ABSTRACT
In recent years, advances in biomedicine have revealed an important role for post-transcriptional mechanisms of gene expression regulation in pathologic conditions. In cancer in general and leukaemia specifically, RNA binding proteins have emerged as important regulator of RNA homoeostasis that are often dysregulated in the disease state. Having established the importance of these pathogenetic mechanisms, there have been a number of efforts to target RNA binding proteins using oligonucleotide-based strategies, as well as with small organic molecules. The field is at an exciting inflection point with the convergence of biomedical knowledge, small molecule screening strategies and improved chemical methods for synthesis and construction of sophisticated small molecules. Here, we review the mechanisms of post-transcriptional gene regulation, specifically in leukaemia, current small-molecule based efforts to target RNA binding proteins, and future prospects.
1. Background
Dysregulation of gene expression is central to many disease states and occurs at several levels of both transcriptional and post-transcriptional control. While transcriptional control and dysregulation have been studied extensively, the role of post-transcriptional mechanisms have only recently come into focus as a significant pathogenetic mechanism. These mechanisms prominently involve the activity of RNA-binding proteins (RBPs), which are a functionally and structurally heterogeneous class of proteins that are only unified by their ability to bind to RNAs. RBPs control a range of processes that are important for the synthesis and homoeostasis of mRNA molecules. Moreover, RBPs have been reported to be dysregulated in multiple disease states, with pathologies typically manifesting due to altered expression levels. This type of dysregulation lends itself to pharmaceutical targeting, particularly in cases where RBPs are overexpressed. A significant amount of effort has been expended in targeting RBPs via oligonucleotide or other nucleic acid-based techniques, with some notable successes particularly in neurological diseases [Citation1,Citation2]. Here we focus on the current state of therapeutically targeting of RBPs with small molecules.
2. RNA-binding proteins regulate a range of RNA homoeostatic mechanisms within the cell
Within the cell, there are three main RNA polymerases that transcribe RNA from DNA [Citation3]. RNA polymerase I is primarily responsible for the transcription of ribosomal RNA, RNA polymerase II is the enzyme that transcribes the bulk of messenger RNA (mRNA) and RNA polymerase III is responsible primarily for transcribing short RNAs such as transfer RNAs and small nucleolar RNAs. Of these polymerases, RNA polymerase II is subject to regulation by DNA-binding proteins, such as transcription factors and epigenetic regulators. This interaction with DNA-binding proteins by RNA polymerase II is the first step in the regulation of gene expression. However, once the pre-mRNA is produced, it undergoes a series of processing steps, including splicing, capping and polyadenylation mediated by RBPs [Citation4–6]. There are a number of RBPs that may participate in ‘folding’ or generation of RNA secondary structures (RNA helicases) [Citation7]. mRNA molecules are subject to modifications, constituting so-called epitranscriptomic marks [Citation8,Citation9]. This latter process appears to be dynamic, with some proteins depositing the modifications (i.e. ‘writers’) and others that remove the modifications (‘erasers’), with N6-methyladenosine (m6A) being the most common epitranscriptomic mark, although more than a hundred have been catalogued [Citation10]. The last step in gene expression is for the mRNAs to be translated in the ribosome into proteins, which themselves are subject to further modification by a variety of covalent modifications.
Mature, modified mRNA molecules can be bound by RBPs that regulate their stability and access to translational machinery within the cell. One important class of such RBPs are the Argonaute proteins, which together with accessory proteins constitute the RNA-induced silencing complex (RISC). The activity of RISC, in turn, is dependent on microRNAs, which are small RNA molecules that bind to mRNAs via sequence homology. These are generated from genes encoding miRNAs by RNA polymerase III-based transcription followed by a series of endoribonucleolytic processing steps. miRNA/RISC-based repression of mRNA stability and translation has been posited as one of the major mechanisms regulating post-transcriptional gene expression.
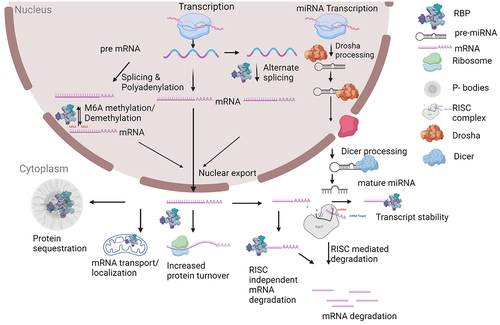
From the preceding discussion, it should be apparent that RBPs can be characterized based on their cell biological function: splicing factors, capping enzymes, polyadenylation factors, RNA helicases, epitranscriptomic writers, erasers and readers. The function of RBPs can also be characterized as occurring via the regulation of RNA stability (RISC-dependent or -independent), or via the localization and transport of mRNA molecules (See for a summary of RNA homoeostasis). These different ways to conceptualize RBP function are overlapping and non-exclusive and are discussed in the chapters below. We also attempt to discuss novel potential therapeutics in light of these different conceptualizations of RBP function.
Figure 1. Schematic of RNA homoeostasis. Gene expression initates with transcription of DNA into a pre-mRNA. Numerous steps of RNA processing occur, many in parallel with transcription. A separate pathway of microRNA (miRNA) processing occurs for primary transcripts that encode miRNAs. Please see text for further details. Schematic created at Biorender.com.
3. RBP dysregulation in cancer and leukaemia
RBPs show a range of dysregulation in cancer, including upregulation and downregulation as well as mutation [Citation11]. These include proteins involved in a range of different RNA-related functions, and many functional studies have now documented a causal or supportive role in driving malignant transformation, including many of the hallmarks of cancer [Citation12]. Splicing factors show altered expression and/or mutation in a range of haematologic malignancies [Citation13], including acute myeloid leukaemia, myelodysplastic syndrome and chronic lymphocytic leukaemia, as well as in solid tumours. Experimentally, the expression of mutant splicing factors has been shown to recapitulate some features of disease, such as with the expression of point-mutated SF3B1 and SRSF2 in myelodysplastic syndrome [Citation14,Citation15]. In addition to splicing factors, RBPs that are involved in epitranscriptomic modifications have been shown to play a role in cancer, such as the m6A writer METTL13 and the m6A eraser FTO, which play functional roles in acute leukaemia [Citation16,Citation17]. Readers of these same modifications, such as the YTHDF2 and IGF2BP2 proteins seem to stabilize specific m6A-modified mRNA transcripts and promote their translation [Citation18,Citation19]. Further, several factors important in microRNA biogenesis, such as the LIN28 proteins, appear to modulate cancer causation [Citation20]. Lastly, a number of proteins that are important in RNA stability are implicated in cancer causation. These include HuR (ELAVL1), which has been extensively studied and has an effect on both RISC-dependent and RISC-independent methods of mRNA stability [Citation21–24]. IGF2BP3, which may also play a role in interpreting m6A signals, acts via an effect on mRNA stability, likely via the RISC [Citation25–27]. However, it should be noted that some conflicting data exist as to the pro- or anti-oncogenic function of IGF2BP3 in specific subtypes of leukaemia [Citation28]. Lastly, components of the translation initiation complex, e.g. eIF4E, have been shown to be overexpressed and functional in cancer progression and metastasis [Citation29]. Together, these functional studies indicate the importance of RBPs and post-transcriptional pathways in driving cancer and indicate that they may represent good targets for cancer therapies.
4. Considerations for designing therapeutics against RBPs
Interestingly, the best functional data for RBPs’ specific role in cancer has been demonstrated for those RBPs which are overexpressed in cancer. In these functional studies, knockdown and/or knockout of the RBPs in a cancer driven by known oncogenes resulted in reduced intensity and/or elimination of various cancer characteristics. Given that chemical antagonism/inhibition may be more readily achievable than chemical agonism, we suggest that targeting RBPs with overexpression in cancer holds high promise [Citation30–32]. The multiplicity of RNA targets bound by RBPs have led to the suggestion of ‘housekeeping’ functions for many of these proteins [Citation33]. While there are clearly RBPs that have high basal expression in many tissues and may truly be ‘housekeeping genes’ with little differential expression in cancer, many RBPs show temporally and spatially restricted expression patterns, including some that demonstrate an oncofetal pattern. A careful analysis of existing data on RBP expression should be carried out to assess for a ‘therapeutic window’. Further, it has been suggested that RBPs that bind RNA in a sequence-independent manner appear to be associated with constitutive or housekeeping function, while those with sequence specific binding appear to have a more specific function [Citation34]. Such sequence specific RBPs with temporal and spatial restricted patterns of expression may therefore be ideal candidates for cancer-specific inhibition. Hence, structure-function and structural analyses of the protein may also assist in understanding the requirements for RNA binding. In addition, high throughput analyses of RNA binding sites, such as those obtained from CLIP-seq, and identification of direct regulatory mRNA targets, by combining binding site data with differential expression analyses, may aid in designing specific assays to measure on-target activity of a therapeutic devised against a specific RBP. Various modalities have been used to target RBPs, including oligonucleotides, peptides, and small molecules. Each of these has distinct advantages and disadvantages; in the current article, we will review small molecule approaches. Small molecule therapeutics are a desirable modality over other strategies for targeting RBPs due to their generally favourable pharmacokinetic properties, the potential to be administered orally and to pass through cell membranes to reach intracellular targets. There are other reviews that more globally consider these alternative approaches [Citation35,Citation36].
