ABSTRACT
A growing body of evidence suggests that RNA interference (RNAi) plays a pivotal role in the communication between plants and pathogenic fungi, where a bi-directional trans-kingdom RNAi is established to the advantage of either the host or the pathogen. Similar mechanisms acting during plant association with non-pathogenic symbiotic microorganisms have been elusive to this date. To determine whether root endophytes can induce systemic RNAi responses to their host plants, we designed an experimental reporter-based system consisting of the root-restricted, beneficial fungal endophyte, Fusarium solani strain K (FsK) and its host Nicotiana benthamiana. Since not all fungi encode the RNAi machinery, we first needed to validate that FsK does so, by identifying its core RNAi enzymes (2 Dicer-like genes, 2 Argonautes and 4 RNA-dependent RNA polymerases) and by showing its susceptibility to in vitro RNAi upon exogenous application of double stranded RNAs (dsRNAs). Upon establishing this, we transformed FsK with a hairpin RNA (hpRNA) construct designed to target a reporter gene in its host N. benthamiana. The hpRNA was processed by FsK RNAi machinery predominantly into 21–24-nt small RNAs that triggered RNA silencing but not DNA methylation in the fungal hyphae. Importantly, when the hpRNA-expressing FsK was used to inoculate N. benthamiana, systemic RNA silencing and DNA methylation of the host reporter gene was recorded. Our data suggest that RNAi signals can be translocated by root endophytes to their hosts and can modulate gene expression during mutualism, which may be translated to beneficial phenotypes.
Introduction
RNA interference (RNAi) is a conserved eukaryotic gene regulatory mechanism that is triggered by small RNAs (sRNAs) of approximately 20–25 nucleotides (nt)[Citation1], [Citation2]. Notwithstanding the diversity of RNAi pathways and the plethora of sRNA classes, there are essentially two types of sRNAs, the small interfering RNAs (siRNAs) and the microRNAs (miRNAs) [Citation3,Citation4]. In general, Dicer and Dicer-like (DCL) endonucleases cleave double stranded RNAs (dsRNAs) and stem loop hairpin RNAs (hpRNAs) into 20–25-nt siRNAs and miRNAs, respectively [Citation5]. The occurring double stranded sRNA is then unzipped in an ATP-dependent reaction so that only one of its two strands will eventually be loaded onto an Argonaute (AGO) protein [Citation6,Citation7]. Then, the AGO-loaded sRNA scans the cytoplasm for complementary mRNA transcripts to cleave them or inhibit their translation [Citation8,Citation9]. At least in plants, AGO-loaded sRNAs may also be transported in the nucleus, where they are involved in RNA-directed DNA methylation (RdDM) of cognate sequences [Citation10,Citation11]. Moreover, in plants, nematodes and some fungi, the presence of RNA-dependent RNA polymerases (RDRs) contributes to the generation of dsRNAs from single stranded transcripts, in a process termed transitivity [Citation12,Citation13].
Fungal RNAi, initially described as ‘quelling’ in Neurospora crassa [Citation14], has essentially a two-fold role. On the one hand, siRNAs generated from (usually RDR-transcribed) dsRNA precursors are involved in genome defence and maintenance of genome integrity as well as fighting against transposons, viruses and transgenes [Citation15,Citation16]. On the other hand, miRNAs (also called miRNA-like, milRNAs), generated by Pol III-transcribed primary miRNA transcripts, fine-tune gene expression during vegetative and sexual development besides responding to various kinds of stresses [Citation15,Citation17]. A growing body of recent evidence suggests that, in addition to the aforementioned roles, RNAi also has a pivotal role in the communication of fungi with their hosts. Indeed, the pathogen Botrytis cinerea delivers sRNAs in Arabidopsis and tomato that target members of the mitogen-activated protein kinases (MAPKs) that function in plant immunity [Citation18]. In reverse, plants fight back; Arabidopsis and tomato deliver sRNAs in B. cinerea targeting the fungal DCL1 and DCL2, to attenuate fungal pathogenicity and growth [Citation19]. Likewise, Fusarium graminearum translocates sRNAs to target defence genes in Hordeum vulgare and Brachypodium distachyon [Citation20], whereas cotton plants, in response to infection with the vascular pathogen Verticillium dahliae, export miR159 and miR166 to silence fungal isotrichodermin C-15 hydroxylase and Ca(2+)-dependent cysteine protease, respectively, both of which are essential for fungal virulence [Citation21]. However, the role of such cross-kingdom RNAi processes in mutualistic interactions remains poorly understood.
Fusarium solani strain K (FsK) is an endophytic, non-pathogenic strain, initially isolated from the roots of tomato plants [Citation22] but other plant species serve as hosts, including legumes [Citation23]. FsK has been shown to protect the host against root and foliar pathogens [Citation22], spider mites [Citation24], zoophytophagous insects [Citation25] and to alleviate drought stress [Citation26]. The beneficial activity of FsK presupposes an intact ethylene signalling pathway, suggesting that the fungus can induce systemic responses to the plant [Citation22]. However, the exact molecular details governing this mutualism remain largely elusive. In this study, we identified the core enzymes of the RNAi machinery in FsK (DCLs, AGOs, RDRs) and provide evidence that the endophyte translocates RNAi signals to its host plant to modulate expression and induce epigenetic modification of a host reporter gene.
Materials and methods
Identification of FsK RNAi genes and phylogenetic analysis
Based on the genome and transcriptome of FsK (upublished, Papadopoulou laboratory) we identified by homology search the FsK DCLs, AGOs and RDRs (Dataset S1), whose sequences are also deposited to Zenodo (https://doi.org/10.5281/zenodo.6088855). The analysed sequences (DCLs, AGOs and RDRs) were aligned with MUSCLE v3.7 [Citation27], and informative sites were selected with Gblocks v0.91b [Citation28]. The aligned selected sites were tested with the Prottest v3.2 software [Citation29] using the Akaike information criterion (AIC) values for optimal residue substitution model matrix selection. The LG [Citation30] residue substitution model matrix scored best for all protein sets. The PhyML v3.0 algorithm [Citation31] using the LG model and bootstrap testing with 100 replicates was used for obtaining the best maximum likelihood tree.
