ABSTRACT
Cervical cancer (CC) is one of the most prevalent malignancies among females. Cytoprotective autophagy could confer cancer cell tolerance to hypoxic stress, promoting cell survival and adaptation. Aspartyl-tRNA synthetase 1 antisense 1 (DARS-AS1) is an oncogenic long non-coding RNA (lncRNA) in various cancers, but how DARS-AS1 regulates cytoprotective autophagy in hypoxic environment in CC remains unclear. Chromatin immunoprecipitation (ChIP) and luciferase reporter assays were conducted to explore the interaction between hypoxia-inducible factor 1 subunit alpha (HIF1α) and DARS-AS1 promoter. Methylated RNA immunoprecipitation (MeRIP) followed by quantitative real-time polymerase-chain reaction (RT-qPCR) detected methylated RNA level. The process of autophagic maturation was monitored by immunofluorescence staining. Higher DARS-AS1 expression was found in CC tissues and cytoprotective. We also uncovered that hypoxic exposure induced cytoprotective autophagy via HIF1α/DARS-AS1/DARS axis. Moreover, DARS-AS1 was validated to facilitate DARS translation via recruiting N6-adenosine-methyltransferase methyltransferase like 3 (METTL3) and methyltransferase like 14 (METTL14), which bound with DARS mRNA DARS mRNA 5’ untranslated region (5ʹUTR) and promoting its translation. The present study demonstrated that the ‘HIF1α/DARS-AS1/DARS/ATG5/ATG3’ pathway regulated the hypoxia-induced cytoprotective autophagy of CC and might be a promising target of therapeutic strategies for patients afflicted with CC.
Introduction
Cervical cancer (CC) is one of the most frequent gynaecological malignancies in the world and has emerged as a prominent public health focus. Based on 2018 statistics, there are over 500,000 new cases and 310,000 deaths globally each year [Citation1,Citation2]. Despite the improvement in diagnostic and therapeutic strategies, the 5-year survival rate of CC patients is still poor [Citation3]. One of the major risk factors for clinically induced CC is high-risk human papillomavirus (HPV) persistence [Citation4]. However, recent evidences have demonstrated that HPV infection alone is insufficient to trigger malignant transformation of normal cervical cells [Citation5]. As a result, a better understanding of the molecular mechanism implicated in CC progression is essential to find more tumour-specific molecular markers.
Hypoxia is a familiar phenomenon in solid tumour microenvironment [Citation6]. Accumulating studies have displayed that hypoxia is involved in a variety of tumour progression, such as angiogenesis and aggressiveness chemotherapy resistance [Citation7]. Hypoxia-inducible factor-1 (HIF-1) is an important modulator, which maintains oxygen balance [Citation8]. Furthermore, hypoxia-inducible factor 1 subunit alpha (HIF1α) is a vital transcription factor in hypoxia and has been widely documented in many human cancers, including colorectal cancer [Citation9], breast cancer [Citation10], and non-small cell lung cancer [Citation11]. Moreover, it has been registered that hypoxia can trigger autophagy through regulating HIF1α, and promote tumour progression.
Autophagy is a crucial intracellular process related to the degradation and recycling of cytosolic material, which is essential for maintaining cellular biosynthesis [Citation12]. A large number of evidences have demonstrated that autophagy plays an important role in protecting cells to survival by holding back apoptosis under multiple metabolic stress microenvironments including hypoxia or oxidative stress [Citation13]. Among the autophagy (ATG)-related genes, the microtubule-associated protein 1A/1B-light chain 3 (LC3), one of the ATG8 homologs in mammals, has been identified as an autophagosome marker and many ATG genes have been knocked out for understanding the physiological role of autophagy in mammals [Citation14]. Despite the extension of our knowledge about autophagy, the mechanisms underlying the abnormal activation of autophagy and the specific effect of autophagy on CC cells under the hypoxia condition remain largely obscure.
N6-Methyladenosine (m6A) is a common message RNA (mRNA) modification that affects gene expression by modulating RNA processing, localization, and translation, all of which can be controlled by ‘writers’, ‘erasers’ and ‘readers’ of epigenetic mark [Citation15]. Methyltransferase like 3 (METTL3) and methyltransferase-like 14 (METTL14) are the important component of ‘writers’ complex, are implicated in the regulation of mRNA stability or translation [Citation16].
Long non-coding RNAs (lncRNAs) are a type of RNAs longer than 200 nucleotides with limited or no protein-coding capacity [Citation17]. LncRNAs can influence gene expression through many ways, such as epigenetic, transcriptional, or post-transcriptional regulations [Citation18]. A growing body of studies has demonstrated that lncRNAs participate in a variety of biological processes including cell proliferation, apoptosis, and autophagy [Citation19]. LncRNA aspartyl-tRNA synthetase 1 antisense RNA 1 (DARS-AS1) has been documented to act as oncogenes in many cancers, such as ovarian cancer [Citation20] and non-small cell lung cancer [Citation21]. However, the role of DARS-AS1 in CC remains largely unclear.
In a nutshell, our study was intended for investigating the function and potential regulatory mechanism of DARS-AS1 in CC. We determined the impacts of DARS-AS1 on CC cell autophagy under hypoxia treatment. Specifically, DARS-AS1 was determined to enhance DARS mRNA stability and facilitate DARS translation. DARS-AS1 modulated DARS translation via METTL3- and METTL14-mediated m6A methylation. Our findings might provide a novel perspective for understanding the molecular mechanism in CC, thus contributing to the discovery of potential effective biomarkers for CC diagnosis and treatment.
Material and methods
Cell culture and treatment
CC cells (SiHa, CaSki, C33A, DoTc2-4510 and HeLa) and human normal cervical epithelial immortalized cells (End1/E6E7) were supplied from American Type Culture Collection (ATCC, Manassas, VA, USA). SiHa, C33A and HeLa cells were grown in Eagle’s Minimum Essential Medium. CaSki cells were grown in Roswell Park Memorial Institute (RPMI) 1640 Medium. DoTc2-4510 cells were grown in Dulbecco’s Modified Eagle’s Medium. End1/E6E7 cells were grown in Keratinocyte Serum Free Medium. All cell mediums were added with 10% foetal bovine serum (FBS, Gibco, USA) in an incubator with 5% CO2 at 37°C. For hypoxia stimulation, the hypoxic incubator (SANYO, Osaka, Japan) including 94% N2, 5% CO2 and 1% O2 was used for culturing HeLa and C33A cells for indicated time periods.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was isolated by TRI reagent (Sigma, USA), and then reversely transcribed using cDNA Synthesis kit (Takara). RT-qPCR was performed using SYBR Green PCR Master Mix (Takara). Gene expression was normalized to Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using 2−ΔΔCt method. All primer sequences were listed in Supplementary file 1.
