ABSTRACT
The small ribosomal subunit protein Rps15/uS19 is involved in early nucleolar ribosome biogenesis and subsequent nuclear export of pre-40S particles to the cytoplasm. In addition, the C-terminal tail of Rps15 was suggested to play a role in mature ribosomes, namely during translation elongation. Here, we show that Rps15 not only functions in nucleolar ribosome assembly but also in cytoplasmic pre-40S maturation, which is indicated by a strong genetic interaction between Rps15 and the 40S assembly factor Ltv1. Specifically, mutations either in the globular or C-terminal domain of Rps15 when combined with the non-essential ltv1 null allele are lethal or display a strong growth defect. However, not only rps15 ltv1 double mutants but also single rps15 C-terminal deletion mutants exhibit an accumulation of the 20S pre-rRNA in the cytoplasm, indicative of a cytoplasmic pre-40S maturation defect. Since in pre-40S particles, the C-terminal tail of Rps15 is positioned between assembly factors Rio2 and Tsr1, we further tested whether Tsr1 is genetically linked to Rps15, which indeed could be demonstrated. Thus, the integrity of the Rps15 C-terminal tail plays an important role during late pre-40S maturation, perhaps in a quality control step to ensure that only 40S ribosomal subunits with functional Rps15 C-terminal tail can efficiently enter translation. As mutations in the C-terminal tail of human RPS15 have been observed in connection with chronic lymphocytic leukaemia, it is possible that apart from defects in translation, an impaired late pre-40S maturation step in the cytoplasm could also be a reason for this disease.
Introduction
Eukaryotic ribosome assembly is a highly complex, hierarchical process that is initiated in the nucleolus upon transcription of a large ribosomal RNA precursor (35S pre-rRNA in yeast) and a pre-5S RNA accompanied by co-transcriptional association of the first ribosome assembly factors and ribosomal proteins (r-proteins). The resulting initial pre-ribosomal particles (90S particles or also called SSU processomes) undergo an intricate maturation pathway, involving more than 200 different assembly factors while separating into precursors for the small 40S and large 60S ribosomal subunits (r-subunits) and transiting to the nucleoplasm and finally to the cytoplasm. Seminal cryo-EM studies of yeast and human pre-ribosomal particles have shed light on the structures of intermediates along this ribosome maturation path, revealing that besides rRNA processing (summarized in Figure S1), also massive rRNA folding and restructuring events take place until rRNAs and r-proteins become compacted into functional mature r-subunits [Citation1–3].
Upon endonucleolytic cleavage of the 35S pre-rRNA at processing site A2 and stepwise release of several structural modules of 90S particles in the nucleolus, the first pre-40S particles emerge [Citation4,Citation5]. These pre-40S particles contain the 20S pre-rRNA, a 3’-extended version of the mature 18S rRNA, as well as already most 40S r-proteins and, in yeast, nine stably bound assembly factors – Rrp12, Nob1, Pno1, Dim1, Tsr1, Rio2, Hrr25, Enp1 and Ltv1 [Citation6]. These pre-40S particles are quickly exported into the cytoplasm by a still poorly understood export mechanism. Several proteins have been suggested to participate in pre-40S export in yeast, among them distinct putative export receptors (Crm1/Xpo1, Rrp12 and Mex67), putative adaptors for export receptor Crm1 (Ltv1, Rio2) and putative auxiliary proteins such as Slx9 as well as the r-protein S15 (Rps15/uS19Footnote1) [Citation7–14].
Freshly exported pre-40S particles are not engaged immediately in translation as important functional sites are blocked by assembly factors [Citation15]. In particular, Dim1, Rio2 and Tsr1 are positioned at the inter-subunit side, and prevent association of 60S and translation initiation factors [Citation16–18]. In the course of cytoplasmic pre-40S maturation, all pre-40S assembly factors are successively released, while, concomitantly, r-proteins which are not yet assembled at their final position are stably accommodated, and in addition, the last missing r-proteins are incorporated into pre-40S particles. For instance, the kinase Hrr25 phosphorylates and releases Ltv1 promoting the stable accommodation of r-protein Rps3/uS3 [Citation19–21]. This maturation event is coordinated with the ATP-hydrolysis-triggered self-release of the ATPase Rio2 [Citation22–24]. Obviously, a recurring theme in ribosome assembly is quality control, in which assembly factors bind to functionally important sites of the nascent r-subunits and probe for correct structure and/or function. In yeast, these maturation steps are believed to occur in a ‘translation-like cycle’, in which an immature pre-40S particle is joined via eukaryotic initiation factor 5B (eIF5B) with an apparently mature 60S subunit [Citation25–27]. Only when this putative ‘translation test-drive’ is successful, the final maturation steps are triggered. The hallmark event of these final 40S maturation steps is the processing of the 20S pre-rRNA into the mature 18S rRNA by the endonuclease Nob1, involving the release of the Nob1-inhibitor Pno1, and finally Nob1, by Rio1 ATPase [Citation28–30].
A key prerequisite for the progression of ribosome biogenesis is the correct and timely binding of r-proteins, as reflected by the fact that the absence of almost any of the 79 yeast r-proteins lead to a specific maturation defect, either in nucleolar ribosome biogenesis steps, in r-subunit export, or maturation steps after export [Citation31–33]. Apart from that, many r-proteins have additional functions that could only be revealed by specific point mutants. The advantage is that such mutant r-protein variants can still assemble onto pre-ribosomal particles and allow maturation to proceed but are disturbed in one of their specific functions. Two well-characterized examples are Rps14/uS11 and Rps5/uS7: while their depletion leads to nucleolar pre-rRNA processing defects [Citation31], Rps14 and Rps5 variants carrying mutations or deletions in the C-terminal unstructured extensions of the proteins cause defects in cytoplasmic 20S pre-rRNA processing, indicating an additional function of these proteins in cytoplasmic pre-40S maturation [Citation34,Citation35].
We are interested in understanding the function of Rps15, an r-protein of the small 40S subunit, which is highly conserved among eukaryotes (Figure S2A). Both in yeast and human cells, depletion of Rps15 was shown to cause retention of pre-40S particles in the nucleus, suggesting that Rps15 assembly onto pre-40S particles is essential for the export of these particles [Citation11,Citation12,Citation31]. Additionally, an earlier, nucleolar function of Rps15 was revealed by a synthetic lethality study with a thermosensitive rps15-1 mutant, uncovering genetic links of RPS15 with NHP2, UTP15, SLX9, and BUD23. Combined mutations of NHP2, UTP15 or SLX9 with rps15-1 led to nucleolar accumulation of pre-40S particles and early pre-rRNA processing defects [Citation36]. In contrast, the rps15-1 Δbud23 double mutant primarily showed nucleoplasmic pre-40S accumulation, and, therefore, defects in nuclear export of pre-40S particles [Citation36].
Besides these functions in 40S subunit biogenesis, Rps15 also fulfils important roles in the mature ribosome. The globular domain of Rps15 is engaged in the B1a inter-subunit bridge by interacting with helix H38 of the 25S rRNA [Citation37,Citation38]. In turn, the largely unstructured C-terminal tail of Rps15 reaches to the decoding site of the 40S subunit, thereby playing an important role in translation elongation [Citation39,Citation40]. Recently, the very C-terminal residues of Rps15 could be visualized in a mature human 80S ribosome, revealing its flexibility during translation [Citation41]. In the post-decoding pre-translocation state of translation elongation, the Rps15 C-terminus interacts with both A- and P-site tRNAs and the mRNA in the decoding site [Citation40]. Based on these structural data, the Rps15-C-terminal domain was suggested to be engaged in the efficient accommodation of tRNAs at the A-site. Notably, mutations in the Rps15 C-terminal tail have been found with a high frequency in patients with relapsing chronic lymphocytic leukaemia (CLL), further substantiating the important function of the Rps15 C-terminus [Citation42–44].
In this study, we identified a specific rps15 mutant, carrying an F138S exchange in the C-terminal tail of the Rps15 r-protein, which shows a synthetically enhanced growth defect together with a Δltv1 chromosomal deletion. A second mutation, F102L, positioned in the region from where the C-terminal tail emerges, further enhanced this defect. In pre-40S particles, both F138 and F102 residues are positioned in close proximity to 40S assembly factor Tsr1. Consequently, tsr1 mutants genetically interact with these different rps15 mutants. Further investigation of these as well as other mutations in Rps15 indicated that, in contrast to the Rps15 depletion, nucleo-cytoplasmic export of pre-40S particles is not impaired, but the final cytoplasmic pre-40S maturation is disturbed. We conclude that the Rps15 C-terminal tail is not only needed for correct translation but already required during the final steps of 40S subunit synthesis.
Results
A synthetic lethality screen links Rps15 to 40S assembly factor Ltv1
Previously, we have performed a synthetic lethality (SL) screen with a deletion mutant of the non-essential 40S subunit assembly factor Ltv1, revealing its functional connection to RNA helicase Prp43, and its G-patch cofactor Pfa1 [Citation30]. The synthetically enhanced growth and 20S pre-rRNA processing defects observed upon LTV1 deletion in prp43 or pfa1 mutants were high-copy-suppressed by NOB1 ( and [Citation30]). These results indicated a role of Prp43 and its cofactor Pfa1 in promoting 20S pre-rRNA processing by the endonuclease Nob1.