Generation of constructs
For the generation of pCS-GFP, a PCR was performed using as template genomic DNA from N. benthamiana line 16C [Citation32] and the primers GFP-F1 and GFP-R1 (Table S1) and the occurring 2017 bp amplicon was cleaved with HpaI/SacI and ligated to a similarly cleaved pSilent-1 (AB303070, Fungal Genetics Stock Center), generating the pSilent-GFP. Next, pSilent-GFP was cleaved with PsiI/SacI and the 6663 bp fragment was ligated into the 7866 bp fragment retrieved upon ZraI/SacI cleavage of pCambia1300, generating the pCS-mGFP. For the generation of pCS-hpGF+GFP, a first PCR was performed using as template genomic DNA from N. benthamiana line 16C and the primers GFP-F2 and GFP-R2 (Table S1) and the occurring 340 bp amplicon was cleaved with XhoI/HindIII and ligated to a similarly cleaved pSilent-1 vector, generating the pSilent-GF. A second PCR was performed using as template genomic DNA from N. benthamiana line 16C and the primers GFP-F3 and GFP-F4 (Table S1) and the occurring 340 bp amplicon was cleaved with KpnI/BglII and ligated to a similarly cleaved pSilent-GF vector, generating the pSilent-hpGF. Next, the 1937 bp fragment emerging upon HpaI/SacI cleavage of the pSilent-GFP was ligated into a similarly cleaved pSilent-hpGF, generating the pSilent-hpGF+GFP. Finally, pSilent-hpGF+GFP was cleaved with PsiI/SacI and the 7277 bp fragment was ligated into the 7866 bp fragment retrieved upon ZraI/SacI cleavage of pCambia1300, generating the pCS-hpGF+GFP.
Agrobacterium-mediated fungal transformation
The binary vectors pCS-GFP and pCS-hpGF+GFP were used to transform Agrobacterium tumefaciens AGL1 strain by electroporation using the MicroPulser Electroporator (www.bio-rad.com) according to the manufacturer’s instructions. The AGL1-pCS-GFP and AGL1-pCS-hpGF+GFP were used to transform FsK conidia as previously described [Citation33].
Isolation of fungal conidia and inoculation
FsK was routinely cultured for 4 days in potato dextrose broth (PDB) (26°C, 160 rpm). Following removal of mycelium fragments by sieving through sterile cheesecloth, conidia were recovered from the filtrate by centrifugation at 6,500 rpm, counted using a haemocytometer and suspended in an appropriate volume of 0.85% NaCl to achieve the desired inoculum concentration. Approximately 100 conidia were used to inoculate N. benthamiana plants at cotyledon stage.
In vitro transcription of sGFP dsRNA
For the generation of the in vitro transcribed sGFP dsRNA, genomic DNA was extracted from FsK-sGFP [Citation34] and used as template for PCR with KAPA Taq DNA Polymerase (www.sigmaaldrich.com) with the T7 promoter-containing primers T7-sGFP-F and T7-sGFP-R (Table S1). The T7 promoter-containing 491 bp amplicon was then used as template in the MEGAscript™ RNAi Kit (www.thermofisher.com) for the generation of a 445 bp sGFP dsRNA.
In vitro RNAi assay
In 24 wells of a 96-well plate, FsK-sGFP conidia were added (in each well, 6 conidia diluted in 100 μl PDB/100). In 12 wells containing these FsK-sGFP conidia, in vitro transcribed sGFP dsRNA was added (100 μl, 1 ng/μl) (dsRNA application samples). In the remaining 12 wells containing FsK-sGFP conidia, 100 μl water was added (control samples). The 96 well was covered with a removable membrane and incubated at 28°C. At timepoints 0–24-48 hpa, the plate was subjected to fluorometric analysis using the Varioskan™ LUX multimode microplate reader (www.thermofisher.com).
Fungal RNA isolation
FsK was routinely cultured for 4 days in potato dextrose broth (PDB) (26°C, 160 rpm). From the occurring mycelium total RNA was isolated with TRIzol™ Reagent (www.thermofisher.com) to be subsequently used in RT-qPCR reactions. For small RNA sequencing, the fraction enriched for small RNAs was isolated from the mycelium using mirVana™ miRNA Isolation Kit (www.thermofisher.com) according to the manufacturer’s instructions.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR)
DNaseI-treated (www.thermofisher.com) RNA isolated from mycelium was quantified with by Qubit 2.0 Fluorometric Quantification (www.thermofisher.com). The DNA-free RNA (10 ng) was then subjected to RT-qPCR using the Luna® Universal Probe One-Step RT-qPCR Kit (www.neb.com) according to the manufacturer’s instructions. Essentially, the total volume of the reaction was reduced to 10 μl and the cycling parameters consisted of incubation at 55°C for 10 min for reverse transcription, 95°C for 1 min followed by 39 cycles of 95°C for 10 sec and 60°C for 30 sec. The relative expression of GFP gene (133 bp amplicon) was calculated from three (for FsK-GFP strain) and two (for the three FsK-hpGF+GFP transformants) technical replicates using as internal controls the ITS (108 bp amplicon) and Tef-1a 1a (120 bp amplicon) genes. Analysis was carried out using the geometric mean of FsK ITS and Tef-1a transcripts [Citation23]. All primers used, TEF-F and TEF-R, ITS-F and ITS-R and GFP-F4 and GFP-R4 are listed in Table S1. Data analysis was performed according to the ΔCT method [Citation35], with all data being expressed as mean ± SD (standard deviation). Data were analysed using the Student’s two-tailed homoscedastic t-test.
Plant and fungal DNA isolation
Genomic DNA from plant and fungal tissue was isolated with DNeasy Plant Pro (/www.qiagen.com) according to the manufacturer’s instructions.
