ABSTRACT
The RNA-targeting CRISPR-Cas13 system has enabled precise engineering of endogenous RNAs, significantly advancing our understanding of RNA regulation and the development of RNA-based diagnostic and therapeutic applications. This review aims to provide a summary of Cas13-based RNA targeting tools and applications, discuss limitations and challenges of existing tools and suggest potential directions for further development of the RNA targeting system.
KEYWORDS:
CRISPR-Cas13 as a programmable RNase
The clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) proteins, are bacterial adaptive immune systems, most of which are potent DNA/RNA endonucleases. In class 2 type VI CRISPR/Cas systems, single-subunit Cas13 system as an RNA-guided RNA endonuclease (RNase) has been reported since 2016. Cas13a (also known as C2c2) is the first subtype of Cas13 family [Citation1–4]. The Cas13 family now comprises 11 subtypes, including Cas13a [Citation1,Citation2], Cas13b [Citation3], Cas13c [Citation4], Cas13d [Citation4], Cas13e [Citation5], Cas13f [Citation5], Cas13g [Citation5], Cas13h [Citation5], Cas13i [Citation5], Cas13x [Citation6] and Cas13y [Citation6], with potentially more to be identified. While these Cas13 systems are all RNA-guided RNases, they differ in size, protein sequence and in their efficiency in eukaryotic cells [Citation5,Citation6]. Among them, Cas13a, Cas13b and Cas13d are the most widely used in mammalian cells.
Cas13 is guided by a CRISPR RNA (crRNA) and is programmed to cleave RNA targets carrying complementary protospacers [Citation1] (). Unlike most DNA-targeting CRISPR/Cas systems that require a protospacer adjacent motif (PAM), Cas13 shows no strong bias on protospacer flanking sequences (PFS) in eukaryotic cells, allowing it to target essentially any sequences. Cas13 proteins typically contain two higher eukaryote and prokaryote nucleotide-binding (HEPN) RNase domains, which form a single catalytic site that cleaves target RNAs [Citation7–10]. Cas13 also possesses another RNase activity that can process its own crRNAs from a pre-crRNA array that consists of multiple repeats of spacers and direct repeats (DRs) [Citation2]. The pre-crRNA processing activity facilitates multiplexed targeting by expressing multiple crRNAs (also called guide RNAs or gRNAs) from a single transcript [Citation11].
Figure 1. CRISPR-Cas13 and its collateral activity.
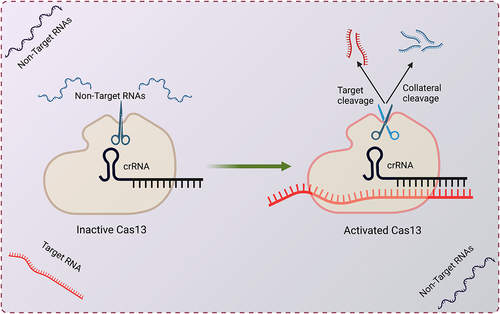
Initial studies of Cas13 in mammalian cells and plant cells have shown comparable levels of knockdown as RNA interference (RNAi) and with improved specificity [Citation12]. While RNAi is less effective against nuclear RNAs, Cas13 efficiently inhibits both nuclear and cytoplasmic RNAs. For instance, Cas13a has been used to knock down nuclear noncoding RNAs such as MALAT1, HOTTIP[Citation11] and enhancer RNAs [Citation13]. Cas13a has also been used to inhibit cancer-associated fusion transcripts, such as the EML4-ALK fusion transcript to inhibit cell viability in lung cancer cells [Citation14]. In addition to linear RNAs, Cas13d can downregulate circular RNAs (circRNAs) by using guide RNAs that target sequences spanning back-splicing junction sites of circRNAs [Citation15]. The flexibility in targeting essentially any sequence without the restriction of PAM/PFS allows most circRNAs to be targeted without affecting the linear transcripts. In addition to endogenous cellular RNAs, Cas13 has been used to degrade viral RNAs, including SARS-CoV-2 [Citation16], influenza virus [Citation16], HIV [Citation17], PRRSV [Citation18] and HPV [Citation19].
Cas13 has also been widely used in vivo, allowing targeted RNA knockdown in xenograft tumours [Citation20,Citation21] and various tissues, including eye [Citation22–24], ear [Citation25,Citation26], brain [Citation22,Citation27,Citation28], lung [Citation29–31], kidney [Citation30], liver [Citation30] and spleen [Citation30]. Cas13 proteins, and especially Cas13d proteins, are significantly smaller than Cas9, allowing efficient package into Adeno-associated viruses (AAVs) along with its gRNA expression cassette for in vivo delivery [Citation11]. AAV-mediated expression of Cas13a/gRNA targeting several oncogenes significantly inhibited tumour growth in xenograft models [Citation20]. Similarly, AAV-Cas13d targeting the oncogenic KRASG12D mRNA significantly inhibited tumour growth in patient-derived xenografts in mice [Citation21]. In addition to tumour, AAV-Cas13d has also been delivered to specific tissues to knock down various targets and achieved phenotypic rescue in mouse models of age-related macular degeneration [Citation23], hearing loss [Citation25], Parkinson’s disease [Citation22], TDP-43 proteinopathy [Citation32] and amyotrophic lateral sclerosis (ALS) [Citation33], respectively. In addition to AAV, non-viral approaches including lipid nanoparticles (LNPs) [Citation31], extracellular vesicles (EVs) [Citation30] and nebulizer-based nanoformulated RNA complex delivery [Citation29] have also been used to deliver Cas13/gRNA, especially to the lung.
Engineering RNA with CRISPR-dCas13
Point mutations in the two HEPN RNase domains inactivate the gRNA-dependent target cleavage activity, allowing target RNAs to be bound without being degraded (). While often called catalytically dead Cas13 (dCas13), the resulting HEPN-deficient Cas13 retains the pre-crRNA processing RNase activity and thus remains capable of multiplexing. dCas13 can be used as a programmable RNA-binding protein (RBP), allowing it to act as a competitive inhibitor of endogenous RBPs on a specific transcript [Citation34]. Similarly, other functional RNA signals such as splice sites, polyadenylation signals and start codons of a specific transcript can be targeted by dCas13 to modulate splicing [Citation11], polyadenylation [Citation35] and translation initiation [Citation36], respectively.
