ABSTRACT
microRNAs are short noncoding RNAs that buffer fluctuations in gene expression in a myriad of physiological conditions. Here, we carried out a screen to identify the role of microRNAs in the maintenance of age-dependent neuronal functions in adult Drosophila. We report that miR-190 acts in the neurons to regulate lifespan, neuronal maintanence and age-related locomotor activity specifically in male flies. miR-190, a highly conserved microRNA, shows higher expression levels in male flies. Our data suggest that miR-190 functions by regulating target genes that are involved in maintaining neuronal activity and lifespan in male flies.
Introduction
The lifespan of organisms is determined by both genetic and environmental factors. Nutrition, insulin signalling and reactive oxygen species signalling have been reported to affect the lifespan of an organism [Citation1]. Most notably, a higher telomere shortening rate has been shown to lead to a decrease in lifespan in mammals [Citation2]. Other factors that regulate lifespan are metabolic status [Citation3,Citation4] and disruptions in the DNA damage repair machinery [Citation5]. Interestingly, hormonal differences [Citation6] as well as the asymmetric inheritance of sex chromosomes between male and females [Citation7] have been linked to an increased lifespan in females.
microRNAs are short noncoding RNAs, which bind to various target mRNAs based on sequence similarity and regulate gene expression post-transcriptionally. Much of the earlier work on microRNAs were focused on deciphering their role in development [Citation8–12] but recently, the focus has shifted to understanding the roles of microRNAs in the adult physiology and ageing [Citation13, Citation14]. Several recent studies demonstrate the role of microRNAs in neuronal maintenance [Citation14–16], and thereby lifespan in Drosophila. Pioneering studies have implicated the critical role of miR-8 [Citation17] and miR-34 [Citation13], in regulating lifespan. In addition, studies have shown that let-7 to miR-125 switch from larva to adult stage is critical for the maintenance of neuronal integrity and regulation of lifespan [Citation18]. Other reports show that miR-969 [Citation19], miR-305 [Citation20], miR-277 [Citation21] and miR-282 [Citation22] have roles in the regulation of lifespan in Drosophila.
Prior screens carried out to decipher the role of microRNAs in flies involved either the use of microRNA knockout flies [Citation23] or tissue-specific modulation of microRNA levels from the early developmental stages [Citation11, Citation24]. We began with a genetic screen to identify microRNAs that play a role in regulating lifespan in the adult stage, mainly to circumvent the effects that originate from developmental disruptions. Here, we show that blocking miR-190 expression in the neurons reduced the life span of male flies and had no effects in female flies. Reducing miR-190 levels affected climbing ability and enhanced neurodegeneration in male flies, suggesting an earlier onset of neuronal ageing. However, these flies appeared to exhibit enhanced locomotor activity, but their metabolic status, hypoxia response, and ROS levels remained largely unaltered. Moreover, we find that miR-190 regulates several genes that are implicated in the regulation of neurotransmitter activity and lifespan specifically in male flies.
Methods
Fly stocks
The w1118 (6326), elavGAL4 (458), UAS-miR-190-sponge (61397), UAS-miR-190OE (59885) and tubGAL80ts (7017) lines were obtained from BDSC. The UAS Scramble-sponge was a kind gift from Dr. Tudor Fulga (Weatherall Institute, Oxford, UK). The UAS-microRNA sponges used are listed as Table S8.
Survival assays
50 larvae were collected into regular food vials and raised on standard media at 18°C. Multiple independent larval collections were performed. Upon eclosion, the flies were allowed to mate at 29°C for 2 days before being separated by gender. Fifteen male/female flies were collected in each vial and the flies were provided with fresh food every alternate day. The number of dead flies was noted every day. Multiple independent sets were carried out on different months, to minimize background effects. A log rank test was carried out on the survival curves using OASIS 2 software.
Activity Recordings
Upon eclosion, virgin flies were collected and entrained for 2 days in an incubator maintained at 29°C with a LD cycle of 12:12. The activity of the flies was recorded using the Drosophila Activity Monitors (DAM) at 29°C with a LD cycle of 12:12. Data from these flies was recorded across time until they died, with both cross-sectional and longitudinal experiments being carried out. The flies were transferred to fresh activity tubes every 7 days. The total activity values generated every week were analysed using the Student’s t-test to their corresponding control.
Climbing assay
Climbing assays were carried out using elongated vials. A mark was made on the vials, 5 cm from the bottom. A maximum of 15 flies were transferred to these elongated vials and were allowed to acclimatize for 10 minutes. Following this the vial was tapped thrice in an arrhythmic fashion, following which a 3 second timer was activated. The number of flies that cross 5cms in 3s were noted. This assay was carried out thrice with a gap of about 3 minutes between each trial.
Bioinformatics
microRNA targets were predicted using the target prediction software, TargetScan Fly 7.2 and DIANA-microT-CDS. The list of human orthologues of the dme-miR-190 targets shortlisted here were obtained using the DRSC Integrative Ortholog Prediction Tool and compared with the list of hsa-miR-190a/b targets predicted by DIANA-microT-CDS (threshold set to 0.5)
Gene Expression analysis
Five-day-old flies were flash frozen, before being stored at −80OC. Total RNA was extracted from 30 fly heads each using the TriZol Reagent as per the manufacturer’s instruction and pelletted using Chloroform and isopropanol. The pellet was washed with 70% ethanol and then centrifuged at 15,000 rpm. The pellet was dried and then dissolved in 40 µl of Molecular biology-grade water (Himedia). Any contaminating DNA was degraded with the help of a QIAGEN RNase-Free DNase set as per the manufacturer’s instructions. Following which the RNA was re-extracted and dissolved in 40 µl of Molecular biology-grade water (Himedia). Takara Perfect Real-time kit was used to convert similar quantities of the RNA to cDNA. The cDNA was used for qPCR quantification using TB Green Premix Ex Taq (Tli RNase H Plus) in a CFX96 machine (Bio-Rad). The level of the genes were normalized to the level of actin. The ΔCt values generated were analysed using the Student’s t-test. The list of primers used is shown as Table S7.
MicroRNA Expression
A microRNA stem loop qPCR was performed to check the levels of miR-190 in male and female flies. Five day and 30-day-old flies were flash frozen, before being stored at −80°C. The total RNA was extracted as per the protocol listed above. The microRNA was reverse transcribed to cDNA using a microRNA-specific stem loop primer and the Takara PrimeScript 1st strand cDNA Synthesis Kit. The resulting cDNA was used for qPCR quantification using TB Green Premix Ex Taq (Tli RNase H Plus) in a CFX96 machine (Bio-Rad). The levels of the microRNA were normalized to the levels of 2S rRNA. The ΔCt values generated were analysed by two-way ANOVA using Tukey’s multiple comparison test.
The TaqMan MicroRNA Assay (ABI) was used for the quantification of microRNA levels in the miR-190 downregulated and overexpressed backgrounds, to show the efficiency of the sponge and overexpression lines, after the RNA was extracted as listed above. cDNA was prepared with High-Capacity cDNA Reverse Transcription Kits (Cat#437496, ABI). The protocol was followed as per the manufacturer’s instructions. For real-time PCR of TaqMan MicroRNA Assay, the 2xTaqMan Universal PCR Master Mix was used, with 2S rRNA TaqMan miRNA control assay being utilized as the endogenous control. The protocol for the same was as per the manufacturer’s instructions. The ΔCt values generated were analysed using the Student’s t-test.
Brain vacuole quantification
Adult fly brains were dissected on day 24 and fixed in 4% PFA solution. The samples were stained with anti-DLG1 antibody (1:100) [4F3 anti-discs large from DSHB) followed by anti-mouse Alexa-fluor 633 secondary antibody. The number of vacuoles and their area was quantified using Fiji software. The largest area of each vacuole in the entire stack was quantified. And only vacuoles with an area larger than 5 μm2 were considered for quantification. A detailed protocol can be found in Citation25.
Results
Here, we investigated the functions of microRNAs that are conserved across all bilaterians, by blocking the expression of these microRNA in the nervous system of adult Drosophila. At the current stage we have completed analysing the neuronal effects of six microRNAs from a total of 30 bilaterian microRNAs on lifespan (Table S1]. This screen was carried out by expressing microRNA-sponges [Citation26], which are transgenically expressed inhibitors of microRNAs, specifically in the adults neurons with the use of an elavGAL4;; tubulinGAL80ts line (see methods section for details). The Gal4 line used here is a pan-neuronal driver and with the use of tubulin-Gal80ts we managed to restrict the expression of various UAS-microRNA sponges to the adult stages.