Quantification of fungal colonization by qPCR
To estimate fungal abundance within plant tissues, absolute quantification of F. solani ITS gene was performed as previously described [Citation23]. Briefly, tissues were washed thoroughly and DNA was extracted using the CTAB method. An external standard curve was generated, using the 108 bp ITS fragment, cloned into pGEM-T Easy vector (www.promega.com); its concentration was determined via Qubit 2.0 Fluorometer. The copy number of the gene was calculated directly from the concentration of the extracted plasmid DNA [Citation36]. Serial 10-fold dilutions of the recombinant plasmid ranging from 6 × 10° to 6 × 107 copies/μl for ITS were subjected in triplicate to qPCR to construct the standard curve. Amplification occurred in a 10 μl reaction mixture containing Kapa SYBR FAST qPCR Master Mix Universal (1x) (www.sigmaaldrich.com), 200 nM of each primer, and 1 μl of DNA, using a thermocycling protocol of 3 min at 95°C; 45 cycles of 15 s at 95°C, 20 s at 58°C, followed by a melting curve to check the specificity of the products. Values are normalized to ng of total DNA isolated. Data are means of two technical replicates for each biological replicate of eight plants per time point and were analysed using the Student’s two-tailed homoscedastic t-test. The experiment was repeated at least twice.
Bisulphite sequencing
Genomic DNA from the fungus (20 ng) or the plant (100 ng) was used for bisulphite sequencing analysis using the EZ DNA Methylation-Gold Kit (www.zymoresearch.com) according to the manufacturer’s instructions and as previously described [Citation37] . Essentially, for the cis-RdDM bisulphite analysis on FsK, the primers Bis-F1 and Bis-R1 (Table S1) were used, whereas for the trans-RdDM bisulphite analysis on FsK and Nb-16C the primers Bis-F2 and Bis-R2 (Table S1) were used in a PCR reaction with ZymoTaq PreMix (www.zymoresearch.com) according to the manufacturer’s instructions. The occurring 262 bp and 311 bp amplicons for cis-RdDM and trans-RdDM, respectively, were cloned into pGEM®-T Easy Vector (www.promega.com) and for each analysis 5 clones were subjected to Sanger sequencing. The data were analysed by CyMATE software [Citation38] and the values (% methylation at CG, CHG at CHH context) obtained through its analysis results were used to produce the methylation bar graphs.
Small RNA sequencing
Sequencing of small RNAs from fungal RNA (small RNA fraction) was performed by GenXPro (https://genxpro.net/) as previously described [Citation37]. Essentially, RNA enriched for the sRNA fraction was extracted from RNA that was isolated with mirVana™ miRNA Isolation Kit (www.thermofisher.com) according to the manufacturer’s instructions. For preparation of sRNA libraries, 300 ng RNA (sRNA fraction) were spiked with 1 fmol Caenorhabditis elegans miR-39 RNA and ligated using the New England Biolabs Next Small RNA Library Prep (www.ned.de) to modified 3’ and 5’ adapters (TrueQuant RNA adaptors, GenXPro). Adapter-ligated RNA was reverse transcribed (First-Strand Synthesis System, www.lifetechnologies.com), purified with SPRI beads (SPRIselect, www.beckmancoulter.com) and PCR-amplified with nine cycles (KAPA HiFi Hot-Start Polymerase). The PCR products were purified (Amicon Utracel-10, www.emdmillipore.com) and size-selected by polyacrylamide gel electrophoresis. The sRNA library was sequenced on an Illumina HiSeq2000 with 1 × 50 bps. SRNA reads from 15 up to 40 nt were considered. The Tablet software was used for reading of the data and sRNA mapping depictions [Citation39].
Results and discussion
FsK colonizes the root system of Nicotiana benthamiana and stimulates plant growth
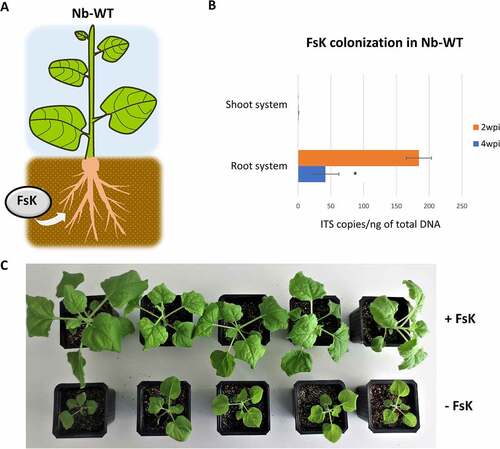
During the colonization process of its host plants, FsK penetrates the root and grows in the root cortex and proliferates even in the vascular system of root system [Citation23]. Notably, although not yet explained for, FsK growth in tomato is restricted to the root system and extends only to the crown and not to the stem and leaf tissues [Citation22]. Here, we investigated the capacity of FsK to colonize another member of the Solanaceae, Nicotiana benthamiana, which is a widely used model plant for RNAi studies [Citation40]. Similar to tomato, upon root-inoculation, the fungal endophyte colonized the root system but failed to expand to the shoot system () and Figure S1). Interestingly, the FsK-colonized plants exhibited considerably stimulated growth, at least up to 4 weeks post inoculation (wpi) when grown in both non-sterile compost and sterile sand () and Figure S2) underpinning the beneficial effect of FsK to this host, at least in terms of growth.
FsK encodes the core RNAi components
Despite being largely conserved among eukaryotes, not all fungi encode the core RNAi pathway; indeed, Saccharomyces cerevisiae lacks DCLs, AGOs and RDRs [Citation41]. Ustilago maydis also lacks DCLs, AGOs and RDRs, in contrast, surprisingly, to its close relative U. hordei [Citation42]. Furthermore, miRNAs have been identified in most fungal species but not in the basal fungus Mucor circinelloides [Citation15]. Interestingly though, whereas RNAi-deficient mutants of most ascomycetes and basidiomycetes are not impaired in vegetative growth and development, sexual differentiation and response to stress, M. circinelloides is severely impaired in these aspects [Citation43]. These being said, the mechanistic details and role of RNAi in fungal kingdom can be unusually diverse.
