Abstract
Background
With the development of liquid biopsy technology, the demand for noninvasive prenatal testing (NIPT) is increasing rapidly. The aim of the study is to evaluate the effects of different blood collection tubes on plasma cfDNA and NIPT quality control.
Methods
We investigated hemolysis, cfDNA concentration, and fragment distribution within blood samples stored in EDTA, ImproGene, and Streck tubes. The effects of ImproGene and Streck tubes on NIPT quality control were evaluated.
Results
The ImproGene tubes prevented the time-dependent increase of cfDNA concentration and preserved the cfDNA fragment size distribution. For NIPT quality control, there is no significant difference in cfDNA, library concentration, and fetal fraction between ImproGene and Streck tubes samples. GC content of the samples in ImproGene tubes was closer to the human genome.
Conclusion
The ImproGene cfDNA tube has excellent performance and is an effective choice for storing blood samples for NIPT testing or other cfDNA analysis.
Introduction
The majority of cell-free DNA (cfDNA) is derived from apoptotic or necrotic cells which release their DNA into the circulation, and has a size distribution which reveals an enrichment in short fragments of about 166 bp [Citation1,Citation2]. At present, studies have found that various biomarkers such as carcinoma antigen 125 can be detected in the body fluids of cancer patients, but such markers have a high false positive rate. Recently, with the rapid development of molecular technology, scientists have realized that cfDNA is a potential biomarker [Citation3–7]. Fetal cfDNA is a special kind of cfDNA released from the placenta into the maternal circulation. The genetic information of the fetus can be detected in fetal cfDNA, so it has been used by many laboratories for genetic diagnosis. cfDNA is generally extracted from maternal plasma through noninvasive prenatal testing, and fetal genetic disease status is obtained through next-generation sequencing (NGS) [Citation8]. Hui sequenced the haplotypes of the parental genome and specific genes in maternal plasma DNA, respectively, and relative haplotype dosage analysis was used to determine the fetal monogenic disease status [Citation9].
Compared with the amniocentesis test method, NIPT has no risk of miscarriage and infection, but it has many requirements for test samples, for example, maternal plasma samples with fetal DNA concentrations below 4% may give rise to false negative results [Citation9–15]. Therefore, it is important to estimate the fetal DNA fraction accurately to make sure that it has passed the quality control (QC) threshold needed to guarantee a correct interpretation of downstream sequencing results.
In order to ensure optimal cfDNA quality, after blood collection, the blood collection tubes need to be turned upside down to mix the blood and additives, then transported smoothly and stored at a temperature of 0–30 °C. To date, a number of specialized cell-stabilizing blood collection tubes have been developed and tested to measure their ability to optimally preserve cfDNA [Citation16–20]. All ethylenediaminetetraacetic acid (EDTA) tubes demonstrated a time-dependent increase of cfDNA concentrations, while cfDNA concentrations remained stable in specialized blood collection tubes such as Streck’s Cell-Free DNA BCT. Our testing also showed the same result (). There is a lack of publicly available data on the use of ImproGene tubes. The purpose of this study is to investigate the effect on the quality of cfDNA in general and of fetal cfDNA in pregnant women under the following pre-analytical conditions: different time intervals before plasma isolation, and the use of different types of preservative in blood collection tubes (EDTA tubes, Streck tubes and ImproGene tubes). For each variable, the amount of cfDNA isolated from plasma was quantitated and the size and fetal fraction were determined. It is theorized that the quality of DNA is improved when blood samples are collected in tubes containing preservatives that will stabilize white blood cells (WBCs), including the ImproGene tubes.