Figure 1. LTV1 is genetically linked to RPS15. (A) Schematic overview of the genetic interactions uncovered in a synthetic lethality (SL) screen with a Δltv1 strain. Genes displayed on the left side were identified in a previous study [Citation30], while genes identified in the current study are shown on the right side. (B) Mutants #SL111 and #SL432 isolated in the SL screen were transformed with LEU2-plasmids carrying the indicated genes or with an empty plasmid (-); transformants were spotted in serial 10-fold dilution steps onto SDC-leu plates (-leu) as well as plates containing 5-fluoroorotic acid (5-FOA), and incubated at 30°C for 5 days. Red colony colour on SDC−leu and slow growth on 5-FOA plates indicate a synthetic growth defect. Red/white sectoring on SDC-leu and cell growth on 5-FOA plates indicate complementation of the synthetic enhancement phenotype. (C) Temperature dependence of the observed genetic interactions. Strains carrying the indicated wild-type and mutant RPS15 alleles on LEU2-plasmids plus HIS3-empty plasmid (Δltv1) or LTV1 wild-type allele on a HIS3-plasmid were spotted onto SDC-his -leu (-his -leu) plates to select for the transformed plasmids and incubated at 23°C, 30°C, and 37°C for 4 days.
![Figure 1. LTV1 is genetically linked to RPS15. (A) Schematic overview of the genetic interactions uncovered in a synthetic lethality (SL) screen with a Δltv1 strain. Genes displayed on the left side were identified in a previous study [Citation30], while genes identified in the current study are shown on the right side. (B) Mutants #SL111 and #SL432 isolated in the SL screen were transformed with LEU2-plasmids carrying the indicated genes or with an empty plasmid (-); transformants were spotted in serial 10-fold dilution steps onto SDC-leu plates (-leu) as well as plates containing 5-fluoroorotic acid (5-FOA), and incubated at 30°C for 5 days. Red colony colour on SDC−leu and slow growth on 5-FOA plates indicate a synthetic growth defect. Red/white sectoring on SDC-leu and cell growth on 5-FOA plates indicate complementation of the synthetic enhancement phenotype. (C) Temperature dependence of the observed genetic interactions. Strains carrying the indicated wild-type and mutant RPS15 alleles on LEU2-plasmids plus HIS3-empty plasmid (Δltv1) or LTV1 wild-type allele on a HIS3-plasmid were spotted onto SDC-his -leu (-his -leu) plates to select for the transformed plasmids and incubated at 23°C, 30°C, and 37°C for 4 days.](/cms/asset/4117bf94-dfda-4afb-9ec2-6b2f44cb124d/krnb_a_2064073_f0001_oc.jpg)
To identify additional players in late pre-40S maturation functionally connected to Ltv1, we set out to investigate seven of the so far uncharacterized mutants found in this SL screen. Considering the previous observations of a synthetically sick phenotype upon combination of certain rio2 mutants with the null or point-mutant ltv1 alleles [Citation23,Citation24], we transformed the remaining mutants with a RIO2 containing plasmid and scored for complementation of synthetic growth defects, as evaluated by a white or red/white sectoring colony colour and the ability to grow on 5-FOA containing plates (see Methods section for details). Indeed, six of these seven mutants could be complemented by RIO2 (), exemplarily shown for mutant #111). This strong genetic link between LTV1 and RIO2 is also in agreement with their joint action during cytoplasmic pre-40S maturation [Citation24].
One remaining mutant (#432) was neither complemented by RIO2, PRP43 and PFA1 nor suppressed by NOB1. To identify the mutation leading to synthetic enhancement in this strain, we transformed a yeast genomic library into the mutant and screened for transformants re-establishing the red/white sectoring phenotype, and hence for the ability to lose the LTV1 containing plasmid.
Using this strategy, we found that the original SL mutant #432 was complemented by the RPS15 gene ()). Sequencing of the chromosomal copy of this gene confirmed a mutation, leading to a phenylalanine to serine exchange (F138S) in the C-terminal unstructured tail of the Rps15 protein (position indicated in Figures S2B and 3A).
To avoid interference by other mutations arising in the strain generated by UV mutagenesis, we constructed a strain in which the chromosomal copy of RPS15 was deleted and complemented by a low-copy plasmid carrying the rps15-F138S allele; then, the growth of this mutant was compared to that of the same strain but carrying the RPS15 wild-type (WT) plasmid. While the rps15-F138S mutant did not display any growth defects by itself at any tested temperature (23°C, 30°C or 37°C), it strongly enhanced the growth defect of a Δltv1 strain, in particular at low temperature, indicating a genetic interaction between LTV1 and the rps15-F138S allele ().
To exclude that this genetic interaction is caused by a reduced level of the Rps15-F138S compared to the wild-type protein, we chromosomally fused the sequence encoding an N-terminal 2xHA-tag to RPS15 and rps15-F138S in the presence and absence of LTV1, respectively. Rps15 protein levels, detected with an α-HA-antibody, were apparently similar in the wild-type and the rps15-F138S mutant strain at any of the temperatures tested (Figure S3A). Based on these results, we conclude that there is a specific functional link between Ltv1 and the C-terminal domain of Rps15. As the chromosomally integrated constructs, however, displayed growth defects due to the N-terminal 2xHA-tag (Figure S3B), we performed all subsequent experiments using untagged Rps15 and variants thereof, encoded from a low-copy plasmid in the background of the knockout of the chromosomal RPS15 copy.
The rps15-F138S mutation enhances the 40S maturation defect of the Δltv1 deletion
Considering the genetic link of the rps15-F138S mutant to the late 40S assembly factor Ltv1, we speculated that the Rps15 C-terminal tail may be important for 40S subunit maturation. Polysome profiling indicated that the rps15-F138S mutant showed a mild 40S subunit shortage, as concluded from the slightly increased free 60S subunit peak (). The Δltv1 mutant showed a decreased free 40S peak and a strongly increased free 60S peak, as observed previously [Citation30]. An increased free 60S peak is characteristic for a 40S synthesis defect, as a deficit of available 40S subunits for subunit joining is leading to a consequent accumulation of free 60S subunits. Importantly, the rps15-F138S mutation clearly enhanced the 40S subunit synthesis defect of the Δltv1 mutant, as obvious from the absence of a free 40S subunit peak, and a massive increase of a combined 60S subunit/80S ribosome peak compared to the Δltv1 mutant alone. Moreover, polysomes were drastically reduced in the rps15-F138S Δltv1 strain at 30°C, and even more so at 23°C, indicating that the 40S subunit synthesis defect results in reduced translation elongation ().
Figure 2. 40S maturation defect of the Δltv1 strain is enhanced by the rps15-F138S allele. (A) Yeast cells of the Δrps15 and Δrps15 Δltv1 strains carrying a LEU2-plasmid encoding wild-type RPS15 or rps15-F138S were grown in liquid SDC-leu medium at 23°C or 30°C, respectively. After addition of cycloheximide and inhibition of translation, cells were lysed and polysome profiles recorded. Peaks corresponding to the free 40S and 60S subunits, 80S ribosomes and polysomes are indicated in the profile of the wild-type strain at 23°C. (B) rps15-F138S enhances the 20S pre-rRNA accumulation occurring in a Δltv1 strain. For northern blotting, cells were grown in liquid SDC-leu at the indicated temperatures. For pre- and mature rRNA detection, the indicated probes were used (the yeast pre-rRNA processing pathway is shown in Figure S1). (C) The mutant rps15-F138S shows no pre-40S export defect but an impairment in cytoplasmic 20S pre-rRNA processing. For fluorescence in situ hybridization (FISH), cells were grown in liquid SDC-leu at 30°C. After cell fixation, spheroplasts were incubated with a probe specific to the D/A2 segment of the ITS1 region to detect 20S pre-rRNA and nuclei were stained with DAPI.
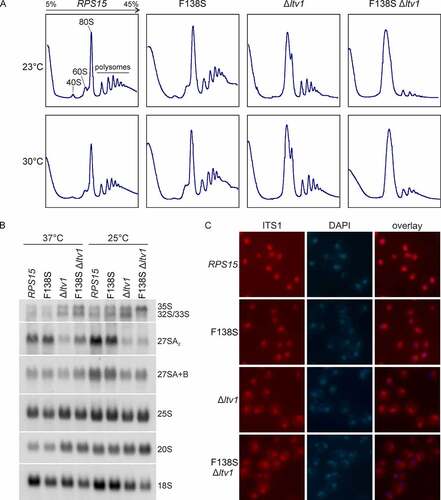
To determine the underlying maturation defects more precisely, we performed northern blotting and detected several intermediates of the pre-rRNA processing pathway (see also the schematic overview of the pathway in Figure S1). While the rps15-F138S mutant alone did not display any apparent defects, this mutation enhanced the defects observed for the Δltv1 strain, especially at 25°C, leading to a slight additional accumulation of 20S pre-rRNA as compared to Δltv1 alone and, even more noticeable, a reduction of mature 18S rRNA levels. Moreover, possibly as a secondary effect, the 35S pre-rRNA accumulated in this double mutant at low temperature ().
Both the Δltv1 deletion and Rps15 depletion were previously observed to result in 40S subunit export defects [Citation12,Citation13,Citation31]. To assess if the cause for the strong growth defect of the rps15-F138S Δltv1 double mutant is a pre-40S export defect or a defect prior to pre-40S export, we examined the localization of the 40S subunit reporter Rps3-GFP (uS3-GFP) in this mutant by fluorescence microscopy (Figure S4). No significant nuclear accumulation of Rps3-GFP was observed in the rps15-F138S Δltv1 double mutant as compared to Δltv1 alone, pointing towards a later, mainly cytoplasmic defect of the double mutant. Fluorescence in situ hybridization (FISH) using a probe specific to the D/A2 segment of ITS1 further indicated that 20S pre-rRNA synthesized in the rps15-F138S Δltv1 mutant can be exported into the cytoplasm, where, similar to the Δltv1 mutant, increased amounts of 20S pre-rRNA are detected; thus, its cytoplasmic processing is impaired (). We conclude that besides the previously reported function of Rps15 in 40S subunit export, Rps15 is also required for cytoplasmic steps of pre-40S maturation.