Cell transfection
Short hairpin RNAs (shRNAs) including sh-HIF1A, sh-DARS-AS1, sh-DARS, sh-METTL3 and sh-METTL14 were generated for knockdown of respective genes. Full sequences of HIF1A, DARS-AS1, ATG3, ATG5, DARS, Fat mass and obesity associated (FTO), and AlkB homolog 5 (ALKBH5) were cloned into the pcDNA3.1 expression vector for overexpression. All these plasmids were purchased from Genepharma (Shanghai, China), and transfected using Lipofectamine 3000 reagent (Invitrogen, CA, USA).
Chromatin immunoprecipitation (ChIP)
ChIP assay was conducted using a ChIP assay kit (Beyotime). Cells were cross-linked and sonicated to about 200–500 basepairs. 1% agarose gel with a 100 bp DNA marker was applied to determine DNA fragment size. Cell lysates were incubated with protein A/G beads coated with the Anti-HIF1α antibody. Immunoglobulin G antibody (Anti-IgG) and RNA polymerase II antibody (anti-RNA Pol II) were used as a negative control and a positive control. Cross-linked DNA was purified and eluted, followed by RT-qPCR. Relevant primer sequences were provided in Supplementary file 1.
Western blot
Protein was extracted using Radioimmunoprecipitation (RIPA) lysis buffer (Thermo Fisher). The concentration of extracted protein was determined by Bicinchoninic Acid (BCA) protein assay kit (Abcam, USA). After being separated by sodium dodecyl sulphate (SDS) gel electrophoresis, the proteins were transmitted onto polyvinylidene fluoride (PVDF) membranes (Roche, Basel, Switzerland). Then, the membranes were blocked with 5% skim milk and incubated with primary antibodies including Anti-HIF1α (Abcam, 1/500), Anti-p62 (Abcam, 1/10000), Anti-LC3 (Abcam, 1/2000), anti-GAPDH (Abcam, 1/1000), Anti-Autophagy Related 16 Like 1 (ATG16L1, Abcam, 1/1000), Anti-ATG16L2 (Abcam, 1/500), Anti-ATG12 (Abcam, 1/1000), Anti-ATG5 (Abcam, 1/1000), Anti-ATG8 (Abcam, 1/4000), Anti-ATG2 (Abcam, 1/500), Anti-Beclin 1 (BECN1, Abcam, 1/1000), Anti-ATG10 (Abcam, 1/1000), Anti-Sirtuin 1 (SIRT1, Abcam, 1/1000), Anti-ATG7 (Abcam, 1/100000), Anti-ATG3 (Abcam, 1/10000), Anti-ATG14 (Abcam, 2 µg/ml), Anti-DARS (Abcam, 1/1000), Anti-METTL3 (Abcam, 1/1000), Anti-METTL14 (Abcam, 1/500), Anti-FTO (Abcam,1/10000) and Anti-ALKBH5 (Abcam, 1/1000) at 4°C overnight. Then, the membranes were washed and incubated with the corresponding secondary antibody for 2 h. Finally, proteins were detected with the enhanced chemiluminescence (ECL) method (Thermo Fisher Scientific, USA). The raw blot data were presented in Supplementary file 4.
Luciferase reporter assay
To confirm the binding between HIF1α and DARS-AS1 promoter, we co-transfected the cells with DARS-AS1 promoter-wide type (Wt), mutant DARS-AS1 promoter-1 (DARS-AS1 promoter-Mut-1), DARS-AS1 promoter-Mut-2 or DARS-AS1 promoter-Mut-3 (1 + 2), pGL3 luciferase reporter constructs harbouring the HIF1α response element target sequence and vector or HIF1A. Similarly, pGL3 luciferase reporters sub-cloned with DARS promoter was co-transfected with sh-NC or sh-DARS-AS1 into hypoxia treated CC cells. After 48 h, the luciferase activities were measured using a luciferase assay system (Promega, WI, USA).
Cell Counting Kit-8 (CCK-8)
Cell viability was detected by CCK-8 assay. Cells were grown in 96-well plates in an incubator (37°C, 5% CO2). Then, 10 μl CCK-8 solutions were added to each well for incubation. The absorbance was measured at 450 nm under a microplate spectrophotometer.
Evaluation of fluorescent LC3 puncta
Autophagic flux was examined by assessing fluorescent LC3 puncta after transfection with red fluorescent protein-green fluorescent protein-LC3 (RFP-GFP-LC3) (Hanbio, Inc., Shanghai, China) into HeLa cells under normoxia or hypoxia treatment. Then, cells were washed and fixed. The distribution of GFP and RFP puncta was detected under a confocal fluorescence microscope and visualized by ImageJ (National Institutes of Health, Bethesda, MD, USA).
Subcellular fractionation
Subcellular fractionation assay was performed using a PARIS™ Kit (Thermo Fisher). The cytoplasmic and nuclear RNA were analysed by RT-qPCR. GAPDH and U6 small nuclear RNA (U6) were used as cytoplasmic and nuclear controls, respectively.
Fluorescent in situ hybridization (FISH) and immunofluorescence
FISH was performed with DARS-AS1 specific probe (Biosearch Technologies). Cells were fixed and then incubated with DARS-AS1 probe overnight at 37°C. After washing and sealing, cells were incubated with DARS antibody for 1 h. Next, cells were washed and incubated with secondary antibodies conjugated with Alexa Fluor® 488. Cells were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) and imaged with a confocal microscope.
RNA binding protein immunoprecipitation (RIP)
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) was used for RIP assays, with the manufacturer’s instructions strictly followed. Cells were lysed and then incubated with magnetic beads conjugated to anti-METTL3 or anti-MRTTL14 or the negative control IgG. The immunoprecipitated RNA was isolated and analysed by RT-qPCR for assessing the enrichment of target genes. Relevant sequences were provided in Supplementary file 1.
Quantification of cellular m6A level
The EpiQuick™ RNA Methylation Quantification Kit (Epigentek, USA) was used to quantify the m6A level in the cellular RNA. The relative m6A amount was measured with a spectrophotometer at 450 nm.
Methylated RNA immunoprecipitation (MeRIP) assay
m6A RNA immunoprecipitation was performed to quantify m6A level in the DARS 5’ untranslated region (5ʹUTR). Cells were cultured with anti-m6A antibody and anti-IgG antibody conjugating to magnetic beads overnight. The eluted RNA was detected using RT-qPCR.