Figure 2. Applications of dCas13.
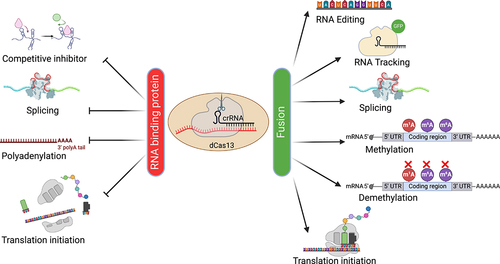
Furthermore, various effector domains have been fused with dCas13 for additional applications. For example, dCas13-GFP has been used to label and track RNAs in live cells [Citation12,Citation37]. Affinity purification of tagged dCas13 loaded with specific gRNAs allows the detection of proteins bound to an RNA of interest [Citation38,Citation39]. Artificial splicing factors fusing dCas13 with domains of splicing factors can modulate alternative splicing of endogenous transcripts [Citation11,Citation40]. Direct insertion or replacement of thousands of nucleotides in endogenous RNAs is now possible with dCas13-mediated trans-splicing [Citation41]. Targeted A-to-I and C-to-U RNA editing have also been developed by fusing dCas13 with either endogenous or evolved deaminase domains, respectively [Citation42,Citation43]. Fusing nucleus- or cytoplasm-localized dCas13 with a methyltransferase domain enables site-specific N6-Methyladenosine (m6A) incorporation within distinct cellular compartments [Citation44]. Conversely, RNA modifications such as m6A and m1A can be removed in site-specific manner by fusing dCas13 with corresponding demethylases, such as ALKBH5 and ALKBH3, respectively [Citation45,Citation46]. Translation of specific mRNAs can be enhanced by dCas13 fused to translation initiation factors such as IF3 in Escherichia coli [Citation47] and PABPC1 in various human cells [Citation48].
In addition to fusing protein effector domains to dCas13, new functions can be generated by fusing functional RNA elements to the gRNA. While similar ideas have successfully been used in dCas9-based applications [Citation49,Citation50], tethering RNA regulatory elements via gRNAs may be particularly suitable for RNA-targeting applications. For example, the SINEB2 RNA element has been fused with gRNA to enhance translation of target mRNAs [Citation51]. Given the large number of functional RNA regulatory elements controlling every step of the RNA life cycle [Citation52], we envision further expansion of the dCas13 RNA toolkit by tethering other RNA elements to the gRNA. Compared to protein effector domains, RNA elements are typically much smaller, making it easier to multiplex and package in adeno-associated virus (AAV) for in vivo delivery.
Target specificity of Cas13: off-targets and mismatch tolerance
In general, CRISPR/Cas13 exhibits tolerance for a single mismatch between the target RNA, with the gRNA spacer nucleotides 15–21 being more sensitive to mismatches in the target site [Citation53]. The presence of more than two mismatches often leads to a reduction in cleavage efficiency [Citation54], although in some cases more mismatches can be tolerated [Citation55]. Moreover, the sensitivity to mismatches is also highly variable across positions within the gRNA/target RNA duplex as well as the nucleotide identity of the mismatch [Citation56,Citation57]. The lack of a set of simple rules for targeting specificity calls for sophisticated machine learning models to be trained using large-scale experimental measurements of mismatch tolerance for each Cas13 system. Encouragingly, at least two approaches have been shown to decrease mismatch tolerance and allow specific detection of SNPs. The first approach is synthetic mismatch, i.e. use gRNAs with one or more mismatches to the desired targets [Citation58,Citation59]. Cas13 will tolerate the limited number of mismatches in the target but not more mismatches found in off-targets. The second approach is to extend the spacer at the 3’ end of the gRNA with a sequence that will form a short stem loop/hairpin with the spacer. By blocking part of the spacer, the hairpin prevents the gRNA from binding to off-target RNA sequences and improves the specificity of the CRISPR/Cas13a system for single nucleotide polymorphism (SNP) identification [Citation60]. Despite extensive research endeavours, enhancing the specificity of Cas13 systems remains an area that warrants further attention and improvement.
Cas13 collateral activity in bacteria and in vitro
Upon target RNA recognition, Cas13 undergoes a conformational change and becomes an activated RNase. Unlike most Cas proteins that cleave within the guide/target duplex, the activated RNase site is exposed on the surface of Cas13 [Citation7,Citation8], allowing it to cleave not only the bound target RNA in cis but also other nearby RNAs in trans (). Such collateral activity allows bacteria to degrade both phage RNAs and host RNAs upon phage infection, resulting in growth arrest of the host cell and the abortion of the infectious cycle [Citation61]. Similar ‘suicide’ responses are also triggered by several other antiviral systems [Citation62], including the type IIIA CRISPR-Cas system employing the RNase Csm6 that indiscriminately degrades both viral and host RNAs.
It is important to keep in mind that collateral activity is fundamentally different from conventional off-target effects. Off-targets of CRISPR or RNAi still directly pair with the guide RNA or siRNA, albeit with one or more mismatches. In contrast, Cas13 collateral activity is indiscriminate, and the degraded substrate does not need to have sequence complementarity to the guide RNA. Moreover, off-target effects are largely independent of whether the intended on-target is present, whereas collateral activity is only activated upon recognition of the on-target RNA that perfectly matches the guide RNA, although in principle conventional off-targets can also activate collateral activity, as long as the mismatch is tolerated.
