ABSTRACT
Understanding how cells sense gases or gaseous solutes is a fundamental question in biology and is pivotal for the evolution of molecular and organismal life. In numerous organisms, gases can diffuse into cells, be transported, generated, and sensed. Controlling gases in the cellular environment is essential to prevent cellular and molecular damage due to interactions with gas-dependent free radicals. Consequently, the mechanisms governing acute gas sensing are evolutionarily conserved and have been experimentally elucidated in various organisms. However, the scientific literature on direct gas sensing is largely based on hemoprotein-based gasoreceptors (or sensors). As RNA-based G-quadruplex (G4) structures can also bind to heme, I propose that some ribozymes that contain or associated with heme-binding G4 can act as gas-sensing riboceptors (ribonucleic acid receptors). Additionally, I present a few other ideas for non-heme metal ion- or metal cluster-based gas-sensing riboceptors. Studying riboceptors can help understand the evolutionary origins of environmental gas sensing, cellular and gasocrine signaling, and interspecies communication.
If there are protein-based gasoreceptors, are there nucleic acid-based gasoreceptors?
Gases are among the evolutionarily oldest molecules, likely preceding the RNA world and potentially contributing to the formation of complex polymeric structures [Citation1–6]. As RNA-based organisms evolved, gaseous solute (gas) sensing must have been one of the earliest mechanisms to develop, playing a pivotal role not only in organism evolution but also in the evolution of molecular machinery. I have recently proposed gasocrine signaling to unify all the gasoreceptor-based cellular signaling events and to relax the gasotransmitter criteria to include oxygen (O2) as an essential gasotransmitter [Citation7,Citation8]. Currently, several protein-based acute gas-sensing sensors or gasoreceptors with diverse signaling domains and activity have been identified across various genera (). However, the identity of O2-sensing gasoreceptors in vertebrates and plants is largely unknown.
Table 1. List of gasoreceptors in diverse organisms.
Similar to proteins, RNA (riboswitches) can also sense and trigger a cellular signaling event. For instance, bacterial nhaA-I and nhaA-II RNA motif riboswitches can sense Li+, while the glmS ribozyme/catalytic riboswitch can sense molecules such as GlcN6P (glucosamine-6-phosphate) and glucose-6-phosphate [Citation9–12]. However, whether gas-sensing riboswitches exist is unknown [Citation13]. Recently, the lncRNA HIPLS (hypoxia-induced lncRNA for polo-like kinase 1 stabilization) was identified as essential for O2 sensing, acting downstream of HIF1α, one of the hypoxia-inducible factor subunits [Citation14]. To the best of my knowledge, there are no scientific literature on RNA-based structures (ribozyme, riboswitch, RNA-based gas sensor, or receptor, etc.) that can directly sense gases. Does this mean that during evolution, the only ‘sensory’ functions of ribozymes or riboswitches were for amino acids, metal ions, metabolites and temperature but not gases? [Citation15]. Or are such gas-sensing ribozymes yet to be experimentally demonstrated, or have they not been described in the literature with a unifying term similar to the parable of the blind men and the elephant? [Citation16,Citation17]
Heme-based protein gasoreceptors
Even before we could debate about the existence of gas-sensing ribozymes or catalytic riboswitches, a question arises: how common are heme (iron protoporphyrin IX)-based gasoreceptors or sensors other than the well-known NO (nitric oxide)-sensing soluble guanylate cyclase? [Citation18] Theoretical arguments for the presence of heme-based gas-sensing proteins have been proposed by others, even for O2 [Citation19,Citation20]. My arguments for mammalian protein-based gasoreceptors are based on the evolutionary conservation of gas-sensing hemoproteins and gas-binding metalloproteins [Citation21–23]. Heme-based gasoreceptors include DosP (Direct sensor of O2), FixL (Oxygen sensor), CooA (CO-sensing transcription factor) in bacteria, E75/Eip75 (ecdysone-induced protein 75) in flies, soluble adenylate cyclase in Leishmania, and soluble guanylate cyclase in worms, mice and human () [Citation7,Citation24–30]. Circadian regulators CLOCK (Clock Circadian Regulator), NPAS2 (Neuronal PAS domain protein 2), and nuclear hormone receptor family protein REV-ERB/NR1D1 are also heme-based gasoreceptors [Citation31–34]. The activity of all these proteins has been experimentally demonstrated to be affected by the binding state of either O2, CO, NO, or CO2. The heme in the pentacoordinate (and even in hexacoordinate in the case of CooA) geometry allows binding of these gases, which is usually further stabilized by additional interactions such as hydrogen bond formation with the distal histidine [Citation30]. Overall, heme-based gasoreceptors or sensors exist at the protein level with diverse signaling domains and activity that include phosphodiesterase, kinase, adenylate cyclase, guanylate cyclase, DNA-binding transcription factor activity (). However, to the best of my knowledge, the identity of O2-, H2S-, C2H4-, CH4-, NH3-, and N2-, sensing gasoreceptors in vertebrates is largely unknown [Citation35–39]. Moreover, it is not clear if heme is the sole co-factor in gasoreceptors, as proteins can bind gases such as O2, CO, H2S, and N2 either via metal ions or metal clusters [Citation22,Citation40–42]. Finally, it is worth noting that not all the hemoproteins may have a gas-sensing role (at least without additional binding partners). For example, O2-binding hemoprotein cytoglobin exhibits nitrite reductase activity and has been proposed as a redox sensor [Citation43,Citation44].