Actinomycin D assay
Cells were treated with actinomycin D at a final concentration of 4 μM. The actinomycin D was added to cells 0 h, 3 h, 6 h, 9 h, 12 h and 15 h prior to RNA extraction. RT-qPCR was used to analyse RNA levels.
RNA pull down assay
DARS-AS1, DARS 3ʹUTR, wild-type or mutant-type DARS 5ʹUTR were in vitro transcribed with Biotin RNA Labelling Mix (Roche, Switzerland), and incubated with cell lysates overnight at 4°C. The eluted proteins were purified and detected by western blot.
Sucrose density gradient assay
Sucrose density gradient analysis was implemented as previously described [Citation22] for detecting the distribution of DARS mRNA in cell extracts. The corresponding cytosolic extracts were sedimented by centrifugation on 10–50% sucrose gradients.
Statistical analysis
The above experiments were repeated at least three times and all data were demonstrated as mean ± standard deviation (SD) and analysed using SPSS17.0 software. The differences were estimated by Student’s t-test or analysis of variance (ANOVA). P value < 0.05 was considered to indicate statistical significance.
Results
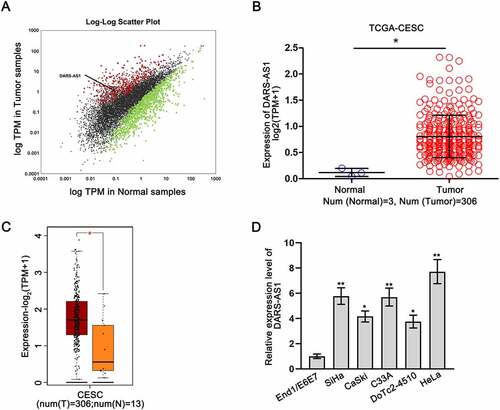
DARS-AS1 is up-regulated in CC
To probe the association between lncRNAs and CC, we utilized RNA sequencing dataset to analyse the expression of numerous lncRNAs in tumourous versus non-tumourous samples from The Cancer Genome Atlas Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA-CESC). A total of 1464 lncRNAs showed differential expression in 306 cancerous tissues compared with 3 normal tissues, among which 365 lncRNAs were up-regulated and 1099 lncRNAs were down-regulated (). We focused on DARS-AS1 (Enzemble ID: ENSG00000231890, NR_110199.1), which exhibited a significant up-regulation in CESC samples () and has been characterized as an oncogene in many cancers, such as thyroid cancer [Citation23] and ovarian cancer [Citation24]. Based on GEPIA 2 (http://gepia2.cancer-pku.cn) database, we found that DARS-AS1 exhibited a high expression level in 306 cases of CESC tissues relative to 13 controls of normal tissues (). Moreover, the expression level of DARS-AS1 was detected by RT-qPCR in CC cells (SiHa, CaSki, C33A, DoTc2-4510, and HeLa) and human normal cervical epithelial immortalized cells (End1/E6E7). Our results indicated that DARS-AS1 expression was higher in CC cells than in End1/E6E7 cells (). All these data suggested that DARS-AS1 was up-regulated in CC.
Figure 1. DARS-AS1 is up-regulated in CC. A Scatter plot illustrated the differential expression of lncRNAs in CESC. B-C DARS-AS1 expression was analysed in CESC and normal tissues from TCGA and GEPIA 2 databases. D DARS-AS1 expression was measured in CC cells via RT-qPCR. Experimental procedures were repeated independently for three times. *P < 0.05, **P < 0.01.
Hypoxia-induced up-regulation of DARS-AS1 is transcriptionally mediated by hypoxia inducible factor HIF1α
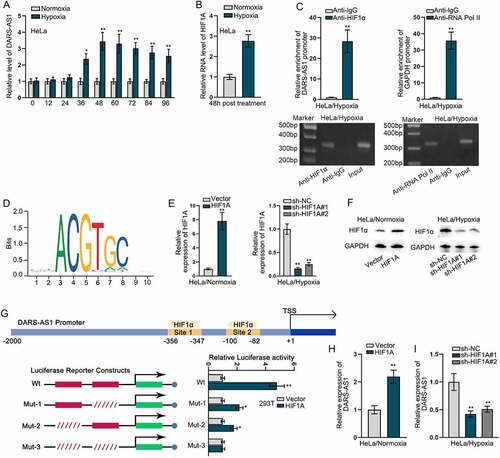
We next explored which factors induced high DARS-AS1 expression in CC. Hypoxia is a pervasive feature of solid tumours and plays critical roles in tumour development [Citation6]. Recent studies have displayed that aberrant expression of some lncRNAs is attributed to hypoxia microenvironment of cancers [Citation25]. Thus, we mimicked a hypoxic cell model with a gradient treatment time, and then measured the expression of DARS-AS1 using RT-qPCR. As revealed in , DARS-AS1 expression reached the peak after 48-hour exposure to hypoxia, but remained unchanged in subsequent 48 hours. Hence, we selected 48 hours for duration of hypoxic exposure. Previous studies have revealed that cellular response to hypoxia is orchestrated by hypoxia-inducible factors (HIFs) that activate transcription of genes involved in pathogenesis of cancers [Citation26,Citation27], among which HIF1α is well known and widely studied in cancers [Citation28–30]. Thus, we supposed that HIF1α might be responsible for the transcriptional induction of DARS-AS1 upon exposure to oxygen-deficient environment. Through RT-qPCR analysis, we found that HIF1A level was obviously elevated upon exposure to hypoxia (). ChIP assay further revealed a marked binding affinity of HIF1α with the promoter of DARS-AS1, using RNA Pol II as a positive control, as the combination between RNA Pol II and gene promoter is an essential step in transcriptional regulation (). As HIFs sequence-specifically bind to consensus HIF1α-response elements (HREs) in the promoter of downstream genes [Citation31], two putative HREs were identified in the promoter region (−2000 to −1) of DARS-AS1 by the use of conserved HIF1α motif () from JASPAR database (http://jaspar.genereg.net). To confirm whether HIF1α transcriptionally regulated DARS-AS1 through these two HREs (binding sites), we stably overexpressed HIF1α in HeLa cells under normoxia and silenced HIF1α in HeLa cells under hypoxia (), and then constructed luciferase activity reporters harbouring the promoter region of DARS-AS1 ( upper). As expected, ectopic HIF1α increased luciferase activity of wild-type reporter, as well as Mut-1 and Mut-2 reporters, but had no effect on that of luciferase reporter with two simultaneously mutated HREs ( lower), validating the binding of HIF1α with both two predicted HREs. At the same time, we observed that DARS-AS1 expression was distinctly promoted by overexpressed HIF1A and was repressed after HIF1A deficiency (). Above data demonstrated that hypoxia-induced up-regulation of DARS-AS1 was dependent on HIF1α at transcriptional level.