To examine whether FsK encodes the core RNAi machinery, we performed transcriptome-validated genome annotation and identified two DCLs (FsKDCL1 and FsKDCL2), two AGOs (FsKAGO1 and FsKAGO2) and four RDRs (FsKRDR1-4) ()). FsKDCL1 and FsKDCL2 contain the Dicer-like protein structures with a Dead-like helicases superfamily domain box (DEXDc) box, a helicase superfamily c-terminal domain (HELICc), and two ribonuclease III domains (RIBOc) responsible for the cleavage of dsRNA precursors into sRNAs [Citation5]. Both FsKAGO1 and FsKAGO2 proteins contain PAZ and PIWI domains; PAZ recognizes the 3’ end of sRNAs while PIWI exhibits an RNaseH-like endonucleolytic activity and mediates target cleavage [Citation44]. All four FsKRDRs contain the RdRP/RDR domain, which is highly conserved in fungi [Citation45]. FsKRDR2 and FsKRDR3 contain the DLDGD motif, which is often encountered in plants, whereas FsKRDR1 and FsKRDR4 contain the DYDGD motif, which is more common in fungi [Citation46]. To explore the molecular evolution of these proteins, we performed phylogenetic analyses of DCL, AGO and RDR proteins including Fusarium graminearum and Neurospora crassa ()). Our analysis showed that FsKDCL1 is related to FgDCL1 and NcDCL1 that function in the meiotic silencing by unpaired DNA (MSUD) pathway ()) [Citation47], whereas FsKDCL2 is closer to FgDCL2 and NcDCL2 which have a prominent role in RNAi by processing of dsRNAs into siRNAs [Citation45,Citation48]. FsKAGO1 is closely related to FgAGO1 and NcQDE2 that are loaded with dsRNA-processed siRNAs during RNAi, whereas FsKAGO2 is closer to FgAGO2 and the N. crassa SMS2 that are involved in MSUD [Citation49]. Of note, FsKRDR4 is closely related to NcQDE1 which is essential for quelling and suggested to be functionally related to plant RDR6 [Citation46].
Figure 2. Identification of FsK RNAi core machinery. (A) Schematic representation of FsK DCL, AGO and RDR proteins using DOG1.0 software [Citation50]. (B) Maximum likelihood phylogenies of the FsK (indicated red), Fusarium graminearum, Neurospora crassa and Arabidopsis thaliana (as an outgroup member) DCL, AGO and RDR proteins using the LG model matrix and 100 bootstrap replicates for assessing branch support.
![Figure 2. Identification of FsK RNAi core machinery. (A) Schematic representation of FsK DCL, AGO and RDR proteins using DOG1.0 software [Citation50]. (B) Maximum likelihood phylogenies of the FsK (indicated red), Fusarium graminearum, Neurospora crassa and Arabidopsis thaliana (as an outgroup member) DCL, AGO and RDR proteins using the LG model matrix and 100 bootstrap replicates for assessing branch support.](/cms/asset/b8f81de6-f2b8-49a3-8674-433409a161ce/krnb_a_2159158_f0002_oc.jpg)
FsK takes up RNAi molecules from its environment
In order to test the functionality of FsK’s RNAi machinery, an in vitro-transcribed 445 bp sGFP dsRNA was applied to a sGFP-expressing FsK ()), sGFP being a GFP variant that contains a serine-to-threonine substitution at amino acid 65, optimized for use in fungi [Citation34] ()). Fluorometric analysis revealed that the sGFP expression levels dropped to almost 50% 24 hours post application (hpa) (). These data suggested that the externally applied dsRNA was processed by fungal DCLs into siRNAs that were loaded onto fungal AGOs to mediate cleavage of the sGFP mRNA. However, no further decrease of sGFP levels could be observed at later timepoints (48 hpa), reminiscent of similar observations in F. asiaticum [Citation51] and implying the absence an active RDR-mediated self-reinforcing mechanism of RNAi that could ensure ongoing RNAi even at the absence/degradation of the initial dsRNA input.
Figure 3. In vitro RNAi in FsK-sGFP. (A) Schematic representation of the sGFP transgene that is present in FsK-sGFP. PToxA: promoter for the promoter from Pyrenophora tritici-repentis ToxA gene; sGFP: GFP variant that contains a serine-to-threonine substitution at amino acid 65; TNOS: terminator for the nopaline synthase gene. The 445 bp fragment chosen for in vitro transcription of dsRNA is depicted. (B) Stereoscopic observation of sGFP fluorescence. (C) Fluorometric analysis for in vitro RNAi in FsK-sGFP. Vertical axis: RFU: relative fluorescence unit, calculated as the ratio of sGFP-indicative fluorescence (excitation 488 nm, emission 515 nm) to growth-indicative absorbance (wavelength 595 nm). Horizontal axis: 1–12: 12 wells containing FsK-sGFP conidia. 13–24: 12 wells containing FsK-sGFP conidia plus 100 ng (each well) sGFP dsRNA.
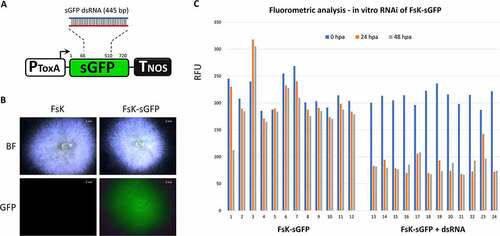
Overall, these data suggest not only that the RNAi machinery in FsK is functional but also that FsK is able to take up RNAi molecules from its environment. Not all fungi are able to take up RNA molecules from their environment; Colletotrichum gloeosporioides, Trichoderma virens and Phytophthora infestans being some notable examples that fail to do so [Citation52]. Of note, fungi that are indeed able to receive RNAi molecules from their environment are not only promising candidates for RNAi-based fungicidal control [Citation53] but also likely partners in an RNAi-based cross-kingdom communication with their host [Citation54].
FsK processes hairpin RNA transcripts into siRNAs that trigger mRNA degradation but not DNA methylation in the fungal hyphae
In order to examine the mode of dsRNA processing in the endophyte, FsK was transformed with a transgene comprising a full length green fluorescent protein (GFP) corresponding to mGFP5-ER [Citation55] and a hairpin (hp) construct of the first 322 bp of GFP (hpGF) (). In this setup, the hpGF locus served as the RNAi-trigger while the GFP locus as the RNAi-target. Small RNA sequencing (sRNA-seq) in three independent FsK-hpGF+GFP transformants (#6, #7, #27) revealed the accumulation of GF siRNAs (perfectly matching the GF region) having variable sizes from 18 to 30 nt but predominantly of 21-nt, 22-nt and 24-nt , and S3). This finding was reminiscent of the situation in plants, where hpRNAs are typically processed by DCLs to 21-, 22- and 24-nt siRNAs [Citation56]. To the best of our knowledge, similar sRNA-seq studies in fungi, aiming to reveal the mode of processing of a specific hpRNA/dsRNA, are absent; yet, genome-wide sRNA-seq studies (identifying siRNAs, miRNAs but also DCL-independent sRNAs) reveal a remarkably diverse pattern, with prominent size classes ranging from 19–22-nt in Penicillium chrysogenum [Citation57], to 22–25-nt in S. pombe [Citation58] and 27–28-nt in F. graminearum [Citation45]. Our analysis does not allow us to identify whether all sRNA size classes are actual DCL products (e.g. they could represent degradation products) or whether they all exhibit biological activity.