5. Structural features of RBPs and considerations for targeting
The binding of RBPs may be dependent on primary sequence, secondary structure, as well as epigenetic modification (e.g. m6A) of the mRNA molecules. Many RBPs are characterized by containing one or more RNA-binding domains (RBDs) that bind to specific RNA sequences and structural motifs. The most well-defined and prevalent RBDs are the RNA recognition motif (RRM), hnRNP K homology (KH), DEAD/DEAH helicase and zinc-finger domains [Citation37]. However, many RBPs lack these domains, and the domains themselves do not necessarily confer a specific mRNA sequence that is bound. Furthermore, many RBPs exist in large multiunit complexes, as has been well demonstrated for the splicing factors that work together in the spliceosome [Citation4]. Classically, RBPs were considered undruggable, as most lack enzymatic activity that provides a handy read-out for measuring on-target activity [Citation30]. Rational design of small molecules was also considered to be difficult, given the lack of highly specific structural motifs for intermolecular interactions or binding pockets within RBPs. Indeed, combinatorial recognition of target RNA sequences may be the rule for many RBPs, as has been demonstrated for IGF2BP3, rendering it difficult to design a small molecule that can target the binding sites [Citation38]. Additionally, in vivo data to demonstrate the anti-cancer effect of RBP loss-of-function was previously lacking for many proteins. These features hindered the development of small molecule-based approaches to targeting RBPs. Recently, however, the advent and refinement of high-throughput screening approaches have allowed for the identification of small molecule inhibitors of RBPs which were once considered undruggable. We first briefly review some of these approaches before discussing specific inhibitors of RBPs.
6. High throughput screening approaches to developing RBP-targeted small molecule therapeutics
Multiple strategies have been used for high throughput screening to discover inhibitors of RBPs. In this section we discuss some commonly used screening techniques in drug discovery.
6.1. F ӧrster resonance energy transfer (FRET) based assay
FRET based methods are powerful tools and a highly efficient platform for drug discovery [Citation39]. The assay is dependent on the overlap of fluorescence emission spectrum of a donor dye, linked to RNA, with the excitation spectrum of an acceptor dye, linked to an RBP, and physical proximity of the RBP and bound RNA [Citation40]. Binding of a small molecule inhibitor with the protein restricts the binding of the RNA ligand; as a result, there is a gain/loss of endpoint fluorescence signal depending on the assay type. For example, a small molecule inhibitor SB1301 (10.4, ) of the LIN28 protein, which regulates miRNA biogenesis, was identified using the FRET-based method [Citation41].
6.2. Fluorescence polarization (FP) assay
This method has been widely used for the high throughput screening of compounds for drug discovery. The method is homogenous, sensitive, robust and relatively insensitive to certain types of technical interference [Citation42]. The principle of FP is that changes in the apparent molecular weight of the probe (in this case, a labelled nucleic acid) alter the polarization of the sample’s emitted light. In the unbound state, rotational diffusion of the probe leads to depolarization of light; when bound (to a RBP in this case), the apparent molecular weight increases and leads to decreased rotational diffusion and a relative increase in emission of polarized light. An example of small molecule inhibitor identified by FP was C902 (10.6; ), again targeting LIN28 [Citation43].
6.3. Amplified Luminescent Proximity Homogeneous Assay (ALPHA) Screen method
ALPHA screen is a bead-based method that is dependent on the interaction of biological molecules. The endpoint fluorescence is dependent on the proximity of the two biological molecules. The singlet oxygen from the donor molecule is excited upon photosensitization and diffuses across to react with thioxene derivative of the acceptor beads, emitting a fluorescent light at 520–620 nm wavelength range. Although amenable to high throughput screening, it has been more extensively used in confirmation of small molecule interference with RNA-RBP binding, as in the case of the poly-A binding protein MSUT1 [Citation44].
6.4. Firefly dual luciferase assay
The dual luciferase reporter assay is widely used in high throughput screening and is considered a ‘workhorse’ for drug discovery. The assay relies on a luciferase reporter fused to a functional RNA element that is under the control of the RBP under study. The reporter construct is introduced into cells, and luciferase activity is measured in the presence and absence of RBP and/or control reporter constructs. The screening strategy is directed at finding small molecules that return the luciferase reporter activity to control levels. This assay has been used to readout splicing and 3’UTR reporters. Specifically, the inhibitors NVS-SM1 and NVS-SM2 were developed against SMN2, an RBP involved in alternative splicing, using the firefly dual luciferase assay [Citation45].
6.5. Catalytic enzyme-linked click-chemistry assay (cat-ELCCA)
The cat-ELCCA is based on click chemistry and has demonstrated applicability in studying biological processes and drug discovery. Based on development of click chemistry [Citation46], this type of assay represents a new class of biochemical assay that involves bioorthogonally tagged analytes and enzymes, in a manner akin to ELISA. This type of assay has been used for HTS to identify small molecule inhibitors against LIN28 [Citation47]. In this assay, a known RNA sequence labelled with 5’-trans-cyclooctene (TCO) interacts with immobilized LIN28 protein. The TCO can then undergo a ‘click’ reaction with methyltetrazine bound to horseradish peroxidase (HRP), bringing LIN28 and HRP into proximity, producing a chemiluminescent signal. In the presence of an RBP inhibitor, the TCO-bound RNA will not bind to LIN28 and chemiluminescence will not be observed. Small molecule inhibitors targeting LIN28, CCG-233094 and CCG-234459 (10.7 and 10.8; ) were identified using this method [Citation47].
Each of these techniques represents a unique method to quantify the strength of the RBP-RNA interaction, with both biochemical and cellular-based methods described. Coupling biochemical and cellular methods in a screen/counter screen strategy might increase the specificity of these HTS assays.
7. Small molecule inhibitors of pre-mRNA splicing
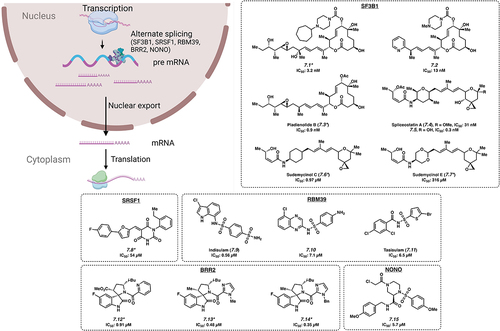
Splicing of pre-mRNA into mRNA is a fundamental process in eukaryotic cells and contributes to normal cellular functioning (). However, aberrant splicing of pre-mRNA is a well-recognized feature of cancer, and one potential mechanism is the upregulation of malignant transcripts [Citation48,Citation49]. The proteins that control splicing, the splicing factor RBPs, are frequently mutated or otherwise dysregulated in cancer [Citation13,Citation50]. This has provided the impetus to develop small molecules to target these RBPs. Here, we highlight the role RBPs have in the increased translation of oncogenic transcripts and the development of small molecules to inhibit the effects of splicing factor RBPs.
Figure 2. Small molecule inhibitors of pre-mRNA splicing. *Mechanism of action for inhibition not reported. Please see text for further details.