A significant decrease in lifespan was found in the miR-190 and miR-375 downregulated flies (Table S1). Further, we focused on miR-190 as miR-375 has been shown to be important for maintenance of neuronal integrity in mammals [Citation27]. miR-190 is a conserved microRNA, reported to play a role in hypoxia responses in Drosophila, by suppressing the levels of the prolyl-hydroxylase fatiga, which leads to activation of Hif1α (hypoxia-inducible factor), sima [Citation28]. Furthermore, deletion of miR-190 led to lethality, with none of the larvae surviving till adulthood [Citation23].
Upon pan-neuronal downregulation of miR-190 in the adult flies, a significantly higher age-associated mortality leading to a lower median lifespan was observed in adult male flies (, Fig S1A). Surprisingly, no changes in median lifespan were observed when miR-190 was downregulated in the neurons of adult female flies (, Fig S1B). Since the effects of reduction of miR-190 levels were observed only in male flies, we hypothesized that miR-190 is differentially expressed in male and female flies, or that the microRNA could regulate a target gene, which is specific to males. To test this, we measured the levels of miR-190 in the head of male and female flies at early [day 5] and late [day 30] adult stages. miR-190 levels were found to be higher in young male flies in comparison to female flies (). Another recent work had reported miR-190 to be expressed at a higher level in the male fly brain in comparison to the female fly brain [Citation29]. While miR-190 levels remained mostly unchanged with age in the head of male flies, a moderate increase in the level of miR-190 with age was seen in the head of the female flies (). Thus, miR-190 is expressed at higher levels in male flies, and plays functional roles in regulating lifespan of male flies.
Figure 1. Effect on lifespan when miR-190 was downregulated in the neurons of (A) adult male (p = 0.0001) (n = 750) and (B) female flies (p = 0.909) (n = 650). (C) miR-190 levels in the heads of 5 and 30 days old male and female flies (p = 0.0408) (n = 5). (D) Rescue of the effect on lifespan (n ≥ 88) (p = 0.3901, for control and rescue conditions) (p < 0.0001, for control and miR-190 sponge expression). Climbing ability in response to miR-190 downregulation in the neurons of (E) adult male (p = 0.002) (n ≥ 34) and (F) female 8 and 24 days old flies (p = 0.374) (n ≥ 11).
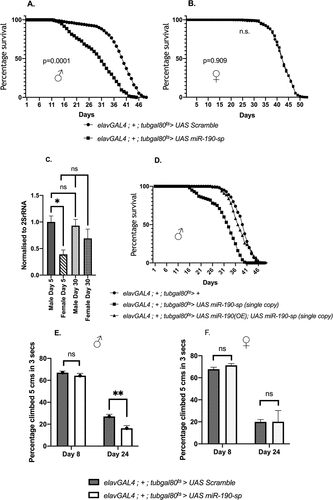
Next, we checked if the effect on lifespan by downregulating miR-190 in the adult male neurons is due to a direct regulation of lifespan by the microRNA. For this we overexpressed miR-190 in both male and female flies. As expected, the overexpression of miR-190 in the neurons of adult flies led to an increase in lifespan in both genders (Fig S2A and S2B). As miR-190 overexpression led to an increase in miR-190 levels to ~4-fold (Fig S1C), the increase in lifespan could be attributed directly to the higher levels of the microRNA in both genders. Moreover, the effects on lifespan in male flies by downregulation of miR-190 in the adult neurons could be rescued by overexpression of miR-190 (), confirming the role of miR-190 in the adult neurons in regulation of lifespan in male flies.
To check if the reduced lifespan in flies that lack miR-190 in the brain is due to premature deterioration of neuronal functions we performed climbing assays. Climbing assays have been widely used in flies as a proxy for neuronal ageing and normal functioning. When miR-190 was downregulated in the neurons, a severe reduction in climbing ability was observed in older male flies [day 24], but not in female flies [day 8 and day 24] and young males [day 8] ( and Table S2). Thus, our findings suggest that miR-190 is crucial for normal age-dependent neuronal functioning and regulates lifespan in adult male flies.