Figure 4. Characterization of FsK RNAi machinery. (A) Schematic representation of the hpGF+GFP transgene. PTrpC: promoter for the Promoter for Aspergillus nidulans trpC gene; GF: 322 bp fragment of the GFP; intron: Magnaporthe grisea cutinase gene intron; TTrpC: promoter for the Promoter for Aspergillus nidulans trpC gene, P35S: Cauliflower mosaic virus 35S promoter; GFP: full-length (792 bp) green fluorescent protein (mGFP-ER version); TNOS: terminator for the nopaline synthase gene. FsK-hpGF+GFP transformants contain the full length hpGF+GFP transgene, whereas FsK-GFP transformants contain only the P35S-GFP-TNOS part of the transgene. Although hpGF+GFP and GFP transgenes correspond to different insertional events, the GFP expression in both transgenes is controlled by the same genetic elements (P35S and TNOS), thus GFP is expected to exhibit similar expression rates in both transgenes. (B) sRNA-seq in FsK-hpGF+GFP #27. All sRNA reads of 18–30-nt fully matching to GFP region are depicted. With light blue the siRNA reads in plus polarity, with dark blue the siRNA reads in minus polarity. The Tablet software [Citation39] was used for visualization of the sRNA reads. (C) Pie graph of the 18–30 nt GFP siRNAs in FsK-hpGF+GFP #27. (D) RT-qPCR for the estimation of GFP mRNA downregulation in FsK-hpGF+GFP compared to FsK-GFP, (E) Bisulphite sequencing for cis RdDM. (F) Bisulphite sequencing for trans RdDM.
![Figure 4. Characterization of FsK RNAi machinery. (A) Schematic representation of the hpGF+GFP transgene. PTrpC: promoter for the Promoter for Aspergillus nidulans trpC gene; GF: 322 bp fragment of the GFP; intron: Magnaporthe grisea cutinase gene intron; TTrpC: promoter for the Promoter for Aspergillus nidulans trpC gene, P35S: Cauliflower mosaic virus 35S promoter; GFP: full-length (792 bp) green fluorescent protein (mGFP-ER version); TNOS: terminator for the nopaline synthase gene. FsK-hpGF+GFP transformants contain the full length hpGF+GFP transgene, whereas FsK-GFP transformants contain only the P35S-GFP-TNOS part of the transgene. Although hpGF+GFP and GFP transgenes correspond to different insertional events, the GFP expression in both transgenes is controlled by the same genetic elements (P35S and TNOS), thus GFP is expected to exhibit similar expression rates in both transgenes. (B) sRNA-seq in FsK-hpGF+GFP #27. All sRNA reads of 18–30-nt fully matching to GFP region are depicted. With light blue the siRNA reads in plus polarity, with dark blue the siRNA reads in minus polarity. The Tablet software [Citation39] was used for visualization of the sRNA reads. (C) Pie graph of the 18–30 nt GFP siRNAs in FsK-hpGF+GFP #27. (D) RT-qPCR for the estimation of GFP mRNA downregulation in FsK-hpGF+GFP compared to FsK-GFP, (E) Bisulphite sequencing for cis RdDM. (F) Bisulphite sequencing for trans RdDM.](/cms/asset/3e3a65a5-7397-49dc-a8f0-d32ee3cb23fe/krnb_a_2159158_f0004_oc.jpg)
To evaluate this RNAi activity, we measured the GF siRNA-mediated downregulation of GFP mRNA in three independent FsK-hpGF+GFP transformants (#6, #7, #27) when compared to FsK-GFP (transformed with a cassette lacking the hpGF transgene) ()). Indeed, GFP expression was virtually eliminated in all FsK-hpGF+GFP transformants ()). Of note, we detected GF siRNAs but no or negligible P siRNAs that could had potentially emerged upon the FsKRDR processing on the GF siRNA-targeted GFP transcript ()). This is in contrast to the situation in plants [Citation13] but in agreement with similar reports in F. asiaticum [Citation51], suggesting the absence of an active RDR-based RNAi mechanism in FsK.
Typically, the onset of RNAi and the accumulation of siRNAs leads to RdDM in plants [Citation59]. DNA methylation also occurs in some, but not all, fungi, and usually in repetitive sequences [Citation60]. Yet, such DNA methylation is considered to be dispensable of RNAi molecules, thus fungi have been considered to lack a bona fide RdDM mechanism [Citation61]. Nevertheless, recent advances challenge this assumption; indeed, sRNA-dependent RdDM-like phenomena has been detected, at least in Pleurotus tuoliensis and P. eryngii var. eryngii (Basiodiomycetes) [Citation62] and Puccinia graminis (Ascomycetes) [Citation63]. Accordingly, and given the abundant accumulation of GF siRNAs in FsK-hpGF+GFP, we were interested to see whether they could trigger RdDM of cognate DNA sequences. To analyse cis-RdDM (at the locus generating the siRNAs), we chose a 262 bp fragment of the hpGF transgene ()). For trans-RdDM (at a locus that does not generate siRNAs but is homologous to them), we chose a 311 bp fragment of the GFP transgene ()). Whereas CG and CHG methylation can be maintained in an RNAi-independent manner [Citation64], CHH methylation is the hallmark of ongoing de novo RdDM [Citation65], and both cis and trans fragments under analysis were rich in asymmetric CHH context (80% for cis and 72% for trans) ()). However, bisulphite sequencing revealed the absence of methylated cytosines in any sequence context (CG, CHG, CHH), at neither cis ()) nor trans ()) loci, suggesting that no RdDM takes place in FsK, at least in our experimental setup. It has been suggested that fungal proteins with de novo methyltransferase (DNMT) and/or helicase-like Snf2 family domains may be involved in RdDM-like pathways in fungi [Citation61]. However, we were unable to detect such genes in the FsK genome, underpinning the conclusions obtained from bisulphite sequencing about the absence of an active RdDM mechanism in FsK.