7.1. SF3B1
SF3B1 is a core component of spliceosomes, the machinery that regulates splicing in eukaryotic cells. In many haematologic malignancies, SF3B1 mutations are thought to be ‘driver mutations’ and result in globally dysregulated splicing patterns. The mechanism is thought to involve alternative splicing of key oncogenic mRNAs, altering mRNA stability and protein isoform expression [Citation51]. Hence, many therapeutic strategies have attempted to specifically inhibit this splicing factor. Small molecules E7107 (7.1) [Citation52,Citation53], H3B–8800 (7.2) [Citation53], Pladienolide B (7.3) [Citation53], Spliceostatin A (7.4) [Citation54], FR901464 (7.5) [Citation54] and Sudemycinol C (7.6) and E (7.7) [Citation55] have been reported to bind to SF3B1. Notably, it was expected that Spliceostatin A (7.4) and 7.5 would bind in a covalent manner via the enone warhead; however, it was found that this binding occurred in a non-covalent manner () [Citation54]. Growth inhibition of non-haematopoietic cell lines with 7.1, 7.3, 7.4 and 7.5 occurred at IC50 values of 3.2 nM, 0.9 nM, 31 nM and 0.3 nM, respectively [Citation53,Citation54]. These four compounds are thought to bind to SF3B1 and interfere with pre-mRNA splicing, both normal and mutant. Of these, 7.1 entered clinical trials, although it was discontinued due to side effects [Citation56]. Another compound, 7.2 was shown to directly inhibit the SF3B1 complex by interfering with the interaction between the SF3b complex and the splicing branchpoint region. 7.2 was active against leukaemia cell lines at an IC50 of 13 nM and showed preferential killing of spliceosome-mutant cells, which the authors attribute to retention of short GC-rich introns, which are found in genes encoding splicing components [Citation57]. This may provide an explanation for H3B–8800’s (7.2) preferential killing of spliceosome-mutant tumour cells, which are already deficient in splicing, and the killing effects were not attributable to the expression of a single target in the mutant cells. This compound is currently in ongoing clinical trials for myeloid malignancies [Citation58]. Most recently, chemical proteomic screens have identified small molecules that can engage the SF3B1 protein, but the specificity for spliceosome mutant cells remains to be established [Citation59]. Other components of the spliceosomal complex, such as SRSF1 have also been targeted by other groups (7.8), see ref [Citation60].
7.2. RBM39
RBM39 was identified as a key member of a network of RBPs that were critical for maintaining RNA splicing and cell survival in acute myeloid leukaemia (AML) [Citation61]. Indisulam (7.9) is an anti-tumour agent that acts as a molecular glue by recruiting the CUL4-DCAF15 ubiquitin ligase, resulting in polyubiquitination and proteasomal degradation of RBM39 [Citation62]. Although it is active in inhibiting cell growth in both haematopoietic and non-haematopoietic cells [Citation63], 7.9 demonstrated limited clinical efficacy in several trials, which were ultimately discontinued [Citation64]. Analogs of 7.9, CQS (7.10) and Tasisulam (7.11), use a similar mechanism to target RBM39 with IC50 values of 7.1 μM and 6.5 μM, respectively [Citation62], but their utility remains to be determined.
7.3. BRR2
Mutations in SNRNP200/BRR2, a Ski2-like RNA helicase that appears to be a key component of the U5 SNRNP complex, are associated with retinitis pigmentosa [Citation65], a form of degenerative blindness. Although its relevance in cancer and leukaemia remain unclear, several compounds have been developed to inhibit Brr2 including 7.12 with an IC50 value of 0.91 μM along with analogs 7.13 and 7.14 with IC50s of 0.48 μM and 0.35 μM, respectively [Citation66].
7.4. NONO
The RBP NONO was discovered to be an androgen receptor-interacting protein which supports its transcriptional activity. The androgen receptor (AR) is a targetable oncogenic driver of prostate cancer; however, aberrant splicing of AR-encoding mRNA can lead to resistance against available treatments. NONO also interacts with a range of other cancer-specific genes, such as SAMHD1, and its expression may be pathogenetic in leukaemia and related to therapy resistance [Citation67–69]. The search for inhibitors that deplete the AR led to the development of small molecule SKBG01 (7.15), with an IC50 value of 5.7 μM in HEK cells. This compound covalently modifies C145 of NONO, altering its binding and regulation of target transcripts, ultimately reducing the quantity of AR. C145 of NONO is located in a hinge region flanked by two RNA recognition motifs, and proximal to two residues important for RNA binding. Furthermore, there is evidence that 7.15 and related compounds act by stabilizing the interaction between NONO and its bound mRNAs. Ultimately, this stabilization effectively stalls transcript processing and maturation, resulting in decreased amount of target mRNA. This represents a unique mechanism of activity (i.e. functional modification rather than inhibition), shown by the authors to be dependent on the presence of wild-type NONO and the covalent modification of the protein by 7.15 [Citation67].
8. Epitranscriptomics: small molecules targeting RNA modifying enzymes
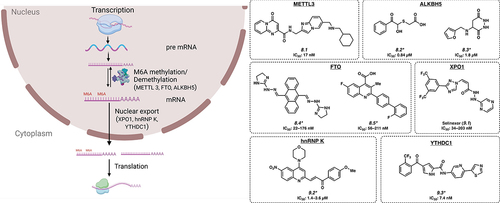
The most prevalent modifications found on RNA molecules are m6A, m1A, m5C and m7G. These modifications play an important role in the gene regulation that affects various biological processes in cells (). Among these, m6A (N6-methyladenosine) is the most abundant modification found on mRNA and has been the most widely studied modification in development and cancer. In acute myeloid leukaemia (AML), m6A modification is more prevalent on RNA and is required for the survival of the cancer cells [Citation70], with a similar pattern noted in both leukaemia as well as in solid tumours [Citation71]. Catalyzing the installation of a methyl group at the 6 position of adenosine to generate m6A are so-called epitranscriptomic writers; a second set of enzymes are known as erasers, which remove this modification.
Figure 3. Inhibitors of RNA modification and nuclear export. *Mechanism of action for inhibition not reported. Please see text for further details.
8.1. METTL3
The RNA methylase enzymes METTL3 and METTL14 install the m6A modification and serve as writers in leukaemia cells. Recent work has led to the identification of STM2457 (8.1) as a small molecule inhibitor of METTL3 with an IC50 of 17 nM, with both in vitro and in vivo activity against AML cells via competitive binding with the SAM co-factor [Citation72]. Many other small molecule inhibitors of METTL3 have been developed, which are mostly analogues of 8.1 and proceed through the same mechanism of action [Citation73].
8.2. FTO/ALKBH5
On the other hand, m6A erasers include FTO and ALKBH5, two RBPs that catalyse the removal of m6A marks and play key roles in AML [Citation17,Citation74]. By removing the m6A marks, these two enzymes alter the epitranscriptome, and modify the stability of a number of crucial mRNA molecules such as TACC3, ASB2 and RARA [Citation17,Citation74]. Compound III and IV (8.2 and 8.3) are small molecule inhibitors of ALKBH5 with IC50 values of 0.84 and 1.8 µM, respectively [Citation75]. CS1 and CS2 (8.4 and 8.5) are small molecule inhibitors of FTO with IC50 values of 22–176 nM and 56–211 nM, respectively, across multiple cell lines [Citation76]. However, the results of these studies are somewhat contradictory, as inhibition of both m6A writers and erasers have been shown to have activity in leukaemia [Citation17,Citation72–74]. Further work is required to clarify how the m6A mark, and its modulation, may represent an effective therapeutic strategy in leukaemia and in cancer.
9. Controlling nuclear export with small molecules
Nuclear export of mRNAs is a key step that allows for the timely and appropriate levels of gene expression to occur in the cytoplasm (). Inhibitors of nuclear transport, specifically of XPO1 have shown clinically relevant activity in several cancers, notably in multiple myeloma [Citation77]. Specifically, Selinexor (9.1) has been approved in combination therapy for multiple myeloma by the FDA and is being currently investigated in this context as well as for other haematologic malignancies. Selinexor (9.1) acts by covalently binding to XPO1 in the nuclear export signalling binding groove at C528, thus inhibiting nuclear export of mRNAs [Citation78]. However, it is thought that additional RBPs may regulate nuclear export in a more nuanced manner. For example, heterogeneous nuclear ribonucleoproteins (hnRNPs) are a class of RBPs that binds to and regulate the nuclear export of the pre-mRNA. There are several different hnRNPs (A-U), and each protein appears to be selectively expressed in specific tissue types. hnRNAP K is overexpressed in human AML and experimental overexpression caused myeloproliferative disease in mice [Citation79]. Compound 5 (9.2) is a small molecule inhibitor developed against hnRNP K with an IC50 of 1.4–3.6 µM [Citation80]. Another RBP involved in mRNA transport, YTHDC1 appears to play a role in cancer [Citation81] and is inhibited by YL-5092 (9.3) with an IC50 of 7.4 nM by binding to the m6A binding site (preprint) [Citation82]. Other proteins such as ELAVL1/HuR and IGF2BP1 have also been reported to play roles in nuclear transport (see section 11.E).