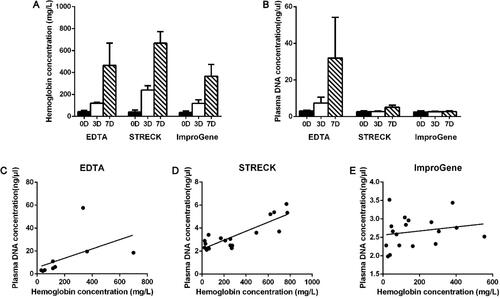
Figure 1. Hemoglobin and plasma DNA concentration of samples at different pre-analytical conditions. Hemoglobin concentration (A) and plasma DNA concentration (B) of samples kept in EDTA, Streck and ImproGene tubes for 0, 3, and 7 days were quantified respectively. The correlation of hemoglobin concentration and plasma DNA concentration of samples in EDTA (C), Streck (D), and ImproGene (E) tubes was analyzed.
Materials and methods
Patient recruitment
Six non-pregnant donors and 38 pregnant donors were included in this study; all patients signed informed consent prior to phlebotomy. Patient ages ranged from 25 to 40 years old; the gestational periods ranged from 12 to 24 weeks. None had fever, blood disease, cardiovascular disease or infectious diseases such as hepatitis B.
Blood collection
10 mL venous blood from each pregnant donor was drawn into Streck Cell-Free DNA BCT tubes (Streck, La Visa, NE; referred to as Streck tubes) and ImproGene Cell-Free DNA tubes (Improve Medical, GUANGZHOU, CHINA; referred to as ImproGene tubes), respectively (20 mL volume total from each patient). 10 mL venous blood from each non-pregnant donor were also in turn drawn into three kinds of tubes (Streck, ImproGene, and EDTA, 30 mL volume total from each patient). All blood samples were collected within one month and tested within 3 and 7 days respectively from the time of blood draw.
Plasma preparation
After the indicated storage time point, the blood in the various collection tubes was centrifuged at 1,600 × g for 10 minutes at 4 °C to prevent the buffy coat from being destroyed. The plasma was transferred to a new 2 mL tube by using a 3 mL pap straw, leaving approximately 3 mm of plasma in the blood collection tube to avoid the transfer of cellular components. The supernatant was centrifuged at 16,000×g for 10minutes at 4 °C to remove residual blood cells. The supernatant was transferred to a new tube and store at −80 °C [Citation17].
Hemolysis
Hemolysis in plasma samples was determined by the absorbance of hemoglobin at 414 nm using a spectrophotometer UV-2700 (SHIMADZU, Kyoto, Japan), averaging three technical replicates per sample.
Extraction of cfDNA
QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) was used to extract cfDNA from plasma. Samples were processed according to the manufacturer’s protocol, with a starting volume of 1 mL of plasma. Purified cfDNA was eluted in 40 µL of AVE buffer and stored at −80 °C.
Analysis of cfDNA concentrations and fragments
cfDNA concentrations were quantified using the Quant-iTdsDNA high sensitivity assay (Invitrogen, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions, using the Qubit fluorometer (Invitrogen) as readout. The High Sensitivity DNA Assay kit on the Agilent 2100 Bioanalyzer was used to analyze cfDNA fragments between 50 and 7,000 bp, according to the manufacturer’s protocol.
Next-generation sequencing and analysis
The sequencing and analysis were performed by Guangzhou Darui Biotechnology Co., Ltd. The cell-free DNA extraction, library construction, sequencing, and bioinformatics analysis were performed according to standard assay protocols. To assess the fetal risk of trisomyies 21, 18, and 13, samples with a T-score ≥3.0 for these chromosomes were classified as positive, whereas samples with a T-score <3.0 were classified as negative for the indicated trisomy.
Statistical analysis
Statistical analysis by using student t-test was performed in GraphPad Prism. p < 0.05 was deemed significant.
Results
Improgene tubes control hemolysis in blood samples for 7 days
It is well known that WBCs start to lyse over time in the commonly used EDTA tubes, which leads to an elevated DNA background [Citation16,Citation20]. The color of the plasma may be used as a rough standard in some cfDNA testing laboratories to decide whether the sample should be further tested [Citation16,Citation21]. In our study, none of the samples from EDTA tubes, Streck tubes, and ImproGene tubes showed obvious plasma turbidity or darkening on days 0, 3, or 7. The hemoglobin concentration of these samples was also quantitatively measured by UV spectrophotometry. shows that among the three kinds of tubes, the hemoglobin concentration using ImproGene tubes had the least amount of change within the 7-day timeframe.