FsK translocates RNAi signals to its host to induce systemic RNAi and epigenetic changes of a reporter gene
Establishment of mutualistic associations between fungi and their host requires genetic and epigenetic reprogramming as well as metabolome modulation of both by the exchange of effector molecules [Citation66]. Indeed, RdDM is essential in Arabidopsis to establish a beneficial relationship with the root-colonizing Trichoderma atroviride while DNA methylation and histone modifications are required for plant priming by the beneficial fungus against B. cinerea [Citation67]. Importantly, it was just recently shown that during the mutualistic interaction of the ectomycorrhizal fungus Pisolithus microcarpus with Eucalyptus grandis, a fungal miRNA, Pmic_miR-8, targets the host NB-ARC domain containing transcripts in a cross-kingdom RNAi manner [Citation68]. Reminiscent of this, an in silico study predicted that the beneficial arbuscular mycorrhizal fungi Rhizophagus irregularis produces sRNAs that have 237 candidate targets in the host plant Medicago truncatula, including specific mRNAs known to be modulated in roots upon AMF colonization [Citation69]. Similarly, a recent study based on transcriptome and sRNA profile change analysis during the onset of the mutualistic interaction between the beneficial root endophyte Serendipita indica with its host Brachypodium distachyon, suggested that interaction-induced sRNAs in both organisms may underlie reciprocal targeting of genes related to plant development and fungal growth and nutrient acquisition [Citation70]. Thus, it is very likely that, similar to fungal pathogens [Citation71], beneficial fungal endophytes also display an RNA-based communication with their hosts. However, clear evidence of actual RNAi molecule translocation and concomitant cross-kingdom RNAi between a beneficial fungal endophyte and its host has been lacking to this date.
In order to address this question, we resorted to the GFP-expressing N. benthamiana plant-line 16C (Nb-GFP) [Citation32], as an RNAi sensor system. Nb-GFP carries a 35S-driven mGF5-ER transgene () and is a well-studied RNAi model plant that allows the monitoring of systemic RNAi (i.e. spreading of RNAi to tissues other than those where RNAi initially occurred) by observation of the presence or abolishment of GFP expression under ultraviolet light. When Nb-GFP plants were inoculated with FsK-hpGF+GFP () and Figure S4), we could record the following outcomes: (i) no visible RNAi (45% of the plants, 6 wpi), (ii) spot-like RNAi (45% of the plants, 4 wpi), (iii) vein-restricted RNAi (5% of the plants, 4 wpi) and (iv) full-tissue RNAi (5% of the plants, 4 wpi) ()). Interestingly, we recorded a positive correlation between the level of endophyte colonization and the efficiency of RNAi in Nb-GFP (Figure S5) suggesting that certain threshold levels (RNAi molecules levels) are associated with the extent of RNAi establishment and spreading. Of note, colonization of Nb-GFP plants with non-transformed FsK and/or FsK-sGFP failed to trigger any visible RNAi phenotype even after 10 wpi, suggesting that it is not the mere presence of the endophyte but the specific RNAi molecules it expresses that are responsible for the induction of RNAi phenotypes in its host.
Figure 5. FsK-hpGF+GFP colonization of Nb-GFP. (A) Schematic overview of the colonization assay. (B) Systemic silencing phenotypes under ultraviolet light 4–6 wpi. (C) Bisulphite sequencing in the host GFP transgene in both roots and leaves in silenced Nb-GFP plants.
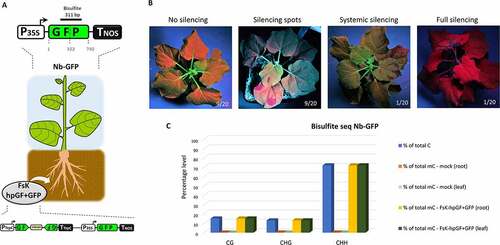
RNAi in plants is tightly coupled to RdDM [Citation59,Citation72]. Accordingly, bisulphite sequencing analysis of leaf and root tissues from the single fully/homogeneously silenced Nb-GFP plant ()) disclosed the dense (100%) onset of DNA methylation in the GFP region ()) in every sequence context: CG, CHG and CHH ()). Overall, these data show that, at least in the case of the fully silenced plant ()), the endophyte triggered not only mRNA degradation but also DNA methylation of a host reporter gene. We favour the scenario that FsK-hpGF+GFP translocated RNAi signals (possibly dsRNAs but most likely siRNAs) to the roots of Nb-GFP initiating local RNAi of the host GFP. Importantly, once present in the plant cells and upon targeting the host GFP transcript for silencing, these endophyte-derived primary siRNAs culminated in the generation of host-derived RDR-mediated secondary siRNAs (as implied by the RdDM pattern, see below). Whether the recorded RdDM in the root tissues was induced by the endophyte-derived primary or the host-derived secondary siRNAs is not clear. Yet, the fact that RdDM could be detected not only in the GF but also in the P region (), 311 bp bisulphite fragment covering both GF and P regions) strongly implies in favour of transitive host-derived secondary siRNAs imposing RdDM. Now, siRNAs are mobile moieties; they can move cell-to-cell through the plasmodesmata and through the vasculature to distant parts of the plant [Citation32,Citation73]. The establishment of systemic silencing in the upper parts of the plant (which FsK fails to colonize, ) suggests that mobile siRNA signals from the root entered the phloem to reach shoot tissues. Most likely both endophyte-derived primary siRNAs and host-derived secondary siRNAs could have played the role of the mobile systemic signal [Citation74]. However, it is unlikely that the mere presence of endophyte-derived primary siRNAs alone could trigger systemic silencing; it rather seems that a certain quantitative siRNA threshold needs to be surpassed for the onset of systemic silencing [Citation75], rendering the abundant presence of host-derived secondary siRNAs indispensable for this process. Importantly, establishment of systemic RNA in the receiving tissues requires RDR6 [Citation76]. Thus, in the receiving tissues, the primary/secondary siRNAs triggered a RDR6-mediated generation of (host-derived) tertiary siRNAs, ensuring the efficient establishment of GFP mRNA degradation and DNA methylation.