10. Small molecule inhibitors of the microRNA biogenesis pathway
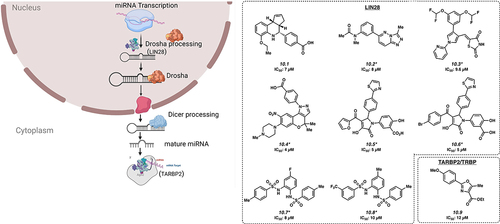
MicroRNA biogenesis is another pathway that is highly regulated by RBPs (). Of note, one of the most studied miRNA regulators is LIN28, which is being targeted for the development small molecules against several cancer types.
Figure 4. Inhibitors of microRNA processing. *Mechanism of action for inhibition not reported. Please see text for further details.
10.1. LIN28
LIN28 consists of two paralogs, LIN28A and LIN28B, which are overexpressed in approximately 15% of human cancers, with overexpression generally associated with a poor prognosis [Citation83]. LIN28 binds to precursor miRNA encoding the tumour suppressive let-7, preventing its maturation [Citation84–86]. Small molecule LI71 (10.1) was discovered to have an IC50 of 7 μM in HeLa cells and targets LIN28 by binding to the CSD domain [Citation87]. Similarly, Compound 1632 and KCB170552 (10.2, 10.3) have shown inhibitory effects by interacting with LIN28 with IC50s of 8 μM and 9.6 μM, respectively [Citation43,Citation88,Citation89]. Other small molecules that inhibit activity of the LIN28A protein include SB1301, PH-43, C902, CCG-233094 and CCG-223095 (10.4 [Citation41], 10.5 [Citation43], 10.6 [Citation43], 10.7 [Citation47], and 10.8 [Citation47] with low micromolar IC50s.
10.2. TARBP2
The RBP TARBP2 is a critical component of the RISC-loading complex, which allows for the loading of microRNAs and miRNAs into RISC, a key step in post-transcriptional gene regulation. This protein has been reported to be overexpressed or otherwise dysregulated in a number of cancer types [Citation90]. CIB-3b (10.9) is a small molecule that binds to TARBP2, inhibiting the RISC-loading complex (IC50 of 12 µM) by disruption of the TRBP/Dicer interaction [Citation91]. Inhibition of the RISC-loading complex ultimately reduces cancer cell proliferation by altering the expression of several different miRNAs processed by Dicer, which presumably result in the de-repression of cell proliferation inhibitory protein expression.
11. Small Molecule targeting of RNA stability modifiers
RBPs can regulate stability of mRNAs, through the interpretation of epitranscriptomic marks, or via regulation of other mechanisms of RNA stability, such as RISC (). These do not reflect mutually exclusive categories, with m6A reader proteins including the IGF2BP proteins and the YTHDF proteins [Citation92]. RBPs reported to interact with and regulate RISC include the IGF2BP proteins, as well as HuR/ELAVL1, and hnRNP family of proteins. In the context of leukaemia, these targets include a number of oncogenic mRNA transcripts, such as LIN28B, HMGA2 and ZFP36L1 [Citation25,Citation93].
Figure 5. Small molecules targeting regulators of mRNA stability. *Mechanism of action for inhibition not reported. Please see text for further details.
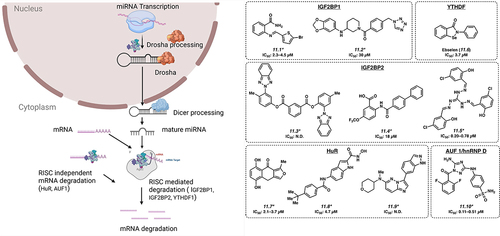
11.1. IGF2BP proteins
There are three proteins in this family: IGF2BP1, IGF2BP2 and IGF2BP3. In polar cells such as fibroblasts and neurons, IGF2BP1 appears to participate in the localization of mRNA transcripts such as ACTB [Citation94]. However, it is also reported to bind to m6A-modified mRNA and participate in post-transcriptional gene regulation at the level of regulating mRNA stability and/or translation, thereby playing a role in oncogenesis [Citation95]. BTYNB and 7773 (11.1 and 11.2) are two small molecule inhibitors developed against IGF2BP1 with IC50s of 2.3–4.5 and 30 µM, respectively, and show activity in a variety of solid tumour cell lines, via regulation of critical oncogenes such as MYC, RAS and E2F [Citation96–98]. The IGF2BP2 protein, also a m6A reader, was recently shown to play an important role in acute myeloid leukaemia pathogenesis by regulating critical mRNA transcripts that in turn control glutamine metabolism [Citation19]. Three compounds have been reported to have activity against IGF2BP2: small molecule inhibitors JX-5 (11.3; IC50: not disclosed) [Citation99], Compound 4 (11.4; IC50: 18 µM) [Citation100] and CWI1–2 (11.5; IC50: 0.20–0.78 µM) [Citation19]. IGF2BP3 is overexpressed in 15% of human cancers, including KMT2A-translocated acute leukaemia [Citation26,Citation27]. An isocorydine derivative, d-ICD is reported to downregulate IGF2BP3 expression, although direct targeting of IGF2BP3 has not been reported [Citation101].
11.2. YTHDF proteins
YTHDF proteins represent another class of RBPs that are cytosolic m6A readers that regulate the degradation and translation of m6A decorated mRNA. YTHDF2 regulates the degradation of m6A modified mRNA [Citation102] and is inhibited by small molecule inhibitor Ebselen (11.6) with an IC50 value of 3.7 µM [Citation103]. 11.6 forms a covalent Se – S bond with YTHDF at C412, nearby the m6A-binding pocket of YTHDF1. While this residue is not in the m6A pocket, binding of 11.6 inhibits the conformational rearrangement of the β4–β5 loop into the m6A binding-competent conformation necessary for interaction with m6A-labelled RNA.
11.3. ELAVL1/HuR
ELAVL1/HuR is reported to be dysregulated in leukaemia, lymphoma and a range of other cancer types, generally showing overexpression [Citation23,Citation104]. This is a multifunctional protein that plays a role in several mechanisms of RNA homoeostasis. In addition to a role in mRNA localization, under stress conditions, HuR interacts with miR-19 to relieve the suppression of RhoB mRNA to proceed for translation [Citation105]. HuR has been reported to function in a RISC independent manner by binding to AU-rich elements (ARE)-containing mRNA, leading to the degradation of the mRNA. Small molecule inhibitors targeting HuR are MS-444 (11.7; IC50 of 2.1–3.7 µM), 1c (11.8; IC50 of 4.7 µM) and SRI-42127 (11.9; IC50: not disclosed) [Citation106–108]. The mechanism of action is not entirely clear for these molecules.
11.4. AUF1/hnRNP D
AUF1/hnRNP D under stress condition binds to ARE-containing mRNA and may play an important role in degradation of oxidized mRNA [Citation109]. JNJ-7706621 (11.10), a small molecule inhibitor targeting AUF1, has been described with an IC50 of 0.11–0.51 µM [Citation110].
12. Targeting protein translation and/or mRNA sequestration
The net effect of transcriptional and post-transcriptional mechanisms discussed thus far is to regulate the amount of mRNA that is accessible to the ribosome for translation into protein (). mRNA may be freely available to the ribosome, or may be sequestered into various cellular compartments, preventing translation. Here, we review selected RBPs that regulate these steps of RNA homoeostasis.
Figure 6. Inhibitors of protein translation and sequestration. *Mechanism of action for inhibition not reported. Please see text for further details.