Expanding the genetic network between Rps15 and late 40S maturation factors
Given the link of the rps15-F138S mutation to LTV1, we wanted to further explore the connections of RPS15 to late 40S maturation. In addition to the rps15-F138S mutant, we included a second mutant into these analyses, which was erroneously generated during PCR amplification of the rps15-F138S allele during cloning. The resulting mutant allele contains a phenylalanine to leucine exchange (F102L) in addition to the F138S exchange. Whereas F138 is positioned only five amino acids from the C-terminal end of Rps15, F102 is positioned in the globular domain in the region from where this C-terminal tail emerges (positions indicated in )). Notably, the rps15-F102L/F138S mutant revealed a slight growth defect at 37°C on its own and was lethal when combined with a Δltv1 null mutant ()).
Figure 3. The rps15-F102L/F138S double mutation is genetically linked to LTV1 and TSR1. (A) The amino acid exchange F138S is located at the C-terminus of Rps15, whereas the second mutation F102L is positioned in the globular domain at which the C-terminus emerges ([Citation45], PDB 6Y7C). (B) Genetic interaction between the Δltv1 null allele and the double mutant rps15-F102L/F138S. The Δrps15 and Δrps15 Δltv1 strains carrying the wild-type plasmid URA3-RPS15 as well as a LEU2-plasmid encoding RPS15, rps15-F138S or rps15-F102L/F138S, respectively, were spotted onto SDC-leu plates and plates containing 5-FOA and were incubated at 30°C for 3 and 6 days. (C) In the Δltv1 strain, the RPS15-mutation F138S leads to cold sensitivity. The shuffled Δrps15 and Δrps15 Δltv1 strains, shown in B, containing only a LEU2-plasmid encoding RPS15, rps15-F138S or rps15-F102L/F138S were spotted onto SDC-leu plates and incubated for 3 days at the indicated temperatures. (D) Genetic interaction between the phosphorylation deficient ltv1 mutant (ltv1-SS>A) and RPS15. The Δrps15 Δltv1 cells carrying URA3-RPS15 and LEU2-LTV1 or LEU2-ltv1-SS>A as well as a TRP1-plasmid with RPS15, rps15-F138S or rps15-F102L/F138S, respectively, were spotted onto SDC-leu-trp plates (-leu -trp) and plates containing 5-FOA and lacking leucine (5-FOA-leu) and were incubated at 30°C for 3 and 6 days. (E) Rps15 is in contact with Rio2 and Tsr1 in the structure of the pre-40S particle ([Citation45], PDB 6Y7C). Rps15 is coloured in teal, Rio2 in blue and Tsr1 in purple. Parts of Tsr1 are more transparent to reveal the C-terminus of Rps15 behind Tsr1. (F) The Rps15 C-terminal tail is positioned between the A- and P-site tRNAs in the post-decoding pre-translocation translating human ribosome ([Citation41], PDB 6Y0G). The indicated position K145 (C-terminal residue of human RPS15) corresponds to yeast K142, the indicated F141 corresponds to yeast F138. (G) Genetic interaction between TSR1 and RPS15. The double knockout strain Δrps15 Δtsr1 carrying URA3-RPS15 and URA3-TSR1 as well as combinations of LEU2-plasmids with TSR1, tsr1-1, tsr1-2 and the HIS3-plasmids with RPS15, rps15 F138S or rps15 F102L/F138S were spotted onto SDC-his-leu plates (-his -leu) and plates containing 5-FOA and were incubated at 23°C or 30°C for 3 days.
![Figure 3. The rps15-F102L/F138S double mutation is genetically linked to LTV1 and TSR1. (A) The amino acid exchange F138S is located at the C-terminus of Rps15, whereas the second mutation F102L is positioned in the globular domain at which the C-terminus emerges ([Citation45], PDB 6Y7C). (B) Genetic interaction between the Δltv1 null allele and the double mutant rps15-F102L/F138S. The Δrps15 and Δrps15 Δltv1 strains carrying the wild-type plasmid URA3-RPS15 as well as a LEU2-plasmid encoding RPS15, rps15-F138S or rps15-F102L/F138S, respectively, were spotted onto SDC-leu plates and plates containing 5-FOA and were incubated at 30°C for 3 and 6 days. (C) In the Δltv1 strain, the RPS15-mutation F138S leads to cold sensitivity. The shuffled Δrps15 and Δrps15 Δltv1 strains, shown in B, containing only a LEU2-plasmid encoding RPS15, rps15-F138S or rps15-F102L/F138S were spotted onto SDC-leu plates and incubated for 3 days at the indicated temperatures. (D) Genetic interaction between the phosphorylation deficient ltv1 mutant (ltv1-SS>A) and RPS15. The Δrps15 Δltv1 cells carrying URA3-RPS15 and LEU2-LTV1 or LEU2-ltv1-SS>A as well as a TRP1-plasmid with RPS15, rps15-F138S or rps15-F102L/F138S, respectively, were spotted onto SDC-leu-trp plates (-leu -trp) and plates containing 5-FOA and lacking leucine (5-FOA-leu) and were incubated at 30°C for 3 and 6 days. (E) Rps15 is in contact with Rio2 and Tsr1 in the structure of the pre-40S particle ([Citation45], PDB 6Y7C). Rps15 is coloured in teal, Rio2 in blue and Tsr1 in purple. Parts of Tsr1 are more transparent to reveal the C-terminus of Rps15 behind Tsr1. (F) The Rps15 C-terminal tail is positioned between the A- and P-site tRNAs in the post-decoding pre-translocation translating human ribosome ([Citation41], PDB 6Y0G). The indicated position K145 (C-terminal residue of human RPS15) corresponds to yeast K142, the indicated F141 corresponds to yeast F138. (G) Genetic interaction between TSR1 and RPS15. The double knockout strain Δrps15 Δtsr1 carrying URA3-RPS15 and URA3-TSR1 as well as combinations of LEU2-plasmids with TSR1, tsr1-1, tsr1-2 and the HIS3-plasmids with RPS15, rps15 F138S or rps15 F102L/F138S were spotted onto SDC-his-leu plates (-his -leu) and plates containing 5-FOA and were incubated at 23°C or 30°C for 3 days.](/cms/asset/6ee08a60-6d07-4bf3-8c94-d892a6dcd631/krnb_a_2064073_f0003_oc.jpg)
Previous studies suggested that LTV1 deletion results in a mixed phenotype, showing both pre-40S export and cytoplasmic pre-40S maturation defects (see, for example, [Citation30]). To further validate our hypothesis that Rps15 has a function in cytoplasmic 40S maturation, we examined ltv1 mutants with no export but only cytoplasmic defects for genetic interactions with rps15 mutants. We have previously generated several ltv1 mutants in which the phosphorylation of the corresponding Ltv1 protein by Hrr25 is reduced, thus, resulting in an inhibition of the release of Ltv1 in the cytoplasm and consequently an exclusively cytoplasmic pre-40S maturation defect; these mutants include the ltv1-S339A/S342A mutant [Citation20]. Indeed, this phosphorylation deficient ltv1 mutant also showed a genetic interaction with both rps15 alleles of this study, with the rps15-F102L/F138S allele showing an almost lethal phenotype in combination with ltv1-S339A/S342A (abbreviated as ltv1-SS>A; ()) and S5A).
Given the genetic links of LTV1 to PFA1 30 and RIO2 23 ()), we next addressed if RPS15 also genetically interacts with these two genes. However, no genetic interaction was observed between our rps15 mutant variants and a Δpfa1 null allele or the rio2-1 point-mutant (Figure S5B and S5C).
We then inspected recent pre-40S cryo-EM structures, to identify assembly factors which might function together with Rps15. Interestingly, while the C-terminal tail of Rps15 is positioned between the A- and P-site tRNAs in the post-decoding pre-translocation state in translation ()) [Citation41], this tail is positioned between 40S assembly factors Rio2 and Tsr1 in pre-40S particles ()) [Citation18,Citation45]. Moreover, domain II of Tsr1 reaches close to the region in which the F102L exchange in the Rps15 mutant variant is positioned. Therefore, we tested for a genetic interaction with the essential TSR1 gene. To this end, we generated tsr1 mutants by random PCR mutagenesis. Two thermo-sensitive tsr1 alleles were obtained, termed tsr1-1 and tsr1-2, leading to three (E588D, Y604C and T704A) and eight (F220L, L239F, R473 G, E481G, F596Y, F710L, T716A, F759L) exchanges in the amino acid sequence of the Tsr1 protein, respectively. Interestingly, both tsr1 mutant alleles caused synthetic lethality in combination with the rps15-F102L/F138S mutation and a synthetically enhanced defect in combination with the rps15-F138S mutation ()) and S5D. These results may be an indication that the contact between Rps15 and Tsr1 is important for cytoplasmic pre-40S subunit maturation.
The C-terminal tail of Rps15 functions in late 40S subunit maturation
The so far obtained rps15 mutants display no (rps15-F138S) or only mild (rps15-F102L/F138S) defects by themselves, and their connection to late 40S maturation becomes only obvious when combined with other mutations (e.g. ltv1 or tsr1). Therefore, we next aimed to generate rps15 mutants displaying late 40S maturation defects on their own.
For this purpose, we created further mutations in proximity to F102 and F138. Residue F102 does not directly contact RNA nor other proteins; however, in the mature 40S and pre-40S structures, it is in close proximity to arginine 77 (R77) and histidine 79 (H79), which are in direct contact with 18S rRNA helices h32 and h33 () [Citation37,Citation45]. Therefore, we speculated that the F102L mutation may affect the positioning of these residues, consequently leading to altered interactions of Rps15 with rRNA. To further evaluate the importance of these residues, we mutated R77 and H79, as well as the residue in between, threonine 78 (T78), alone and in combinations (). However, although most of them (except the R77A mutant) were genetically linked to LTV1 (Figure S6A and S6B), many of these mutants did not display a growth defect on their own, except for the triple mutant rps15-R77A/T78A/H79A (further referred to as rps15-RTH(77–79)>A), which showed a slight growth defect at 37°C ().