The collateral activity of Cas13 has found diverse applications across numerous fields (). For example, the ability to sense a specific sequence and induce self-destruction of bacterial cells has been used for sequence-specific bacteria killing. By packaging programmed CRISPR-Cas13a recognizing resistance genes into bacteriophage capsids, Kiga and colleagues created a new type of antibacterial agent called ‘CapsidCas13a(s)’ that inhibited the growth of carbapenem-resistant Escherichia coli and methicillin-resistant Staphylococcus aureus, two types of bacteria known for their resistance to common antibiotics [Citation63]. Supporting a role of Cas13 collateral activity in causing the observed growth arrest, targeting the same gene with CRISPR/Cas9 did not result in the same defect, which is not known to have collateral activity.
Figure 3. Applications of Cas13 collateral activity.
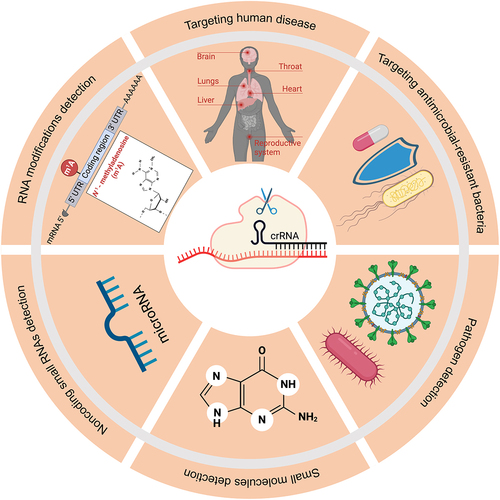
Cas13 collateral activity has also been leveraged for sensitive detection of RNA or DNA (when coupled with in vitro transcription). In the presence of a target RNA, activated Cas13a cleaves a self-quenched reporter RNA, which results in the emission of fluorescence. The amount of fluorescence emitted is proportional to the concentration of the target RNA. The sensitivity can be further increased by isothermal amplification of the target and by incorporating Csm6, an auxiliary CRISPR-associated enzyme [Citation64]. The resulting SHERLOCK platform allows ultrasensitive detection of pathogenic virus (e.g. Zika and Dengue viruses) and bacteria and is also highly specific in distinguishing specific viral strains, human SNPs and tumour mutations [Citation58]. Technical improvements in several related platforms, including HUDSON [Citation65], SHINE [Citation66], CASCADE [Citation67], RT-LAMP-CRISPR-Cas13a [Citation68], have allowed rapid, instrument-free and extraction-free detection of RNAs including SARS-CoV-2 RNA from unextracted samples. Integrated with the CARMEN platform that can simultaneously analyse over 4,500 crRNA-target pairs within a single array, CARMEN-Cas13 can detect all 169 viruses associated with humans in a single test [Citation69]. Cas13 has also been used to detect microRNAs [Citation70,Citation71], RNA modifications [Citation72], as well as small molecules when coupled with riboswitches [Citation73].
Cas13 collateral activity in eukaryotic cells
While evidence supporting Cas13 collateral activity in vitro and in bacterial systems has been abundant and clear, the extent of such activity in eukaryotic cells remains a subject of inconsistency in various published papers. Early studies of Cas13a and Cas13d did not report any off-target or collateral effects in mammalian cells [Citation11,Citation12,Citation42]. Instead, these studies show that Cas13-mediated transcript knockdown is remarkably specific, with zero off-targets compared to hundreds for shRNA-mediated knockdown [Citation11,Citation12] when assayed with RNA-seq. In Drosophila transgenic cell line Sg4_CD, a number of Cas13 variants can efficiently knock down an eCFP reporter without affecting a co-expressed DsRed reporter [Citation74]. Another study tested Cas13d-mediated transcript knockdown in zebrafish embryos and observed no toxic effects, off-target impacts or collateral activity [Citation75]. Similarly, Cas13a downregulated endogenous PPIB and KRAS mRNAs in human lung epithelial cells A549 with no significant changes in the expression of 11 housekeeping genes and 18S rRNA [Citation29].
More recent studies, however, have found substantial collateral activity of various Cas13 systems in mammalian and Drosophila cells using a variety of assays [Citation6,Citation76–79]. For example, we and others have shown that targeting either reporter mRNAs (e.g. EGFP) or endogenous RNAs (e.g. ACTG1) can result in the degradation of co-expressed non-target reporter RNAs (e.g. mCherry), Cas13 mRNAs and other endogenous RNAs, as measured by qRT-PCR, Northern blotting, Western blotting and fluorescence imaging. Multiple studies have also reported a loss of RNA integrity, as indicated by rRNA cleavage and fragmentation in denaturing RNA gel or BioAnalyzer runs [Citation78]. Supporting global RNA degradation caused by Cas13 collateral activity, we further showed a 46% decrease in total RNA extracted from Cas13 targeted cells [Citation77]. Moreover, using polyA+ RNA-seq with external spike-in, we have shown that almost the entire transcriptome is downregulated by Cas13 collateral activity, including GAPDH, ACTB and other housekeeping genes [Citation77].
How do we reconcile the contradictory observations regarding Cas13 collateral activity in eukaryotic cells? The difficulty for internal control-based normalization has probably contributed to the failure to detect collateral activity in some studies, because commonly used control such as GAPDH and even rRNA are also down-regulated by Cas13 collateral activity [Citation77]. Moreover, recent studies have shown that the extent of collateral activity is strongly correlated with target RNA abundance [Citation76,Citation77,Citation79], and earlier studies reporting a lack of collateral activity were mostly targeting low abundance RNAs [Citation11]. There is also a substantial variation of collateral activity observed for different subtypes of Cas13 proteins, with Cas13d showing the strongest effect, followed by Cas13a and then Cas13b [Citation79]. Intriguingly, the extent of collateral activity also varies across cell types. For example, it has been shown that Cas13a exhibits a stronger collateral activity in U87 cells but not in HEK293T cells [Citation78], although we have observed very strong collateral activity in HEK293T cells using Cas13d [Citation77]. Such substantial variations could be due to variation in the expression level of Cas13, gRNA and the target RNA in those cells, or even variation in cellular pH [Citation80–84], as RNase activities are sensitive to pH [Citation85]. Therefore, the extent of collateral activity observed depends on a variety of factors, potentially underlying the contradictory results reported in the literature.