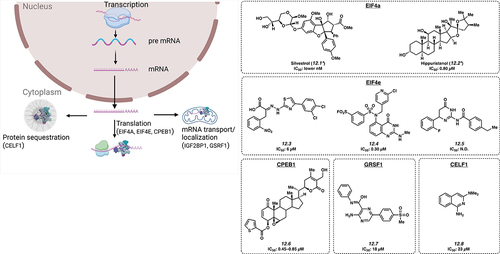
12.1. EIF4A
eIF4A is a member of the DEAD-box family on RNA helicases. In combination with translation factors, it readies mRNA templates during translation initiation. High levels of eIF4A are seen in cancers such as T-cell acute lymphoblastic leukaemia (T-ALL), gastric cancer and cervical cancer with a proposed role via promotion of oncogenic mRNA translation [Citation111,Citation112]. The first small molecule inhibitor of eIF4A reported to have activity in T-cell ALL was natural product Silvestrol (12.1), with a low nanomolar IC50 value [Citation111]. Another small molecule inhibitor of eIF4A, Hippuristanol (12.2) has been found to inhibit eIF4A with an IC50 of about 0.80 µM in HeLa cells [Citation113].
12.2. EIF4E
Increased levels of eIF4E correlate with the growth of many types of tumours. The eIF4E/eIF4G complex is important in regulating gene expression via transcription initiation. The complex is regulated by 4E-BPs, known for their tumour-suppressor activity. By mimicking 4E-BP activity, small molecule 4EGI–1 (12.3) binds to eIF4E and interrupts eIF4E/eIF4G interaction with an IC50 of 6 μM in A549 cells by inhibiting cap-dependent translation [Citation114]. Another small molecule inhibitor of EIF4E is Compound 12 (12.4) with an IC50 of 0.30 µM [Citation115], which binds covalently between the sulphonyl group and K162, also inhibiting cap-dependent translation. Notably, 12.4 was identified using HTS in silico docking. Bn7GxP-based PROTACs (see section 13 for discussion of chemical proximity induced agents) also inhibited eIF4E at high micromolar concentrations [Citation116]; however, these compounds showed minimal effect in cellular assays likely due to limited cell permeability. Small molecule 094 (12.5; IC50: not disclosed) was also found to inhibit the EIF4E-RM38 complex via inhibition of the protein-protein interaction [Citation117].
12.3. CPEB1
CPEB1 is a translation regulator linked to increased metastatic potential in various cancer types including bladder cancer by downregulating TWIST1 and inducing EMT. A variant in the gene is thought to influence risk of chronic lymphocytic leukaemia [Citation118]. Small molecule ASR488 (12.6) has been shown to bind with CPEB1 leading to cell growth arrest and cell death with an IC50 of 0.45–0.85 µM in TCCSUP cells [Citation119].
12.4. GRSF1
GRSF1 is important in most steps of post-transcriptional gene regulation. It has an important part in cancer progression, and is thought to cause metastasis of cervical cancer through the PI3K/AKT/NF-κB and TIMP3/MMP9 pathways. Levels are also observed to be increased in patients with hepatocellular carcinoma (HCC). Studies using VE-821 (12.7), a small molecule, developed to inhibit GRSF1, yielded results showing an IC50 of 18 μM in Hep3B cells. It appears to act by decreasing GRSF1-dependent YY1 stability, a transcriptional activator of oncogenes, potentially by competing with miR-30e-5p microRNA-mediated repression [Citation120].
12.5. CELF1
The RNA binding protein, CUGBP Elav-like family member 1 (CELF1) binds to mRNAs to regulate splicing, translation, and decay and may thereby have a role in translational activation of epithelial-mesenchymal transition genes that drive tumour progression. A small molecule, Compound 27 (12.8), competitively binds CELF1, preventing GU-rich RNA complexation with an IC50 of 23 μM [Citation121].
13. Proximity-based approaches to small molecule development
In recent years, the use of chemical induced proximity via chimeric small molecules has been hailed as a potentially ground-breaking approach in drug development. The first of these, protein- or proteolysis-targeting chimeras (PROTACs), represent hetero-bifunctional molecules that degrade proteins by targeting a specific protein for ubiquitin-proteasome mediated degradation, which is a highly conserved and central pathway for protein degradation. This was first described to target the methionine aminopeptidase MetAP-2, with the chimera consisting of a small molecule linked to a peptide in this early work [Citation122]. Since then, numerous refinements of this technology have been reported, allowing for the tethering of two chemical moieties together, one targeting a protein of interest, and the other targeting an E3 ubiquitin ligase enzyme [Citation123]. The recruitment of the E3 ubiquitin ligase results in the deposition of ubiquitin side chains on the protein, which is then targeted to the proteasome where it is degraded. In the RBP field, a recent report showed that this approach could be used to degrade SF3B1 [Citation124], and as previously mentioned to target EIF4E [Citation116]. For the latter strategy, the investigators designed 7-benzylguanosine analogs of the m7G cap, normally found on the 5’ end of mRNA molecules, linked to a E3 ubiquitin ligase recruiting small molecule. Another variation on this strategy is so-called RNA-PROTAC, where an RNA sequence is linked to a compound that binds to the ubiquitin ligase enzyme [Citation125]. For sequence-specific RBPs (which may include several of the m6A readers), this strategy might be very useful. Lastly, recent work has led to the design of so-called RIBOTACs, where one portion of the chimeric molecule targets RNA molecules and the other portion recruits an RNA-degrading enzyme, such as RNAse L or RNAse H [Citation126,Citation127]. This latter approach was recently reported to be successful in degrading a microRNA, miR-155. The further development of RBP-binding small molecules could enable the recruitment of RNA-degrading enzymes as a targeted approach to degrade specific RNA ligands of RBPs. These chimeric approaches are likely to expand as the number of biological molecules with defined functions, other than ubiquitin ligases and RNAses, are targeted with small molecules. For example, a recent study reported a chimeric molecule (termed a transcriptional/epigenetic chemical inducer of proximity) capable of rewiring cancer-specific transcription by replacing epigenetic repression with transcriptional activation at specific genetic loci [Citation128]. Such approaches hold promise to rationally design chimeric approaches against cancer-specific post-transcriptional gene regulation, as the biological mechanisms are better understood and small molecules binding specific RBPs are developed.
14. Conclusions
In the last 20 years, a remarkable amount of progress has been made in understanding the biology and pathology of RBPs. In haematologic malignancies, a number of important discoveries have led to targeting efforts, particularly with mutant splicing factors, and at least one small molecule remains in clinical trials. The recognition of m6A modifiers and readers and their importance in gene regulation is another major recent development in the field, and there are several new small molecules reported to target m6A writers and readers. A number of other promising small molecules are at various stages of development, and developments in screening and downstream development strategies promise to enrich the pipeline of small molecules. For example, small molecules can be transformed from binders to degraders of RBPs by the construction of heterobifunctional molecules. This field promises to expand rapidly, as it promises to bring novel biologically informed approaches to target proteins that were once deemed undruggable.
Author contributions
All authors contributed to literature review and drafting of the manuscript. AKJ, MLT, DSR were responsible for reviewing the biological aspects of the work, while GMS, JPS, NKG were responsible for reviewing the organic and medicinal chemistry content.
Supplementary_Table_Structures.pdf
Download PDF (201.7 KB)Acknowledgments
We acknowledge that there may be other investigators and authors in the field whose very important contributions to the field were not included in the current manuscript due to space limitations. We acknowledge Tasha Lin, Gunjan Sharma, Tiffany Tran and Jaspal Bassi for their contributions to understanding the function of RBPs in acute leukemia.