Conclusion
Overall, we provide solid evidence that FsK translocates RNAi signals to its host to trigger systemic silencing and epigenetic modifications. To prove the concept, we have employed an artificial RNAi sensor system; future studies coupling sRNAome, degradome and methylome analysis will be required to pinpoint the nature of the endogenous fungal sRNAs (siRNAs and/or miRNAs) that are translocated to the host, which host genes are targeted for transcriptional and/or post-transcriptional silencing and how this process is ultimately translated into a beneficial phenotype. One may even envisage that the elucidation of these small RNAs and their incorporation in RNA-based agricultural products [Citation77] may prove useful for the development of sustainable RNA-based solutions to improve plant productivity and/or plant health. Our data may well reflect a so far unrecognized pathway according to which endophytes establish the mutualism and/or impose their beneficial impact by translocating RNA molecules that modulate host gene expression and affect the epigenome’s plasticity. Overall, it seems that RNAi-mediated communication between plants and their interacting organisms is much more widespread than previously thought and may account for the improved plant performance often observed in the presence of certain associated microbiota.
Author contributions
A.D. and K.K.P. designed research; A.D., A.K., M.A., E.D., A.M. M.G. and E.D., performed research; A.D., O.T. S.V. and K.K.P analysed data; A.D. and K.K.P. wrote the paper. All authors reviewed and approved the manuscript.
Data availability
All sequencing data supporting the findings of this study are deposited to Zenodo (https://doi.org/10.5281/zenodo.6088855).
Supplemental Material
Download MS Word (4.5 MB)Disclosure statement
Fusarium solani FsK is patented (20,070,100,563/1,006,119, issued by the Industrial Property Organization to KKP).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2022.2159158.
Additional information
Funding
References
- Hung Y-H, Slotkin RK. The initiation of RNA interference (RNAi) in plants. Curr Opin Plant Biol. 2021;61:102014.
- Baulcombe D. RNA silencing in plants. Nature. 2004;431(7006):356–363.
- Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16(12):727–741.
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20(7):759–771.
- Paturi S, Deshmukh MV. A glimpse of “Dicer Biology” through the structural and functional perspective. Front Mol Biosci. 2021;8:643657.
- Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13(7):350–358.
- Iwakawa H-O, Tomari Y. Life of RISC: formation, action, and degradation of RNA-induced silencing complex. Mol Cell. 2022;82(1):30–43.
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952.
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320(5880):1185–1190.
- Wassenegger M, Dalakouras A. Viroids as a tool to study RNA-directed DNA methylation in plants. Cells. 2021;10(5):1187.
- Wassenegger M, Heimes S, Riedel L, et al. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76(3):567–576.
- Sakurai Y, Baeg K, Lam AYW, et al. Cell-free reconstitution reveals the molecular mechanisms for the initiation of secondary siRNA biogenesis in plants. Proceedings of the National Academy of Sciences 2021; 118:e2102889118.
- Yuan, de Felippes Ff R, Waterhouse PM. The whys and wherefores of transitivity in plants. Front Plant Sci. 2020;11:11.
- Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6(22):3343–3353.
- Torres-Martinez S, Ruiz-Vazquez RM. The RNAi universe in fungi: a varied landscape of small RNAs and biological functions. Annu Rev Microbiol. 2017;71(1):371–391.
- Lax C, Tahiri G, Patiño-Medina JA, et al. The evolutionary significance of RNAi in the Fungal Kingdom. Int J Mol Sci. 2020;22(1):21.
- Li L, Chang SS, Liu Y. RNA interference pathways in filamentous fungi. Cell Mol Life Sci. 2010;67(22):3849–3863.
- Weiberg A, Wang M, Lin FM, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342(6154):118–123.
- Wang M, Weiberg A, Lin FM, et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants. 2016;2(10):16151.
- Werner B, Koch A, Šečić E, et al. Fusarium graminearum DICER-like-dependent sRNAs are required for the suppression of host immune genes and full virulence. bioRxiv 2021:2021.05.17.444440.
- Zhang T, Zhao YL, Zhao JH, et al. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants. 2016;2(10):16153.
- Kavroulakis N, Ntougias S, Zervakis GI, et al. Role of ethylene in the protection of tomato plants against soil-borne fungal pathogens conferred by an endophytic Fusarium solani strain. J Exp Bot. 2007;58(14):3853–3864.
- Skiada V, Faccio A, Kavroulakis N, et al. Colonization of legumes by an endophytic Fusarium solani strain FsK reveals common features to symbionts or pathogens. Fungal Genet Biol. 2019;127:60–74.
- Pappas ML, Liapoura M, Papantoniou D, et al. The beneficial endophytic fungus Fusarium solani Strain K alters tomato responses against spider mites to the benefit of the plant. Front Plant Sci. 2018;9:1603.
- Garantonakis N, Pappas ML, Varikou K, et al. Tomato inoculation with the endophytic strain fusarium solani K results in reduced feeding damage by the zoophytophagous predator nesidiocoris tenuis. Front Ecol Evol. 2018; 6
- Kavroulakis N, Doupis G, Papadakis IE, et al. Tolerance of tomato plants to water stress is improved by the root endophyte Fusarium solani FsK. Rhizosphere. 2018;6:77–85.
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5(1):113.
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–577.
- Darriba D, Taboada GL, Doallo R, et al. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165.
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–1320.
- Guindon S, Gascuel O, Rannala B. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704.
- Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature. 1997;389(6651):553.
- Zhang T, Ren P, Chaturvedi V, et al. Development of an Agrobacterium-mediated transformation system for the cold-adapted fungi Pseudogymnoascus destructans and P. pannorum. Fungal Genet Biol. 2015;81:73–81.
- Sesma A, Osbourn AE. The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature. 2004;431(7008):582–586.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Whelan JA, Russell NB, Whelan MA. A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods. 2003;278(1–2):261–269.