The unintended indiscriminate degradation of cellular RNAs is a major concern for using Cas13 to knock down specific target RNAs in cells, especially for abundant targets. While reducing Cas13/gRNA abundance [Citation86] or stability [Citation76] can potentially reduce off-target binding and thus off-target-activated collateral activity, it is inherently challenging to reduce on-target-activated collateral activity without compromising on-target knockdown efficiency, as the same active site is responsible for both activities. Nonetheless, the collateral activity varies dramatically across Cas13 subtypes (e.g. PspCas13b vs RfxCas13d) and novel Cas13 orthologs (e.g. DjCas13d) with minimal cellular toxicity have recently been identified [Citation87]. Moreover, high-fidelity variants of Cas13d and Cas13X have also been engineered [Citation88], and in particular the RfxCas13d-N2V7 variant has been independently shown to be more specific than Cas7–11, another RNA-targeting CRISPR system not known to have collateral activity [Citation32,Citation89]. It remains unclear mechanistically how collateral activity is modulated in those Cas13 variants/orthologs. Possibilities include structural changes that position the active site closer to the target region and less exposed on the surface, or decrease the stability of the active Cas13-gRNA-target ternary complex, such that it stays active long enough to cleave the target RNA which is in close proximity, but not long enough to cleave a larger number of other RNAs in cells.
Sequence-specific cell targeting with CRISPR-Cas13
The activation of Cas13 collateral activity arrests the growth of infected bacteria. Similarly in human cells, Cas13-mediated targeting of reporter mRNAs or non-essential endogenous mRNAs substantially reduces the viability and proliferation of cells [Citation77]. Similar to collateral RNA degradation, the extent of growth defect is positively correlated with target RNA abundance [Citation77]. While apoptosis is observed in some studies [Citation76], others reported no increase in cell death, only a reduction in cellular metabolic activity and DNA replication [Citation77]. Interestingly, similar to RNase treatment of nuclei [Citation90,Citation91], Cas13 collateral activity also results in the collapse of chromatin, potentially contributing to global mRNA downregulation and inhibition of DNA replication. The toxicity of Cas13 collateral activity has also been observed in vivo. Strikingly, Cas13d-mediated knockdown of several non-essential mRNAs in adult mouse brain resulted in animal death [Citation92]. The lethality strictly depends on simultaneous expression of Cas13, gRNA and the target, suggesting the toxicity is driven by collateral activity rather than off-target effects [Citation92].
The ability to inhibit cell growth or even induce cell death by sensing a marker RNA opens the door for sequence-specific cell targeting. Programmable elimination of pathogenic cells such as cancer cells, activated fibroblasts and senescent cells will facilitate the development of therapies against cancer, fibrosis and ageing, respectively. Such a technology platform will also allow functional studies of novel cell types or cell states uncovered by single-cell RNA sequencing. As a proof-of-principle, we have previously demonstrated in a competitive growth assay that a subset of cells can be selectively depleted by Cas13-mediated sensing of a dispensable reporter RNA uniquely expressed in the target cell population [Citation77]. The same concept has been applied in vivo for selective elimination of cancer cells by targeting cancer-specific oncogenic mRNAs. For instance, Kang and colleagues showed that Cas13a targeting of oncogenic EGFRvIII mRNA significantly inhibited tumour growth in vivo [Citation78] (although it remains to be shown that Cas13 collateral activity, rather than the loss of the oncogenic target mRNA alone, drove tumour regression). Further enhancing Cas13 collateral activity will enable more potent cell killing. Encouragingly, Yang and colleagues have shown that the collateral activity of Cas13a can be enhanced by inserting an extra RNA-binding domain into a unique active-site-proximal loop within its HEPN domains [Citation93].
Outstanding questions
Given the numerous applications of the Cas13 system in biomedical research, clinical diagnostics and potential therapeutic applications, it is critical to understand its limitations and avoid potential pitfalls. While high-fidelity variants of Cas13 alleviate the concern of unintended collateral RNA degradation for targeted RNA knockdown, recent studies have uncovered unexpected effect of expressing either Cas13 or gRNA alone. A deeper understanding of these observations and the underlying mechanisms will facilitate the development of next-generation high-fidelity Cas13 tools for various applications.
Cas13-independent effects of guide RNAs. While gRNAs are thought to be loaded into Cas13 first and subsequently bind target RNAs via sequence complementarity, the gRNA/target duplex is long (20–30 bp) and can be stably formed in cells even in the absence of Cas13. Binding by the gRNA alone can potentially block regulatory sites on the target mRNA or recruit double-strand RNA (dsRNA) binding proteins. For example, when used in plants, Cas13 gRNAs with 28-nt but not 20-nt spacers down-regulated target RNAs via endogenous RNAi machinery in the absence of Cas13a [Citation94]. Such effect was also seen with Cas9 gRNAs with 28-nt spacers. A similar observation was also made in human A459 cells: one out of three gRNAs tested decreased CXCR4 mRNA by over 90% in the absence of Cas13a [Citation29], although in this case it was unclear whether a long spacer was used and whether the RNAi machinery is involved. Similarly, expressing Cas13b gRNAs with a 31-nt spacer degraded a viral RNA in mosquito cells in the absence of Cas13b protein [Citation95]. Such Cas13-independent RNAi-like target silencing could potentially result in more off-target effects for targeted knockdown and complicate the interpretation of dCas13-based applications such as imaging and splicing modulation. It has also been shown that when the spacer region is longer than 40-nt, the long duplex formed with the target RNA will trigger efficient target RNA editing by recruiting endogenous RNA editing enzyme ADAR1 [Citation96]. The edited target RNA may no longer trigger Cas13-mediated cleavage, potentially explaining why gRNAs longer than 30-nt results in less efficient knockdown [Citation53]. These Cas13-independent effects of gRNAs highlight the caveat of using Cas13 gRNAs with long spacers and underscore the importance of proper controls. More systematic studies are needed to understand the prevalence of Cas13-independent effect and how to avoid it, for both the widely used Cas13 orthologs and the high-fidelity variants.