Can heme-binding G4-based ribozymes act as RNA-based gasoreceptors?
RNA G4 (G-quadruplex) structures have been described not only in mRNA but also in small and long ncRNAs (non-coding RNAs), TERRA (telomeric repeat-containing RNA), and even in mitochondria RNA [Citation45–48]. RNA G4 structures are widespread and enriched in the 5’ UTR of ribosomal protein-coding mRNAs, regulating ribosomal protein translation [Citation49,Citation50]. RNA G4 has been shown to exhibit ribozyme activity, catalysing oxygen transfer reactions and acting as peroxidases and/or peroxygenases both in vitro and in vivo [Citation51–54]. Such RNA G4-based ribozymes have also been demonstrated in the C9orf72 (chromosome 9 open reading frame 72) gene, implicated in neurodegenerative diseases such as ALS (Amyotrophic lateral sclerosis) [Citation55–58].
RNA-based G4 structures, characterized by stacked G-tetrads, have the capability to scavenge heme, which in its freeform is cytotoxic [Citation51,Citation52,Citation59–62]. Even human rRNA and ribosomes have been shown to bind heme in vivo [Citation63]. Functional RNA G4 heme-based ribozymes (e.g. rPS2.M/heme holoenzyme) has been reported to catalyze chlorination of organic substrates [Citation64]. However, the question arises whether all or some RNA G4 associated with heme (heme B) can also bind and sense in vivo under physiological conditions. In my opinion, if soluble guanylate cyclase can be activated by NO binding to its heme and this binding affects its structure and enzymatic activity, then why not consider that at least some RNA G4 structures with bound heme could similarly respond to gas binding? [Citation65] If gas binding to the heme of RNA G4 triggers a change in the activity of G4-containing or G4-associated ribozymes, then these ribozymes could be considered RNA-based gas-sensing receptors ().
Figure 1. Model of putative gas-sensing riboceptors. (A) a riboceptor can consist of a ribozyme or deoxyribozyme associated with sequences capable of binding diverse gas-binding cofactors. (B) a riboceptor can consist of a ribozyme or deoxyribozyme containing sequences capable of binding diverse gas-binding cofactors. The gas-binding must modulate the ribozyme activity.
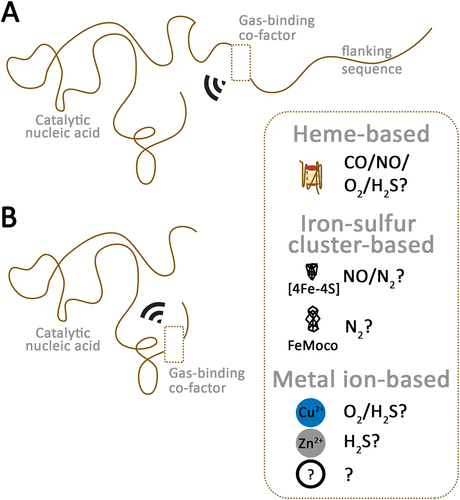
Overall, heme-binding RNA G4 structures found within or associated with RNA-based signaling components (such as ribozymes, riboswitches, RNA-based transcription factors), besides their role as heme scavenging, may also function as gas-sensing riboceptors (ribonucleic acid receptors) [Citation63,Citation66]. I propose ‘riboceptors’ as a unifying term to encompass all classes of RNA molecules capable of sensing not only for gas, ions, metal ions, and metabolites, but also temperature and water.
How to identify heme-based gas-sensing riboceptors?
To identify putative gas-sensing riboceptors, we first need to systemically identify all RNA structures with catalytic (or transcription factor) activity that contain or are associated with heme-binding G4-forming sequences and structures. Next, we should determine which heme-based ribozymes can bind to well-studied gases such as CO, CO2, NO, or O2 and exhibit a change in ribozyme activity upon binding. The binding of the gases may be stabilized through additional interactions with the RNA, such as hydrogen bonding via guanine or cytosine, similar to distal histidine-based hydrogen bonding seen in O2-binding hemoproteins. If binding of the gas does not alter ribozyme activity, then heme-binding could serve a catalytic, scavenging, or transport role for the gas. However, if binding of the gas alters ribozyme structure and activity in vivo, then these ribozymes function as heme-based gas-sensing riboceptors. Mutational studies disrupting heme-binding (or additional interactions with the gas) should confirm loss of gas-binding activity and modulation of ribozyme activity.
Binding partner, co-riboceptor, or riboceptor activity-modifying RNA?
Formation of protein-protein or protein-lipid-based functional homodimers, heterodimers, and heteromultimers/oligomers can alter or enhance the protein activity [Citation67–71]. For example, the Wnt/β-catenin pathway requires interaction of the Wnt ligand with the receptor Frizzled, as well as the co-receptor LRP5/6 (lipoprotein receptor-related protein 5 or 6) [Citation72]. Similarly, ligand-selectivity can be regulated depending on the identity of RAMPs (receptor activity-modifying proteins) and their interaction with receptors [Citation73,Citation74]. In another extreme example, gas-sensing bacterial protein CooA, CO-binding is important for CooA homodimerization and its transcriptional activity [Citation75]. Finally, the RNA-binding protein DGCR8 (DiGeorge Critical Region 8), which interacts with DROSHA to form a pri-miRNA (primary miRNA) processing complex, is also a gas-binding hemoprotein [Citation76–80]. We should consider whether DGCR8 or the pri-miRNA processing factor can be classified as gasoreceptor, given that CO and NO has been shown to inhibit pri-miRNA activity. This suggests that CO and NO act as antagonists of the pri-miRNA processing factor. Debating about it may allow us to consider riboceptors based on dimerization of ribozymes with other gas-sensing modules (). Overall, drawing parallels with protein-based signaling and gas-sensing mechanisms, it remains unknown whether RNA-based gas sensing also requires dimerization of ribozymes or other RNA structures, co-riboceptors, or riboceptor activity-modifying RNAs (RAMRs).
Non-heme-based gas-sensing riboceptors
In proteins, gas-binding occurs not only via heme, but also via metal ions and even metal clusters. For instance, arthropod hemocyanin bind O2 and CO via Cu2+ ions, while hemoglobin in vestimentiferan tubeworms can bind H2S via Zn2+ ions [Citation41,Citation42,Citation81]. Ethylene-sensing gasoreceptors in Arabidopsis and cyanobacteria require Cu2+ ions [Citation82,Citation83] Metal cluster-based gas binding occurs via FeS (Iron-sulphur), FeMoco (Iron molybdenum cofactor), etc [Citation40,Citation84]. In bacteria, FeS cluster-containing transcription factors FNR and NsrR has been proposed as O2- and NO-sensing proteins, respectively [Citation84–87]. However, apart from proteins, nucleic acids can also interact with metal ions [Citation88–92]. Whether metal ion-binding ribozymes or other RNA structures act as gas-sensing riboceptors is currently unknown (). Theoretically, FeS clusters can interact with RNA via coordination bonds, but their stability and role in gas sensing require experimental verification.
Additionally, besides RNA, G-rich DNA can also form G4 structures that bind heme, and DNA (deoxyribozymes or DNAzyme) can exhibit catalytic activity [Citation93–96]. Therefore, it is theoretically plausible that heme- or non-heme-based gas-sensing deoxyriboceptors may exist. Overall, until all putative heme- and non-heme-based nucleic acids have been systemically characterized for their ability to bind and sense gases, it remains challenging to dismiss the possibility of gas-sensing riboceptors. If gas-sensing riboceptors don’t exist, it suggests that the origin of gasocrine signaling and environmental gas-sensing is only via protein-based gasoreceptors, which I think is very unlikely if the RNA world theory is true, since gases preceded the RNA world. Finally, as I recently proposed that some of the heme-based gasoreceptors may also function as water-sensing aquareceptors, it remains to be determined whether gas-sensing riboceptors may also have a dual role as water-sensing riboceptors [Citation97].
Author contributions
Savani Anbalagan: conceptualization, writing of the original draft, and review and editing.
Acknowledgments
The author thank Zofia Szweykowska-Kulinska (Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University, Poznan, Poland) for allowing him to attend her inspiring lectures on molecular evolution. The author also thank Agnieszka Chacinska (Past affiliation: Centre of New Technologies, University of Warsaw, Regenerative Mechanisms for Health - International Research Agendas Programme and current affiliation: International Institute of Molecular Machines and Mechanisms, Polish Academy of Science, Warsaw, Poland) for suggesting him to attend the 44th FEBS Congress meeting.
Disclosure statement
The funding agency and institution S.A. is affiliated with was not involved in the contents of the manuscript.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Gesteland RF, Cech T, Atkins JF. The RNA world: the nature of modern RNA suggests a prebiotic RNA world. United States of America: CSHL Press; 2006.
- Higgs PG, Lehman N. The RNA world: molecular cooperation at the origins of life. Nat Rev Genet. 2015;16(1):7–17. doi: 10.1038/nrg3841
- Saito H. The RNA world ‘hypothesis’. Nat Rev Mol Cell Biol. 2022;23(9):582. doi: 10.1038/s41580-022-00514-6
- Lazcano A, Miller SL. The origin and early evolution of life: prebiotic chemistry, the pre-RNA world, and time. Cell. 1996;85(6):793–798. doi: 10.1016/S0092-8674(00)81263-5
- Pearce BKD, Pudritz RE, Semenov DA, et al. Origin of the RNA world: the fate of nucleobases in warm little ponds. Proc Natl Acad Sci USA. 2017;114(43):11327–11332. doi: 10.1073/pnas.1710339114
- Morasch M, Liu J, Dirscherl CF, et al. Heated gas bubbles enrich, crystallize, dry, phosphorylate and encapsulate prebiotic molecules. Nat Chem. 2019;11(9):779–788. doi: 10.1038/s41557-019-0299-5
- Anbalagan S. Heme-based oxygen gasoreceptors. Am J Physiol Endocrinol Metab. 2024;326(2):E178–81. doi: 10.1152/ajpendo.00004.2024
- Anbalagan S. Oxygen is an essential gasotransmitter directly sensed via protein gasoreceptors. Anim Model Exp Med. 2024;7(2):189–193. doi: 10.1002/ame2.12400
- White N, Sadeeshkumar H, Sun A, et al. Na+ riboswitches regulate genes for diverse physiological processes in bacteria. Nat Chem Biol. 2022;18(8):878–885. doi: 10.1038/s41589-022-01086-4
- Klein DJ, Ar F-D. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313(5794):1752–1756. doi: 10.1126/science.1129666
- Roth A, Nahvi A, Lee M, et al. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA. 2006;12(4):607–619. doi: 10.1261/rna.2266506
- Soukup J. The glmS ribozyme and its multifunctional coenzyme glucosamine-6-phosphate [internet]. In: Ribozymes. John Wiley & Sons, Ltd; 2021 [cited 2024 Jun 14]. p. 91–115. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/9783527814527.ch4
- Kavita K, Breaker RR. Discovering riboswitches: the past and the future. Trends Biochem Sci. 2023;48(2):119–141. doi: 10.1016/j.tibs.2022.08.009
- Chen Z, Chen C, Xiao L, et al. HILPS, a long noncoding RNA essential for global oxygen sensing in humans. Sci Adv. 2023;9: eadi1867. 47). doi: 10.1126/sciadv.adi1867
- Anbalagan S. Temperature-sensing riboceptors. RNA Biol. 2024. doi: 10.1080/15476286.2024.2379118
- Anbalagan S. “Blind men and an elephant”: the need for animals in research, drug safety studies, and understanding civilizational diseases. Anim Model Exp Med. 2023;6(6):627–633. doi: 10.1002/ame2.12364
- Backstein K. The blind men and the elephant. United States of America: Scholastic Incorporated; 1992.
- Ignarro LJ, Freeman B. Nitric oxide: biology and pathobiology. United States of America: Academic Press; 2017.
- Goldberg MA, Dunning SP, Bunn HF. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science. 1988;242(4884):1412–1415. doi: 10.1126/science.2849206
- Hochachka PW, Buck LT, Doll CJ, et al. Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA. 1996;93(18):9493–9498. doi: 10.1073/pnas.93.18.9493
- Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J Exp Biol. 1998;201(8):1099–1117. doi: 10.1242/jeb.201.8.1099
- Burmester T. Origin and evolution of arthropod hemocyanins and related proteins. J Comp Physiol B. 2002;172(2):95–107. doi: 10.1007/s00360-001-0247-7
- Vinogradov SN, Hoogewijs D, Bailly X, et al. A phylogenomic profile of globins. BMC Evol Biol. 2006;6(1):31. doi: 10.1186/1471-2148-6-31
- Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 2000;39(10):2685–2691. doi: 10.1021/bi991911s
- Monson EK, Weinstein M, Ditta GS, et al. The FixL protein of rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc Natl Acad Sci USA. 1992;89(10):4280–4284. doi: 10.1073/pnas.89.10.4280
- Lanzilotta WN, Schuller DJ, Thorsteinsson MV, et al. Structure of the CO sensing transcription activator CooA. Nat Struct Mol Biol. 2000;7(10):876–880. doi: 10.1038/82820
- Reinking J, Lam MMS, Pardee K, et al. The drosophila nuclear receptor E75 contains heme and is gas responsive. Cell. 2005;122(2):195–207. doi: 10.1016/j.cell.2005.07.005
- Sen Santara S, Roy J, Mukherjee S, et al. Globin-coupled heme containing oxygen sensor soluble adenylate cyclase in Leishmania prevents cell death during hypoxia. Proc Natl Acad Sci USA. 2013;110(42):16790–16795. doi: 10.1073/pnas.1304145110
- Gray JM, Karow DS, Lu H, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430(6997):317–322. doi: 10.1038/nature02714
- Liu R, Kang Y, Chen L. Activation mechanism of human soluble guanylate cyclase by stimulators and activators. Nat Commun. 2021;12(1):5492. doi: 10.1038/s41467-021-25617-0
- Freeman SL, Kwon H, Portolano N, et al. Heme binding to human CLOCK affects interactions with the E-box. Proc Natl Acad Sci USA. 2019;116:19911–19916. doi: 10.1073/pnas.1905216116
- Sarkar A, Carter EL, Harland JB, et al. Ferric heme as a CO/NO sensor in the nuclear receptor Rev-erbß by coupling gas binding to electron transfer. Proc Natl Acad Sci USA. 2021;118(3):e2016717118. doi: 10.1073/pnas.2016717118
- Dioum EM, Rutter J, Tuckerman JR, et al. NPAS2: a gas-responsive transcription factor. Science. 2002;298(5602):2385–2387. doi: 10.1126/science.1078456
- Uchida T, Sato E, Sato A, et al. CO-dependent activity-controlling mechanism of heme-containing CO-sensor protein, neuronal PAS domain protein 2. J Biol Chem. 2005;280(22):21358–21368. doi: 10.1074/jbc.M412350200
- Bishop T, Ratcliffe PJ. Genetic basis of oxygen sensing in the carotid body: HIF2α and an isoform switch in cytochrome c oxidase subunit 4. Sci Signal. 2020;13: eaba1302. 615). doi: 10.1126/scisignal.aba1302
- Ernst L, Steinfeld B, Barayeu U, et al. Methane formation driven by reactive oxygen species across all living organisms. Nature. 2022;603(7901):482–487. doi: 10.1038/s41586-022-04511-9
- Carlew TS, Allen CJ, Binder BM. Ethylene receptors in nonplant species. Small Methods. 2020;4(8):1900266. doi: 10.1002/smtd.201900266
- Lieberman M, Hochstein P. Ethylene formation in rat liver microsomes. Science. 1966;152(3719):213–214. doi: 10.1126/science.152.3719.213
- Hird FJ, Marginson MA. Formation of ammonia from glutamate by mitochondria. Nature. 1964;201(4925):1224–1225. doi: 10.1038/2011224a0
- Dance I. Calculating the chemical mechanism of nitrogenase: new working hypotheses. Dalton Trans. 2022;51(33):12717–12728. doi: 10.1039/D2DT01920E
- Flores JF, Fisher CR, Carney SL, et al. Sulfide binding is mediated by zinc ions discovered in the crystal structure of a hydrothermal vent tubeworm hemoglobin. Proc Natl Acad Sci USA. 2005;102:2713–2718. doi:10.1073/pnas.0407455102
- Richey B, Decker H, Gill SJ. Binding of oxygen and carbon monoxide to arthropod hemocyanin: an allosteric analysis. Biochemistry. 1985;24(1):109–117. doi: 10.1021/bi00322a016
- Kaliszuk SJ, Morgan NI, Ayers TN, et al. Regulation of nitrite reductase and lipid binding properties of cytoglobin by surface and distal histidine mutations. Nitric Oxide. 2022;125–126:12–22. doi: 10.1016/j.niox.2022.06.001
- DeMartino AW, Amdahl MB, Bocian K, et al. Redox sensor properties of human cytoglobin allosterically regulate heme pocket reactivity. Free Radic Biol Med. 2021;162:423–434. doi: 10.1016/j.freeradbiomed.2020.10.321
- Figueiredo J, Santos T, Miranda A, et al. Ligands as stabilizers of G-Quadruplexes in non-coding RNAs. Molecules. 2021;26(20):6164. doi: 10.3390/molecules26206164
- Mou X, Liew SW, Kwok CK. Identification and targeting of G-quadruplex structures in MALAT1 long non-coding RNA. Nucleic Acids Res. 2022;50(1):397–410. doi: 10.1093/nar/gkab1208
- Sahayasheela VJ, Yu Z, Hidaka T, et al. Mitochondria and G-quadruplex evolution: an intertwined relationship. Trends Genet. 2023;39(1):15–30. doi: 10.1016/j.tig.2022.10.006
- Hirashima K, Seimiya H. Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Res. 2015;43(4):2022–2032. doi: 10.1093/nar/gkv063
- Varshney D, Cuesta SM, Herdy B, et al. RNA G-quadruplex structures control ribosomal protein production. Sci Rep. 2021;11(1):22735. doi: 10.1038/s41598-021-01847-6
- Yang SY, Lejault P, Chevrier S, et al. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat Commun. 2018;9(1):4730. doi: 10.1038/s41467-018-07224-8
- Poon L-H, Methot SP, Morabi-Pazooki W, et al. Guanine-rich RNAs and DNAs that bind heme robustly catalyze oxygen transfer reactions. J Am Chem Soc. 2011;133(6):1877–1884. doi: 10.1021/ja108571a
- Sen D, Poon LCH. RNA and DNA complexes with hemin [Fe(iii) heme] are efficient peroxidases and peroxygenases: how do they do it and what does it mean? Crit Rev Biochem Mol Biol. 2011;46(6):478–492. doi: 10.3109/10409238.2011.618220
- Grigg JC, Shumayrikh N, Sen D, et al. G-quadruplex structures formed by expanded hexanucleotide repeat RNA and DNA from the neurodegenerative disease-linked C9orf72 gene efficiently sequester and activate heme. PLOS ONE. 2014;9(9):e106449. doi: 10.1371/journal.pone.0106449
- Lat PK, Liu K, Kumar DN, et al. High specificity and tight spatial restriction of self-biotinylation by DNA and RNA G-Quadruplexes complexed in vitro and in vivo with heme. Nucleic Acids Res. 2020;48(10):5254–5267. doi: 10.1093/nar/gkaa281
- Reddy K, Zamiri B, Stanley SYR, et al. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288(14):9860–9866. doi: 10.1074/jbc.C113.452532
- Fratta P, Mizielinska S, Nicoll AJ, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2(1):1016. doi: 10.1038/srep01016
- Geng Y, Cai Q. Role of C9orf72 hexanucleotide repeat expansions in ALS/FTD pathogenesis. Front Mol Neurosci. 2024;17:1322720. doi: 10.3389/fnmol.2024.1322720
- Kharel P, Becker G, Tsvetkov V, et al. Properties and biological impact of RNA G-quadruplexes: from order to turmoil and back. Nucleic Acids Res. 2020;48(22):12534–12555. doi: 10.1093/nar/gkaa1126
- Kharel P, Fay M, Manasova EV, et al. Stress promotes RNA G-quadruplex folding in human cells. Nat Commun. 2023;14(1):205. doi: 10.1038/s41467-023-35811-x
- Shao X, Zhang W, Umar MI, et al. RNA G-Quadruplex structures mediate gene regulation in bacteria. MBio. 2020;11(1):e02926–19. doi: 10.1128/mBio.02926-19
- Balla J, Vercellotti GM, Jeney V, et al. Heme, heme oxygenase, and ferritin: how the vascular endothelium survives (and dies) in an iron-rich environment. Antioxid Redox Signal. 2007;9(12):2119–2138. doi: 10.1089/ars.2007.1787
- Gozzelino R, Jeney V, Soares MP. Mechanisms of cell protection by heme oxygenase-1. Annu Rev Pharmacol Toxicol. 2010;50(1):323–354. doi: 10.1146/annurev.pharmtox.010909.105600
- Mestre-Fos S, Ito C, Moore CM, et al. Human ribosomal G-quadruplexes regulate heme bioavailability. J Biol Chem. 2020;295(44):14855–14865. doi: 10.1074/jbc.RA120.014332
- Hoog TG, Pawlak MR, Gaut NJ, et al. Emergent ribozyme behaviors in oxychlorine brines indicate a unique niche for molecular evolution on mars. Nat Commun. 2024;15(1):3863. doi: 10.1038/s41467-024-48037-2
- Horst BG, Yokom AL, Rosenberg DJ, et al. Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy. Elife. 2019;8:e50634. doi: 10.7554/eLife.50634
- Gray LT, Puig Lombardi E, Verga D, et al. G-quadruplexes sequester free heme in living cells. Cell Chem Biol. 2019;26(12):1681–1691.e5. doi: 10.1016/j.chembiol.2019.10.003
- Villaverde A, Corchero JL, Seras-Franzoso J, et al. Functional protein aggregates: just the tip of the iceberg. Nanomed (Lond). 2015;10(18):2881–2891. doi: 10.2217/nnm.15.125
- Kumari K, Sharma GS, Gupta A, et al. Functionally active cross-linked protein oligomers formed by homocysteine thiolactone. Sci Rep. 2023;13(1):5620. doi: 10.1038/s41598-023-32694-2
- Wang F, Liu K, Han L, et al. Function of a p24 heterodimer in Morphogenesis and protein transport in penicillium oxalicum. Sci Rep. 2015;5(1):11875. doi: 10.1038/srep11875
- Schwenk J, Metz M, Zolles G, et al. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010;465(7295):231–235. doi: 10.1038/nature08964
- Pyle E, Kalli AC, Amillis S, et al. Structural lipids enable the formation of functional oligomers of the eukaryotic purine symporter UapA. Cell Chem Biol. 2018;25(7):840–848.e4. doi: 10.1016/j.chembiol.2018.03.011
- MacDonald BT, He X. Frizzled and LRP5/6 receptors for wnt/ -catenin signaling. Cold Spring Harb Perspect Biol. 2012;4(12):a007880. doi: 10.1101/cshperspect.a007880
- Hay DL, Pioszak AA. RAMPs (receptor-activity modifying proteins): new insights and roles. Annu Rev Pharmacol Toxicol. 2016;56(1):469–487. doi: 10.1146/annurev-pharmtox-010715-103120
- Barbash S, Lorenzen E, Persson T, et al. GPCRs globally coevolved with receptor activity-modifying proteins, RAMPs. Proc Natl Acad Sci USA. 2017;114:12015–12020. doi:10.1073/pnas.1713074114.
- Roberts GP, Thorsteinsson MV, Kerby RL, et al. CooA: a heme-containing regulatory protein that serves as a specific sensor of both carbon monoxide and redox state. Prog Nucleic Acid Res Mol Biol. 2001;67:35–63.
- Barr I, Smith AT, Chen Y, et al. Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc Natl Acad Sci USA. 2012;109:1919–1924. doi: 10.1073/pnas.1114514109
- Faller M, Matsunaga M, Yin S, et al. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14(1):23–29. doi: 10.1038/nsmb1182
- Hines JP, Smith AT, Jacob JP, et al. CO and NO bind to Fe(II) DiGeorge critical region 8 heme but do not restore primary microRNA processing activity. J Biol Inorg Chem. 2016;21(8):1021–1035. doi: 10.1007/s00775-016-1398-z
- Partin AC, Ngo TD, Herrell E, et al. Heme enables proper positioning of drosha and DGCR8 on primary microRNAs. Nat Commun. 2017;8(1):1737. doi: 10.1038/s41467-017-01713-y
- Weitz SH, Gong M, Barr I, et al. Processing of microRNA primary transcripts requires heme in mammalian cells. Proc Natl Acad Sci USA. 2014;111:1861–1866. doi: 10.1073/pnas.1309915111
- Connelly PR, Johnson CR, Robert CH, et al. Binding of oxygen and carbon monoxide to the hemocyanin from the spiny lobster. J Mol Biol. 1989;207(4):829–832. doi: 10.1016/0022-2836(89)90248-9
- Lacey RF, Binder BM. Ethylene Regulates the Physiology of the Cyanobacterium Synechocystis sp. PCC 6803 via an Ethylene Receptor. Plant Physiol. 2016;171(4):2798–809. doi: 10.1104/pp.16.00602
- Lacey RF, Binder BM. How plants sense ethylene gas — The ethylene receptors. J Inorg Biochem. 2014;133:58–62. doi: 10.1016/j.jinorgbio.2014.01.006
- Tucker NP, Hicks MG, Clarke TA, et al. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One. 2008;3(11):e3623. doi: 10.1371/journal.pone.0003623
- Khoroshilova N, Popescu C, Münck E, et al. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94(12):6087–6092. doi: 10.1073/pnas.94.12.6087
- Gruner I, Frädrich C, Böttger LH, et al. Aspartate 141 is the fourth ligand of the oxygen-sensing [4Fe-4S]2+ cluster of Bacillus subtilis transcriptional regulator fnr *. J Biol Chem. 2011;286(3):2017–2021. doi: 10.1074/jbc.M110.191940
- Volbeda A, Dodd EL, Darnault C, et al. Crystal structures of the NO sensor NsrR reveal how its iron-sulfur cluster modulates DNA binding. Nat Commun. 2017;8(1):15052. doi: 10.1038/ncomms15052
- Sagripanti J-L, Goering PL, Lamanna A. Interaction of copper with DNA and antagonism by other metals. Toxicol Appl Pharmacol. 1991;110(3):477–485. doi: 10.1016/0041-008X(91)90048-J
- Simpson RB. Association constants of methylmercuric and mercuric ions with Nucleosides. J Am Chem Soc. 1964;86(10):2059–2065. doi: 10.1021/ja01064a029
- Joshi CK, Jamasb AR, ViñViñAs R, et al. gRnade: geometric deep learning for 3D RNA inverse design. bioRxiv. 2024:2024.03.31.587283.
- Philips A, G Ł, Bujnicki JM. Computational methods for prediction of RNA interactions with metal ions and small organic ligands. Methods Enzymol. 2015;553:261–285.
- Zhao Y, Wang J, Chang F, et al. Identification of metal ion-binding sites in RNA structures using deep learning method. Brief Bioinform. 2023;24(2):bbad049. doi: 10.1093/bib/bbad049
- Ponce-Salvatierra A, Wawrzyniak-Turek K, Steuerwald U, et al. Crystal structure of a DNA catalyst. Nature. 2016;529(7585):231–234. doi: 10.1038/nature16471
- Dong X, Qiu Z, Wang Z, et al. Efficient Silver(I)-containing i-motif DNA hybrid catalyst for enantioselective diels-alder reactions. Angew Chem Int Ed Engl. 2024:e202407838. doi: 10.1002/anie.202407838
- Cozma I, Em M, Brennan JD, et al. DNAzymes as key components of biosensing systems for the detection of biological targets. Biosens Bioelectron. 2021;177:112972. doi: 10.1016/j.bios.2021.112972
- Okamoto C, Momotake A, Yamamoto Y. Structural and functional characterization of complexes between heme and dimeric parallel G-quadruplex DNAs. J Inorg Biochem. 2021;216:111336. doi: 10.1016/j.jinorgbio.2020.111336
- Anbalagan, S. Heme-based aquareceptors. Postepy Biochem. 2024. doi: 10.18388/pb.2021_551