Figure 4. C-terminal Rps15-truncation results in growth defects. (A) The F102L exchange in Rps15 is in close proximity to the amino acids R77 and H79. In the pre-40S particle ([Citation45], PDB 6Y7C), R77 is in contact with helix h33 of the 18S rRNA, and H79 is in contact with helix h32. The third amino acid in this globular region of Rps15, that was exchanged in this study, is threonine 78 (T78). (B) The triple mutation RTH(77–79)>A of RPS15 leads to a slight growth defect at high temperatures. The shuffled Δrps15 strains, containing a LEU2-RPS15 plasmid or LEU2-rps15 variant as indicated were spotted onto SDC-leu plates (-leu) and incubated for 2 days at the indicated temperatures. (C) C-terminal deletions, investigated in this study, are indicated in the pre-40S particle ([Citation45], PDB 6Y7C). (D) RPS15 C-terminal deletions lead to growth defects. The shuffled Δrps15 strains, containing only a LEU2-RPS15 plasmid or C-terminal deleted rps15 variants as indicated were spotted onto SDC-leu plates and incubated for 3 days at the indicated temperatures.
![Figure 4. C-terminal Rps15-truncation results in growth defects. (A) The F102L exchange in Rps15 is in close proximity to the amino acids R77 and H79. In the pre-40S particle ([Citation45], PDB 6Y7C), R77 is in contact with helix h33 of the 18S rRNA, and H79 is in contact with helix h32. The third amino acid in this globular region of Rps15, that was exchanged in this study, is threonine 78 (T78). (B) The triple mutation RTH(77–79)>A of RPS15 leads to a slight growth defect at high temperatures. The shuffled Δrps15 strains, containing a LEU2-RPS15 plasmid or LEU2-rps15 variant as indicated were spotted onto SDC-leu plates (-leu) and incubated for 2 days at the indicated temperatures. (C) C-terminal deletions, investigated in this study, are indicated in the pre-40S particle ([Citation45], PDB 6Y7C). (D) RPS15 C-terminal deletions lead to growth defects. The shuffled Δrps15 strains, containing only a LEU2-RPS15 plasmid or C-terminal deleted rps15 variants as indicated were spotted onto SDC-leu plates and incubated for 3 days at the indicated temperatures.](/cms/asset/b04f1af7-c5b5-430c-8db7-811fe0aaa699/krnb_a_2064073_f0004_oc.jpg)
Next, as F138 is very close to the C-terminal end of Rps15, we generated stepwise C-terminal truncations (depicted in ) and compared the growth of the different mutants to that of the F138S mutant. Removal of the last seven amino acids (rps15-1-135) led to a slight growth defect, deletion of the last eight residues (rps15-1-134) to a strong growth defect, which became even more severe when the last 12 (rps15-1-130) amino acids were removed (). Notably, all tested C-terminally truncated Rps15 variants, including a variant lacking only the very last amino acid (lysine, K142), led to a synthetic enhanced defect (rps15-1-141) or even a synthetic lethal phenotype (all other C-terminally truncated mutants) in combination with the Δltv1 null allele (Figure S6C and S6D).
To address whether the novel rps15 mutants also display late 40S maturation defects on their own, we investigated the phenotypes of the newly generated rps15-RTH(77–79)>A and rps15-1-134 mutants. Polysome profiles showed a slight (rps15-RTH(77–79)>A) or severe (rps15-1-134) increase in the free 60S peak, together with a reduction of the free 40S peak, indicating a 40S subunit maturation defect in these mutants ().
Figure 5. The C-terminus of RPS15 functions in late pre-40S maturation. (A) Δrps15 cells carrying the LEU2-plasmid encoding the wild-type RPS15, rps15-1-134 or rps15-RTH(77–79)>A, respectively, were grown in liquid SDC-leu medium at 30°C. After inhibition of translation by cycloheximide, cells were lysed and polysome profiles were recorded. Peaks corresponding to the 40S and 60S subunit, 80S ribosomes and polysomes are indicated in the profile of the wild-type strain. (B) For northern blotting, rps15 deletion strains carrying LEU2-plasmids with wild-type RPS15 or the indicated rps15 variants were grown in liquid SDC-leu medium at 30°C to an OD600 of 0.1–0.2 and shifted for 3 h to 25°C, 30°C or 37°C, respectively. Blots were probed as indicated in the methods section (the yeast pre-rRNA processing pathway is shown in Figure S1). (C) Eluates from TAP purification of Tsr1-particles from the wild-type RPS15 or rps15-1-134 or rps15-RTH(77–79)>A mutant backgrounds were analysed via SDS-PAGE, followed by western blotting. Tsr1-CBP was detected via α-CBP antibody; Rps3 and Nob1 were detected with antibodies specific to the respective proteins. (D) The rps15-1-134 and rps15-RTH(77–79)>A mutants show cytoplasmic 20S pre-rRNA accumulation. For fluorescence in situ hybridization (FISH), cells were grown in liquid SDC-leu at 30°C. After cell fixation, spheroplasts were incubated with a probe specific to the D/A2 segment of the ITS1 region to detect 20S pre-rRNA and nuclei were stained with DAPI. (E) After fractionation of polysomes from the RPS15 strain and rps15-1-134 or rps15-RTH(77–79)>A, respectively, northern blots were performed. Blots were probed as described in the methods section.
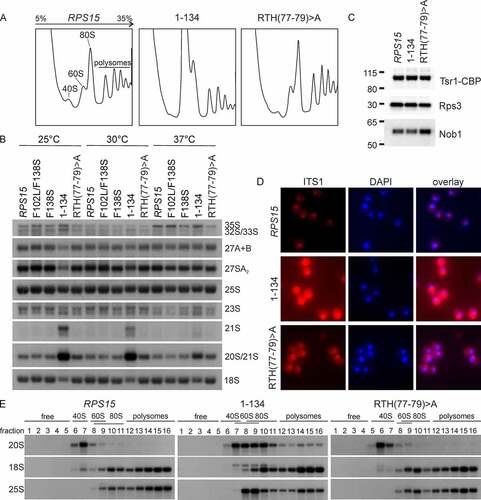
To detect potential rRNA processing defects in these new rps15 mutants, we performed northern blotting and included the rps15-F138S and rps15-F102L/F138S mutants in the analysis for comparison. Indeed, we observed a slight (rps15-RTH(77–79)>A) or strong (rps15-1-134) accumulation of 20S pre-rRNA, which was, in the case of the rps15-1-134 mutant also accompanied by a reduction of mature 18S rRNA, especially at low temperatures. In addition, in the rps15-1-134 mutant, a 21S pre-rRNA was also detected at 30°C and to a higher extent at 25°C (). This aberrant precursor arises when cleavage of the 32S pre-rRNA intermediate at site A2 is skipped and the precursor is cleaved at site A3 instead (see Figure S1). In line with this, a mild accumulation of 32S (and also 35S) pre-rRNA and a reduction of 27SA2 pre-rRNA was also observed under the same conditions. These data suggest that the rps15-1-134 mutant also shows a defect in early nucleolar ribosome biogenesis, next to the late defect in 20S pre-rRNA processing. As 20S pre-rRNA is cleaved by Nob1, a potential explanation for 20S pre-rRNA accumulation in the rps15 mutants could be the failure to recruit Nob1 to pre-40S particles. However, Nob1 was present in late nuclear and cytoplasmic pre-40S particles purified from the rps15-1-134 and rps15-RTH(77–79)>A strains, indicating that a step after Nob1 recruitment has to be affected (). In addition, to determine, in which cellular compartment the 20S pre-rRNA accumulates, we assessed the localization of 20S pre-rRNA in the mutants by FISH (). While the rps15-RTH(77–79)>A mutant showed only slight cytoplasmic accumulation of 20S pre-rRNA, the C-terminal truncation mutant rps15-1-134 accumulated significant amounts of 20S pre-rRNA in the cytoplasm, indicating a strong cytoplasmic 40S maturation defect before site D is cleaved by Nob1.
Although cytoplasmic 20S pre-rRNA accumulation was the strongest phenotype observed in the two rps15 mutants, the rps15-1-134 mutant also appeared to have an increased nuclear ITS1 signal () and showed some accumulation of nuclear pre-rRNAs, i.e. 21S, 23S, 32S and 35S pre-rRNAs (). Therefore, we sought for further confirmation that the cytoplasmic 20S pre-rRNA accumulation is the main defect in these mutants. To this end, we scored for potential pre-40S export defects in these two rps15 mutants, by examining the localization of the 40S subunit export reporter Rps3-GFP via fluorescence microscopy (Figure S7A). As a control, also the 60S subunit export reporter Rpl25-GFP was examined in these strains. Importantly, neither the rps15-RTH(77–79)>A nor the rps15-1-134 mutant showed an r-subunit export defect (Figure S7A), further supporting our model that they mainly display cytoplasmic pre-40S maturation defects. In our previous study, we observed that large amounts of the 20S pre-rRNA accumulating in Δltv1 Δpfa1 as well as Δltv1 prp43 double mutants are incorporated into polysomes [Citation30], suggesting that in these mutants, 20S pre-rRNA containing 40S subunits can escape the quality control exerted by the 'translation-like cycle' and engage in translation. To address if such a phenotype is also observable in the rps15-1-134 and rps15-RTH(77–79)>A mutants, we analysed fractions collected from polysome profiles of these strains by northern blotting (). In the rps15-RTH(77–79)>A mutant, the 20S pre-rRNA sedimented with the 40S peak comparable to the wild-type strain. In contrast, the rps15-1-134 mutant showed small amounts of 20S pre-rRNA in polysomes; however, in contrast to the Δltv1 Δpfa1 and Δltv1 prp43 double mutants investigated previously [Citation30], most of the 20S pre-rRNA signal was found in the 80S peak, suggesting that pre-40S particles from the rps15-1-134 mutant can form 80S ribosomes but engage only inefficiently in translation. Taken together, we conclude that the C-terminal domain of Rps15 is important for the final cytoplasmic pre-40S subunit maturation events prior to 20S pre-rRNA processing.