Guide RNA-independent Cas13 activity and toxicity. Several studies have reported unexpected toxicity when certain variants of Cas13 are expressed alone without corresponding gRNAs. For example, plasmid-based expression of LwaCas13a or PspCas13b but not RfxCas13d (CasRx) alone inhibited neurite and dendrite growth in primary cultures of mouse neurons [Citation97,Citation98]. Similarly, injecting PguCas13b and PspCas13b but not RfxCas13d proteins impaired zebrafish embryonic development [Citation75]. While RfxCas13d/CasRx is less toxic in cultured mouse neurons, ubiquitously expressed CasRx caused embryonic lethality in flies, as flies homozygote for CasRx cannot be generated [Citation99]. Intriguingly, even dCasRx homozygous flies cannot be generated. This observation suggests that the observed toxicity is likely caused by the pre-crRNA processing RNase activity, which remains intact in dCas13 [Citation100,Citation101]. Cas13/dCas13 proteins recognize the direct repeat region in pre-crRNAs, which forms a short hairpin with moderate sequence specificity. Similar hairpin structures could potentially be found in many endogenous RNAs in eukaryotic cells, especially in neurons that are known to enrich for dsRNAs [Citation102]. The recognition of these direct repeat-like structures will result in the cleavage and degradation of endogenous transcripts, potentially causing cellular toxicity. Supporting this idea, Li and colleagues discovered that LwCas13a, PspCas13b and RfxCas13d all bind thousands of endogenous mRNAs in HEK293T cells and are capable of cleaving endogenous RNAs in vitro without corresponding gRNAs [Citation103]. Future studies will delineate whether Cas13/dCas13 processes endogenous RNAs into crRNAs/gRNAs, and whether inactivating the pre-crRNA processing activity will abolish gRNA-independent toxicity.
Conclusions
Since its initial discovery in 2016, the RNA-targeting CRISPR/Cas13 system has not only transformed the basic research of RNA biology but has also significantly advanced RNA-based diagnostics and therapeutics. The ability to target essentially any sequences in RNA, nuclear or cytoplasmic, in a highly efficient and specific manner, makes Cas13 a powerful tool for gene targeting. Its compact size, coupled with its inherent multiplexibility, further enhances its potential as a therapeutic platform. The ever-expanding dCas13 toolkit facilitates a diverse range of RNA manipulations with single-nucleotide precision and specificity. The capability to precisely edit endogenous RNAs, including multi-kilobase replacements or insertions, without altering DNA, opens the door for developing safer therapies across a spectrum of diseases [Citation41]. The collateral activity of Cas13, while initially perceived as a constraint, has proven immensely successful in pathogen detection, and has the potential to become a powerful platform for precision cell targeting. We envision continued development of Cas13 technologies to enable more exciting applications in biomedical research and RNA-based diagnostics and RNA-targeting therapeutics.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573
- East-Seletsky A, O’Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273. doi: 10.1038/nature19802
- Smargon AA, Cox DBT, Pyzocha NK, et al. Cas13b is a type VI-B CRISPR-Associated RNA-Guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618–30 e7. doi: 10.1016/j.molcel.2016.12.023
- Yan WX, Chong S, Zhang H, et al. Cas13d is a compact RNA-Targeting type VI CRISPR effector positively modulated by a WYL-Domain-containing accessory protein. Mol Cell. 2018;70(2):327–39 e5. doi: 10.1016/j.molcel.2018.02.028
- Hu Y, Chen Y, Xu J, et al. Metagenomic discovery of novel CRISPR-Cas13 systems. Cell Discov. 2022;8(1):107. doi: 10.1038/s41421-022-00464-5
- Xu C, Zhou Y, Xiao Q, et al. Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat Methods. 2021;18(5):499–506. doi: 10.1038/s41592-021-01124-4
- Knott GJ, East-Seletsky A, Cofsky JC, et al. Guide-bound structures of an RNA-targeting A-cleaving CRISPR–Cas13a enzyme. Nat Struct Mol Biol. 2017;24(10):825–833. doi: 10.1038/nsmb.3466
- Liu L, Li X, Ma J, et al. The molecular architecture for RNA-Guided RNA cleavage by Cas13a. Cell. 2017;170(4):714–26 e10. doi: 10.1016/j.cell.2017.06.050
- Liu L, Li X, Wang J, et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168(1–2):121–34 e12. doi: 10.1016/j.cell.2016.12.031
- Zhang B, Ye Y, Ye W, et al. Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d. Nat Commun. 2019;10(1):2544. doi: 10.1038/s41467-019-10507-3
- Konermann S, Lotfy P, Brideau NJ, et al. Transcriptome engineering with RNA-Targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–76 e14. doi: 10.1016/j.cell.2018.02.033
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR–Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049
- Che W, Ye S, Cai A, et al. CRISPR-Cas13a targeting the enhancer RNA-SMAD7e inhibits bladder cancer development both in vitro and in vivo. Front Mol Biosci. 2020;7:607740. doi: 10.3389/fmolb.2020.607740
- Saifullah SM, Suzuki T, Yano S, et al. Effective RNA knockdown using CRISPR-Cas13a and Molecular Targeting of the EML4-ALK Transcript in H3122 lung cancer Cells. Int J Mol Sci. 2020;21(23):21. doi: 10.3390/ijms21238904
- Li S, Li X, Xue W, et al. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat Methods. 2021;18(1):51–59. doi: 10.1038/s41592-020-01011-4
- Abbott TR, Dhamdhere G, Liu YX, et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181(4):865–876.e12. doi: 10.1016/j.cell.2020.04.020
- Yin L, Zhao F, Sun H, et al. CRISPR-Cas13a inhibits HIV-1 infection. Mol Ther Nucleic Acids. 2020;21:147–155. doi: 10.1016/j.omtn.2020.05.030
- Cui J, Techakriengkrai N, Nedumpun T, et al. Abrogation of PRRSV infectivity by CRISPR-Cas13b-mediated viral RNA cleavage in mammalian cells. Sci Rep. 2020;10(1):9617. doi: 10.1038/s41598-020-66775-3
- Chen Y, Jiang H, Wang T, et al. In vitro and in vivo growth inhibition of human cervical cancer cells via human papillomavirus E6/E7 mRnas’ cleavage by CRISPR/Cas13a system. Antiviral Res. 2020;178:104794. doi: 10.1016/j.antiviral.2020.104794
- Gao J, Luo T, Lin N, et al. A new tool for CRISPR-Cas13a-based cancer gene therapy. Mol Ther Oncolytics. 2020;19:79–92. doi: 10.1016/j.omto.2020.09.004
- Jiang W, Li H, Liu X, et al. Precise and efficient silencing of mutant kras G12D by CRISPR-CasRx controls pancreatic cancer progression. Theranostics. 2020;10(25):11507–11519. doi: 10.7150/thno.46642
- Zhou H, Su J, Hu X, et al. Glia-to-neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell. 2020;181(3):590–603 e16. doi: 10.1016/j.cell.2020.03.024
- Zhou C, Hu X, Tang C, et al. CasRx-mediated RNA targeting prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Natl Sci Rev. 2020;7(5):835–837. doi: 10.1093/nsr/nwaa033
- Yan Z, Yao Y, Li L, et al. Treatment of autosomal dominant retinitis pigmentosa caused by RHO-P23H mutation with high-fidelity Cas13X in mice. Mol Ther Nucleic Acids. 2023;33:750–761. doi: 10.1016/j.omtn.2023.08.002
- Guo Y, Han L, Han S, et al. Specific knockdown of Htra2 by CRISPR-CasRx prevents acquired sensorineural hearing loss in mice. Mol Ther Nucleic Acids. 2022;28:643–655. doi: 10.1016/j.omtn.2022.04.014
- Zheng Z, Li G, Cui C, et al. Preventing autosomal-dominant hearing loss in bth mice with CRISPR/CasRx-based RNA editing. Sig Transduct Target Ther. 2022;7(1):79. doi: 10.1038/s41392-022-00893-4
- Li J, Shen Z, Liu Y, et al. A high-fidelity RNA-targeting Cas13 restores paternal Ube3a expression and improves motor functions in Angelman syndrome mice. Mol Ther. 2023;31(7):2286–2295. doi: 10.1016/j.ymthe.2023.02.015
- Morelli KH, Wu Q, Gosztyla ML, et al. An RNA-targeting CRISPR–Cas13d system alleviates disease-related phenotypes in Huntington’s disease models. Nat Neurosci. 2023;26(1):27–38. doi: 10.1038/s41593-022-01207-1
- Blanchard EL, Vanover D, Bawage SS, et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat Biotechnol. 2021;39(6):717–726. doi: 10.1038/s41587-021-00822-w
- Li T, Zhang L, Lu T, et al. Engineered extracellular vesicle-delivered CRISPR/CasRx as a novel RNA editing tool. Adv Sci. 2023;10(10):e2206517. doi: 10.1002/advs.202206517
- Cui Z, Zeng C, Huang F, et al. Cas13d knockdown of lung protease ctsl prevents and treats SARS-CoV-2 infection. Nat Chem Biol. 2022;18(10):1056–1064. doi: 10.1038/s41589-022-01094-4
- Zeballos CM, Moore HJ, Smith TJ, et al. Mitigating a TDP-43 proteinopathy by targeting ataxin-2 using RNA-targeting CRISPR effector proteins. Nat Commun. 2023;14(1):6492. doi: 10.1038/s41467-023-42147-z
- Powell JE, Lim CKW, Krishnan R, et al. Targeted gene silencing in the nervous system with CRISPR-Cas13. Sci Adv. 2022;8(3):eabk2485. doi: 10.1126/sciadv.abk2485
- Li M, Li D, Lin L, et al. Precise interference of RNA–protein interaction by CRISPR-Cas13-mediated peptide competition. ACS Synth Biol. 2023;12(10):2827–2833. doi: 10.1021/acssynbio.3c00287
- Tian S, Zhang B, He Y, et al. CRISPR-iPAS: a novel dCAS13-based method for alternative polyadenylation interference. Nucleic Acids Res. 2022;50(5):e26. doi: 10.1093/nar/gkac108
- Montagud-Martinez R, Marquez-Costa R, Rodrigo G. Programmable regulation of translation by harnessing the CRISPR-Cas13 system. Chem Commun (Camb). 2023;59(18):2616–2619. doi: 10.1039/D3CC00058C
- Yang LZ, Wang Y, Li SQ, et al. Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol Cell. 2019;76(6):981–97 e7. doi: 10.1016/j.molcel.2019.10.024
- Han S, Zhao BS, Myers SA, et al. RNA–protein interaction mapping via MS2- or Cas13-based APEX targeting. Proc Natl Acad Sci U S A. 2020;117(36):22068–22079. doi: 10.1073/pnas.2006617117
- Tang T, Han Y, Wang Y, et al. Programmable system of Cas13-mediated RNA modification and its biological and biomedical applications. Front Cell Dev Biol. 2021;9:677587. doi: 10.3389/fcell.2021.677587
- Du M, Jillette N, Zhu JJ, et al. CRISPR artificial splicing factors. Nat Commun. 2020;11(1):2973. doi: 10.1038/s41467-020-16806-4
- Jacob B, Kamyab J, James G, et al. Programmable multi-kilobase RNA editing using CRISPR-mediated trans-splicing. Preprint at bioRxiv. 2023. doi: 10.1101/2023.08.18.553620
- Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–1027. doi: 10.1126/science.aaq0180