Disclosure statement
D.S.R. serves on advisory boards and has consulted for AbbVie, Inc., a pharmaceutical company that markets drugs for hematologic malignancies. The other authors declare no potential competing interests.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2024.2303558
Additional information
Funding
References
- Korobeynikov VA, Lyashchenko AK, Blanco-Redondo B, et al. Antisense oligonucleotide silencing of FUS expression as a therapeutic approach in amyotrophic lateral sclerosis. Nat Med. 2022;28(1): 104–16. doi: 10.1038/s41591-021-01615-z
- Nussbacher JK, Tabet R, Yeo GW, et al. Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron. 2019;102(2): 294–320. doi: 10.1016/j.neuron.2019.03.014
- Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12(1): 89–97. doi: 10.1016/S0959-440X(02)00294-4
- Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15(2): 108–21. doi: 10.1038/nrm3742
- Herzel L, Ottoz DSM, Alpert T, et al. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol. 2017;18(10): 637–50. doi: 10.1038/nrm.2017.63
- Rodríguez-Molina JB, Turtola M. Birth of a poly(A) tail: mechanisms and control of mRNA polyadenylation. FEBS Open Bio. 2023;13(7): 1140–53. doi: 10.1002/2211-5463.13528
- Ranji A, Boris-Lawrie K. RNA helicases: emerging roles in viral replication and the host innate response. RNA Biol. 2010;7(6): 775–87. doi: 10.4161/rna.7.6.14249
- Wiener D, Schwartz S. The epitranscriptome beyond m6A. Nat Rev Genet. 2021;22(2): 119–31. doi: 10.1038/s41576-020-00295-8
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10): 608–24. doi: 10.1038/s41580-019-0168-5
- Kadumuri RV, Janga SC. Epitranscriptomic Code and its alterations in human disease. Trends Mol Med. 2018;24(10): 886–903. doi: 10.1016/j.molmed.2018.07.010
- Choi PS, Thomas-Tikhonenko A. RNA-binding proteins of COSMIC importance in cancer. J Clin Investig. 2021;131(18): 131. doi: 10.1172/JCI151627
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5): 646–74. doi: 10.1016/j.cell.2011.02.013
- Chen S, Benbarche S, Abdel-Wahab O. Splicing factor mutations in hematologic malignancies. Blood. 2021;138(8): 599–612. doi: 10.1182/blood.2019004260
- Obeng EA, Chappell RJ, Seiler M, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30: 404–417. doi: 10.1016/j.ccell.2016.08.006
- Kim E, Ilagan JO, Liang Y, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27: 617–630. doi: 10.1016/j.ccell.2015.04.006
- Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22: 191–205.e9. doi: 10.1016/j.stem.2017.11.016
- Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31: 127–141. doi: 10.1016/j.ccell.2016.11.017
- Paris J, Morgan M, Campos J, et al. Targeting the RNA m(6)A Reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25: 137–48.e6. doi: 10.1016/j.stem.2019.03.021
- Weng H, Huang F, Yu Z, et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. 2022;40: 1566–82.e10. doi: 10.1016/j.ccell.2022.10.004
- Zhou J, Bi C, Ching YQ, et al. Inhibition of LIN28B impairs leukemia cell growth and metabolism in acute myeloid leukemia. J Hematol Oncol. 2017;10: 138. doi: 10.1186/s13045-017-0507-y
- Ishimaru D, Ramalingam S, Sengupta TK, et al. Regulation of bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol Cancer Res. 2009;7(8): 1354. doi: 10.1158/1541-7786.MCR-08-0476
- Kim HH, Kuwano Y, Srikantan S, et al. HuR recruits let-7/RISC to repress c-myc expression. Genes Dev. 2009;23(15): 1743–8. doi: 10.1101/gad.1812509
- Topisirovic I, Siddiqui N, Orolicki S, et al. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol. 2009;29(5): 1152–62. doi: 10.1128/MCB.01532-08
- Ishimaru D, Zuraw L, Ramalingam S, et al. Mechanism of regulation of bcl-2 mRNA by nucleolin and A+U-rich element-binding factor 1 (AUF1). J Biol Chem. 2010;285(35): 27182–91. doi: 10.1074/jbc.M109.098830
- Ennajdaoui H, Howard Jonathan M, Sterne-Weiler T, et al. IGF2BP3 modulates the interaction of invasion-associated transcripts with RISC. Cell Rep. 2016;15: 1876–1883. doi: 10.1016/j.celrep.2016.04.083
- Palanichamy JK, Tran TM, Howard JM, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Invest. 2016;126(4): 1495–511. doi: 10.1172/JCI80046
- Tran TM, Philipp J, Bassi JS, et al. The RNA-binding protein IGF2BP3 is critical for MLL-AF4-mediated leukemogenesis. Leukemia. 2022;36(1): 68–79. doi: 10.1038/s41375-021-01346-7
- Okuda H, Miyamoto R, Takahashi S, et al. RNA-binding proteins of KHDRBS and IGF2BP families control the oncogenic activity of MLL-AF4. Nat Commun. 2022;13(1): 6688. doi: 10.1038/s41467-022-34558-1
- De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23(18): 3189–99. doi: 10.1038/sj.onc.1207545
- Song P, Yang F, Jin H, et al. The regulation of protein translation and its implications for cancer. Sig Transduct Target Ther. 2021;6: 68. doi: 10.1038/s41392-020-00444-9
- Fabbri L, Chakraborty A, Robert C, et al. The plasticity of mRNA translation during cancer progression and therapy resistance. Nat Rev Cancer. 2021;21(9): 558–77. doi: 10.1038/s41568-021-00380-y
- Lee LJ, Papadopoli D, Jewer M, et al. Cancer plasticity: the role of mRNA translation. Trends Cancer. 2021;7(2): 134–45. doi: 10.1016/j.trecan.2020.09.005
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15(12): 829–45. doi: 10.1038/nrg3813
- Bertoldo JB, Müller S, Hüttelmaier S. RNA-binding proteins in cancer drug discovery. Drug Discovery Today. 2023;28(6): 103580. doi: 10.1016/j.drudis.2023.103580
- Mohibi S, Chen X, Zhang J. Cancer the‘RBP’eutics–rNA-binding proteins as therapeutic targets for cancer. Pharmacol Ther. 2019;203: 107390. doi: 10.1016/j.pharmthera.2019.07.001
- Hong S. RNA binding protein as an emerging therapeutic target for cancer prevention and treatment. J Cancer Prev. 2017;22(4): 203–10. doi: 10.15430/JCP.2017.22.4.203
- Gebauer F, Schwarzl T, Valcárcel J, et al. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22(3): 185–98. doi: 10.1038/s41576-020-00302-y
- Schneider T, Hung L-H, Aziz M, et al. Combinatorial recognition of clustered RNA elements by the multidomain RNA-binding protein IMP3. Nat Commun. 2019;10(1): 2266. doi: 10.1038/s41467-019-09769-8
- Tian H, Ip L, Luo H, et al. A high throughput drug screen based on fluorescence resonance energy transfer (FRET) for anticancer activity of compounds from herbal medicine. Br J Pharmacol. 2007;150(3): 321–34. doi: 10.1038/sj.bjp.0706988
- Rogers MS, Cryan LM, Habeshian KA, et al. A FRET-based high throughput screening assay to identify inhibitors of anthrax protective antigen binding to capillary morphogenesis gene 2 protein. PloS One. 2012;7(6): e39911. doi: 10.1371/journal.pone.0039911
- Lim D, Byun WG, Koo JY, et al. Discovery of a small-molecule inhibitor of protein-MicroRNA interaction using binding assay with a site-specifically labeled Lin28. J Am Chem Soc. 2016;138: 13630–13638. doi: 10.1021/jacs.6b06965
- Hall MD, Yasgar A, Peryea T, et al. Fluorescence polarization assays in high-throughput screening and drug discovery: a review. Methods Appl Fluoresc. 2016;4(2): 022001. doi: 10.1088/2050-6120/4/2/022001
- Borgelt L, Li F, Hommen P, et al. Trisubstituted pyrrolinones as small-molecule inhibitors disrupting the protein-RNA interaction of LIN28 and let-7. ACS Med Chem Lett. 2021;12: 893–898. doi: 10.1021/acsmedchemlett.0c00546
- Baker JD, Uhrich RL, Strovas TJ, et al. Targeting pathological tau by small molecule inhibition of the Poly(A): MSUT2 RNA-Protein interaction. ACS Chem Neurosci. 2020;11: 2277–2285. doi: 10.1021/acschemneuro.0c00214