Figure 2. Hypoxia-induced up-regulation of DARS-AS1 is transcriptionally mediated by hypoxia inducible factor HIF1α. A DARS-AS1 expression was detected in HeLa cells under hypoxia at different times using RT-qPCR. B HIF1A expression was detected in HeLa cells after 48-h hypoxia treatment using RT-qPCR. C ChIP-AGE assays detected the enrichment of DARS-AS1 promoter by HIF1α in HeLa cells under hypoxia. D JASPAR predicted the binding sites between DARS-AS1 promoter and HIF1α. E-F HIF1A overexpression and silencing efficiencies were verified by RT-qPCR and western blot. G Luciferase reporter assays detected the effectiveness of two binding sites between DARS-AS1 promoter and HIF1α by calculating the relative luciferase activity of DARS-AS1 promoter Wt/Mut-1/Mut-2/Mut-3 in cells with transfection of vector or HIF1A overexpression plasmids. H-I Impact of HIF1A overexpression or silence on DARS-AS1 expression was detected using RT-qPCR. Experimental procedures were repeated independently for three times. *P < 0.05, **P < 0.01.
Hypoxia-induced autophagy of CC cells is mediated by DARS-AS1/ATG5/ATG3 signalling
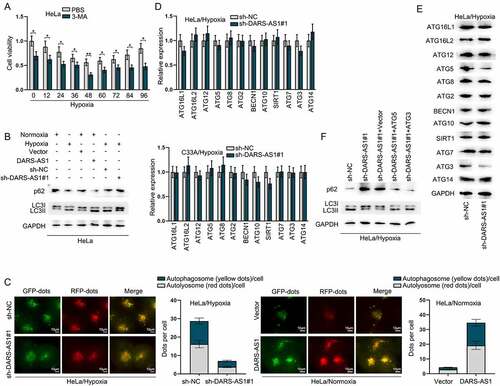
Hypoxia is able to trigger cytoprotective autophagy in tumour cells, thereby promoting cell survival and resistance to antitumor therapy, as evidenced by multiple published papers [Citation32–34]. We speculated that hypoxic treatment might induce CC cell protective autophagy via DARS-AS1 up-regulation. Firstly, we treated 1 mM autophagy inhibitor 3-MA into HeLa cells under hypoxia, followed by the detection of cell viability. CCK-8 results indicated that 3-MA significantly decreased the cell viability (), which confirmed that hypoxic treatment could induce CC cell protective autophagy. Then, we carried out western blot to analyse the protein levels of classical autophagic markers, namely, LC3II and p62 [Citation35]. As presented in , LC3II expression was higher while p62 expression was lower in hypoxia treated cells compared with cells under normoxia treatment, and DARS-AS1 overexpression led to increase in LC3II expression but decrease in p62 expression. Moreover, LC3II expression was reduced and p62 expression was enhanced in hypoxia treated cells after DARS-AS1 depletion. To further dynamically monitor the maturation of autophagy flux and clarify the impacts of hypoxia and DARS-AS1 on the autophagy of cells, we next transfected a tandem labelled GFP-mRFP-LC3 adenovirus into HeLa cells as described in previous autophagy studies [Citation36]. As shown in , clearly accumulated detectable yellow autophagic LC3 puncta (GFP+RFP+) was observed in HeLa cells under hypoxia treatment, which was lessened after sh-DARS-AS1 transfection. On the contrary, the yellow autophagic LC3 puncta was slightly found in HeLa cells under normoxia treatment, and then much enhanced by DARS-AS1 elevation. Herein, DARS-AS1 was validated to affect the autophagy of CC cells. It was learnt from previous research that lncRNA could regulate the expression of autophagy-related genes to affect autophagy [Citation37]. Thus, we utilized RT-qPCR and western blot to analyse the mRNA and protein levels of several autophagy-related genes in cells before and after DARS-AS1 depletion under hypoxia treatment. Intriguingly, we found that DARS-AS1 knockdown barely affected the mRNA levels of these autophagy-related genes, but could reduce the protein levels of ATG5 and ATG3 (), implying that DARS-AS1 could modulate the translation of ATG5 and ATG3. To check whether DARS-AS1 was truly involved in the autophagy via regulating ATG5 and ATG3 translation, we further used western blot to examine the protein levels of LC3II and p62. Data revealed that overexpressed ATG5 or ATG3 reversed the enhanced level of p62 as well as the inhibited LC3II protein level in DARS-AS1-depleted HeLa cells with hypoxia treatment (). Overall, hypoxia-induced autophagy of CC cells was mediated by DARS-AS1/ATG5/ATG3 signalling.
Figure 3. Hypoxia-induced autophagy of CC cell is mediated by DARS-AS1/ATG5/ATG3 signalling. A HeLa cell viability was detected at different times after hypoxia treatment in the presence of 3-MA via CCK-8. B Impacts of DARS-AS1 up-regulation or down-regulation on the protein levels of p62 and LC3 in HeLa cells treated under normoxia or hypoxia treatment were investigated via western blot. C Fluorescence images of GFP-mRFP-LC3 stably expressed in DARS-AS1-silenced HeLa cells under hypoxia or DARS-AS1-overexpressed HeLa cells under normoxia were observed. The puncta visible due to GFP (green) and RFP (red) revealed the formation of autophagosomes. D-E RT-qPCR and western blot measured the mRNA and protein levels of autophagy-related genes in DARS-AS1-silenced cells under hypoxia. F Western blot assay detected the protein levels of p62 and LC3II in hypoxia-treated HeLa cells after transfection of sh-DARS-AS1 and ATG5/3 on under hypoxia. Experimental procedures were repeated independently for three times. *P < 0.05, **P < 0.01.