- Abudayyeh OO, Gootenberg JS, Franklin B, et al. A cytosine deaminase for programmable single-base RNA editing. Science. 2019;365(6451):382–386. doi: 10.1126/science.aax7063
- Wilson C, Chen PJ, Miao Z, et al. Programmable m(6)A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat Biotechnol. 2020;38(12):1431–1440. doi: 10.1038/s41587-020-0572-6
- Xie S, Jin H, Yang F, et al. Programmable RNA N 1 -methyladenosine demethylation by a Cas13d-directed demethylase. Angew Chem Int Ed Engl. 2021;60(36):19592–19597. doi: 10.1002/anie.202105253
- Li J, Chen Z, Chen F, et al. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res. 2020;48(10):5684–5694. doi: 10.1093/nar/gkaa269
- Otoupal PB, Cress BF, Doudna JA, et al. CRISPR-RNAa: targeted activation of translation using dCas13 fusions to translation initiation factors. Nucleic Acids Res. 2022;50(15):8986–8998. doi: 10.1093/nar/gkac680
- Torkzaban B, Kawalerski R, Coller J. Development of a tethered mRNA amplifier to increase protein expression. Biotechnol J. 2022;17(10):e2200214. doi: 10.1002/biot.202200214
- Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136
- Shechner DM, Hacisuleyman E, Younger ST, et al. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12(7):664–670. doi: 10.1038/nmeth.3433
- Cao C, Li A, Xu C, et al. Enhancement of protein translation by CRISPR/dCasRx coupled with SINEB2 repeat of noncoding RNAs. Nucleic Acids Res. 2023;51(6):e33. doi: 10.1093/nar/gkad010
- Ahmed F, Benedito VA, Zhao PX. Mining functional elements in Messenger RNAs: overview, challenges, and perspectives. Front Plant Sci. 2011;2:84. doi: 10.3389/fpls.2011.00084
- Wessels HH, Mendez-Mancilla A, Guo X, et al. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat Biotechnol. 2020;38(6):722–727. doi: 10.1038/s41587-020-0456-9
- Zhao X, Liu L, Lang J, et al. A CRISPR-Cas13a system for efficient and specific therapeutic targeting of mutant KRAS for pancreatic cancer treatment. Cancer Lett. 2018;431:171–181. doi: 10.1016/j.canlet.2018.05.042
- Mayes CM, Santarpia J. Evaluating the impact of gRNA SNPs in CasRx activity for reducing viral RNA in HCoV-OC43. Cells. 2022;11(12):11. doi: 10.3390/cells11121859
- Molina Vargas AM, Sinha S, Osborn R, et al. New design strategies for ultra-specific CRISPR-Cas13a-based RNA detection with single-nucleotide mismatch sensitivity. Nucleic Acids Res. 2024;52(2):921–939. doi: 10.1093/nar/gkad1132
- Wessels HH, Stirn A, Mendez-Mancilla A, et al. Prediction of on-target and off-target activity of CRISPR–Cas13d guide RNAs using deep learning. Nat Biotechnol. 2023;42(4):628–637. doi: 10.1038/s41587-023-01830-8
- Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321
- Casati B, Verdi JP, Hempelmann A, et al. Rapid, adaptable and sensitive Cas13-based COVID-19 diagnostics using ADESSO. Nat Commun. 2022;13(1):3308. doi: 10.1038/s41467-022-30862-y
- Ke Y, Huang S, Ghalandari B, et al. Hairpin-spacer crRNA-enhanced CRISPR/Cas13a system promotes the specificity of single nucleotide polymorphism (SNP) identification. Adv Sci. 2021;8(6):2003611. doi: 10.1002/advs.202003611
- Meeske AJ, Nakandakari-Higa S, Marraffini LA. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature. 2019;570(7760):241–245. doi: 10.1038/s41586-019-1257-5
- Lopatina A, Tal N, Sorek R. Abortive infection: bacterial suicide as an antiviral immune strategy. Annu Rev Virol. 2020;7(1):371–384. doi: 10.1146/annurev-virology-011620-040628
- Kiga K, Tan XE, Ibarra-Chavez R, et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat Commun. 2020;11(1):2934. doi: 10.1038/s41467-020-16731-6
- Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179
- Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836
- Arizti-Sanz J, Freije CA, Stanton AC, et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020;11(1):5921. doi: 10.1038/s41467-020-19097-x
- Lopez-Valls M, Escalona-Noguero C, Rodriguez-Diaz C, et al. CASCADE: naked eye-detection of SARS-CoV-2 using Cas13a and gold nanoparticles. Anal Chim Acta. 2022;1205:339749. doi: 10.1016/j.aca.2022.339749
- Ortiz-Cartagena C, Fernandez-Garcia L, Blasco L, et al. Reverse transcription-loop-mediated isothermal amplification-CRISPR-Cas13a technology as a promising diagnostic tool for SARS-CoV-2. Microbiol Spectr. 2022;10(5):e0239822. doi: 10.1128/spectrum.02398-22
- Ackerman CM, Myhrvold C, Thakku SG, et al. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582(7811):277–282. doi: 10.1038/s41586-020-2279-8
- Bruch R, Baaske J, Chatelle C, et al. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv Mater. 2019;31(51):e1905311. doi: 10.1002/adma.201905311
- Zhou T, Huang R, Huang M, et al. CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and Specific MiRNA Detection. Adv Sci. 2020;7(13):1903661. doi: 10.1002/advs.201903661
- Chen Y, Yang S, Peng S, et al. N1-methyladenosine detection with CRISPR-Cas13a/C2c2. Chem Sci. 2019;10(10):2975–2979. doi: 10.1039/C8SC03408G
- Iwasaki RS, Batey RT. SPRINT: a Cas13a-based platform for detection of small molecules. Nucleic Acids Res. 2020;48(17):e101. doi: 10.1093/nar/gkaa673