The C-terminal domain of Rps15 acts together with the N-terminal extension of Rps31 in ensuring translational fidelity
Recent findings suggested an important function of the Rps15 C-domain in translational decoding [Citation39,Citation41], while our data indicate an additional role of Rps15 in late pre-40S maturation. These phenotypes are reminiscent of those described for r-protein Rps31/eS31, for which a dual role in late 40S maturation and decoding was also described [Citation46–48], and a ∆rps31 (also known as ∆ubi3) mutant was also found to be synthetic lethal with ∆ltv1 [Citation47].
Moreover, Rps31 is positioned at the beak of the 40S subunit and has an unstructured N-terminal extension, which reaches into close proximity to the C-terminal domain of Rps15 () [Citation37]. Similar to C-terminal deletions of Rps15, the deletion of the N-terminal domain of Rps31 (ubi3ΔN allele, herein rps31ΔN) or complete deletion of the non-essential RPS31 gene leads to late pre-40S maturation defects, including a huge 20S pre-rRNA accumulation in the cytoplasm. Moreover, in ubi3Δ cells, 20S pre-rRNA mainly co-sediments with 80S ribosomes, with small amounts of 20S pre-rRNA entering into polysomes [Citation47], similar as we observed for the rps15-1-134 mutant. Besides, cells lacking Rps31 show an increased rate of amino acid misincorporation during translation, indicating that Rps31 ensures optimal translational fidelity [Citation47]. In order to find out if Rps15 and Rps31 could cooperate in their functions, we first tested for a genetic interaction between rps15 and rps31 N-terminal mutants. Indeed, the rps31ΔN mutant was synthetically lethal in combination with the rps15-1-134, rps15-F102L/F138S and rps15-RTH(77–79)>A alleles (). Moreover, rps15-F138S, as well as rps15-1-141 showed a mild synthetically enhanced growth defect in combination with the rps31ΔN allele (Figure S8).
Figure 6. Rps31 and Rps15 are functionally linked and ensure translational fidelity. (A) The Rps15 C-terminal tail is in close proximity to the Rps31 N-terminal tail in mature 40S subunits (PDB 4V88 [Citation37]). (B) Genetic interaction between RPS15 and RPS31. A Δrps15 rps31ΔN [URA3-RPS15] shuffle strain carrying the indicated wild-type and mutant alleles was spotted onto SDC-leu-ura plates (-leu -ura) and 5-FOA containing plates. The rps15-1-134 rps31ΔN, the rps15-F102L/F138S rps31ΔN and rps15-RTH(77–79)>A rps31ΔN mutants were inviable on 5-FOA, indicating synthetic lethality. (C) Measurement of misincorporation frequencies in the indicated mutants. Strains were transformed with URA3-plasmids pDB688 and pDB868 to measure misreading (Arg245(GGC) to His245(CAC)). Transformants were grown in liquid SDC-ura medium to mid-log phase at 30°C and then Renilla and firefly luciferase activity was measured. Assays were done in quadruplicate, and the data were expressed as the mean ± the standard deviation. The percentage of misreading was expressed as the firefly (H245R)/Renilla luciferase activity divided by the firefly (wild-type)/Renilla luciferase activity multiplied by 100. Significance levels were determined by Student’s t-test (*, p < 0.05).
![Figure 6. Rps31 and Rps15 are functionally linked and ensure translational fidelity. (A) The Rps15 C-terminal tail is in close proximity to the Rps31 N-terminal tail in mature 40S subunits (PDB 4V88 [Citation37]). (B) Genetic interaction between RPS15 and RPS31. A Δrps15 rps31ΔN [URA3-RPS15] shuffle strain carrying the indicated wild-type and mutant alleles was spotted onto SDC-leu-ura plates (-leu -ura) and 5-FOA containing plates. The rps15-1-134 rps31ΔN, the rps15-F102L/F138S rps31ΔN and rps15-RTH(77–79)>A rps31ΔN mutants were inviable on 5-FOA, indicating synthetic lethality. (C) Measurement of misincorporation frequencies in the indicated mutants. Strains were transformed with URA3-plasmids pDB688 and pDB868 to measure misreading (Arg245(GGC) to His245(CAC)). Transformants were grown in liquid SDC-ura medium to mid-log phase at 30°C and then Renilla and firefly luciferase activity was measured. Assays were done in quadruplicate, and the data were expressed as the mean ± the standard deviation. The percentage of misreading was expressed as the firefly (H245R)/Renilla luciferase activity divided by the firefly (wild-type)/Renilla luciferase activity multiplied by 100. Significance levels were determined by Student’s t-test (*, p < 0.05).](/cms/asset/f1972eb3-def1-4b97-b7d1-0985a852cd86/krnb_a_2064073_f0006_oc.jpg)
To assess if these synthetic growth defects go along with an enhancement of the translational error rate upon combination of rps15 and rps31 mutations, we analysed amino acid misincorporation in the viable rps15 rps31ΔN double mutants (). Additionally, we also analysed the rps15 single mutants F138S, 1–141, 1–134, F102L/F102S and RTH(77–79)>A. To this end, we made use of a plasmid-borne tandem Renilla and firefly luciferase reporter system [Citation49], where the CAC (His) codon at position 245, essential to form functional firefly luciferase, has been substituted by a CGC (Arg) codon, resulting in only residual firefly luciferase enzymatic activity. An increased firefly luciferase activity is indicative of a misreading event, leading to misincorporation of histidine at this critical position, and consequently to reconstitution of the enzymatic activity of the protein.
Our results indicated that the single rps31∆N and the rps15-1-141 mutation leads to a modest but statistically significant increase in the misreading rate (). Although we observed the tendency that the combined mutation of rps31∆N and rps15-1-141 further increased the misincorporation rate compared to the single mutations, while the combination of rps31∆N and rps15-F138S reduced the misreading rate compared to rps31∆N alone, no significance was detected for these combinations compared to the single mutations. Similarly, single rps15 mutants F138S, F102L/F138S, and RTH(77–79)>A repeatedly showed slightly increased misreading rates in our assays, however without significance. In contrast, no difference compared to the wild-type was observed for the rps15-1-134 mutant. Interestingly, the ∆ltv1 knockout mutation also significantly increased the translation error rate compared to the wild-type strain, but only tendencies for increased misreading without significance were detected for the double ∆ltv1 rps15-1-141 mutant, and for decreased misreading in the double ∆ltv1 rps15-F138S mutant. Altogether, these results suggest that slight alterations in the C-terminal tail of Rps15 can lead to increased misreading. Moreover, our data support a model where the N-terminal tail of Rps31 and the tip of the C-terminal end of Rps15 (missing in the rps15-1-141 mutant) cooperate in ensuring translational fidelity. Ltv1 is a crucial factor for final maturation of cytoplasmic pre-40S particles, including correct positioning of different r-proteins [Citation20,Citation21,Citation24]; thus, as our results showed, it is not surprising that 40S subunits are also prone to make more misreading in the absence of Ltv1.
Discussion
Many eukaryotic r-proteins contain, in addition to globular domains, long unstructured N- or C-terminal extensions [Citation50]. Interestingly, in several instances, such extensions were described to fulfil important functions during ribosome biogenesis. For example, the C-terminal tails of Rps14 and Rps5 and the N-terminal tail of Rps31 were shown to be required for 20S pre-rRNA processing in the cytoplasm [Citation34,Citation35,Citation48]. Interestingly however, Rps5, Rps14 and Rps31 are assembled to pre-ribosomes early on, and the absence of the essential Rps5 and Rps14 r-proteins stalls the ribosome biogenesis pathway in the nucleus [Citation31]. Hence, while their physical presence is required early in maturation, these r-proteins fulfil additional important functions in late 40S subunit maturation steps. Here, we show that Rps15 is yet another r-protein with more than one function in ribosome biogenesis, being not only involved in early nucleolar maturation events and required for 40S export, as previously reported [Citation11,Citation12,Citation31,Citation36], but also in cytoplasmic pre-40S maturation.
In particular, the mutant in which the seven C-terminal amino-acids of Rps15 are missing, rps15-1-134, accumulated 20S pre-rRNA in the cytoplasm, suggesting a defect in the final 40S maturation steps prior to D-site cleavage. The molecular reason for this phenotype is not yet clear, however, we consider it unlikely that the Rps15 C-terminal tail directly regulates 20S pre-rRNA processing for several reasons: (1) Nob1 is present in pre-40S particles purified from the rps15-1-134 mutant, excluding a defect in Nob1 recruitment. (2) Our rps15 mutants did not exacerbate the mild slow-growth phenotype of a NOB1-TAP strain (data not shown), in contrast to other mutants more directly connected to D-site cleavage, like rpl3-W255C [Citation27]. (3) In pre-40S structures, the Rps15 C-terminal tail is ~50 Å apart from Nob1 and more than 80 Å apart from the 3’ end of the 18S rRNA [Citation29,Citation45,Citation51], making a direct engagement of the Rps15 C-terminal tail in D-site cleavage unlikely.