DARS-AS1 regulates the expression of ATG5 and ATG3 via modulating DARS to affect CC cell autophagy
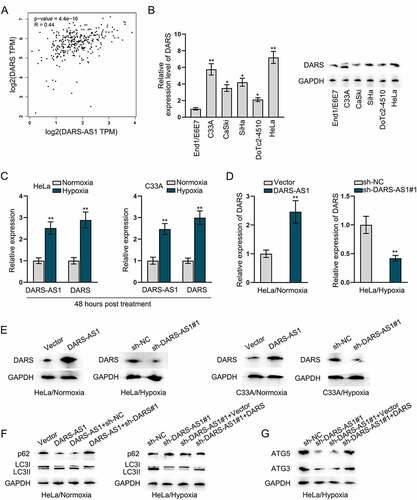
LncRNAs are proposed to play essential roles in regulating the expression of the nearby loci [Citation38]. To probe the interaction between DARS-AS1 and its nearby gene DARS in CC, we first employed GEPIA 2 database to analyse the correlation between their expression in CC tissues. As illustrated in , we found a positive correlation between DARS-AS1 and DARS expression in CC tissues (p-value = 4.4e-16, R = 0.44). RT-qPCR and western blot results further uncovered that the expression profile of DARS in CC cells was in line with that of DARS-AS1, as evidenced that DARS was up-regulated in 5 CC cell lines (). Moreover, we found that both DARS-AS1 and DARS expression was markedly promoted in HeLa and C33A cells after 48-h hypoxia treatment (), and we validated that DARS-AS1 could regulate DARS at both mRNA level and protein level (). In order to determine whether DARS-AS1 regulated DARS to affect CC cell autophagy, we applied western blot to detect the protein levels of LC3II and p62. We observed that in HeLa cells under normoxia treatment, the increase in LC3II protein level and decline in p62 protein level induced by DARS-AS1 overexpression was reversed after co-transfection of sh-DARS, while in HeLa cells under hypoxia treatment, the increased level of p62 and decreased LC3II due to DARS-AS1 knockdown was countered by DARS up-regulation (). DARS is a cytoplasmic aspartyl-tRNA synthetase gene [Citation39], and ATG5 and ATG3 possess a higher proportion of aspartic acid than other autophagy proteins [Citation40]. Thus, we assumed that DARS-AS1 might regulate the expression of ATG5 and ATG3 to affect CC cell autophagy via modulating DARS. Through western blot analysis, we found that DARS increase could neutralize the repressive effects of DARS-AS1 silence on ATG5 and ATG3 protein levels (). Additionally, the increased protein levels of ATG5 and ATG3 in response to DARS-AS1 augment were reversed by DARS knockdown (Figure S1A). Taken together, DARS-AS1 regulated the expression of ATG5 and ATG3 to affect CC cells autophagy via modulating DARS expression.
Figure 4. DARS-AS1 regulates the expression of ATG5 and ATG3 to affect CC cytoprotective autophagy via modulating DARS translation. A Correlation between DARS-AS1 and DARS expressions in CC tissues was obtained from GEPIA 2 database. B DARS expression in CC cells was analysed by RT-qPCR and western blot. C Expressions of DARS-AS1 and DARS were detected in HeLa and C33A cells treated with 48-h hypoxia. D-E DARS mRNA and protein levels were examined when DARS-AS1 was overexpressed or silenced in HeLa and C33A cells treated with normoxia or hypoxia. F LC3II and p62 protein levels were detected in different transfection groups. G Impacts of DARS-AS1 depletion and DARS overexpression on the protein levels of ATG3 and ATG5 in HeLa cells under hypoxia was analysed. Experimental procedures were repeated independently for three times. *P < 0.05, **P < 0.01.
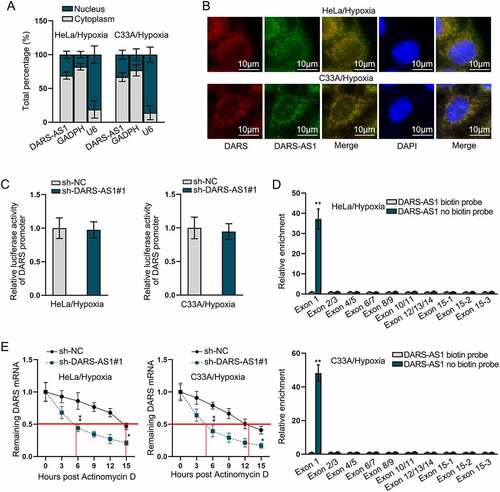
DARS-AS1 binds to DARS 5ʹUTR to regulate DARS mRNA stability and translation
To further explore the potential mechanism of DARS expression regulated by DARS-AS1, we first performed subcellular fractionation assay to identify the cellular location of DARS-AS1. As revealed in , DARS-AS1 was distributed both in nucleus and cytoplasm. In FISH assay, it was visually manifested that both DARS-AS1 and DARS were located in the nucleus and cytoplasm (). In the meantime, we found that DARS-AS1 could not influence the luciferase activity of DARS promoter, which excluded transcriptional regulation of DARS-AS1 on DARS (). Then, the binding between DARS-AS1 and DARS 5ʹUTR was displayed in Supplementary file 2. We also obtained all the positions of DARS exons, which were demonstrated in Supplementary file 3. RNA pull-down assay further validated that DARS-AS1 could bind to DARS at Exon 1 (). Previous study has revealed that 5ʹUTR plays a role in modulating mRNA stability and translation [Citation41], thus we supposed that DARS-AS1 might post-transcriptionally regulate DARS mRNA stability and translation. Subsequently, Actinomycin D assay validated that DARS-AS1 silence significantly reduced the stability of DARS mRNA (). From Figure S1B-C, sucrose density gradient assay was implemented for analysing DARS-AS1 influence on DARS translation, and based on the results, increases in DARS-AS1 level resulted in the changes of the distribution of DARS mRNA from light polysomes to heavy polysomes, indicating that DARS translation was enhanced by DARS-AS1 up-regulation, while DARS-AS1 depletion led to the opposite consequences. Collectively, DARS-AS1 bound to DARS 5ʹUTR to regulate DARS mRNA stability and translation.
Figure 5. DARS-AS1 binds to DARS 5ʹUTR to regulate DARS mRNA stability. A Subcellular fractionation assays identified the cellular location of DARS-AS1 in HeLa and C33A cells treated with hypoxia. B FISH assays detected the co-existence of DARS-AS1 and DARS in HeLa and C33A cells treated with hypoxia. C Luciferase activity of DARS promoter in DARS-AS1-silenced HeLa and C33A cells treated with hypoxia was analysed. D Combination between DARS 5ʹUTR and DARS-AS1 in hypoxia-treated cells was confirmed using RNA pull down assay. E Stability of DARS mRNA in DARS-AS1-silenced cells treated with hypoxia in the presence of actinomycin D was analysed. Experimental procedures were repeated independently for three times. *P < 0.05, **P < 0.01.