- Huynh N, Depner N, Larson R, et al. A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol. 2020;21(1):279. doi: 10.1186/s13059-020-02193-y
- Kushawah G, Hernandez-Huertas L, Abugattas-Nunez Del Prado J, et al. CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev Cell. 2020;54(6):805–17 e7. doi: 10.1016/j.devcel.2020.07.013
- Kelley CP, Haerle MC, Wang ET. Negative autoregulation mitigates collateral RNase activity of repeat-targeting CRISPR-Cas13d in mammalian cells. Cell Rep. 2022;40(7):111226. doi: 10.1016/j.celrep.2022.111226
- Shi P, Murphy MR, Aparicio AO, et al. Collateral activity of the CRISPR/RfxCas13d system in human cells. Commun Biol. 2023;6(1):334. doi: 10.1038/s42003-023-04708-2
- Wang Q, Liu X, Zhou J, et al. The CRISPR-Cas13a gene-editing system induces collateral cleavage of RNA in glioma cells. Adv Sci. 2019;6(20):1901299. doi: 10.1002/advs.201901299
- Ai Y, Liang D, Wilusz JE. CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells. Nucleic Acids Res. 2022;50(11):e65. doi: 10.1093/nar/gkac159
- Webb BA, Chimenti M, Jacobson MP, et al. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11(9):671–677. doi: 10.1038/nrc3110
- Persi E, Duran-Frigola M, Damaghi M, et al. Systems analysis of intracellular pH vulnerabilities for cancer therapy. Nat Commun. 2018;9(1):2997. doi: 10.1038/s41467-018-05261-x
- White KA, Grillo-Hill BK, Barber DL. Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J Cell Sci. 2017;130(4):663–669. doi: 10.1242/jcs.195297
- Parks SK, Chiche J, Pouyssegur J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat Rev Cancer. 2013;13(9):611–623. doi: 10.1038/nrc3579
- Swietach P. What is pH regulation, and why do cancer cells need it? Cancer Metast Rev. 2019;38(1–2):5–15. doi: 10.1007/s10555-018-09778-x
- Baba M, Kojima K, Nakase R, et al. Effects of neutral salts and pH on the activity and stability of human RNase H2. J Biochem. 2017;162(3):211–219. doi: 10.1093/jb/mvx021
- Wessels HH, Mendez-Mancilla A, Hao Y, et al. Efficient combinatorial targeting of RNA transcripts in single cells with Cas13 RNA perturb-seq. Nat Methods. 2023;20(1):86–94. doi: 10.1038/s41592-022-01705-x
- Wei J, Lotfy P, Faizi K, et al. Deep learning and CRISPR-Cas13d ortholog discovery for optimized RNA targeting. Cell Syst. 2023;14(12):1087–102 e13. doi: 10.1016/j.cels.2023.11.006
- Tong H, Huang J, Xiao Q, et al. High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effects. Nat Biotechnol. 2023;41(1):108–119. doi: 10.1038/s41587-022-01419-7
- Ozcan A, Krajeski R, Ioannidi E, et al. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature. 2021;597(7878):720–725. doi: 10.1038/s41586-021-03886-5
- Caudron-Herger M, Muller-Ott K, Mallm JP, et al. Coding RNAs with a non-coding function: maintenance of open chromatin structure. Nucleus. 2011;2(5):410–424. doi: 10.4161/nucl.2.5.17736
- Hall LL, Carone DM, Gomez AV, et al. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156(5):907–919. doi: 10.1016/j.cell.2014.01.042
- Li Y, Xu J, Guo X, et al. The collateral activity of RfxCas13d can induce lethality in a RfxCas13d knock-in mouse model. Genome Biol. 2023;24(1):20. doi: 10.1186/s13059-023-02860-w
- Yang J, Song Y, Deng X, et al. Engineered LwaCas13a with enhanced collateral activity for nucleic acid detection. Nat Chem Biol. 2023;19(1):45–54. doi: 10.1038/s41589-022-01135-y
- Sharma VK, Marla S, Zheng W, et al. CRISPR guides induce gene silencing in plants in the absence of Cas. Genome Biol. 2022;23(1):6. doi: 10.1186/s13059-021-02586-7
- Tng PYL, Carabajal Paladino L, Verkuijl SAN, et al. Cas13b-dependent and Cas13b-independent RNA knockdown of viral sequences in mosquito cells following guide RNA expression. Commun Biol. 2020;3(1):413. doi: 10.1038/s42003-020-01142-6
- Qu L, Yi Z, Zhu S, et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat Biotechnol. 2019;37(9):1059–1069. doi: 10.1038/s41587-019-0178-z
- Wu QW, Kapfhammer JP. The bacterial enzyme Cas13 interferes with neurite outgrowth from cultured cortical neurons. Toxins (Basel). 2021;13(4):262. doi: 10.3390/toxins13040262
- Wu QW, Kapfhammer JP. The Bacterial Enzyme RfxCas13d is less neurotoxic than PspCas13b and could Be a promising RNA editing and interference tool in the nervous system. Brain Sci. 2021;11(8):11. doi: 10.3390/brainsci11081054
- Buchman AB, Brogan DJ, Sun R, et al. Programmable RNA targeting using CasRx in flies. Crispr J. 2020;3(3):164–176. doi: 10.1089/crispr.2020.0018
- Slaymaker IM, Mesa P, Kellner MJ, et al. High-resolution structure of Cas13b and biochemical characterization of RNA targeting and cleavage. Cell Rep. 2019;26(13):3741–51 e5. doi: 10.1016/j.celrep.2019.02.094
- Zhang C, Konermann S, Brideau N, et al. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell. 2018;175(1):212–223.e17. doi: 10.1016/j.cell.2018.09.001
- Dorrity TJ, Shin H, Wiegand KA, et al. Long 3′UTRs predispose neurons to inflammation by promoting immunostimulatory double-stranded RNA formation. Sci Immunol. 2023;8(88):eadg2979. doi: 10.1126/sciimmunol.adg2979
- Li Z, Li Z, Cheng X, et al. Intrinsic targeting of host RNA by Cas13 constrains its utility. Nat Biomed Eng. 2023;8(2):177–192. doi: 10.1038/s41551-023-01109-y
