3):e0194150.
- Agrawal A, Rajamani V, Reddy VS, et al. Transgenic plants over-expressing insect-specific microRNA acquire insecticidal activity against Helicoverpa armigera: an alternative to Bt-toxin technology. Transgenic Res. 2015;24(5):791–801.
- Jiang S, Wu H, Liu H, et al. The overexpression of insect endogenous small RNAs in transgenic rice inhibits growth and delays pupation of striped stem borer (chilo suppressalis). Pest Management Science. 2016;73(7):1453–1461.
- Miozzi L, Gambino G, Burgyan J, et al. Genome-wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera. Mol Plant Pathol. 2013;14(1):30–43.
- Wang XB, Wu Q, Ito T, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107(1):484–489.
- Donaire L, Barajas D, Martínez-García B, et al. Structural and genetic requirements for the biogenesis of tobacco rattle Virus -derived small interfering RNAs. Journal of Virology. 2008;82(11):5167–5177.
- Shahid S, Kim G, Johnson NR, et al. MicroRNAs from the parasitic plant cuscuta campestris target host messenger RNAs. Nature. 2018;553(7686):82–85.
- Hudzik C, Hou Y, Ma W, et al. Exchange of small regulatory rnas between plants and their pests. Plant Physiology. 2020;182(1):51–62.