Genomic DNA release is significantly reduced in ImproGene tubes
We measured cfDNA concentrations of plasma collected in EDTA tubes, Streck tubes and ImproGene tubes respectively at different intervals after blood withdrawal. cfDNA concentrations remained constant in the ImproGene tubes at three different time points, but cfDNA concentrations increased in both the EDTA and Streck tubes in a time-dependent manner as shown in .
We compared the correlation between the concentrations of hemoglobin and cfDNA. The results show that the concentration of cfDNA had almost no linear correlation with hemoglobin in the ImproGene tubes () and a slightly linear increasing relationship in the Streck tubes (), but the cfDNA concentration in the EDTA tubes increased significantly with the FHb (free hemoglobin) concentration ().
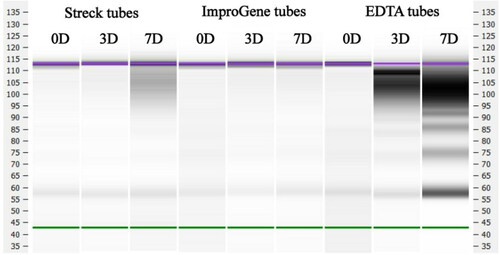
No differences in size distribution of the DNA fragment in the ImproGene tubes were observed as shown in , but degradation of DNA occurred in the Streck tubes within 7 days, and the size distribution of the DNA fragments changed drastically in the EDTA tubes.
Figure 2. cfDNA fragment distribution of samples at different pre-analytical conditions. cfDNA fragment distribution of samples kept in EDTA, Streck, and ImproGene tubes for 0, 3, and 7 days were analyzed respectively. The High Sensitivity DNA Assay kit on the Agilent 2100 Bioanalyzer was used to analyze cfDNA fragments between 50 and 7,000 bp, according to the manufacturer’s protocol.
Performance of ImproGene tubes in NIPT quality control steps
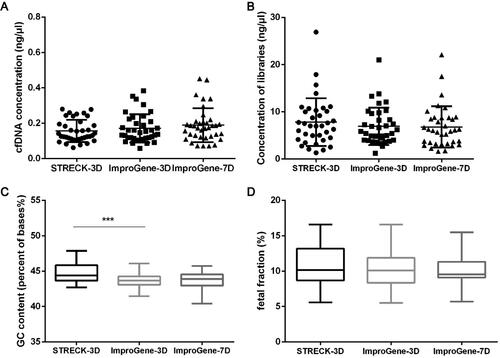
As shown in , blood samples from 36 pregnant donors were collected in Streck tubes and ImproGene tubes respectively and processed for QC testing for Next-generation sequencing in NIPT. We compared the performances of ImproGene and Streck tubes in QC steps as shown in . There were no significant differences in the cfDNA and library concentrations between the samples in the Streck tubes and ImproGene tubes (, b)). As shown in , the GC content of the samples in the ImproGene tubes was 43.8% (average value; 3 days), while the GC content in the Streck tubes was 44.8% (average value; 3 days). The fetal fraction of all samples in the ImproGene and Streck tubes were more than 4%, and there was no significant difference between them. The results also showed that no significant differences were found in the test results of 3-day and 7-day samples in the ImproGene tubes.
Figure 3. Experimental scheme. Whole blood from 38 pregnant donors was collected into Streck tubes and ImproGene tubes, respectively, 10 mL for each tube, and transported to Guangzhou Darui Biotechnology Co., Ltd by S.F. Express Company. The blood in Streck tubes was tested at day 3 after blood draw, while the blood in ImproGene tubes was processed at day 3 and day 7 after blood draw. All of the tubes were stored at room temperature with 10 times inversion, twice a day.