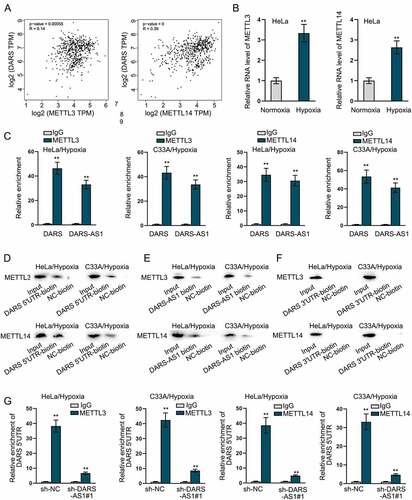
DARS-AS1 recruits METTL3 and METTL14 to enhance DARS translation
CeRNA network is one of the common post-transcriptional regulations. Through the StarBase (http://starbase.sysu.edu.cn) database, we found no miRNA combining with DARS-AS1 and DARS in common. However, 33 RBPs binding to DARS-AS1 were found out on starBase (Table S1). Of note, METTL3 and METTL14, RNA methyltransferases, have been suggested to mediate m6A modification which can modulate RNA translation and stability [Citation42]. Then, we first used GEPIA 2 database to analyse the correlations between METTL3 and DARS expression as well as METTL14 and DARS expression in CC tissues. As shown in , METTL3 and METTL14 expression was positively correlated with DARS expression, respectively (p-value = 0.00055, R = 0.14; p-value = 0, R = 0.39). Then, we found that both METTL3 and METTL14 were significantly up-regulated in HeLa cells treated with hypoxia (). In addition, RIP and RNA pull-down assays further validated the combination of METTL3 and METTL14 with DARS-AS1 as well as METTL3 and METTL14 with DARS (). More importantly, METTL3 and METTL14 bound to DARS 5ʹUTR rather than DARS 3ʹUTR (). However, these affinities were attenuated when DARS-AS1 was knocked down (). Additionally, we divided the sequence length of DARS and conducted RIP assay, after which we found that both METTL3 and METTL14 bound to DARS at exon 1 (Figure S1D). Then, we stably down-regulated METTL3 and METTL14 (Figure S2A-S2B) and utilized RT-qPCR and western blot to measure the mRNA and protein levels of DARS in hypoxia-treated cells. Data showed that METTL3 and METTL14 had no effects on DARS mRNA level, but significantly reduced the protein level of DARS (Figure S2C-S2E). Then, sucrose density gradient assay was implemented, and based on the experimental results, increases in METTL3 or METTL14 level contributed to changed distribution of DARS mRNA from light polysomes to heavy polysomes, indicating that DARS translation was enhanced by METTL3 or METTL14 up-regulation (Figure S2F). All these data suggested that DARS-AS1 recruited METTL3 and METTL14 to enhance DARS translation.
Figure 6. DARS-AS1 and DARS bind to METTL3 and METTL14. A GEPIA 2 database analysed the correlation between METTL3 and DARS expression and between METTL14 and DARS expression in CC tissues. B Expressions of METTL3 and METTL14 were detected in HeLa cells treated with normoxia or hypoxia. C-D RIP and RNA pull down assays assessed the combination of DARS-AS1/DARS with METTL3/METTL14. E RNA pull down experiments were carried out for evaluating the combination of DARS-AS1 and METTL3/MATTL14. F RNA pull down experiments were carried out for evaluating the combination of DARS 3ʹUTR and METTL3/MATTL14. G RIP experiments were performed for investigation into impact of DARS-AS1 knockdown on binding between DARS 5ʹUTR and METTL3 or MATTL14. Experimental procedures were repeated independently for three times. **P < 0.01.
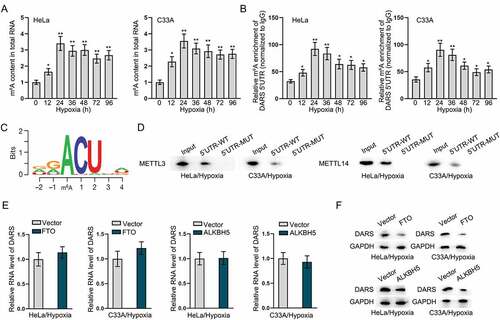
DARS translation is regulated by METTL3- and METTL14- mediated m6A methylation
To further determine whether DARS translation is mediated by m6A methylated modification, we first examined the m6A level of total RNA in HeLa and C33A cells under hypoxia treatment. Data indicated that the content of m6A in total RNA was significantly increased after hypoxia treatment (). Then, RIP assay measured the m6A enrichment of DARS 5ʹUTR in HeLa and C33A cells treated with hypoxia, and it was found that DARS 5ʹUTR bound to m6A (). Through JASPAR and starBase databases, we predicted the methylated conserved sequences and the binding sites between DARS 5ʹUTR and m6A modification (). RNA pull-down assay further confirmed that METTL3 and METTL14 bound to the DARS 5ʹUTR-WT rather than DARS 5ʹUTR-Mut (). Moreover, we overexpressed two famous m6A methylated modification erasers, namely, FTO and ALKBH5 (Figure S3A-S3D), and found that overexpression of FTO and ALKBH5 had no changes on DARS mRNA level but decreased DARS protein level (). Conclusively, these data suggested that DARS translation was regulated by METTL3- and METTL14-mediated m6A methylation.
Figure 7. DARS translation is regulated by METTL3- and METTL14- mediated m6A methylation. A Colorimetric method detected the m6A content in total RNA of HeLa and C33A cells treated with hypoxia. B RIP assays measured the m6A enrichment of DARS 5ʹUTR in HeLa and C33A cells treated with hypoxia. C JASPAR predicted the methylated conserved sequences and the binding sites between DARS 5ʹUTR and m6A modification. D RNA pull down assays verified the combination of METTL3/METTL14 and DARS 5ʹUTR-WT/Mut. E-F RT-qPCR and western blot examined the mRNA and protein levels of DARS in FTO- or ALKBH5-overexpressed HeLa and C33A cells treated with hypoxia. Experimental procedures were repeated independently for three times. *P < 0.05, **P < 0.01.
Discussion
Recent evidences have suggested that lncRNAs are important modulators in cell biology and lncRNA dysregulation is involved in a variety of human cancers, including CC [Citation43]. Previous study has demonstrated that DARS-AS1 functions as an oncogene in CC cell growth via sponging miR-188-5p to regulate HMGB1 expression [Citation44]. In our study, we not only identified that DARS-AS1 was significantly up-regulated in CC, but also unveiled the potential mechanism of DARS-AS1 up-regulation in CC cells.