Notably, the Rps15 C-terminal tail alters its positioning throughout the translational cycle. In the post-decoding pre-translocation state of translation elongation, it is wedged between the A- and P-site tRNAs [Citation41]. In line with this, deletion of the last 15 residues from the C-terminal tail of human RPS15 leads to defects in translation elongation, as was concluded from the reduction of polysomes and the accumulation of 80S particles [Citation39]. Moreover, some of the mutations in the RPS15 C-terminus linked to CLL showed reduced overall translation, while others showed increased misreading rates or increased stop-codon readthrough (Figure S2C) [Citation43].
Here, we show that the deletion of the very C-terminal residue of Rps15 (K141) results in increased misreading rates. This result is in compliance with structural data suggesting that this terminal lysine residue (K145 in human RPS15) is in direct contact with mRNA during decoding [Citation41]. Moreover, the genetic interaction between rps15 and rps31 mutants, as well as the enhanced misreading rates in the rps31ΔN rps15-1-141 double mutant suggest that the Rps31 N-terminal extension and the Rps15 C-terminal tail might act in concert in ensuring not only the correct execution of final pre-40S maturation events but also correct decoding. Interestingly, RPS31 is non-essential, and also our rps15- mutants are viable, indicating that although these mutants display growth defects, they tolerate a certain extent of translational error. Consistently, the misreading rates we detected were quite modest, especially when compared to the values obtained for mutants of other r-proteins (e.g. rps9B-D94N) or the wild-type strain treated with low doses of paromomycin [Citation49]. Moreover, we did not observe increased misreading in the rps15-1-134 mutant, which showed the strongest defects in cytoplasmic pre-40S maturation. Together with the previous results that only few of the human RPS15 C-terminal tail mutants show increased misreading (Figure S2C) [Citation43], these data suggest that mild positional alterations at some critical residues of the tail can alter translational fidelity, potentially due to altered communication with mRNA or tRNAs, while other mutations or the complete absence of this tail do not have such an effect. However, the main defect of C-terminal tail mutants is likely not the altered misreading, as it occurs only in some mutants and the increase in misreading is only subtle.
Strikingly, the strategic positioning of the Rps15 C-terminal tail between the A- and P-site tRNAs resembles the orientation of the Rps15 C-terminal tail in pre-40S particles, which is clamped between assembly factors Tsr1 and Rio2 instead of tRNA (compare PDB 6Y0G [Citation41] and PDB 6Y7C [Citation45], ). Considering this positioning, we speculate that Rio2 and Tsr1 might quality-check for the integrity of the Rps15 C-terminal tail. In the absence of an interaction of the C-terminal tail with Rio2 and/or Tsr1, further maturation (including 20S pre-rRNA processing) and engagement of faulty pre-40S particles lacking the Rps15 C-terminal tail in translation might be prevented, resulting in the accumulation of these immature 40S particles in 80S(-like) particles. An alternative possibility would be that the rps15-1-134 mutant might cause an earlier cytoplasmic pre-40S maturation defect (e.g. an altered rRNA structure or protein positioning) and that not the absence of the Rps15 C-terminal tail directly, but the defect caused by its absence is responsible for the blocking of subsequent pre-40S maturation steps. Last but not least, it might be possible that the Rps15 C-terminal tail is itself part of a quality control mechanism to test the functionality of 40S subunits, and that in its absence, a particular quality control step cannot take place, consequently trapping pre-40S particles in an inactive form.
While the functional connection of the Rps15 C-terminal tail to Tsr1 can be explained by the structural data demonstrating a direct interaction, the precise connection of the Rps15 C-terminal tail to Ltv1 is more puzzling. Ltv1 interacts with Enp1 and Rps3 [Citation19,Citation20], which are both positioned in the head domain of the pre-40S particle, as is the globular domain of Rps15. So far, the flexible and poorly structured protein Ltv1 has only been partly resolved on pre-40S particles [Citation17,Citation18,Citation45,Citation52]. However, biochemical interaction data indicate a direct interaction between Rps15 and Ltv1 [Citation53]. Moreover, a human pre-40S particle cryo-EM analysis succeeded in visualizing a C-terminal α-helix of Ltv1 that is in direct contact with the globular domain of Rps15 [Citation29]. In lack of further structural information, it is not possible to judge whether additional contacts are formed between Ltv1 and Rps15 or whether Ltv1 even interacts with the Rps15 C-terminal domain directly. The observed defects in the absence of Ltv1 might either be a consequence of the missing interaction with the Rps15 globular domain (potentially leading to an altered positioning of Rps15) or might be due to the effect of the missing Ltv1 on other proteins linked to Rps15 like Rio2 or Tsr1. The third possibility we envisage is an RNA folding problem in these mutants. ∆ltv1 mutants are cold-sensitive, and the genetic interaction between LTV1 and RPS15 is also most pronounced at low temperatures. Cold-sensitive phenotypes are frequently observed upon RNA folding problems [Citation54,Citation55], hence we speculate that an altered rRNA structure occurring in the absence of Ltv1 might be the reason for the genetic interactions with Rps15.
Evidence for a cytoplasmic pre-40S maturation function of Rps15 also comes from a recent study, in which several different mutants including rps15 mutants were investigated for defects in the cytoplasmic release of Ltv1, Rio2 and Tsr1 from pre-40S particles [Citation56]. Mutants in the C-terminal tail of Rps15 showed defects in the release of Rio2 from pre-40S particles in the mentioned study [Citation56]. In our experiments, however, we did not see Rio2 release defects in rps15 C-terminal mutants (data not shown). The reason for these divergent results could be related to the fact that, in the study by Huang et al. [Citation55], the phenotypes were investigated in a strain depleted for Fap7, a condition normally leading to the accumulation of 80S-like particles [Citation25,Citation57]. Hence, Rio2 release defects might only be visible when rps15 mutations are combined with the depletion of Fap7.
In recent years, a strong connection between mutations clustering in the evolutionary-conserved C-terminal tail of human RPS15 (131-PGIGATHSSR-140) and aggressive, chemo-refractory CLL has been uncovered, and the molecular reason has been attributed to translational defects [Citation42–44]. Although a mutation of F141, the residue corresponding to yeast F138, has not yet been observed in this context, F141 is positioned directly after a cluster of amino-acids found to be mutated in CLL (Figure S2B). Additionally, C-terminal truncation mutants tested in our study lack at least one (rps15-1-141), or several (rps15-1-134) of the equivalent CLL-linked residues. In human cells, it was shown that the respective mutated RPS15 variants are incorporated into ribosomes and negatively affect translation fidelity and global protein synthesis (Figure S3C) [Citation43]. Our study revealed that in yeast, Rps15 C-terminal truncation mutants already show defects in the course of 40S subunit maturation before 40S particles are even joined with 60S subunits forming translation-competent ribosomes. Thus, a plausible alternative possibility to explain the disease relapse could be related to the 40S biogenesis defects, likely at the late cytoplasmic maturation steps and, therefore, should be considered in the future.
Materials and Methods
Yeast strains and genetic methods
The S. cerevisiae strains used in this study are W303 derivatives generated by integration at the genomic locus and are listed in Table S1. Yeast plasmids were constructed using standard recombinant DNA techniques and are listed in Table S2. All DNA fragments amplified by PCR were verified by sequencing. The tsr1-1 and tsr1-2 mutants were generated by random PCR mutagenesis as described previously [Citation58,Citation59].
Identification of mutants from the SL screen
Mutants showing synthetic growth defects or synthetic lethality in combination with Δltv1 were previously generated in a synthetic lethality (SL) screen [Citation30]. The screen is based on a combination of the ade2/ade3 red/white colony-sectoring assay and counter-selection on 5-FOA (5-fluoro orotic acid, Thermo Scientific)-containing plates and scores for the inability to lose a plasmid carrying an LTV1 wild-type copy, resulting in a red non-sectoring, 5-FOA-sensitive phenotype (for more details, see [Citation60]). The seven mutants that were not further characterized in the previous study [Citation30] were first transformed with LEU2-plasmids containing genes already known to be genetically linked to LTV1 (i.e. PRP43, PFA1, NOB1 and RIO2). While mutants remained red upon transformation with non-complementing plasmids, the mutants carrying complementing plasmids recovered the ability to lose the LTV1 wild-type plasmid, and hence showed red/white sectoring colonies. Using this strategy, we identified six of the seven mutants to be complemented by RIO2. Sequencing of the chromosomal RIO2 copy confirmed mutation of this gene in these six mutants. The remaining SL mutant (#432) was transformed with a genomic LEU2-plasmid based library. Colonies showing a red/white sectoring phenotype were re-streaked onto plates lacking leucine (SDC-leu) and then on 5-FOA containing plates. Complementing plasmids resulting in red/white sectoring colonies on SDC-leu and growth on 5-FOA were isolated, and LTV1 containing plasmids were identified by PCR. All other plasmids were subjected to DNA sequencing, revealing that all of them contained RPS15. Mutation of RPS15 in the SL mutant #432 was subsequently confirmed by DNA sequencing at the genomic locus.
Plasmid shuffle assays
The Δrps15 shuffle strain (Δrps15 [pRS316-RPS15]) was constructed by chromosomal deletion of RPS15 in a diploid yeast strain, followed by transformation with the URA3-plasmid [pRS316-RPS15]. After tetrad dissection, the spores harbouring the gene knockout and the complementing URA3 plasmid were recovered. A similar strategy was used to generate the RPS15 shuffle Δltv1 strain (Δrps15 Δltv1 [pRS316-RPS15]).
The ‘double shuffle’ strain Δrps15 Δtsr1 [pRS316-RPS15] [pRS316-TSR1] was generated by crossing of the respective single shuffle strains and subsequent sporulation.
The rps31∆N-HA ∆rps15 [pRS316-RPS15] shuffle strain was generated by crossing of the ∆rps15 [pRS316-RPS15] shuffle strain with a rps31∆N-HA strain (also known as ubi3∆N-HA [Citation48]). The resulting diploid was sporulated, tetrads were dissected and a representative strain harbouring the appropriate alleles selected.