- Palacino J, Swalley SE, Song C, et al. SMN2 splice modulators enhance U1–pre-mRNA association and rescue SMA mice. Nat Chem Biol. 2015;11: 511–517. doi: 10.1038/nchembio.1837
- Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48(28): 6571–84. doi: 10.1021/bi9007726
- Lorenz DA, Kaur T, Kerk SA, et al. Expansion of cat-ELCCA for the discovery of small molecule inhibitors of the pre-let-7-Lin28 RNA-Protein interaction. ACS Med Chem Lett. 2018;9: 517–521. doi: 10.1021/acsmedchemlett.8b00126
- Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer — implications for care. Nat Rev Clin Oncol. 2020;17(8): 457–74. doi: 10.1038/s41571-020-0350-x
- Hershberger CE, Moyer DC, Adema V, et al. Complex landscape of alternative splicing in myeloid neoplasms. Leukemia. 2021;35(4): 1108–20. doi: 10.1038/s41375-020-1002-y
- Seiler M, Peng S, Agrawal AA, et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018;23(1): 282–96.e4. doi: 10.1016/j.celrep.2018.01.088
- Zhou Z, Gong Q, Wang Y, et al. The biological function and clinical significance of SF3B1 mutations in cancer. Biomark Res. 2020;8(1): 38. doi: 10.1186/s40364-020-00220-5
- Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25(5): 440–4. doi: 10.1101/gad.2009411
- Kotake Y, Sagane K, Owa T, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3(9): 570–5. doi: 10.1038/nchembio.2007.16
- Kaida D, Motoyoshi H, Tashiro E, et al. Spliceostatin a targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9): 576–83. doi: 10.1038/nchembio.2007.18
- Shi Y, Bray W, Smith AJ, et al. An exon skipping screen identifies antitumor drugs that are potent modulators of pre-mRNA splicing, suggesting new therapeutic applications. PloS One. 2020;15(5): e0233672. doi: 10.1371/journal.pone.0233672
- Eskens FA, Ramos FJ, Burger H, et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin Cancer Res. 2013;19: 6296–6304. doi: 10.1158/1078-0432.CCR-13-0485
- Seiler M, Yoshimi A, Darman R, et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nature Med. 2018;24(4): 497–504. doi: 10.1038/nm.4493
- Steensma DP, Wermke M, Klimek VM, et al. Phase I first-in-human dose escalation study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. Leukemia. 2021;35(12): 3542–50. doi: 10.1038/s41375-021-01328-9
- Lazear MR, Remsberg JR, Jaeger MG, et al. Proteomic discovery of chemical probes that perturb protein complexes in human cells. Molecular Cell. 2023;83: 1725–42.e12. doi: 10.1016/j.molcel.2023.03.026
- Sidarovich A, Will CL, Anokhina MM, et al. Identification of a small molecule inhibitor that stalls splicing at an early step of spliceosome activation. Elife. 2017;6: e23533. doi: 10.7554/eLife.23533
- Wang E, Lu SX, Pastore A, et al. Targeting an RNA-Binding Protein Network in acute myeloid leukemia. Cancer Cell. 2019;35(3): 369–84.e7. doi: 10.1016/j.ccell.2019.01.010
- Han T, Goralski M, Gaskill N, et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356. doi: 10.1126/science.aal3755
- Ozawa Y, Sugi NH, Nagasu T, et al. E7070, a novel sulphonamide agent with potent antitumour activity in vitro and in vivo. Eur J Cancer. 2001;37(17): 2275–82. doi: 10.1016/S0959-8049(01)00275-1
- Pogacar Z, Groot K, Jochems F, et al. Genetic and compound screens uncover factors modulating cancer cell response to indisulam. Life Sci Alliance. 2022;5(9): e202101348. doi: 10.26508/lsa.202101348
- Ledoux S, Guthrie C. Retinitis Pigmentosa mutations in bad response to refrigeration 2 (Brr2) impair ATPase and helicase activity. J Biol Chem. 2016;291(23): 11954–65. doi: 10.1074/jbc.M115.710848
- Ito M, Iwatani M, Yamamoto T, et al. Discovery of spiro[indole-3,2’-pyrrolidin]-2(1H)-one based inhibitors targeting Brr2, a core component of the U5 snRNP. Bioorg Med Chem. 2017;25: 4753–4767. doi: 10.1016/j.bmc.2017.07.017
- Kathman SG, Koo SJ, Lindsey GL, et al. Remodeling oncogenic transcriptomes by small molecules targeting NONO. Nat Chem Biol. 2023;19(7): 825–36. doi: 10.1038/s41589-023-01270-0
- Zhang X, Wu C, Xiong W, et al. Knockdown of p54nrb inhibits migration, invasion and TNF-α release of human acute monocytic leukemia THP1 cells. Oncol Rep. 2016;35(6): 3742–8. doi: 10.3892/or.2016.4756
- Zhang F, Sun J, Tang X, et al. Stabilization of SAMHD1 by NONO is crucial for ara-C resistance in AML. Cell Death Dis. 2022;13(7): 590. doi: 10.1038/s41419-022-05023-0
- Vu LP, Pickering BF, Cheng Y, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23: 1369–1376. doi: 10.1038/nm.4416
- Vu LP, Cheng Y, Kharas MG. The biology of m(6)A RNA methylation in normal and malignant hematopoiesis. Cancer Discovery. 2019;9: 25–33. doi: 10.1158/2159-8290.CD-18-0959
- Yankova E, Blackaby W, Albertella M, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593(7860): 597–601. doi: 10.1038/s41586-021-03536-w
- Fiorentino F, Menna M, Rotili D, et al. METTL3 from target validation to the first Small-molecule inhibitors: a medicinal chemistry journey. J Med Chem. 2023;66(3): 1654–77. doi: 10.1021/acs.jmedchem.2c01601
- Shen C, Sheng Y, Zhu AC, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. 2020;27(1): 64–80.e9. doi: 10.1016/j.stem.2020.04.009
- Sabnis RW. Novel small molecule RNA m6A demethylase AlkBH5 inhibitors for treating cancer. ACS Med Chem Lett. 2021;12(6): 856–7. doi: 10.1021/acsmedchemlett.1c00102
- Su R, Dong L, Li Y, et al. Targeting FTO suppresses cancer stem Cell maintenance and immune evasion. Cancer Cell. 2020;38(1): 79–96.e11. doi: 10.1016/j.ccell.2020.04.017
- Chari A, Vogl DT, Gavriatopoulou M, et al. Oral selinexor-dexamethasone for triple-class refractory multiple myeloma. N Engl J Med. 2019;381: 727–738. doi: 10.1056/NEJMoa1903455
- Kashyap T, Argueta C, Aboukameel A, et al. Selinexor, a selective inhibitor of nuclear export (SINE) compound, acts through NF-κB deactivation and combines with proteasome inhibitors to synergistically induce tumor cell death. Oncotarget. 2016;7(48): 78883–95. doi: 10.18632/oncotarget.12428
- Aitken MJL, Malaney P, Zhang X, et al. Heterogeneous nuclear ribonucleoprotein K is overexpressed in acute myeloid leukemia and causes myeloproliferation in mice via altered Runx1 splicing. NAR Cancer. 2022;4: zcac039. doi: 10.1093/narcan/zcac039
- Shu B, Zeng P, Kang S, et al. Syntheses and evaluation of new quinoline derivatives for inhibition of hnRNP K in regulating oncogene c-myc transcription. Bioorg Chem. 2019;85: 1–17. doi: 10.1016/j.bioorg.2018.12.020
- Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRnas. Elife. 2017;6: 6. doi: 10.7554/eLife.31311
- Yang S, Zhang H, Li Y, et al. Discovery of a selective YTHDC1 inhibitor that targets acute myeloid leukemia. Res Square. 2023. doi: 10.21203/rs.3.rs-2644364/v1
- Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nature Genet. 2009;41(7): 843–8. doi: 10.1038/ng.392
- Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRnas in mouse embryonic stem cells. RNA. 2009;15(3): 357–61. doi: 10.1261/rna.1368009
- Yuan J, Nguyen CK, Liu X, et al. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate Fetal-Like Lymphopoiesis. Science. 2012;335: 1195. doi: 10.1126/science.1216557
- Wang S, Chim B, Su Y, et al. Enhancement of LIN28B-induced hematopoietic reprogramming by IGF2BP3. Genes Dev. 2019;33(15–16): 1048–68. doi: 10.1101/gad.325100.119
- Wang L, Rowe RG, Jaimes A, et al. Small-molecule inhibitors disrupt let-7 oligouridylation and release the selective blockade of let-7 processing by LIN28. Cell Rep. 2018;23(10): 3091–101. doi: 10.1016/j.celrep.2018.04.116