Hypoxia is familiar in solid tumours including CC, and it exerts important functions in tumour progression along with therapeutic responses [Citation45]. HIF1α, a primary regulator in hypoxia, can serve as a transcription factor to induce the transcription of lncRNAs. As reported previously, HIF1α induces the transcription of lncRNA RAET1K to affect hypoxia-induced glycolysis in hepatocellular carcinoma cells via through miR-100-5p [Citation46]. Coincidently, our study displayed that HIF1α transcriptionally enhanced DARS-AS1 expression by binding to DARS-AS1 promoter during hypoxia. Moreover, Jia Tong et al. have proposed that hypoxia-induced DARS-AS1 contributes to myeloma tumorigenesis [Citation47]. Consequently, all these results suggested hypoxia-induced DARS-AS1 increase was dependent on HIF1α in CC cells.
Autophagy, as a degradation pathway vital for energy and cellular homoeostasis, acts majorly as a protective role that prevents cell death under stressful conditions like hypoxia [Citation48]. In this study, we selected 3-MA as an autophagic inhibitor. The results showed that 3-MA can effectively inhibit hypoxia-induced cell viability in CC. Besides, our study also found that hypoxia induced the autophagy via up-regulating DARS-AS1 expression through detecting the expression of autophagosome markers, LC3II and p62. DARS-AS1 silence reversed the increased the expression of LC3II as well as the decreased that of p62 in HeLa cells under hypoxia. It has been reported that lncRNAs can regulate autophagy-related genes to control autophagy activation [Citation49]. ATG5 and ATG3, as autophagy proteins, have been widely documented to participate in the autophagosome formation [Citation50]. Consistently, our study further demonstrated that DARS-AS1 regulated ATG5 and ATG3 expression to affect hypoxia-induced autophagy.
Current study has disclosed that DARS-AS1 positively regulates its nearby gene DARS to promote clear cell renal cell carcinoma progression [Citation51]. Congruously, our study also confirmed the interaction between DARS-AS1 and DARS in CC. We have shown that DARS expression was positively correlated with DARS-AS1 expression in CC, and DARS was highly expressed in CC cells. DARS gene polymorphisms have been discovered to associate with the risk of isolated ventricular septal defects [Citation52]. From previous literature, DARS has been testified to serve as an oncogene in clear cell renal cell carcinoma (ccRCC) and participate in the regulatory mechanism of DARS-AS1, which exerted promoting impacts in ccRCC malignancy [Citation53]. In current study, it was confirmed that DARS-AS1 affected ATG5 and ATG3 expression to control hypoxia-induced autophagy via modulating DARS translation.
As one of the most famous RNA modifications, m6A methylation plays a crucial role in mRNA metabolism processes, such as splicing and stability, and translation [Citation54]. In our study, we discovered that DARS-AS1 regulated DARS mRNA stability and translation via METTL3- and METTL14-mediated m6A modification. Based on previous reports, RNA m6A modification mediated by METTL3 and METTL14 is detected to affect various mRNA in many cancers, such as gastric cancer [Citation55], triple-negative breast cancer [Citation56] and hepatocellular carcinoma [Citation57,Citation58]. Our study further used two famous m6A methylated modification erasers FTO and ALKBH5 to verify whether DARS translation was truly induced by m6A methylation.
In summary, our study elucidated a novel mechanism of which HIF1α-activated up-regulation of DARS-AS1 regulated hypoxia-induced cytoprotective autophagy in CC cells via METTL3- and METTL14-mediated m6A modification to promote DARS translation. Our study might provide an effective therapeutic strategy for CC.
The limitations of our study were presented as follows. First, the detailed regulatory mechanism of DARS-AS1 transcription induced by HIF1α was not investigated. Second, we searched many RBPs of DARS-AS1, but didn’t probe their functions as well as the relation between them and DARS-AS1. Last, whether DARS-AS1 regulates hypoxia-induced autophagy via the signalling pathways was not discussed. All these deficiencies will be improved in the near future.
Supplemental Material
Download Zip (2.6 MB)Acknowledgments
Thank you for all authors.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2022.2079889.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Tsikouras P, Zervoudis S, Manav B, et al. Cervical cancer: screening, diagnosis and staging. J BUON. 2016;21:320–325.
- Nuranna L, Fahrudin A. Survival rate of cervical cancer in national referral hospital in 2012 - 2014. Acta Med Indones. 2019;51:145–150.
- Wang R, Pan W, Jin L, et al. Human papillomavirus vaccine against cervical cancer: opportunity and challenge. Cancer Lett. 2020;471:88–102.
- Hemmat N, Bannazadeh Baghi H. Association of human papillomavirus infection and inflammation in cervical cancer. Pathog Dis. 2019;77(5).
- Mennerich D, Kubaichuk K, Kietzmann T. DUBs, hypoxia, and cancer. Trends Cancer. 2019;5:632–653.
- Wigerup C, Påhlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol Ther. 2016;164:152–169.
- Pezzuto A, Carico E. Role of HIF-1 in cancer progression: novel insights. A review. Curr mol med. 2018;18:343–351.
- Zhang W, Yuan W, Song J, et al. LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α. Biochimie. 2018;144:21–27.
- Liu J, Zhang C, Zhao Y, et al. Parkin targets HIF-1α for ubiquitination and degradation to inhibit breast tumor progression. Nat Commun. 2017;8:1823.
- Yin X, Xia J, Sun Y, et al. CHCHD2 is a potential prognostic factor for NSCLC and is associated with HIF-1a expression. BMC Pulm Med. 2020;20:40.
- Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42.
- Huang HY, Wang WC, Lin PY, et al. The roles of autophagy and hypoxia in human inflammatory periapical lesions. Int Endod J. 2018;51(Suppl 2):e125–e45.
- Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13:1619–1628.
- Wang T, Kong S, Tao M, et al. The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer. 2020;19:88.
- Tao Z, Zhao Y, Chen X. Role of methyltransferase-like enzyme 3 and methyltransferase-like enzyme 14 in urological cancers. Peer J. 2020;8:e9589.
- Chi Y, Wang J, Wang J, et al. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8:1015.
- Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772.
- Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509.
- Huang K, Fan WS, Fu XY, et al. Long noncoding RNA DARS-AS1 acts as an oncogene by targeting miR-532-3p in ovarian cancer. Eur Rev Med Pharmacol Sci. 2020;24:7211.
- Liu D, Liu H, Jiang Z, et al. Long non-coding RNA DARS-AS1 promotes tumorigenesis of non-small cell lung cancer via targeting miR-532-3p. Minerva Med. 2019;1121: 408–409 .
- Gandin V, Sikström K, Alain T, et al. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J vis exp. 2014. DOI:10.3791/51455.
- Zheng W, Tian X, Cai L, et al. LncRNA DARS-AS1 regulates microRNA-129 to promote malignant progression of thyroid cancer. Eur Rev Med Pharmacol Sci. 2019;23:10443–10452.
- Huang K, Fan WS, Fu XY, et al. Long noncoding RNA DARS-AS1 acts as an oncogene by targeting miR-532-3p in ovarian cancer. Eur Rev Med Pharmacol Sci. 2019;23:2353–2359.
- Zhang J, Jin HY, Wu Y, et al. Hypoxia-induced LncRNA PCGEM1 promotes invasion and metastasis of gastric cancer through regulating SNAI1. Clin Transl Oncol. 2019;21:1142–1151.
- Gonzalez FJ, Xie C, Jiang C. The role of hypoxia-inducible factors in metabolic diseases. Nat Rev Endocrinol. 2018;15:21–32.
- Wu D, Potluri N, Lu J, et al. Structural integration in hypoxia-inducible factors. Nature. 2015;524:303–308.
- Sun S, Ning X, Zhang Y, et al. Hypoxia-inducible factor-1alpha induces Twist expression in tubular epithelial cells subjected to hypoxia, leading to epithelial-to-mesenchymal transition. Kidney Int. 2009;75:1278–1287.
- Lim JH, Chun YS, Park JW. Hypoxia-inducible factor-1alpha obstructs a Wnt signaling pathway by inhibiting the hARD1-mediated activation of beta-catenin. Cancer Res. 2008;68:5177–5184.
- Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature. 2014;508:103–107.
- Ghoshal P, Teng Y, Lesoon LA, et al. HIF1A induces expression of the WASF3 metastasis-associated gene under hypoxic conditions. Int J Cancer. 2012;131:E905–E15.
- Xue H, Yuan G, Guo X, et al. A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3-MIR155-3p-CREBRF pathway. Autophagy. 2016;12:1129–1152.
- Hu YL, Jahangiri A, De Lay M, et al. Hypoxia-induced tumor cell autophagy mediates resistance to anti-angiogenic therapy. Autophagy. 2012;8:979–981.
- Hu YL, DeLay M, Jahangiri A, et al. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res. 2012;72:1773–1783.
- Chai W, Ye F, Zeng L, et al. HMGB1-mediated autophagy regulates sodium/iodide symporter protein degradation in thyroid cancer cells. J Exp Clin Cancer Res. 2019;38:325.
- Guo FX, Wu Q, Li P, et al. The role of the LncRNA-FA2H-2-MLKL pathway in atherosclerosis by regulation of autophagy flux and inflammation through mTOR-dependent signaling. Cell Death Differ. 2019;26:1670–1687.
- Song S, Tan J, Miao Y, et al. Crosstalk of autophagy and apoptosis: involvement of the dual role of autophagy under ER stress. J Cell Physiol. 2017;232:2977–2984.
- Liu Z, Dai J, Shen H. Dataset for regulation between lncRNAs and their nearby protein-coding genes in human cancers. Data Brief. 2018;19:1902–1906.
- Fröhlich D, Suchowerska AK, Spencer ZH, et al. In vivo characterization of the aspartyl-tRNA synthetase DARS: homing in on the leukodystrophy HBSL. Neurobiol Dis. 2017;97:24–35.
- Katsumata K, Nishiyama J, Inoue T, et al. Dynein- and activity-dependent retrograde transport of autophagosomes in neuronal axons. Autophagy. 2010;6:378–385.
- Jia L, Mao Y, Ji Q, et al. Decoding mRNA translatability and stability from the 5’ UTR. Nat Struct Mol Biol. 2020;27:814–821.
- Wang X, Feng J, Xue Y, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578.
- Ma S, Deng X, Yang Y, et al. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/β-catenin signaling in cervical cancer. Biomed Pharmacothe. 2018;108:1686–1693.
- Zhu J, Han S. DARS-AS1 knockdown inhibits the growth of cervical cancer cells via downregulating HMGB1 via sponging miR-188-5p. Technol Cancer Res Treat. 2020;19:1533033820971669.
- Chen XJ, Deng YR, Wang ZC, et al. Hypoxia-induced ZEB1 promotes cervical cancer progression via CCL8-dependent tumour-associated macrophage recruitment. Cell Death Dis. 2019;10:508.
- Zhou Y, Huang Y, Hu K, et al. HIF1A activates the transcription of lncRNA RAET1K to modulate hypoxia-induced glycolysis in hepatocellular carcinoma cells via miR-100-5p. Cell Death Dis. 2020;11:176.
- Tong J, Xu X, Zhang Z, et al. Hypoxia-induced long non-coding RNA DARS-AS1 regulates RBM39 stability to promote myeloma malignancy. Haematologica. 2020;105:1630–1640.
- Ravanan P, Srikumar IF, Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci. 2017;188:53–67.
- Gu Z, Hou Z, Zheng L, et al. LncRNA DICER1-AS1 promotes the proliferation, invasion and autophagy of osteosarcoma cells via miR-30b/ATG5. Biomed Pharmacothe. 2018;104:110–118.
- Zhou B, Liu J, Kang R, et al. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2020;66:89–100.
- Jiao M, Guo H, Chen Y, et al. DARS-AS1 promotes clear cell renal cell carcinoma by sequestering miR-194-5p to up-regulate DARS. Biomed Pharmacothe. 2020;128:110323.
- Feng Y, Chen R, Mo X. Association of DARS gene polymorphisms with the risk of isolated ventricular septal defects in the Chinese Han population. Ital J Pediatr. 2016;42:102.
- Jiao M, Guo H, Chen Y, et al. DARS-AS1 promotes clear cell renal cell carcinoma by sequestering miR-194-5p to up-regulate DARS. Biomed Pharmacothe. 2020;128:110323.
- Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacothe. 2019;112:108613.
- Yue B, Song C, Yang L, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142.
- Shi Y, Zheng C, Jin Y, et al. Reduced expression of METTL3 promotes metastasis of triple-negative breast cancer by m6A methylation-mediated COL3A1 up-regulation. Front Oncol. 2020;10:1126.
- Liu X, Qin J, Gao T, et al. Analysis of METTL3 and METTL14 in hepatocellular carcinoma. Aging (Albany NY). 2020;12:21638–21659.
- Yang N, Wang T, Li Q, et al. HBXIP drives metabolic reprogramming in hepatocellular carcinoma cells via METTL3-mediated m6A modification of HIF-1α. J Cell Physiol. 2020;236(5):3863–3880 .