- Dalakouras A, Dadami E, Wassenegger M, et al. RNA-directed DNA methylation efficiency depends on trigger and target sequence identity. Plant J. 2016;87(2):202–214.
- Hetzl J, Foerster AM, Raidl G, et al. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51(3):526–536.
- Milne I, Stephen G, Bayer M, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14(2):193–202.
- Philips JG, Naim F, Lorenc MT, et al. The widely used Nicotiana benthamiana 16c line has an unusual T-DNA integration pattern including a transposon sequence. PLoS One. 2017;12(2):e0171311.
- Drinnenberg IA, Weinberg DE, Xie KT, et al. RNAi in budding yeast. Science. 2009;326:544–550.
- Laurie JD, Linning R, Bakkeren G. Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis. Curr Genet. 2008;53(1):49–58.
- Ruiz-Vazquez RM, Nicolas FE, Torres-Martinez S, et al. Distinct RNAi pathways in the regulation of physiology and development in the fungus Mucor circinelloides. Adv Genet. 2015;91:55–102.
- Wu J, Yang J, Cho WC, et al. Argonaute proteins: structural features, functions and emerging roles. J Adv Res. 2020;24:317–324.
- Chen Y, Gao Q, Huang M, et al. Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci Rep. 2015;5(1):12500.
- Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11(3):142–151.
- Alexander WG, Raju NB, Xiao H, et al. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet Biol FG & B. 2008;45(5):719–727.
- Carreras-Villaseñor N, Esquivel-Naranjo EU, Villalobos-Escobedo JM, et al. The RNAi machinery regulates growth and development in the filamentous fungus Trichoderma atroviride. Mol Microbiol. 2013;89(1):96–112.
- Lee DW, Pratt RJ, McLaughlin M, et al. An argonaute-like protein is required for meiotic silencing. Genetics. 2003;164(2):821–828.
- Ren J, Wen L, Gao X, et al. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19(2):271–273.
- Song X-S, Gu K-X, Duan -X-X, et al. Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol Plant Pathol. 2018;19(12):2543–2560.
- Qiao L, Lan C, Capriotti L, et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol J. 2021;19(9):1756–1768.
- Šečić E, Kogel K-H. Requirements for fungal uptake of dsRNA and gene silencing in RNAi-based crop protection strategies. Curr Opin Biotechnol. 2021;70:136–142.
- He B, Hamby R, Jin H. Plant extracellular vesicles: trojan horses of cross-kingdom warfare. FASEB Bioadv. 2021;3(9):657–664.
- Haseloff J, Siemering KR. The uses of green fluorescent protein in plants. Methods Biochem Anal. 2006;47:259–284.
- Fusaro AF, Matthew L, Smith NA, et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7(11):1168–1175.
- Dahlmann TA, Kück U, Herrera-Estrella A. Dicer-dependent biogenesis of small RNAs and evidence for MicroRNA-like RNAs in the penicillin producing fungus penicillium chrysogenum. PLOS ONE. 2015;10(5):e0125989.
- Djupedal I, Kos-Braun IC, Mosher RA, et al. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J. 2009;28(24):3832–3844.
- Dalakouras A, Vlachostergios D, Manavella P. Epigenetic approaches to crop breeding: current status and perspectives. J Exp Bot. 2021;72(15):5356–5371.
- Bewick AJ, Hofmeister BT, Powers RA, et al. Diversity of cytosine methylation across the fungal tree of life. Nat Ecol Evol. 2019;3(3):479–490.
- Nai Y-S, Huang Y-C, Yen M-R, et al. Diversity of fungal DNA methyltransferases and their association with DNA methylation patterns. Front Microbiol. 2021;11:616922.
- Zhang Z, Wen J, Li J, et al. The evolution of genomic and epigenomic features in two pleurotus fungi. Sci Rep. 2018;8(1):8313.
- Sperschneider J, Jones AW, Nasim J, et al. The stem rust fungus Puccinia graminis f. sp. tritici induces centromeric small RNAs during late infection that are associated with genome-wide DNA methylation. BMC Biol. 2021;19(1):203.
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220.
- Pelissier T. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999;27(7):1625–1634.
- Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21(14):1204–1209.
- Rebolledo-Prudencio OG, Estrada-Rivera M, Dautt-Castro M, et al. The small RNA-mediated gene silencing machinery is required in Arabidopsis for stimulation of growth, systemic disease resistance, and suppression of the nitrile-specifier gene NSP4 by Trichoderma atroviride. Plant J. 2022;109(4):873–890.
- Wong-Bajracharya J, Singan VR, Monti R, et al. The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc Natl Acad Sci U S A. 2022 Jan 18;119(3):e2103527119.
- Silvestri A, Fiorilli V, Miozzi L, et al. In silico analysis of fungal small RNA accumulation reveals putative plant mRNA targets in the symbiosis between an arbuscular mycorrhizal fungus and its host plant. BMC Genomics. 2019;20(1):169.
- Secic E, Zanini S, Wibberg D, et al. A novel plant-fungal association reveals fundamental sRNA and gene expression reprogramming at the onset of symbiosis. BMC Biol. 2021;19(1):171.
- Cai Q, He B, Jin H. A safe ride in extracellular vesicles – small RNA trafficking between plant hosts and pathogens. Curr Opin Plant Biol. 2019;52:140–148.
- Jones L, Hamilton AJ, Voinnet O, et al. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11(12):2291–2301.
- Voinnet O. Revisiting small RNA movement in plants. Nat Rev Mol Cell Biol. 2022;23(3):163–164.
- Devers EA, Brosnan CA, Sarazin A, et al. Movement and differential consumption of short interfering RNA duplexes underlie mobile RNA interference. Nat Plants. 2020;6(7):789–799.
- Kalantidis K, Tsagris M, Tabler M. Spontaneous short-range silencing of a GFP transgene in Nicotiana benthamiana is possibly mediated by small quantities of siRNA that do not trigger systemic silencing. Plant J. 2006;45(6):1006–1016.
- Schwach F, Vaistij FE, Jones L, et al. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138(4):1842–1852.
- Dalakouras A, Wassenegger M, Dadami E, et al. Genetically modified organism-Free RNA interference: exogenous application of RNA molecules in plants. Plant Physiol. 2020;182(1):38–50.