To analyse the growth phenotypes conferred by mutant alleles of RPS15 either alone or in combination with Δltv1 and tsr1- or rps31-mutations, we transformed the strains with the respective plasmids. Thereafter, transformants were spotted in 10-fold serial dilutions on 5-FOA containing plates to evaluate the phenotypes caused after loss of the URA3-RPS15 plasmid or both the URA3-RPS15 and URA3-TSR1 plasmids. Subsequently, strains that were viable on 5-FOA containing plates were re-streaked on plates selecting for the transformed plasmids. Subsequently, these strains were spotted in 10-fold serial dilutions onto the respective plates and incubated at different temperatures.
Fluorescence in situ hybridization (FISH), fluorescence microscopy
For fluorescence in situ hybridization, cells were grown in 50 ml SDC-leu medium at 30°C to an OD600 of ~0.5 and fixated with formaldehyde with a final concentration of 4% for 1 h at room temperature. After fixation, cells were washed twice with buffer containing 0.1 M KPO4 buffer (K2HPO4 and KH2PO4 mixed in the appropriate ratio to obtain a pH of 6.4) and washed once with washing buffer containing 0.1 M KPO4 and 1.2 M sorbitol (pH of 6.4). For cell wall lysis, cells were incubated with 1 ml washing buffer containing 500 µg/ml Zymolyase 100 T (Amsbio) for 60 min at room temperature, followed by one washing step with the washing buffer. Finally, the spheroplasts were resuspended in ~1.5-fold pellet volume, applied to adhesive coated 10-well diagnostic microscope slides (Thermo Scientific, LOT #381,613) and incubated for 10 min. For equilibration and to remove non-adhering cells by aspiration, spheroplasts were washed with a 2x SSC buffer (pH 7) and, afterwards, incubated in a humid chamber overnight at 37°C with hybridization buffer containing 50% formamide, 10% dextran sulphate sodium salt from Leuconostoc ssp. (Fluka), 125 µg/ml E. coli MRE600 tRNA (Boehring Mannheim GmbH), 500 µg/ml salmon sperm DNA sodium salt (AppliChem), 4x SSC, 1x Denhardt solution (Invitrogen) and approximately 0.8 pmoles of a Cy3-labelled ITS1-specific probe (5′-Cy3-ATGCTCTTGCCAAAACAAAAAAATCCATTTTCAAAATTATTAAATTTCTT-3′). After probing, spheroplasts were washed once with 200 ml 2x SSC, with 1x SSC and, finally, incubated with 200 ml 0.5x SSC containing 5 µg DAPI. After nuclear staining with DAPI, spheroplasts were washed twice with 0.5x SSC and the wells were dried and layered with Mowiol before the microscopy slides were covered with coverslips. Cells were imaged by fluorescence microscopy using either an Imager Z1 microscope (Carl Zeiss) with Plan-Apo-Chromat 100 oil immersion lens and a DICIII, 4’,6-diamidino-2-phenylindole, and a HECy3 filter, or a Leica DM6 B microscope, equipped with a DFC 9000 GT camera, using the PLAN APO 100x objective and narrow band TXR and LDA filters and the LasX software.
Sucrose gradient analysis
Cells were grown in 70 ml SDC-leu medium at 30°C to an OD600 of ~0.5–0.7 (log-phase). Cycloheximide (CHX) was added to 50 ml culture in a final concentration of 100 µg/ml, and cells were incubated on ice for 5 min. After harvesting, cells were resuspended in lysis buffer containing 10 mM HCl-Tris (pH 7.5), 100 mM NaCl, 30 mM MgCl2 and 100 µg/ml CHX. After mechanical cell lysis using glass beads, 7 A260 units of the cell extracts were loaded onto 5–35% sucrose gradients containing 50 mM HCl-Tris (pH 7.5), 50 mM NaCl and 10 mM MgCl2 and centrifuged at 38,000 rpm at 4°C for 2 h 45 min using a Beckman OptimaTM LE-80 K Ultracentrifuge. Gradients were analysed using a UA-6 system (Teledyne Isco) with continuous monitoring at A254 nm.
Northern blotting
For the analysis of polysome profile fractions by northern blotting, RNA from the fractions (approximately 500 µl) was extracted by mixing three times with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1). RNA was precipitated as described below.
For the analysis of total RNA by northern blotting, cells were grown in 50 ml SDC-leu medium at 30°C to an OD600 of 0.1–0.2 and shifted for 3 h to 25°C, 30°C or 37°C, respectively. Cells were resuspended in 200 µl lysis buffer containing 10 mM HCl-Tris (pH 7.5), 10 mM EDTA, and 0.5% SDS and mechanically lysed with 200 µl glass beads (0.5 mm diameter) for 3 min. After the addition of 450 µl lysis buffer, RNA was extracted by adding four times phenol-chloroform-isoamyl alcohol (25:24:1) and once chloroform-isoamyl alcohol (24:1) to the samples. RNA was precipitated by the addition of 1/10 volumes of 3 M sodium acetate (pH 5.2), 2.5 volumes of 100% ethanol and 1 µl GlycoBlueTM co-precipitant (Invitrogen) and, after drying, dissolved in nuclease-free water. Three µg of the isolated RNA were separated on 1.6% MOPS-agarose gels containing 20 mM 3-(N-morpholino)-propanesulfonic acid (MOPS), 5 mM sodium acetate, 1 mM EDTA, 0.75% formaldehyde and ethidium bromide (pH 7.0), transferred overnight onto Hybond N+ nylon membranes (Amersham Biosciences) by capillary transfer and UV cross-linked to the membrane. Except for the probe E/C2 (27S A + B): 5′-GGCCAGCAATTTCAAGTTA-3′, which was hybridized at 37°C, hybridization with the following 5′-32P-radiolabeled oligonucleotides was performed at 42°C overnight in buffer containing 0.5 M Na2HPO4, pH 7.2, 7% SDS, and 1 mM EDTA: probe D/A2 (20S/21S): 5′-GACTCTCCATCTCTTGTCTTCTTG-3′, probe A2A3 (35S, 32S/33S, 27SA2, 23S, 21S): 5′-TGTTACCTCTGGGCCC-3′, probe 18S: 5′-GCATGGCTTAATCTTTGAGAC-3′, probe 25S: 5′-CTCCGCTTATTGATATGC-3′). After three subsequent washing steps with a buffer containing 40 mM Na2HPO4, pH 7.2, 1% SDS, signals were detected by exposing X-ray films. Membranes were regenerated by washing in 1% SDS prior to hybridization.
Tsr1-TAP purification
The Tsr1-TAP Δrps15 strains carrying the LEU2-plasmids expressing RPS15, rps15-1-134 or rps15-RTH/77-79)>A, respectively, were grown at 30°C in 4 l YPD each to an OD600 of 2. TAP purifications were performed in a lysis buffer containing 50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 1.5 mM MgCl2, 0.075% NP-40 and 1 mM dithiothreitol (DTT). Prior to use, 1× Protease Inhibitor Mix FY (Serva) was added freshly to the lysis buffer. Cells were lysed by mechanical disruption using glass beads and the lysate was incubated with 300 µL IgG Sepharose™ 6 Fast Flow (GE Healthcare) at 4°C for 60 min. After incubation, beads were transferred into Mobicol columns (MoBiTec) and washed with buffer. Elution from IgG Sepharose™ beads was performed via TEV protease under rotation at room temperature for 70 min. A final concentration of 2 mM CaCl2 was added to the TEV eluates and they were then incubated with 300 µL Calmodulin Sepharose™ 4B (GE Healthcare) at 4°C for 60 min, washed with lysis buffer containing 2 mM CaCl2 and finally eluted with 5 mM EGTA. Protein samples were TCA-precipitated and dissolved in an SDS sample buffer, separated on NuPAGE™ 4–12% Bis–Tris gels (Invitrogen) and analysed via western blotting.
Western blotting
Western blot analysis was performed using the following antibodies: α-CBP antibody (1:5000; Merck–Millipore, cat. no. 07–482), α-Rps3 antibody (1:50,000; provided by Matthias Seedorf), α-Nob1 antibody (1:3000; provided by David Tollervey), horseradish peroxidase conjugated α-HA antibody (1:5000; Roche, cat. no. 12,013,819,001), α-GAPDH antibody (1:40,000; Cell Signalling Technology, cat. no. 2118S), secondary α-rabbit horseradish peroxidase-conjugated antibody (1:15,000; Sigma, cat. no. A0545). Protein signals were visualized using the Clarity™ Western ECL Substrate Kit (Bio-Rad) and captured by Chemi-Doc™ Touch Imaging System (Bio-Rad).
Quantification of translation accuracy
To measure the rate of amino acid misincorporation, the appropriate strains were transformed with a dual-luciferase reporter plasmid generously provided by David M. Bedwell (see Table S2). Luciferase activities were measured as previously described [Citation49] using the Dual-Glo® Luciferase Assay System (Promega). Cells from each strain were grown in liquid SDC-ura medium to mid-log phase at 30°C and firefly and Renilla luciferase luminescence levels were measured at room temperature with a CLARIOstar 1.20 microplate reader (BMG Labtech, Germany) adjusted to endpoint read-type and default settings. Assays were repeated four times (biological replicas), each of these was replicated three times (technical replicas), and the data were expressed as the mean ± the standard deviation. Error rates for each strain were calculated as the percentage of the firefly/Renilla luciferase activity (mutant plasmid) divided by the firefly/Renilla luciferase activity (wild-type plasmid).