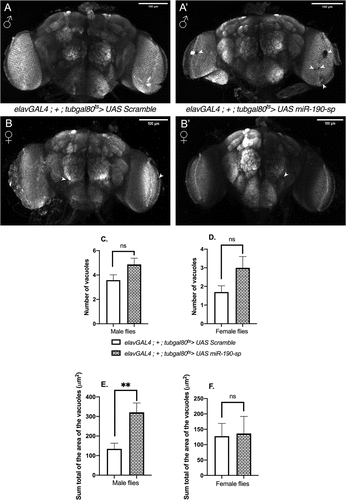
Next, we checked if the reduction in climbing ability in male flies in response to blocking miR-190 was due to a decrease in general locomotor activity. Surprisingly, lowering miR-190 levels in neurons led to an increase in locomotor activity in older male flies [day 24–28], but not in young male flies [day 3 to 7] and female flies [day 3 to 7 and day 24 to 28] ( B). This is in contrast to the previous data where a significant decrease in the percent activity in adult flies was observed when miR-190 was downregulated in the whole larvae from early development [Citation24]. Despite an increase in general motor activity, the reduced climbing ability suggests an early onset of neuronal ageing in the miR-190 loss-of-function male flies. Next, we checked if lowering miR-190 levels would lead to enhanced neurodegeneration by whole-brain imaging to visualize vacuoles. We saw an increase in brain vacuole size in 24-day-old adult male flies that express miR-190 sponge (, A’, E), no substantial changes were observed for the brain vacuole size in females (, B’, F). However, we noticed an increase in brain vacuole numbers, though not significant, in both genders in response to reduced miR-190 levels (, D). Thus, miR-190 function is crucial for maintenance of neuronal integrity in male flies.
Figure 2. Locomotor activity when miR-190 was downregulated in the neurons of (A) adult male flies, tested at day 3–7 (p > 0.9999); day 24–28 (p < 0.0001) (n ≥ 96) and (B) female flies tested at day 3–7 (p > 0.9999); day 24–28 (p > 0.9999) (n ≥ 63).
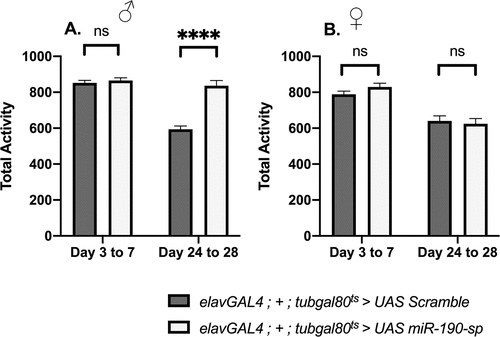
Figure 3. Vacuoles in the brain on day 24 when miR-190 was downregulated in the neurons of adult male flies (A, A’) and adult female flies (B, B’). Shown are representative images, vacuoles are marked by an arrow head. Number of vacuoles on day 24 when miR-190 was downregulated in the neurons of adult male flies (C) (n ≥ 23)(p = 0.062) and female flies (D) (n ≥ 10)(p = 0.104). Sum total of the area of the vacuoles on day 24 when miR-190 was downregulated in the neurons of adult male flies (E) (n ≥ 23) (p = 0.0019) and female flies (F) (n ≥ 10) (p = 0.902).
Lifespan has a strong correlation to metabolic status, with diet also being shown to affect the ageing process in Drosophila [Citation4, [Citation30]. Next, we examined if the levels of miR-190 alters the metabolic status of flies, and influences lifespan. We measured the levels of energy stores in the form of triglycerides and glycogen in adult flies [day 5] that express low miR-190 in the neurons. We also subjected miR-190 sponge flies [day 5] to chronic food withdrawal. In addition, we measured circulating levels of glucose, as another metabolic readout. Any deviations in these readouts would indicate a change in metabolic status. miR-190 downregulation in the neurons of adult flies [day 5] did not affect various metabolic parameters measured (Fig S3A-D, G, H), but led to a slight significant change in the ability of flies to resist starvation (Fig S3E, F) . We also observed a gender-specific difference in the response of flies with lower miR-190 levels. However, we believe that such a small effect on starvation responses, may not have contributed to the lifespan and climbing defects. Surprisingly, an increase in feeding in male flies but not in females flies [day 5] was observed in response to expression of miR-190 sponge in the adult nervous system (Fig S3I, J); however, this seems to have had no effects on the metabolic status of these flies. Thus, the role of miR-190 on lifespan and neuronal maintenance in male Drosophila could be independent of any changes in their metabolic status.