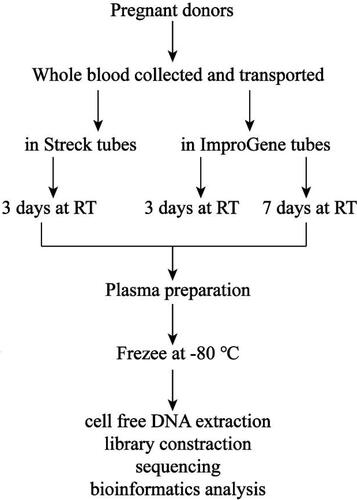
Figure 4. Quality control steps of Next-generation sequencing in NIPT. Concentrations of cfDNA (A) and libraries (B), GC content (C), and fetal fraction (D) were quantified for different pre-analytical conditions. Paired student’s t-test was used to compare the differences between matched Streck-3D (3 day) and ImproGene-3D samples, ImproGene-3D and ImproGene-7D (7 day) samples. ***p < 0.05.
Discussion
cfDNA is widely used in the noninvasive prenatal diagnosis of trisomy [Citation8,Citation9,Citation12,Citation22]. The newest findings also show that cfDNA derived from tumors can be used as biomarkers for cancer diagnosis [Citation1,Citation6]. One of the major technical challenges to use cfDNA as biomarkers is the ability to quantify targets at low concentrations. Any additional DNA released from nucleated cells before the separation of plasma decreases the proportion of fetal or tumor derived cfDNA in the blood [Citation21]. Previous studies have shown that increasing the time interval between the blood draw and the separation of plasma leads to an increase in the total DNA concentration in EDTA tubes [Citation16,Citation18,Citation20]. In order to maintain the proportion of the target cfDNA in blood samples, and keep background “noise” to a minimum, it is generally recommended that blood samples in EDTA tubes be centrifuged to separate plasma immediately after the blood draw, or at most, within a few hours [Citation16,Citation18]. It is also necessary to minimize WBC disruption, so specialized cell-stabilizing blood collection tubes have been developed.
FHb, the content of hemoglobin in plasma, is a regular indicator of blood preservation quality. In the process of blood collection, transportation, and storage, FHb will noticeably increase when improper operation occurs and/or there is a longer preservation time. Some researchers have shown that the increased FHb concentration can reflect the destruction of red blood cells, which is not synchronized with the release of DNA from white blood cell lysis [Citation23,Citation24]. In our study, samples collected in ImproGene tubes showed the results consistent with the conclusion mentioned above while Streck tubes had only a weak correlation; the cfDNA concentration in EDTA tubes markedly increased with FHb concentration (). This suggests that fewer nucleated cells were destroyed in samples collected in the ImproGene tubes. The preservatives in the ImproGene tubes prevented the time-dependent increase in cfDNA concentration observed in the EDTA tubes, and even in the Streck tubes (), as well as maintained the size distribution of the original cfDNA fragments (). These results also indicate that using plasma color to determine whether a sample is qualified for clinical use may not be accurate, because hemolysis or other interfering substances may not be grossly recognized. Storage time is not unlimited, even for specialized cell-stabilizing blood collection tubes, indicating that an optimized and standardized pre-analytical protocol is needed for various clinical applications.cfDNA is the target of NIPT, so its quality is an important factor in determining the accuracy of detection. In recent years Streck tubes have become commonly used blood collection tubes for NIPT. The results of our research indicated that there were no significant differences in the cfDNA and library concentrations between ImproGene tubes and Streck tubes (, b)). The GC content of all samples was greater than 40%, and the testing company considered that they all met the QC requirements. The results also showed that the GC content of the samples in ImproGene tubes was close to the average 40% content of the human genome [Citation25] (). The test result of the fetal fraction showed that both the Streck tubes and the ImproGene tubes resulted in the fetal fraction being above 4%, in addition, there were no significant differences in the fetal fraction between the samples of 3 days and 7 days in the ImproGene tubes, indicating that the ImproGene tubes adequately preserves the samples well, reducing a potential source of preanalytic error.