- Roos M, Pradère U, Ngondo RP, et al. A Small-molecule inhibitor of Lin28. ACS Chem Biol. 2016;11(10): 2773–81. doi: 10.1021/acschembio.6b00232
- Byun WG, Lim D, Park SB. Discovery of small-molecule modulators of protein-RNA interactions by fluorescence intensity-based binding assay. Chembiochem. 2020;21: 818–824. doi: 10.1002/cbic.201900467
- Yu X, Li Z. The role of TARBP2 in the development and progression of cancers. Tumour Biol. 2016;37: 57–60. doi: 10.1007/s13277-015-4273-6
- Peng T, He Y, Wang T, et al. Discovery of a Novel Small-Molecule Inhibitor Disrupting TRBP-Dicer Interaction against Hepatocellular Carcinoma via the Modulation of microRNA Biogenesis. J Med Chem. 2022;65: 11010–11033. doi: 10.1021/acs.jmedchem.2c00189
- Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20: 285–295. doi: 10.1038/s41556-018-0045-z
- Müller S, Bley N, Glaß M, et al. IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 2018;46(12): 6285–303. doi: 10.1093/nar/gky229
- Huttelmaier S, Zenklusen D, Lederer M, et al. Spatial regulation of beta-actin translation by src-dependent phosphorylation of ZBP1. Nature. 2005;438: 512–515. doi: 10.1038/nature04115
- Bell JL, Wachter K, Muhleck B, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70(15): 2657–75. doi: 10.1007/s00018-012-1186-z
- Mahapatra L, Andruska N, Mao C, et al. A novel IMP1 inhibitor, BTYNB, targets c-myc and inhibits melanoma and ovarian cancer Cell proliferation. Transl Oncol. 2017;10(5): 818–27. doi: 10.1016/j.tranon.2017.07.008
- Wallis N, Oberman F, Shurrush K, et al. Small molecule inhibitor of Igf2bp1 represses Kras and a pro-oncogenic phenotype in cancer cells. RNA Biol. 2022;19(1): 26–43. doi: 10.1080/15476286.2021.2010983
- Müller S, Bley N, Busch B, et al. The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res. 2020;48(15): 8576–90. doi: 10.1093/nar/gkaa653
- Feng P, Chen D, Wang X, et al. Inhibition of the m6A reader IGF2BP2 as a strategy against T-cell acute lymphoblastic leukemia. Leukemia. 2022;36(9): 2180–8. doi: 10.1038/s41375-022-01651-9
- Dahlem C, Abuhaliema A, Kessler SM, et al. First small-molecule inhibitors targeting the RNA-Binding protein IGF2BP2/IMP2 for cancer therapy. ACS Chem Biol. 2022;17(2): 361–75. doi: 10.1021/acschembio.1c00833
- Li M, Zhang L, Ge C, et al. An isocorydine derivative (d-ICD) inhibits drug resistance by downregulating IGF2BP3 expression in hepatocellular carcinoma. Oncotarget. 2015;6(28): 25149–60. doi: 10.18632/oncotarget.4438
- Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates Messenger RNA translation efficiency. Cell. 2015;161: 1388–1399. doi: 10.1016/j.cell.2015.05.014
- Micaelli M, Dalle Vedove A, Cerofolini L, et al. Small-molecule ebselen binds to YTHDF proteins interfering with the Recognition of N (6)-methyladenosine-modified RNAs. ACS Pharmacol Transl Sci. 2022;5: 872–891. doi: 10.1021/acsptsci.2c00008
- Prieto C, Kharas MG. RNA regulators in leukemia and lymphoma. Cold Spring Harb Perspect Med. 2020;10: 10. doi: 10.1101/cshperspect.a034967
- Glorian V, Maillot G, Polès S, et al. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Diff. 2011;18(11): 1692–701. doi: 10.1038/cdd.2011.35
- Meisner NC, Hintersteiner M, Mueller K, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol. 2007;3(8): 508–15. doi: 10.1038/nchembio.2007.14
- Wu X, Ramesh R, Wang J, et al. Small molecules targeting the RNA-Binding protein HuR inhibit tumor growth in xenografts. J Med Chem. 2023;66(3): 2032–53. doi: 10.1021/acs.jmedchem.2c01723
- Chellappan R, Guha A, Si Y, et al. SRI-42127, a novel small molecule inhibitor of the RNA regulator HuR, potently attenuates glial activation in a model of lipopolysaccharide-induced neuroinflammation. Glia. 2022;70(1): 155–72. doi: 10.1002/glia.24094
- Ishii T, Hayakawa H, Sekiguchi T, et al. Role of Auf1 in elimination of oxidatively damaged messenger RNA in human cells. Free Radic Biol Med. 2015;79: 109–16. doi: 10.1016/j.freeradbiomed.2014.11.018
- Li KT, Wu XZ, Sun ZY, et al. A novel strategy for regulating mRNA’s degradation via interfering the AUF1’s binding to mRNA. Molecules. 2022;27: 27. doi: 10.3390/molecules27103182
- Wolfe AL, Singh K, Zhong Y, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513(7516): 65–70. doi: 10.1038/nature13485
- Xue C, Gu X, Li G, et al. Expression and functional roles of eukaryotic initiation factor 4A family proteins in human cancers. Front Cell Dev Biol. 2021;9: 711965. doi: 10.3389/fcell.2021.711965
- Bordeleau ME, Mori A, Oberer M, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2(4): 213–20. doi: 10.1038/nchembio776
- Moerke NJ, Aktas H, Chen H, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128(2): 257–67. doi: 10.1016/j.cell.2006.11.046
- Wan X, Yang T, Cuesta A, et al. Discovery of lysine-targeted eIF4E inhibitors through covalent docking. J Am Chem Soc. 2020;142(11): 4960–4. doi: 10.1021/jacs.9b10377
- Kaur T, Menon A, Garner AL. Synthesis of 7-benzylguanosine cap-analogue conjugates for eIF4E targeted degradation. Eur J Med Chem. 2019;166: 339–50. doi: 10.1016/j.ejmech.2019.01.080
- Lucchesi CA, Zhang J, Gao M, et al. Identification of a first-in-class small-molecule inhibitor of the EIF4E-RBM38 complex that enhances wild-type TP53 protein translation for tumor growth suppression. Mol Cancer Ther. 2023;22(6): 726–36. doi: 10.1158/1535-7163.MCT-22-0627
- Crowther-Swanepoel D, Broderick P, Di Bernardo MC, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42(2): 132–6. doi: 10.1038/ng.510
- Tyagi A, Kolluru V, Chandrasekaran B, et al. ASR488, a novel small molecule, activates an mRNA binding protein, CPEB1, and inhibits the growth of bladder cancer. Oncol Lett. 2020;20(1): 850–60. doi: 10.3892/ol.2020.11593
- Han L, Huang C, Wang X, et al. The RNA-binding protein GRSF1 promotes hepatocarcinogenesis via competitively binding to YY1 mRNA with miR-30e-5p. J Exp Clin Cancer Res. 2022;41(1): 17. doi: 10.1186/s13046-021-02217-w
- Tan Y, Sun X, Xu Y, et al. Small molecule targeting CELF1 RNA-binding activity to control HSC activation and liver fibrosis. Nucleic Acids Res. 2022;50(5): 2440–51. doi: 10.1093/nar/gkac139
- Sakamoto KM, Kim KB, Kumagai A, et al. Protacs: chimeric molecules that target proteins to the Skp1-cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA. 2001;98: 8554–8559. doi: 10.1073/pnas.141230798
- Chirnomas D, Hornberger KR, Crews CM. Protein degraders enter the clinic — a new approach to cancer therapy. Nat Rev Clin Oncol. 2023;20(4): 265–78. doi: 10.1038/s41571-023-00736-3
- Gama-Brambila RA, Chen J, Zhou J, et al. A PROTAC targets splicing factor 3B1. Cell Chem Biol. 2021;28(11): 1616–27.e8. doi: 10.1016/j.chembiol.2021.04.018
- Ghidini A, Cléry A, Halloy F, et al. RNA-PROTACs: degraders of RNA-Binding proteins. Angew Chem Int Ed Engl. 2021;60: 3163–3169. doi: 10.1002/anie.202012330
- Costales MG, Matsumoto Y, Velagapudi SP, et al. Small Molecule Targeted Recruitment of a Nuclease to RNA. J Am Chem Soc. 2018;140(22): 6741–4. doi: 10.1021/jacs.8b01233
- Tong Y, Lee Y, Liu X, et al. Programming inactive RNA-binding small molecules into bioactive degraders. Nature. 2023;618(7963): 169–79. doi: 10.1038/s41586-023-06091-8
- Gourisankar S, Krokhotin A, Ji W, et al. Rewiring cancer drivers to activate apoptosis. Nature. 2023;620(7973): 417–25. doi: 10.1038/s41586-023-06348-2