Author contributions:
B.P., E.H., J.d.l.C. and I.R. designed the experiments and interpreted the results. I.R., S.W., B.P., D.S., J.F.-F. and S.M.-V. constructed strains and plasmids. I.R., S.W., J.F.-F., S.M.-V. and B.P. analysed genetic interactions. I.R. and B.P. performed northern blotting and microscopy and I.R., B.P. and S.W. performed FISH experiments. Sucrose gradients were analysed by I.R., S.W. and B.P. Translational misreading assays were performed by J.F.-F., and S.M.-V. I.R. and B.P. wrote the initial manuscript. All authors commented on the manuscript.
Supplemental Material
Download PDF (11.9 MB)Acknowledgments
We thank Selene Cordeiro and Matthias Diepold for their experimental help in the initial phase of the project. We thank D. M. Bedwell for plasmids pDB688 and pDB868.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
Notes
1. Recently, a new nomenclature for ribosomal proteins was introduced (Ban et al., 2014). In this publication, the standard nomenclature is used, and the new nomenclature is additionally indicated upon first mentioning of a r-protein.
References
- Klinge S, Woolford JL. Ribosome assembly coming into focus. Nat Rev Mol Cell Biol. 2019;20(2):116–131.
- Kressler D, Hurt E, Baßler J. A Puzzle of Life: crafting Ribosomal Subunits. Trends Biochem Sci. 2017;42(8):640–654.
- Baßler J, Hurt E. Eukaryotic Ribosome Assembly. Annu Rev Biochem. 2019;88(1):281–306.
- Du Y, An W, Zhu X, et al. Cryo-EM structure of 90S small ribosomal subunit precursors in transition states. Science. 2020;369(6510):1477–1481.
- Cheng J, Lau B, La Venuta G, et al. 90S pre-ribosome transformation into the primordial 40S subunit. Science. 2020;369(6510):1470–1476.
- Cerezo E, Plisson-Chastang C, Henras AK, et al. Maturation of pre-40S particles in yeast and humans. Wiley Interdiscip Rev RNA. 2019;10(1):e1516.
- Moy TI, Silver PA. Requirements for the nuclear export of the small ribosomal subunit. J Cell Sci. 2002;14(14):2985–2995.
- Faza MB, Chang Y, Occhipinti L, et al. Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet. 2012;8(8):e1002915.
- Fischer U, Schäuble N, Schütz S, et al. A non-canonical mechanism for Crm1-export cargo complex assembly. Elife. 2015;4:e05745.
- Peña C, Schütz S, Fischer U, et al. Prefabrication of a ribosomal protein subcomplex essential for eukaryotic ribosome formation. Elife. 2016;5. DOI:10.7554/eLife.21755.
- Léger-Silvestre I, Caffrey JM, Dawaliby R, et al. Specific Role for Yeast Homologs of the Diamond Blackfan Anemia-associated Rps19 Protein in Ribosome Synthesis. J Biol Chem. 2005;280(46):38177–38185.
- Léger-Silvestre I, Milkereit P, Ferreira-Cerca S, et al. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 2004;23(12):2336–2347.
- Seiser RM, Sundberg AE, Wollam BJ, et al. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics. 2006;174(2):679–691.
- Oeffinger M, Dlakić M, Tollervey D. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 2004;18(2):196–209.
- Strunk BS, Loucks CR, Su M, et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333(6048):1449–1453.
- McCaughan UM, Jayachandran U, Shchepachev V, et al. Pre-40S ribosome biogenesis factor Tsr1 is an inactive structural mimic of translational GTPases. Nat Commun. 2016;7(1):11789.
- Heuer A, Thomson E, Schmidt C, et al. Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae. Elife. 2017;6:e30189.
- Scaiola A, Peña C, Weisser M, et al. Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. EMBO J. 2018;37(7). DOI:10.15252/embj.201798499.
- Schäfer T, Maco B, Petfalski E, et al. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441(7093):651–655.
- Mitterer V, Murat G, Réty S, et al. Sequential domain assembly of ribosomal protein S3 drives 40S subunit maturation. Nat Commun. 2016;7(1):10336.
- Pertschy B. When a ribosomal protein grows up - the ribosome assembly path of Rps3. Microb Cell. 2017;4(5):140–143.
- Knüppel R, Christensen RH, Gray FC, et al. Insights into the evolutionary conserved regulation of Rio ATPase activity. Nucleic Acids Res. 2018;46(3):1441–1456.
- Ferreira-Cerca S, Sagar V, Schäfer T, et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat Struct Mol Biol. 2012;19(12):1316–1323.
- Mitterer V, Shayan R, Ferreira-Cerca S, et al. Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism. Nat Commun. 2019;10(1):2754.
- Strunk BS, Novak MN, Young CL, et al. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150(1):111–121.
- Lebaron S, Schneider C, van Nues RW, et al. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol. 2012;19(8):744–753.
- García-Gómez JJ, Fernández-Pevida A, Lebaron S, et al. Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3. PLoS Genet. 2014;10(3):e1004205.
- Parker MD, Collins JC, Korona B, et al. A kinase-dependent checkpoint prevents escape of immature ribosomes into the translating pool. PLoS Biol. 2019;17(12):e3000329.
- Ameismeier M, Cheng J, Berninghausen O, et al. Visualizing late states of human 40S ribosomal subunit maturation. Nature. 2018;558(7709):249–253.
- Pertschy B, Schneider C, Gnädig M, et al. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284(50):35079–35091.
- Ferreira-Cerca S, Pöll G, Gleizes P-E, et al. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell. 2005;20(2):263–275.
- Pöll G, Braun T, Jakovljevic J, et al. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PloS One. 2009;4(12):e8249.
- de la Cruz J, Karbstein K, Woolford JL. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem. 2015;84(1):93–129.
- Jakovljevic J, de MPA, Miles TD, et al. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol Cell. 2004;14(3):331–342.
- Neueder A, Jakob S, Pöll G, et al. A local role for the small ribosomal subunit primary binder rpS5 in final 18S rRNA processing in yeast. PloS One. 2010;5(4):e10194.
- Bellemer C, Chabosseau P, Gallardo F, et al. Genetic interactions show the importance of rRNA modification machinery for the role of Rps15p during ribosome biogenesis in S. cerevisiae. PloS One. 2010;5(5):e10472.
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529.
- Bowen AM, Musalgaonkar S, Moomau CA, et al. Ribosomal protein uS19 mutants reveal its role in coordinating ribosome structure and function. Translation (Austin). 2015;3(2):e1117703.
- Bulygin K, Malygin A, Gopanenko A, et al. The functional role of the C-terminal tail of the human ribosomal protein uS19. Biochim Biophys Acta Gene Regul Mech. 2020;3(3):194490.
- Babaylova ES, Gopanenko AV, Bulygin KN, et al. mRNA regions where 80S ribosomes pause during translation elongation in vivo interact with protein uS19, a component of the decoding site. Nucleic Acids Res. 2020;48(2):912–923.
- Bhaskar V, Graff-Meyer A, Schenk AD, et al. Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes. Cell Rep. 2020;31(1):107473.
- Ljungström V, Cortese D, Young E, et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood. 2016;127(8):1007–1016.
- Bretones G, Álvarez MG, Arango JR, et al. Altered patterns of global protein synthesis and translational fidelity in RPS15-mutated chronic lymphocytic leukemia. Blood. 2018;132(22):2375–2388.
- Yu L, Kim HT, Kasar S, et al. Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation. Clin Cancer Res. 2017;23(3):735–745.
- Shayan R, Rinaldi D, Larburu N, et al. Good Vibrations: structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy. Molecules. 2020;25(5):1125.
- Lacombe T, García-Gómez JJ, de la Cruz J, et al. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol. 2009;72(1):69–84.
- Martín-Villanueva S, Fernández-Fernández J, Rodríguez-Galán O, et al. Role of the 40S beak ribosomal protein eS12 in ribosome biogenesis and function in Saccharomyces cerevisiae. RNA Biol. 2020;17(9):1261–1276.
- Fernández-Pevida A, Martín-Villanueva S, Murat G, et al. The eukaryote-specific N-terminal extension of ribosomal protein S31 contributes to the assembly and function of 40S ribosomal subunits. Nucleic Acids Res. 2016;44(16):7777–7791.
- Salas-Marco J, Bedwell DM. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol. 2005;348(4):801–815.
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, et al. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19(6):560–567.
- Ameismeier M, Zemp I, van den Heuvel J, et al. Structural basis for the final steps of human 40S ribosome maturation. Nature. 2020;587(7835):683–687.
- Sun Q, Zhu X, Qi J, et al. Molecular architecture of the 90S small subunit pre-ribosome. Elife. 2017;6:e22086.
- Campbell MG, Karbstein K, Kobe B. Protein-protein interactions within late pre-40S ribosomes. PloS One. 2011;6(1):e16194.
- Gerhardy S, Oborská-Oplová M, Gillet L, et al. Puf6 primes 60S pre-ribosome nuclear export at low temperature. Nat Commun. 2021;12(1):4696.
- Stokes JM, Davis JH, Mangat CS, et al. Discovery of a small molecule that inhibits bacterial ribosome biogenesis. Elife. 2014;3:e03574.
- Huang H, Ghalei H, Karbstein K. Quality control of 40S ribosome head assembly ensures scanning competence. J Cell Biol. 2020;219(11). DOI:10.1083/jcb.202004161
- Ghalei H, Trepreau J, Collins JC, et al. The ATPase Fap7 Tests the Ability to Carry Out Translocation-like Conformational Changes and Releases Dim1 during 40S Ribosome Maturation. Mol Cell. 2017;67(6):990‐1000.e3.
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8(2):79–82.
- Santos-Rosa H, Moreno H, Simos G, et al. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18(11):6826–6838.
- Kressler D, Doère M, Rojo M, et al. Synthetic lethality with conditional dbp6 alleles identifies Rsa1p, a nucleoplasmic protein involved in the assembly of 60S ribosomal subunits. Mol Cell Biol. 1999;19(12):8633–8645.