There is a well-documented relationship between hypoxia and ageing. Exposure to hypoxia during adult and larval stages leads to a decrease in lifespan [Citation31,Citation32], which suggests that activation of hypoxia signalling pathway could be responsible for the short lifespan when miR-190 levels are lowered in male neurons. Moreover, fatiga, a gene that encodes for a key component of the hypoxia response pathway, is a target of miR-190 [Citation28]. We measured the levels of hypoxia response genes ldh (lactate dehydrogenase) and fatiga, and did not observe any changes in their expression levels (Fig S4). We also measured the levels of ROS activity, another key regulator of lifespan, in male flies that express miR-190 sponge. The expression levels of MnSOD, thioredoxin1 and catalase genes, indicators of ROS activity, did not differ (Fig S4). Thus, the neuronal function of miR-190 in the regulating lifespan in male flies is not due to any alterations in their hypoxia and ROS status.
Several target genes for miR-190 are predicted by bioinformatic tools (TargetScan Fly 7.2 and DIANA-microT-CDS), so we decided to focus on common genes that are picked up by both target prediction programs. A notable number of these genes are linked to neuronal activity including the synthesis and regulation of the neurotransmitter acetylcholine. An increase in the levels of neural activity-related targets, mostly acetylcholine signalling related genes, was observed when miR-190 was downregulated in the brain of adult male flies [day 5] in comparison to the control UAS-scramble flies (). The levels of miR-190 target genes were affected less strongly in females, when compared to the respective gender control (). A decrease in the levels of some of these predicted target genes was seen in flies [day 5] when miR-190 was overexpressed in the neurons of both genders, as expected (Fig S5A, B). Thus, we show that endogenous levels of miR-190 regulates the levels of various genes that maintain acetylcholine neurotransmitter levels in male flies. The sexually dimorphic differences in the regulation of these target genes by miR-190 could be responsible for the lack of effects on lifespan and climbing behaviour in the miR-190 downregulated female flies.
Figure 4. Expression of putative target genes involved in acetylcholine regulation in (A) adult male flies (n = 10) and (B) female flies (n = 10) or involved in regulating lifespan in (C) adult male flies (n = 10) and (D) female flies when miR-190 is downregulated in the neurons of 5 day old flies (n = 10).
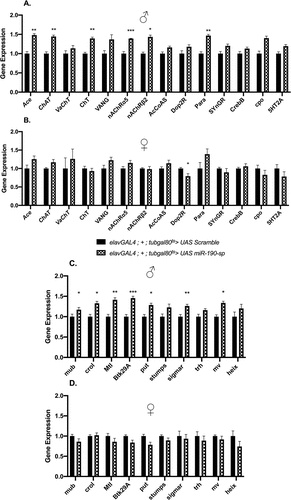
In addition to acetylcholine regulatory genes, miR-190 is also predicted to target genes identified by computational approaches implicated in regulating longevity in Drosophila [Citation33]. Few of these genes, mub, crol and Btk29A have already been shown by experimental approaches to regulate Drosophila lifespan [Citation34,Citation35, Citation36]. We measured the levels of these genes to confirm if they are regulated by miR-190, and therefore may contribute to the ageing phenotype observed when miR-190 is downregulated in the neurons of adult male flies. An increase in the expression of several of these target genes was observed when miR-190 was downregulated in the neurons of adult male flies [day 5] compared to its respective gender control (). As expected from the lack of phenotypes we did not observe a change in these target gene levels in female flies, when compared to the UAS-scramble control flies (). Also, overexpression of miR-190 in the neurons of both genders [day 5] led to a reduction in the levels of several of these genes (Fig S5C, D). Thus, miR-190 also targets genes that are crucial for regulation of the ageing process in males.