Conclusion
In the early years, amniocentesis was widely used in the field of prenatal diagnosis, which was a typical liquid biopsy. With the continuous development of liquid biopsy, the technology behind NIPT has gradually improved, and it has become the most advanced prenatal testing technology. Compared with amniocentesis, the noninvasive nature of NIPT detection technology avoids the risk of infection and miscarriage secondary to invasive diagnostic procedures, especially for women at advanced maternal age [Citation26]. NIPT is affected by various factors, and includes differing preservation of cfDNA in the different types of blood collecting tubes.
We evaluated the effects of plasma separation time intervals and different blood collection tubes on plasma cfDNA and NIPT quality control, and confirmed that ImproGene cfDNA tubes prevent the release of gDNA and maintain the original cfDNA proportions. This research provides the foundation of using ImproGene cfDNA tubes to preserve blood samples for NIPT detection or other cfDNA analysis.
Acknowledgments
The authors would like to thank Professor Jiancheng Tu (Department of Clinical Laboratory Medicine, Zhongnan Hospital of Wuhan University) for helping improve the article.
Disclosure statement
The authors declared that they have no conflicts of interest to this work.
Funding
The author(s) reported there is no funding associated with the work featured in this article.
References
- Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–65.
- Lo YMD, Chan KCA, Sun H, Chen EZ, Jiang P, Lun FMF, Zheng YW, Leung TY, Lau TK, Cantor CR, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2(61):61ra91. doi:10.1126/scitranslmed.3001720.
- Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46(5):318–22. doi:10.1159/000226740.
- Lo YMD, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. The Lancet. 1997;350(9076):485–7. doi:10.1016/S0140-6736(97)02174-0.
- Duvvuri B, Lood C. Cell-free DNA as a biomarker in autoimmune rheumatic diseases. Front Immunol. 2019;10:502. doi:10.3389/fimmu.2019.00502.
- Salvi S, Gurioli G, De Giorgi U, Conteduca V, Tedaldi G, Calistri D, Casadio V. Cell-free DNA as a diagnostic marker for cancer: current insights. Onco Targets Ther. 2016;9:6549–59. doi:10.2147/OTT.S100901.
- Hampson P, Dinsdale RJ, Wearn CM, Bamford AL, Bishop JRB, Hazeldine J, Moiemen NS, Harrison P, Lord JM. Neutrophil dysfunction, immature granulocytes, and cell-free DNA are early biomarkers of sepsis in burn-injured patients: a prospective observational cohort study. Ann Surg. 2017;265(6):1241–9. doi:10.1097/SLA.0000000000001807.
- Zhang J, Li J, Saucier JB, Feng Y, Jiang Y, Sinson J, McCombs AK, Schmitt ES, Peacock S, Chen S, et al. Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA. Nat Med. 2019;25(3):439–47. doi:10.1038/s41591-018-0334-x.
- Peng XL, Jiang P. Bioinformatics approaches for fetal DNA fraction estimation in noninvasive prenatal testing. Int J Mol Sci. 2017;18(2):453.
- Luo Y, Jia B, Yan K, Liu S, Song X, Chen M, Jin F, Du Y, Wang J, Hong Y, et al. Pilot study of a novel multi-functional noninvasive prenatal test on fetus aneuploidy, copy number variation, and single-gene disorder screening. Mol Genet Genomic Med. 2019;7(4):e00597. doi:10.1002/mgg3.597.
- Song Y, Zhou X, Huang S, Li X, Qi Q, Jiang Y, Liu Y, Ma C, Li Z, Xu M, et al. Quantitation of fetal DNA fraction in maternal plasma using circulating single molecule amplification and re-sequencing technology (cSMART). Clin Chim Acta. 2016;456:151–6. doi:10.1016/j.cca.2016.03.005.
- Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2017;50(3):302–14. doi:10.1002/uog.17484.
- Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, Klugman S, Watson MS. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2016;18(10):1056–65. doi:10.1038/gim.2016.97.
- Dahl F, Ericsson O, Karlberg O, Karlsson F, Howell M, Persson F, Roos F, Stenberg J, Ahola T, Alftrén I, et al. Imaging single DNA molecules for high precision NIPT. Sci Rep. 2018;8(1):4549. doi:10.1038/s41598-018-22606-0.
- Hu P, Liang D, Chen Y, Lin Y, Qiao F, Li H, Wang T, Peng C, Luo D, Liu H, et al. An enrichment method to increase cell-free fetal DNA fraction and significantly reduce false negatives and test failures for non-invasive prenatal screening: a feasibility study. J Transl Med. 2019;17(1):124. doi:10.1186/s12967-019-1871-x.
- Sorber L, Zwaenepoel K, Jacobs J, De Winne K, Van Casteren K, Augustus E, Lardon F, Prenen H, Peeters M, Van Meerbeeck J, et al. Specialized blood collection tubes for liquid biopsy: improving the pre-analytical conditions. Mol Diagn Ther. 2020;24(1):113–24. doi:10.1007/s40291-019-00442-w.
- Medina Diaz I, Nocon A, Mehnert DH, Fredebohm J, Diehl F, Holtrup F. Performance of streck cfDNA blood collection tubes for liquid biopsy testing. PLoS One. 2016;11(11):e0166354. doi:10.1371/journal.pone.0166354.
- Toro PV, Erlanger B, Beaver JA, Cochran RL, VanDenBerg DA, Yakim E, Cravero K, Chu D, Zabransky DJ, Wong HY, et al. Comparison of cell stabilizing blood collection tubes for circulating plasma tumor DNA. Clin Biochem. 2015;48(15):993–8. doi:10.1016/j.clinbiochem.2015.07.097.
- Enko D, Halwachs-Baumann G, Kriegshäuser G. Plasma free DNA: evaluation of temperature-associated storage effects observed for Roche Cell-Free DNA collection tubes. Biochem Med (Zagreb). 2019;29(1):010904. doi:10.11613/BM.2019.010904.
- Norton SE, Lechner JM, Williams T, Fernando MR. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem. 2013;46(15):1561–5. doi:10.1016/j.clinbiochem.2013.06.002.
- Nishimura F, Uno N, Chiang P-C, Kaku N, Morinaga Y, Hasegawa H, Yanagihara K. The effect of in vitro hemolysis on measurement of cell-free DNA. J Appl Lab Med. 2019;4(2):235–40. doi:10.1373/jalm.2018.027953.
- Langlois S, Brock J-A, Douglas Wilson R, GENETICS COMMITTEE. Current status in non-invasive prenatal detection of down syndrome, trisomy 18, and trisomy 13 using cell-free DNA in maternal plasma. J Obstet Gynaecol Can. 2013;35(2):177–81. doi:10.1016/S1701-2163(15)31025-2.
- Stokowski R, White K, Hacker C, Doshi J, Schmid M. Hemolysis and fetal fraction in cell-free DNA blood collection tubes for noninvasive prenatal testing. Mol Diagn Ther. 2020;24(2):185–90. doi:10.1007/s40291-020-00446-x.
- Zhao Y, Li Y, Chen P, Li S, Luo J, Xia H. Performance comparison of blood collection tubes as liquid biopsy storage system for minimizing cfDNA contamination from genomic DNA. J Clin Lab Anal. 2019;33(2):e22670. doi:10.1002/jcla.22670.
- Li W. On parameters of the human genome. J Theor Biol. 2011;288:92–104. doi:10.1016/j.jtbi.2011.07.021.
- