Abstract
Circular RNA (circRNA) are novel types of non-coding RNA that may be used as non-invasive noninvasive biomarkers in clinical plasma samples. However, the role of circRNA in plasma samples from patients with new-onset systemic lupus erythematosus (SLE) has not been extensively investigated. In the present study, reverse transcription-quantitative PCR was used to screen differentially-expressed circRNA (hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367, hsa_circ_0002316, hsa_circ_0104871, hsa_circ_0001947, hsa_circ_0001481, hsa_circ_0008675, hsa_circ_0082689 and hsa_circ_0082688) in plasma samples isolated from 22 patients with new-onset SLE and 22 healthy control (HC). The results indicated hsa_circ_0000175, hsa_circ_ 0044235, hsa_circ_0068367, and hsa_circ_0001947 expression levels were significantly lower in plasma samples from new-onset SLE patients compared with corresponding levels in HC subjects and patients with new-onset rheumatoid arthritis (RA). Multivariate analysis indicated expression levels of hsa_circ_0044235 and hsa_circ_0001947 in plasma were independent risk factors for SLE. ROC curve analysis suggested that the combination of hsa_circ_0044235 and hsa_circ_0001947 indicated significant value in discriminating new-onset SLE from HC subjects and patients with RA. Moreover, the levels of hsa_circ_0044235 in plasma samples from patients with new-onset SLE were associated with platelet count, platelet-crit, and platelet distribution width; the expression of hsa_circ_0001947 in plasma from patients with SLE was associated with treatment. Thus, the present study demonstrated a promise for the combination of plasma hsa_circ_0044235 and hsa_circ_0001947 expression as potential diagnostic and prognostic biomarkers in patients with new-onset SLE.
Introduction
Systemic lupus erythematosus (SLE) is described as a prototype autoimmune syndrome accompanied by autoantibody production and immune complex formation. This is promoted by the binding of self-antigen and antibodies, which may potentially lead to life-threatening renal, cardiac or brain damage (Di Battista et al. Citation2018). Since patients with SLE have heterogeneous clinical manifestations and unpredictable disease courses, accurate diagnosis is urgent for appropriate treatment and optimal disease prognosis. However, the diverse manifestations and disease heterogeneity increase the difficulty of successful SLE diagnosis (Lech et al. Citation2011; Chang et al. Citation2015; O’Gorman et al. Citation2015). The current diagnostic methods for SLE lack sensitivity and specificity. Therefore, the identification of appropriate new biomarkers for the early diagnosis of SLE is urgently required.
Circular RNA (circRNA) is a type of non-coding RNA that form covalently-closed RNA circles (Chen and Yang Citation2015). In contrast to linear RNA, circRNA are more stable and resistant to exonuclease-mediated degradation due to the lack of 5' or 3' ends (Jeck et al. Citation2013). Moreover, circRNA often shows tissue or developmental stage-specific expression and is widely distributed in a variety of body fluids, such as blood (Memczak et al. Citation2013; Memczak et al. Citation2015). Given the high abundance, stability, tissue-specific expression and availability of circRNA, these RNA molecules can be used as biomarkers for the diagnosis of various diseases, such as cancer, infectious and autoimmune diseases (Huang et al. Citation2018; Ouyang et al. Citation2018; Yu et al. Citation2020).
Plasma circRNA possesses diagnostic value for certain diseases. Huang et al. (Citation2018) demonstrated that the expression levels of hsa_circ_0001953 and hsa_circ_0009024 in plasma could be used to diagnose active tuberculosis, whereas Zhu et al. (Citation2020) showed that plasma hsa_circ_0027089 levels could be employed as a new marker for the diagnosis of hepatitis B virus-related hepatocellular carcinoma. Furthermore, Wu et al. (Citation2019) reported three specific plasma circRNA (i.e., hsa_circRNA_004183, hsa_circRNA_079265, hsa_circRNA_105039) could be used as diagnostic biomarkers for congenital heart diseases.
Although increasing studies have demonstrated that circRNA are widely used to diagnose human diseases, the diagnostic role of plasma circRNA in patients with SLE is still unclear. Our previous study revealed that certain circRNA were differentially expressed in peripheral blood mononuclear cells (PBMC) and peripheral blood samples between patients with SLE and healthy control (HC) subjects (Luo et al. Citation2019b, Citation2020a,Citationb). In the present study, the potential for a wide array of differentially-expressed circRNA (i.e., hsa_circ_0000175, _0044235, _0068367, _0002316, _0104871, _0001947, _0001481, _0008675, _0082689, and _0082688) to be useful as novel diagnostic biomarkers for new-onset SLE was investigated in plasma samples from patients with SLE and healthy counterparts. It was hoped that the results might demonstrate the promise of the combination of plasma circRNA expressions as potential diagnostic and prognostic biomarkers in patients with new-onset SLE.
Materials and methods
Patient variables
The present study recruited 65 patients with new-onset SLE, who fulfilled the SLICC criteria for SLE (Petri et al. Citation2012), 51 patients with new-onset rheumatoid arthritis (RA) who fulfilled the revised ACR 2010 criteria for RA (Aletaha et al. Citation2010), and 51 healthy control (HC) subjects with no history of autoimmune diseases. Patient enrollment was performed in The First Affiliated Hospital of Nanchang University between May 2019 and July 2020. All patients were new-onset that was diagnosed for the first time and received no treatment(s) with immunosuppressants or corticosteroids prior to recruitment. Subsequently, 22 new-onset SLE cases were recruited following the administration of therapeutic regimens for at least 15 days. Patients with RA were recruited as an autoimmune disease control sample. Exclusion criteria used for the selection process included those patients accompanied by other autoimmune, inflammatory or hormonal diseases, cancers, or mental disorders. All study protocols were approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University (approval no. (2022) CDYFYYLK(06-005)).
Collection of plasma and total RNA extraction
A total of 5 ml peripheral blood from each subject was obtained into a K2-EDTA tube. Each sample was then centrifuged first at 2000 × g for 10 min at 4°C, followed by a second centrifugation at 12,000 × g for 10 min at 4°C. The resultant plasma (supernatant) samples were extracted; visual examination of the samples indicated an absence of hemolysis (also confirmed by the absence of free hemoglobin in the plasma; data not shown). Thereafter, aliquots of each plasma sample were incubated with TRIzol® reagent (Invitrogen; ThermoFisher Scientific, Waltham, MA) according to manufacturer protocols. Total RNA was then extracted using a miRNeasy Mini kit (Qiagen AB, Hilden, Germany) and a NanoDrop ND-1000 system (Agilent Technologies, Santa Clara, CA) was used to evaluate RNA integrity and quantity. The final isolates were each then stored at −80°C until PCR analyses.
Reverse transcription-quantitative PCR analysis
The cDNA samples were acquired by a reverse transcriptase reaction from total RNA using a PrimeScript RT kit (Takara Bio Inc., Kyoto, Japan). Thereafter, the protocol for the Reverse transcription-quantitative PCR (RT-qPCR) assays was as follows: initial denaturation step at 95°C for 5 min, followed by 40 cycles of 15 sec at 95°C (denaturation), 1 min at 60°C (annealing and elongation), and 2 min at 72°C (final extension). Each singular reaction with each given primer was performed on an ABI 7500 Real-Time PCR system (Applied Biosystems) with SYBR Premix Ex TaqTM II kit (Takara). The primers (shown in ) used in the present study were designed by the Circinteractome Divergent Primers web, verified by primer-BLAST and synthesized by Shanghai Shenggong. In each case, GADPH was used as an internal control. For all samples, relative gene expression was calculated using the 2-ΔΔCt comparative threshold cycle method (Livak and Schmittgen Citation2001).
Table 1. Specific circRNA primers used for RT-qPCR analysis.
Statistical analysis
Quantile normalization was performed using SPSS software (v. 17.0, SPSS, Inc., Cary, NC) and subsequent data processing was performed using Prism software (v.5, GraphPad. San Diego, CA) or SPSS 17.0. The equation n = Z2 × б2/d2 (wherein values used were 1.282 × [0.52/0.12] = 40.96) was used to calculate the minimum sample size needed for the current study to prove its hypothesis. A Student’s t-test and the Mann–Whitney U-test were employed to compare normally distributed parameters and those with skewed distribution, respectively. Multivariate analysis (logistic regression) was performed to analyze the risk factors. The changes noted following different treatment conditions were analyzed by the paired t-test, whereas the correlation analysis was evaluated by Spearman’s method. Receiver operating characteristic (ROC) curves were used to evaluate the diagnostic efficiency of plasma circRNA. The validated plasma biomarkers were enrolled by the binary logistic regression models and the model selection was performed to determine the final combinations of the biomarkers. A p-value < 0.05 was considered to indicate a significant difference.
Results
Characteristics of the study subjects
A total of 167 subjects were recruited in the present study, including 65 patients with SLE, 51 patients with RA and 51 HC subjects. Patients with SLE were classified into screening and validation cohorts. The screening cohort included 22 patients with SLE and 22 HC subjects. An independent cohort was used consisting of 43 patients with SLE and 39 HC subjects. Regarding sample size, use of an N = 65 was deemed sufficient for this study. One reason for this determination was that sample sizes of ≤60 have been used in many studies similar to the current one (see Li Let al. Citation2018; Wang et al. Citation2018; Miao et al. Citation2019); compared to these, the current sample size is not small. Further, when the minimal sample size needed for this study was calculated, i.e., using n = Z2 × σ2/d2 (1.28 2 × [0.52/0.12] = 40.96), the N = 65 sample size ultimately was larger than the minimum needed.
The patients were enrolled in the validation set for the evaluation of abnormal plasma circRNA levels. Disease activity was assessed by the SLE disease activity index (SLEDAI) (Bombardier et al. Citation2010). The characteristics of patients with SLE and RA and HC subjects are shown in . No significant differences were noted between patients with SLE and HC subjects regarding age or sex. No significant differences were noted between patients with SLE and RA with regard to sex. Patients with SLE and RA were not age-matched due to the difference in age of onset of the two diseases (the incidence of RA was high among women 50–60 years of age, while that of SLE was high among women of 20–40 years of age). No association was noted between plasma circRNA levels and age or sex in patients with SLE or HC subjects, as well as with regard to sex in patients with RA (data not shown). The plasma expression levels of hsa_circ_0044235 and hsa_circ_0068367 in patients with RA correlated with age (Figure S1), while the expression levels of the other plasma circRNA did not correlate with age (data not shown).
Figure 1. Screening of differentially-expressed circRNA in plasma samples from 22 patients with SLE and 22 HC subjects. Expression levels of (A) hsa_circ_0000175 [Student’s t-test], (B) hsa_circ_0044235 [Mann–Whitney U-test], and (C) hsa_circ_0068367 [Mann–Whitney] were significantly down-regulated in patients with SLE (compared with those noted in HC subjects). (D) Expression levels of hsa_circ_0002316 exhibited no significant differences between patients with SLE and HC subjects (Mann–Whitney). (E) Expression levels of hsa_circ_0104871 exhibited no significant differences between patients with SLE and HC subjects (Mann–Whitney test). (F) Expression levels of hsa_circ_0001947 were significantly down-regulated in patients with SLE compared with those noted in HC subjects (Mann–Whitney). (G) No differences in hsa_circ_0001481 expression levels between SLE and HC subjects (Student’s t-test). circRNA/circ = circular RNA. HC = healthy control. SLE = systemic lupus erythematosus.
![Figure 1. Screening of differentially-expressed circRNA in plasma samples from 22 patients with SLE and 22 HC subjects. Expression levels of (A) hsa_circ_0000175 [Student’s t-test], (B) hsa_circ_0044235 [Mann–Whitney U-test], and (C) hsa_circ_0068367 [Mann–Whitney] were significantly down-regulated in patients with SLE (compared with those noted in HC subjects). (D) Expression levels of hsa_circ_0002316 exhibited no significant differences between patients with SLE and HC subjects (Mann–Whitney). (E) Expression levels of hsa_circ_0104871 exhibited no significant differences between patients with SLE and HC subjects (Mann–Whitney test). (F) Expression levels of hsa_circ_0001947 were significantly down-regulated in patients with SLE compared with those noted in HC subjects (Mann–Whitney). (G) No differences in hsa_circ_0001481 expression levels between SLE and HC subjects (Student’s t-test). circRNA/circ = circular RNA. HC = healthy control. SLE = systemic lupus erythematosus.](/cms/asset/2f058e7e-21be-416b-a368-b5c966c7201a/iimt_a_2196453_f0001_b.jpg)
Table 2. Clinical parameters in the patients in this study.
Expression levels of hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367 and hsa_circ_0001947 in plasma samples of patients with SLE (screening stage)
Because plasma samples were easy to collect and the expression levels of circRNA in plasma were generally lower than in peripheral blood and peripheral blood mononuclear cells (PBMC) (data not shown), expression levels of certain circRNA in the study subjects’ plasma were evaluated. The RT-qPCR analyses found that expression of hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367, hsa_circ_0002316, hsa_circ_0104871, hsa_circ_0001947, hsa_circ_0001481, hsa_circ_0008675, hsa_circ_0082689 and hsa_circ_0082688 was initially detected in the plasma isolated from the 22 SLE patients and 22 HC subjects. Expression levels of hsa_circ_0000175 (p = 0.0031; ), hsa_circ_0044235 (p < 0.0001; ), hsa_circ_0068367 (p = 0.0376; ) and hsa_circ_0001947 (p = 0.0124; ) were significantly decreased in the plasma of SLE patients compared with those of HC subjects. The expression levels of hsa_circ_0002316 (p = 0.8235; ), hsa_circ_0104871 (p = 0.1927; ) and hsa_circ_0001481 (p = 0.5233; ) did not significantly differ between the two groups. This was also true for the expression of hsa_circ_0008675, hsa_circ_0082689 and hsa_circ_0082688 in plasma (data not shown).
Validation of the differentially-expressed circRNA in plasma samples from patients with SLE and RA, as well as HC subjects
To validate the aforementioned results noted during the screening stage, a further study was conducted in an independent cohort of 43 SLE patients and 29 HC subjects. The analyses found that expression levels of hsa_circ_0000175 (p = 0.0006; ), hsa_circ_0044235 (p < 0.0001; ), hsa_circ_0068367 (p = 0.0314; ) and hsa_circ_0001947 (p = 0.0175; ) in plasma samples of the SLE patients (when data pooled with the earlier 22 subjects; n = 65) were significantly decreased compared with those in the plasma from the now n = 51 HC subjects. Independent comparisons of just the 43 and 29 subjects showed the same trends for hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367, and the expression level of hsa_circ_0001947 in plasma samples of the 43 SLE patients were decreased compared with those in the plasma from the 29 HC subjects, but the difference was not significant (data not shown).
Figure 2. Validation of expression levels of hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367 and hsa_circ_0001947 in plasma samples during the second stage. (A) In plasma samples from 65 patients with SLE. expression levels of (A) hsa_circ_0000175 [Student’s t-test], (B) hsa_circ_0044235 [Mann–Whitney U-test], (C) hsa_circ_0068367 [Mann–Whitney], and (D) hsa_circ_0001947 [Student’s t-test] were significantly down-regulated compared with those in samples from 51 HC subjects. In plasma samples from 65 patients with SLE, expression levels of (E) hsa_circ_0000175 [Mann–Whitney], (F) hsa_circ_0044235 [Mann–Whitney], (G) hsa_circ_0068367 [Mann–Whitney], and (H) hsa_circ_0001947 [Mann–Whitney] in plasma samples from 65 patients with SLE were significantly down-regulated compared with those in samples from 51 patients with RA. SLE = systemic lupus erythematosus. HC = healthy control. RA = rheumatoid arthritis. circ = circular RNA.
![Figure 2. Validation of expression levels of hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367 and hsa_circ_0001947 in plasma samples during the second stage. (A) In plasma samples from 65 patients with SLE. expression levels of (A) hsa_circ_0000175 [Student’s t-test], (B) hsa_circ_0044235 [Mann–Whitney U-test], (C) hsa_circ_0068367 [Mann–Whitney], and (D) hsa_circ_0001947 [Student’s t-test] were significantly down-regulated compared with those in samples from 51 HC subjects. In plasma samples from 65 patients with SLE, expression levels of (E) hsa_circ_0000175 [Mann–Whitney], (F) hsa_circ_0044235 [Mann–Whitney], (G) hsa_circ_0068367 [Mann–Whitney], and (H) hsa_circ_0001947 [Mann–Whitney] in plasma samples from 65 patients with SLE were significantly down-regulated compared with those in samples from 51 patients with RA. SLE = systemic lupus erythematosus. HC = healthy control. RA = rheumatoid arthritis. circ = circular RNA.](/cms/asset/f1ca3d84-de31-4b0a-b7e5-20d6ea948e53/iimt_a_2196453_f0002_b.jpg)
In addition, the expression levels of this specific four circRNA were compared in plasma samples from patients with SLE and those with RA. The results indicated that the plasma expression levels of hsa_circ_0000175 (p < 0.0001; ), hsa_circ_0044235 (p = 0.0002; ), hsa_circ_0068367 (p = 0.0006; ) and hsa_circ_0001947 (p < 0.0001; ) in the pool of 65 patients with SLE were significantly down-regulated as compared to in the plasma samples from the 51 RA patients. In a comparison of outcomes with the HC vs. RA groups, it was seen that there was a trend toward elevated hsa_circ_0000175, elevated hsa_circ_0068367, and decreased hsa_cir0044235 in the RA patients’ plasma in comparison to samples from the HC group; however, the actual differences in each measured values were ultimately seen to not be statistically significant. Further, plasma expression levels of hsa_circ_0001947 in the 51 RA patients with RA were significantly up-regulated as compared to the plasma samples from the 51 HC (data not shown).
Plasma expression levels of hsa_circ_0044235 and hsa_circ_0001947 are risk factors for SLE
The above data indicated that plasma expression of hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367 and hsa_circ_0001947 in the SLE patients differed from those of the HC subjects and RA cases. Thus, to explore whether plasma expression of hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367 and hsa_circ_0001947 could potentially be considered risk factors for SLE, the ‘enter method’ of logistic regression was used.
The equation for the expression of this four circRNA in plasma was found to be: Y = −0.640 x [hsa_circ_0000175-1.507 x hsa_circ_0044235-0.177 x hsa_circ_0068367-0.925 x hsa_circ_0001947] + 2.846 (). From these analyses, the decrease in plasma levels of hsa_circ_0044235 and hsa_circ_0001947 was each identified as positive risk factors for SLE (both p < 0.0001), while expression levels of hsa_circ_0000175 and hsa_circ_0068367 were not (p = 0.0520, p = 0.3550, respectively).
Table 3. Expression of plasma hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367 and hsa_circ_0001947 in the equation.
Diagnostic value of plasma hsa_circ_0044235 and hsa_circ_0001947 in patients with SLE
Given that decreased plasma expressions of hsa_circ_0044235 and hsa_circ_0001947 were now determined to be positive risk factors for SLE, ROC (receiver operating characteristics) curves were used to evaluate the diagnostic efficiency of using plasma hsa_circ_0044235 and _0001947 expressions to determine SLE. Of the two, hsa_circ_0044235 exhibited the higher under the curve (AUC: 0.789, 95% CI: 0.708–0.869; p < 0.0001; ; ) value; hsa_circ_0001947 had an AUC = 0.641 (95% CI: 0.541–0.742; p = 0.0092; ; ). When logistic regression analysis was performed, the data indicated that the combination of expressions of both this plasma circRNA exhibited an improved potential to distinguish patients with SLE from HC subjects (AUC = 0.800, 95% CI: 0.723–0.878; p < 0.0001; ; ) when compared to that of using hsa_circ_0044235 (AUC: 0.789) or hsa_circ_0001947 (AUC = 0.641) alone as a marker alone.
Figure 3. ROC curve analysis of plasma hsa_circ_0044235 and hsa_circ_0001947 expression levels in patients with SLE and HC. (A) ROC curve analyses of plasma (A) hsa_circ_0044235 and (B) hsa_circ_0001947 expression levels in SLE patients and HC subjects. (C) ROC curve analysis using combined plasma hsa_circ_0044235, and hsa_circ_0001947 expression levels in SLE patients and HC subjects. ROC = receiver operating characteristics. SLE = systemic lupus erythematosus. HC = healthy control. circ = circular RNA.
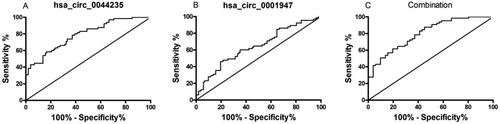
Table 4. ROC curve analysis of circRNA as SLE diagnosis from hosts in the RA and RA + HC groups.
The ROC curves constructed based on plasma hsa_circ_0044235 and hsa_circ_0001947 levels were also analyzed in patients with SLE and RA. The analyses found that hsa_circ_0001947 exhibited the higher AUC (0.854, 95% CI: 0.785–0.923; p < 0.0001; ; ); hsa_circ_0044235 had an AUC = 0.701 (95% CI: 0.607–0.795; p = 0.0002; ; ). Logistic regression analysis used to assess the diagnostic efficacy found that the combination of plasma hsa_circ_0044235 and hsa_circ_0001947 expressions reached an AUC = 0.883, which was superior to the AUC of either of this circRNA alone (95% CI: 0.823-0.943; p < 0.0001; ; ).
Figure 4. ROC curve analysis of plasma hsa_circ_0044235 and hsa_circ_0001947 expression levels in patients with SLE and RA, RA + HC. ROC curve analysis of plasma (A) hsa_circ_0001947 and (B) hsa_circ_0044235 expression levels in patients with SLE and RA. (C) ROC curve analysis of combined plasma hsa_circ_0044235 and hsa_circ_0001947 expression levels in patients with SLE and RA. ROC curve analysis of plasma (D) hsa_circ_0001947 and (E) hsa_circ_0044235 expression levels in SLE and control subjects (patients with RA + HC subjects). (F) ROC curve analysis of combined plasma hsa_circ_0044235 and hsa_circ_0001947 expression levels in SLE and control subjects (patients with RA + HC subjects). ROC : receiver operating characteristics; SLE : systemic lupus erythematosus; RA : rheumatoid arthritis; HC : healthy control; circ : circular RNA.
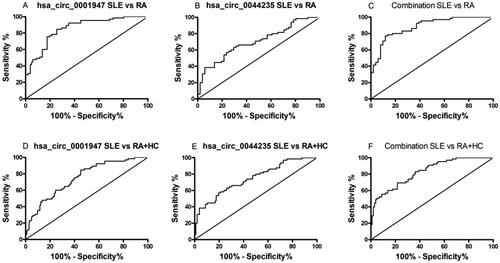
In addition, the question was raised as to whether plasma expression levels of each of this circRNA could be used to distinguish patients with SLE from all “controls” (i.e., HC and RA). The analyses to address this found that levels of hsa_circ_0001947 exhibited a higher AUC (0.748, 95% CI: 0.673-0.822; p < 0.0001; ; ), followed by hsa_circ_0044235 (AUC: 0.745, 95% CI: 0.669-0.821; p < 0.0001; ; ). The logistic regression model revealed that using a combination of expression levels of this two circRNA resulted in an improved ability to distinguish patients with SLE from all controls (HC and RA) (AUC = 0.829, 95% CI: 0.767-0.890; p < 0.0001; ; ) which was improved compared with the AUC associated with each circRNA alone.
Correlation of confirmed plasma circRNA expression with clinical characteristics of SLE
To explore whether plasma expression levels of hsa_circ_0044235 and hsa_circ_0001947 in SLE patients could be used to assess disease severity, a Spearman’s analysis was used to investigate associations between circRNA expression and clinical characteristics, including SLEDAI, white blood count, lymphocyte count, lymphocyte percentage, monocyte count (M), monocyte percentage (M%), neutrophil count, neutrophil percentage (N), neutrophil percentage (N%), red blood cell count, hemoglobin, hematocrit, platelet count (PLT), mean platelet volume (MPV), platelet-crit (PCT), platelet distribution width (PDW), neutrophil-lymphocyte ratio, platelet-lymphocyte ratio, lymphocyte-monocyte ratio, erythrocyte sedimentation rate, C-reactive protein and presence of autoantibodies. Plasma expression of hsa_circ_0044235 in SLE patients was found to correlate with PLT (rs = 0.3357, p = 0.0066), PCT (rs = 0.3114, p = 0.0261), and PDW (rs = −0.3118, p = 0.0259); changes in values for this three marker reflected the activity and severity of the SLE itself in the study subjects (). In contrast, no correlation was found between plasma hsa_circ_0001947 levels and these (or any other measured) clinical characteristics.
Figure 5. Correlation of plasma hsa_circ_0044235 and hsa_circ_0001947 expression levels with clinical manifestations of SLE. (A) Plasma expression levels of hsa_circ_0044235 in patients with SLE were positively associated with PLT (Spearman’s method). (B) Plasma expression levels of hsa_circ_0044235 in patients with SLE were positively associated with PCT (Spearman’s method). (C) Plasma expression levels of hsa_circ_0044235 in patients with SLE were negatively associated with PDW (Spearman’s method). (D) No significant difference was noted in the plasma expression levels of hsa_circ_0044235 prior to and following treatment (Student’s t-test, paired t-test). (E) Plasma expression levels of hsa_circ_0001947 were increased in these patients with SLE following treatment (Mann-Whitney test, Wilcoxon matched-pairs test). SLE : systemic lupus erythematosus; PCT : platelet crit; PDW : platelet distribution width; PLT : platelet count.
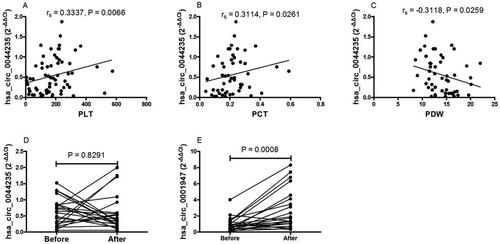
To see if such measures have application in monitoring treatments for SLE, the plasma expression levels of hsa_circ_0044235, and hsa_circ_0001947 were evaluated again in 22 SLE cases both prior to and following pharmacologic treatments (i.e., corticosteroids and immunosuppressants, 15 days). The analyses found that hsa_circ_0001947 plasma levels were significantly increased in the SLE patients following the treatment regimen (p = 0.0008; . However, no impact on levels of hsa_circ_0044235 due to treatment was noted ().
Discussion
It is increasingly believed that circular RNA (circRNA) might play an important role in many pathologies. Indeed, many studies have shown that various circRNA impact the functions of a variety of critical immune system cells during these health events (in re: macrophages and neutrophils [Maass et al. Citation2017; Ye et al. Citation2018; Duanet al. Citation2021; Feng et al. Citation2021; Song et al. Citation2021; Liang et al. Citation2022; Cao et al. Citation2022; Wan et al. Citation2022; Zhou et al. Citation2022; Tofigh et al. Citation2023] and lymphocytes [Liang et al. Citation2020; Chen et al. Citation2022; Cheng et al. Citation2022; Jiang et al. Citation2022]. Beyond determining the impact of circRNA on immune cell functions, the above-noted and several other studies have begun to examine the potential use of plasma circRNA expression levels as biomarkers for the presence of different pathologies, including autoimmune- and inflammation-based diseases. The results from this current study build upon this existing database of information about circRNA and autoimmune diseases in humans.
Indeed, Ouyang et al. (Citation2018) found that circRNA_002453 expression was up-regulated in the plasma of lupus nephritis (LN) patients and as this was associated with the severity of renal involvement, this suggested that circRNA_002453 might serve as a potential biomarker for the diagnosis of LN. With respect to SLE, the differential expression of a variety of circRNA in the plasma of SLE patients was studied using microarrays and RT-qPCR (Li H et al. Citation2018); those investigators suggested that the comprehensive expression profiles of various circRNA in SLE patients plasma and of hsa_circ_101471, hsa_circ_102584, hsa_circ_400011, hsa_circ_100226 may be useful as novel noninvasive biomarkers for SLE disease in the future. In addition, Zhang et al. (Citation2018) found that expression of hsa_circRNA_407176 and hsa_circRNA_001308 were decreased in the plasma of SLE patients and so each of this circRNA could be potential biomarker to detect SLE. Even those these early studies provided credible data supporting the potential use of certain circRNA as biomarkers, the results from microarrays over different studies were inconsistent. Although the reason for this remains unclear, it is possible SLE patient heterogeneity and diversity in specimen collection gave rise to these disparities.
Given that the reproducibility, convenience, and economy of examining biomarkers to detect autoimmune diseases (including SLE) is clear, the present studies were undertaken to focus specifically on 10 circRNA (e.g., hsa_circ_0000175, hsa_circ_0044235, hsa_circ_0068367, hsa_circ_0002316, hsa_circ_0104871, hsa_circ_0001947, hsa_circ_0001481, hsa_circ_0008675, hsa_circ_0082689, and hsa_circ_0082688). The main reason for the selection of this 10 circRNA for analyses was based on pilot study analyses of their levels in the peripheral blood and associated blood cells from SLE patients.
The study here showed that expression of hsa_circ_0000175, _0044235, _0068367, and _0001947 were significantly decreased in plasma from SLE patients, whereas expression of hsa_circ_0002316, _0104871, and _0001481 did not differ from levels in healthy controls. These findings were consistent with the results from previous studies (Li et al. Citation2018; Ouyang et al. Citation2018; Zhang et al. Citation2018), suggesting that the expression of circRNA in the plasma of SLE patients were dysregulated. Interestingly, though previous studies demonstrated that hsa_circ_0008675, hsa_circ_0082689, and hsa_circ_0082688 levels were elevated in the peripheral blood/blood cells of SLE patients (Luo et al. Citation2019b, Citation2020a,Citationb), such findings were not reproduced here in the plasma samples.
The present results showed that expressions of plasma hsa_circ_0000175, _0044235, _0068367, and _0001947 were significantly decreased in SLE patients compared with HC and RA ‘controls’. Multivariate analysis indicated that the expression levels of hsa_circ_0044235 and hsa_circ_0001947 in plasma were independent risk factors for SLE. Thus, hsa_circ_0044235 and hsa_circ_0001947 were selected for ROC curve analysis to ascertain whether this plasma circRNA could potentially be used as diagnostic biomarkers for SLE. The results here suggested that plasma hsa_circ_0044235 could be used to differentiate patients with SLE from HC subjects, an outcome consistent with findings reported previously for hsa_circ_0044235 in other types of samples (Luo et al. Citation2019b, Citation2020a). Moreover, the results demonstrated that a ROC curve analysis of a combination of plasma hsa_circ_0044235 and hsa_circ_0001947 expressions could be used to differentiate SLE from RA. Thus, this study became the first to demonstrate an improved diagnostic role for a combination of this two plasma circRNA for SLE diagnosis. Such an outcome was consistent with previous findings on potential diagnostic roles for monitoring circRNA, such as hsa_circ_0000479 and hsa_circ_0057762, when evaluating SLE patients (Guo et al. Citation2019; Li et al. Citation2019).
As the data revealed the potential for select circRNA to be used to diagnose the presence at all of SLE, the question was raised as to if they could also be useful as markers of disease severity. In fact, the data indicated that plasma expression levels of hsa_circ_0044235 in patients with SLE correlated with clinical values for PLT, PCT and PDW, themselves markers of SLE severity (Paradowska-Gorycka et al. Citation2016; Wang et al. Citation2019; Lopez et al. Citation2020). Moreover, the analyses here also found that plasma expression levels of hsa_circ_0001947 in SLE patients could be modulated in tandem with patient treatment regimens. Taken together, these results indicate that monitoring of plasma hsa_circ_0044235 and hsa_circ_0001947 (alone or in combination) could provide improved diagnostic accuracy for detecting a presence of SLE, as well as any worsening/progression of the disease and/or host responses to known treatments.
As circRNA regulate the expression of target microRNA (miRNA) via sponging effects, which in turn promotes degradation of the corresponding target genes of these miRNA, the next series of studies need to address the mechanism of action that each of the above-analyzed circRNA might have in SLE (Zhang et al. Citation2018). To begin to better define functions for the two circRNA here that showed the most significant relationships with the various endpoints evaluated in the study groups (i.e., hsa_circ_0044235 and hsa_circ_0001947), Arraystar’s prediction software was used to predict possible miRNA targets for this circRNA. As before, a total of five putative miRNA targets of hsa_circ_0044235, and hsa_circ_0001947 were identified (Luo et al. Citation2019a; Luo et al. Citation2019b; Luo et al. Citation2020a) (Table SI). Accordingly, expression levels of these putative miRNA in plasma will be investigated in more detail in future studies in order to assess the impact of changes in the expression of this two circRNA during the course of SLE.
Conclusions
In the present study, the potential association of select plasma circRNA with SLE was examined. The data revealed that plasma expression levels of hsa_circ_0000175, _0044235, _0068367, and _0001947 were differentially-expressed between patients with SLE and RA (or even with HC subjects). Further, the analyses here indicated that decreased levels of plasma hsa_circ_0044235 and hsa_circ_0001947 each could be seen as positive risk factors for SLE. Moreover, the studies also found that combinational analyses of both plasma hsa_circ_0044235 and hsa_circ_0001947 expression might be useful as a novel biomarker assay for the diagnosis of new-onset SLE and to monitor disease progression/patient response to treatment regimens against SLE.
Study limitations
The present study contains certain limitations. For example, the sample size for the new-onset patients who were enrolled at The First Affiliated Hospital of Nanchang University was relatively small and the patients were recruited from only one hospital, which may restrict result validity. Therefore, a larger cohort study and additional cases from different medical centres or hospitals are required to validate the potential clinical application of this circRNA.
Author contributions
QL, YTY and LZ performed the experiments. YJG, JYR, YG, ZKH and JML analyzed and interpreted the data. QL and JML made substantial contributions to the design/supervision of the present study and wrote the manuscript. All authors have reviewed the results and approved the final version of the manuscript.
Supplemental Material
Download MS Word (59.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of the present study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Aletaha D, Neogi T, Silman A, Funovits J, Felson D, Bingham C, Birnbaum N, Burmester G, Bykerk V, Cohen M, et al. 2010. The 2010 rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 69:1580–1588.
- Bombardier C, Gladman D, Urowitz M, Caron D, Chang C. 2010. Derivation of the SLEDAI. A disease activity index for lupus patients. Committee on Prognosis Studies in SLE. Arthritis Rheum. 35:630–640.
- Cao S, Zeng Y, Chen M, Ouyang W. 2022. Integrated analysis of immune-related circRNA-miRNA-mRNA regulatory network in ischemic stroke. Front Neurol. 13:889855.
- Chang N, Li T, Kim J, Landolt-Marticorena C, Fortin P, Gladman D, Urowitz M, Wither J. 2015. Interferon-α induces altered transitional B-cell signaling and function in systemic lupus erythematosus. J Autoimmun. 58:100–110.
- Chen J, Jia G, Lv X, Li S. 2022. Type 1 diabetes mellitus-related circRNA regulate CD4+ T-cell functions. Bio Med Res Intl. 2022:4625183.
- Chen L, Yang L. 2015. Regulation of circRNA biogenesis. RNA Biol. 12:381–388.
- Cheng S, Tang Q, Xie S, Wen S, Zhang H, Xie Z, Jiang W. 2022. Role of non-coding RNA in airway allergic diseases through regulation of T-cell subsets. Mediat Inflamm. 2022:6125698.
- Di Battista M, Marcucci E, Elefante E, Tripoli A, Governato G, Zucchi D, Tani C, Alunno A. 2018. One year in review 2018: Systemic lupus erythematosus. Clin Exp Rheumatol. 36(5):763–777.
- Duan S, Wang S, Huang T, Wang J, Yuan X. 2021. circRNAs: Insight into their role in tumor-associated macrophages. Front Oncol. 11:780744.
- Feng Z, Li L, Tu Y, Shu X, Zhang Y, Zeng Q, Luo L, Wu A, Chen W, Cao Y, et al. 2021. Identification of circular RNA-based immunomodulatory networks in colorectal cancer. Front Oncol. 11:779706.
- Guo G, Wang H, Ye L, Shi X, Yan K, Lin K, Huang Q, Li B, Lin Q, Zhu L, et al. 2019. Hsa_circ_0000479 as a novel diagnostic biomarker of systemic lupus erythematosus. Front Immunol. 10:2281.
- Huang Z, Su R, Qing C, Peng Y, Luo Q, Li J. 2018. Plasma ccircular RNAs hsa_circ_0001953 and hsa_circ_0009024 as diagnostic biomarkers for active tuberculosis. Front Microbiol. 9:2010.
- Jeck W, Sorrentino J, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 19:141–157.
- Jiang Z, Huang L, Chen L, Zhou J, Liang B, Bai X, Wu L, Huang H. 2022. Circular RNA profile in Graves’ Dsease and potential function of hsa_circ_0090364. Endocrine Connect. 11:e220030.
- Lech M, Kantner C, Kulkarni O, Ryu M, Vlasova E, Heesemann J, Anz D, Endres S, Kobayashi K, Flavell R, et al. 2011. Interleukin-1 receptor-associated kinase-M suppresses systemic lupus erythematosus. Ann Rheum Dis. 70:2207–2217.
- Li H, Li K, Lai W, Li X, Wang H, Yang J, Chu S, Wang H, Kang C, Qiu Y. 2018. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta. 480:17–25.
- Li L, Zhu Z, Zhao W, Tao S, Li B, Xu S, Wang J, Zhang M, Wu J, Leng R, et al. 2018. Circular RNA expression profile and potential function of hsa_circ_0045272 in systemic lupus erythematosus. Immunology. 155:137–149.
- Li S, Zhang J, Tan X, Deng J, Li Y, Piao Y, Li C, Yang W, Mo W, Sun J, et al. 2019. Microarray expression profile of circular RNAs and mRNAs in children with systemic lupus erythematosus. Clin Rheumatol. 38:1339–1350.
- Liang B, Li M, Deng Q, Wang C, Rong J, He S, Xiang Y, Zheng F. 2020. CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease. Ann Translat Med. 8:741.
- Liang Q, Fu J, Wang X, Liu L, Xiao W, Gao Y, Yang L, Yu H, Xie X, Tu Z, et al. 2022. Circs100a11 enhances M2a macrophage activation and lung inflammation in children with asthma. Allergy. Sep 14. doi: 10.1111/all.15515.Online ahead of print.
- Livak K, Schmittgen T. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 25:402–408.
- Lopez P, Rodriguez-Carrio J, Martinez-Zapico A, Pérez-Álvarez A, Suorez-Diaz S, Mozo L, Benavente L, Caminal-Montero L, Suorez A. 2020. Low-density granulocytes and monocytes as biomarkers of cardiovascular risk in systemic lupus erythematosus. Rheumatology. 59:1752–1764.
- Luo Q, Liu J, Fu B, Zhang L, Guo Y, Huang Z, Li J. 2019a. Circular RNAs Hsa_circ_0002715 and Hsa_circ_0035197 in peripheral blood are novel potential biomarkers for new-onset rheumatoid arthritis. Dis Markers. 2019:2073139.
- Luo Q, Zhang L, Fang L, Fu B, Guo Y, Huang Z, Li J. 2020a. Circular RNAs hsa_circ_0000479 in peripheral blood mononuclear cells as novel biomarkers for systemic lupus erythematosus. Autoimmunity. 53:167–176.
- Luo Q, Zhang L, Li X, Fu B, Guo Y, Huang Z, Li J. 2019b. Identifcation of circular RNA hsa_circ_0044235 and hsa_circ_0068367 as novel biomarkers for systemic lupus erythematosus. Intl J Mol Med. 44:1462–1472.
- Luo Q, Zhang L, Xiong L, Fu B, Guo Y, Huang, ZLJ 2020b. Peripheral blood circular RNA hsa_circ_0082688-hsa circ_0008675 can be used as a candidate biomarker of systemic lupus erythematosus with renal involvement. Clin Exp Rheumatol. 38:822–833.
- Maass P, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer A, Toka O, Aiuti A, Luft F, et al. 2017. A map of human circular RNAs in clinically relevant tissues. J Mol Med. 95:1179–1189.
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak S, Gregersen L, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNA with regulatory potency. Nature. 495:333–338.
- Memczak S, Papavasileiou P, Peters O, Rajewsky N. 2015. Identification and characterization of circular RNA as a new class of putative biomarkers in human blood. PLoS One. 10(10):e0141214.
- Miao Q, Zhong Z, Jiang Z, Lin Y, Ni B, Tang W, Tang J. 2019. RNA-seq of circular RNA identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. Lupus. 28:520–528.
- O’Gorman W, Hsieh E, Savig E, Gherardini P, Hernandez J, Hansmann L, Balboni I, Utz P, Bendall S, Fantl W, et al. 2015. Single-cell systems-level analysis of human toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J Allergy Clin Immunol. 136:1326–1336.
- Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi G, Yang M. 2018. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol Immunol. 101:531–538.
- Paradowska-Gorycka A, Sowinska A, Stypinska B, Grobelna MK, Walczyk M, Olesinska M, Piotrowski P, Jagodzinski PP. 2016. Genetic variants in IL-12B and IL-27 in Polish patients with systemic lupus erythematosus. Scand J Immunol. 84:49–60.
- Petri M, Orbai A, Alarcón G, Gordon C, Merrill J, Fortin P, Bruce IN, Isenberg D, Wallace D, Nived O, et al. 2012. Derivation and validation of the systemic lupus international collaborating clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 64:2677–2686.
- Song M, Gao J, Yan T, Bi E, An T, Wang X, Jiang W, Wang T, Chen Z, Shi Z, et al. 2021. Hsa_circ_0000652 aggravates inflammation by activation of macrophages and enhancement of OX40/OX40L interaction in ankylosing spondylitis. Front Cell Devel Biol. 9:737599.
- Tofigh R, Hosseinpourfeizi M, Baradaran B, Teimourian S, Safaralizadeh R. 2023. Rheumatoid arthritis and non-coding RNAs: How to trigger inflammation. Life Sci. 315:121367.
- Wan L, Liu J, Huang C, Zhu Z, Li F, Sun G, Wang K, Li S, Ma XCX, Yuan W. 2022. Role of m6A modification and novel circ_0066715/miR-486-5p/ETS1 axis in rheumatoid arthritis macrophage polarization progression. Aging. 14:10009–10026.
- Wang J, Niu R, Jiang L, Wang Y, Shao X, Wu M, Ma Y. 2019. The diagnostic values of C-reactive protein and procalcitonin in identifying systemic lupus erythematosus infection and disease activity. Medicine. 98:e16798.
- Wang X, Zhang C, Wu Z, Chen Y, Shi W. 2018. CircIBTK inhibits DNA demethylation and activation of AKT signaling pathway via miR-29b in peripheral blood mononuclear cells in systemic lupus erythematosus. Arthritis Res Ther. 20:118.
- Wu J, Li J, Liu H, Yin J, Zhang M, Yu Z, Miao H. 2019. Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children. J Clin Lab Anal. 2019; 33:e22998.
- Ye Z, Liu X, Yang Y, Zhang X, Yu T, Li S, Feng Y, Luo G. 2018. The differential expression of novel circular RNAs in an acute lung injury rat model caused by smoke inhalation. J Physiol Biochem. 74:25–33.
- Yu J, Ding W, Wang M, Guo X, Xu J, Xu Q, Yang Y, Sun S, Liu J, Qin L, et al. 2020. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: A large-scale, multi-center study. Intl J Cancer. 146:1754–1763.
- Zhang C, Wang X, Chen Y, Wu Z, Zhang C, Shi W. 2018. Down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4+ T-cells of systemic lupus erythematous. Clin Sci. 132:2285–2298.
- Zhang M, Wang J, Zhu Z, Li LJ, Liu R, Yang X, Leng R, Li X, Pan H, Ye D. 2018. Differentially-expressed circular RNA in systemic lupus erythematosus and their clinical significance. Biomed Pharmacother. 107:1720–1727.
- Zhou R, Shi Z, Shan K, Zhang S, Zhang Y, Liang Y, Yan B, Zhao C. 2022. Comparative analysis of differentially-expressed circular RNAs in polarized macrophages. Front Genet. 13:823517.
- Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia H, Dai Z, Tang Z, Fan J, Zhou J. 2020. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. 41:296–302.