Discussion
Ageing is a process characterized by a progressive decline in physiological processes needed for the survival of the organism, which is mainly attributed to the senescence of the individual cells of the organism. A growing litany of articles have demonstrated the role of microRNAs in CNS maintenance either directly through their role in neurons [Citation16,Citation17, Citation13, Citation14] or indirectly via the glia [Citation15]. Here, we show that in male flies miR-190 aids in maintaining normal lifespan (. In addition, miR-190 seems to be required for neuronal maintenance, normal negative geotactic responses and locomotor activity in an age-dependent manner in male flies (, , , A’). It is interesting to note that lowering miR-190 levels led to increased locomotor activity in older male flies, whereas, these flies showed a reduced response in negative geotaxis assays. Increased locomotion is indicative of hyperactivity of neurons, expected from a possible enhanced acetylcholine neurotransmitter activity or due to upregulation of other target genes like para, known to play a role in generation of action potentials [Citation37]. Neuronal hyperactivation is known to result in neurodegeneration, which could explain the larger brain vacuoles and climbing defects seen in these flies Citation38,Citation39). More experimental work needs to be done to verify if the miR-190 downregulation causing hyperactivity is the cause of the neurodegeneration.
In this study, we show that the miR-190 expression is higher in male flies (). Despite the age-dependent function of miR-190, the levels of miR-190 in male flies are not affected by age. As miR-190 is located in an intron of gene rhea, we checked the levels of rhea mRNA, which was also found to be higher in males (Fig S6A). The higher expression of miR-190 in male flies seems to be linked to the regulation of gene expression of rhea. However, the phenotypes that we observe by expressing miR-190 sponge are independent of any effects on rhea transcript levels (Fig S6B).
In agreement with the male-specific phenotypes of lowering miR-190 expression, various miR-190 target genes, which are implicated in the regulation of neurotransmission and longevity, were expressed at a higher level in male flies with lower levels of miR-190 in the neurons (). In comparison to males the levels of these genes were not affected much in females in response to lowering adult neuronal miR-190 levels (). Overexpression of miR-190, which extended lifespan, reduced the expression of some of the target genes in male flies (Fig S2 and S5). However, among the miR-190 target genes tested in this study only Ace, ChAT, para and crol levels changed significantly in response to sponge treatment and overexpression as well in male flies. Of these genes ChAT and Ace regulate acetylcholine synthesis and activity in the synaptic cleft, respectively [Citation40]. The product of para, a subunit of the voltage gated sodium channels plays a key role in regulation of neuronal activity [Citation37]. The crol gene encodes for a zinc finger transcription factor that has been reported to regulate lifespan in Drosophila [Citation34]. The changes in the levels of these target genes in miR-190 downregulation and overexpression strongly suggests their roles in the phenotypes observed. However, other genes that change significantly only in either overexpression or downregulation of the microRNA could also contribute to the phenotypes in these individual conditions (; Fig S5). The difference in the regulatory capacity of target genes that are implicated in neurotransmission and longevity could be attributed to the higher levels of miR-190 in male flies. Female flies may have other microRNAs that act as genetic buffers by which optimal neuronal activity and lifespan is maintained. In the adult Drosophila head the number of female biased microRNAs are higher in comparison to male biased microRNAs, hence miR-190 mediated regulation of lifespan may not be crucial in females [Citation29]. However, the levels of miR-190 increase in the head during later stages in female flies (), which might be due to other yet unidentified functions, and this would be an interesting aspect to pursue in future.
Several miR-190 target genes that were shown to be regulated by this study (, S5) have also been identified in efforts that looked at genome-wide microRNA-target interaction [Citation41, Citation42, Citation43]. Remarkably the orthologues of a large number of the miR-190 targets tested by this study were also predicted to be targets of miR-190a/b in the human genome (Tables S3 and S4), which raises several future possibilities. miR-190 was also recently discovered to have eight differentially expressed isoforms on account of being a microRNA targeted by the Nibbler exonuclease [41]. The role of these isoforms in the context of the lifespan would also be an interesting aspect to pursue. Thus, we report a microRNA with a male bias in expression, which is crucial for normal neuronal functioning and ageing process in males.
Author contributions
JLF: conceived, designed, acquired data, analyzed data, drafted the manuscript. JV: conceived, designed, supervised the work, received funding, prepared the final draft of the manuscript.
Supplemental Material
Download PDF (14.5 MB)Acknowledgments
We thank Dr. Nisha Kannan for the use of the activity monitors. We thank Dr. Smitha Vishnu, Dr. Nisha Kannan, Reshma Menon, Dr. Himani Pathak and Swetha Gopal for their critical comments. We thank Dr. Tudor Fulga for the UAS-scrambled sponge lines. We thank Dr. Shamprasad Varija Raghu for the anti-DLG1 antibody. We thank IISER TVM Institutional Support; Extra-Mural Grant from Science and Engineering Research Board, Department of Science and Technology, Government of India; Ramanujan Fellowship from Science and Engineering Research Board, Department of Science and Technology, Government of India and Junior/Senior Research Fellowship from University Grants Commission, Government of India to JF for funding.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2022.2127544
Additional information
Funding
References
- Iliadi KG, Knight D, Boulianne GL. Healthy aging -insights from Drosophila. Front Physiol. 2012;3. DOI:10.3389/fphys.2012.00106.
- Vera E, Bernardes de Jesus B, Foronda M, et al. The rate of increase of short telomeres predicts longevity in mammals. Cell Rep. 2012;2(4):732–737.
- Piper MDW, Skorupa D, Partridge L. Diet, metabolism and lifespan in Drosophila. Exp Gerontol. 2005;40(11):857–862.
- Strilbytska O, Strutynska T, Semaniuk U, et al. Dietary sucrose defines lifespan and metabolism in drosophila. Ukrainian Biochemical Journal. 2020; DOI:10.15407/ubj92.05.097
- Flurkey K, Currer JM, and Harrison DE. Mouse Models in Aging Research.The Mouse in Biomedical Research (Second Edition). 2007;637–672. DOI:10.1016/B978-012369454-6/50074-1
- Russell S J, Kahn C Ronald. (2007). Endocrine regulation of ageing. Nat Rev Mol Cell Biol, 8(9), 681–91. DOI:10.1038/nrm2234
- Tower J. Sex-specific regulation of aging and apoptosis. Mech Ageing Dev. 2006;127(9):705–718.
- Boulan L, Martín D, Milán M. Bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr Biol. 2013;23(6):473–478.
- Chawla G, Sokol N S. (2011). MicroRNAs in Drosophila development. Int Rev Cell Mol Biol, 286 1–65. DOI:10.1016/B978-0-12-385859-7.00001-X
- Redmond W, Allen D, Christian Elledge M, et al. Screening of microRNAs controlling body fat in Drosophila melanogaster and identification of miR-969 and its target, Gr47b. PLoS ONE. 2019;14(7):e0219707.
- Suh YS, Bhat S, Hong S-H, et al. Genome-wide microRNA screening reveals that the evolutionary conserved miR-9a regulates body growth by targeting sNPFR1/NPYR. Nat Commun. 2015;6:7693.
- Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21(18):2277–2282.
- Liu N, Landreh M, Cao K, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482(7386):519–523.
- Weigelt CM, Hahn O, Arlt K, et al. Loss of miR-210 leads to progressive retinal degeneration in Drosophila melanogaster. Life Science Alliance. 2019; DOI:10.26508/lsa.201800149
- Aw SS, Lim IKH, Tang MXM, et al. A glio-protective role of mir-263a by tuning sensitivity to glutamate. Cell Rep. 2017;19(9):1783–1793.
- Chawla G, Deosthale P, Childress S, et al. A let-7-to-miR-125 MicroRNA Switch regulates neuronal integrity and lifespan in Drosophila. PLoS Genet. 2016a;12:8.
- Karres JS, Hilgers V, Carrera I, et al. The conserved microRNA MiR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131(1):136–145.
- Chawla G, Deosthale P, Childress S, et al. A let-7-to-miR-125 MicroRNA switch regulates neuronal integrity and lifespan in Drosophila. PLoS Genet. 2016b;12(8):e1006247.
- Zhang X, Lyu J, Jin X, et al. A motor neuron protective role of miR-969 mediated by the transcription factor kay. RNA Biol. 2020;17(9):1277–1283.
- Ueda M, Sato T, Ohkawa Y, et al. Identification of miR-305, a microRNA that promotes aging, and its target mRNAs in Drosophila. Genes Cells. 2018;23(2):80–93.
- Esslinger SM, Schwalb B, Helfer S, et al. Drosophila miR-277 controls branched-chain amino acid catabolism and affects lifespan. RNA Biol. 2013;10(6):1042–1056.
- Vilmos P, Bujna Á, Szuperák M, et al. Viability, longevity, and egg production of drosophila melanogaster are regulated by the miR-282 microRNA. Genetics. 2013;195(2):469–480.
- Chen YW, Song S, Weng R, et al. Systematic study of Drosophila MicroRNA functions using a collection of targeted knockout mutations. Dev Cell. 2014;31(6):784–800.
- Donelson NC, Dixit R, Pichardo-Casas I, et al. MicroRNAs regulate multiple aspects of locomotor behavior in Drosophila. G3 (Bethesda). 2020;10(1):43–55.
- Behnke JA, Ye C, Moberg KH, et al. A protocol to detect neurodegeneration in Drosophila melanogaster whole-brain mounts using advanced microscopy. STAR Protocols. 2021;2(3):100689.
- Fulga TA, McNeill EM, Binari R, et al. A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs. Nat Commun. 2015;6(1). DOI:10.1038/ncomms8279
- Cai LJ, Tu L, Li T, et al. Up-regulation of microRNA-375 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease by inhibiting SP1. Aging (Albany NY). 2020. DOI:10.18632/aging.102649
- De Lella Ezcurra AL, Bertolin AP, Kim K, et al. miR-190 enhances HIF-dependent responses to hypoxia in drosophila by inhibiting the prolyl-4-hydroxylase fatiga. PLoS Genet. 2016;12(5):e1006073.
- Fagegaltier D, Fagegaltier D, Gordon A, et al. A genome-wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics. 2014;198(2):647–668.
- Lee K Pum, Simpson S J, Clissold F J, Brooks R, Ballard J William, Taylor P W, Soran N, Raubenheimer D. (2008). Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc Natl Acad Sci U S A, 105(7), 2498–503. DOI:10.1073/pnas.0710787105
- Malacrida S, De Lazzari F, Mrakic-Sposta S, et al. Lifespan and ROS levels in different Drosophila melanogaster strains after 24 h hypoxia exposure. Biol Open. 2022;11(6). DOI:10.1242/bio.059386
- Polan DM, Alansari M, Lee B, et al. Early-life hypoxia alters adult physiology and reduces stress resistance and lifespan in Drosophila. J Exp Biol. 2020. DOI:10.1242/jeb.226027
- Hall BS, Barnett YA, Crofts JJ, et al. Identification of novel genes associated with longevity in Drosophila melanogaster - a computational approach. Aging (Albany NY). 2019;11(23):11244–11267.
- Magwire MM, Yamamoto A, Carbone MA, et al. Quantitative and molecular genetic analyses of mutations increasing Drosophila life span. PLoS Genet. 2010;6(7):e1001037.
- Merzetti E M, Staveley B E. (2016). Identifying potential PARIS homologs in D. melanogaster. Genet Mol Res, 15(4). DOI:10.4238/gmr15048934
- Hamada N, Bäckesjö C, Smith C Edvard and Yamamoto D. (2005). Functional replacement of Drosophila Btk29A with human Btk in male genital development and survival. FEBS Letters, 579(19), 4131–4137. DOI:10.1016/j.febslet.2005.06.042
- O’Dowd DK, Germeraad SE, Aldrich RW. Alterations in the expression and gating of Drosophila sodium channels by mutations in the para gene. Neuron. 1989;2(4):1301–1311.
- Rival T, Soustelle L, Strambi C, Besson M, Iché M, Birman S. (2004). Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr Biol, 14(7), 599–605. DOI:10.1016/j.cub.2004.03.039
- Palladino M J, Bower J E, Kreber R, Ganetzky B. (2003). Neural dysfunction and neurodegeneration in Drosophila Na+/K+ ATPase alpha subunit mutants. J Neurosci, 23(4), 1276–86.
- Parsons SM, Prior C, Marshal IG. Acetylcholine trans-port, storage and release. Int Rev Neurol. 1993; DOI:10.1016/S0074-7742(08)
- Wessels HH, Lebedeva S, Hirsekorn A, et al. Global identification of functional microRNA-mRNA interactions in Drosophila. Nat Commun. 2019;10(1). DOI:10.1038/s41467-019-09586-z
- Xia X, Fu X, Wu B, et al. Circadian regulation of microRNA-target chimeras in Drosophila. BioRxiv. 2019. DOI:10.1101/622183
- Liu N, Abe M, Sabin LR, et al. The exoribonuclease nibbler controls 3′ end processing of microRNAs in drosophila. Curr Biol. 2011;21(22):1888–1893.
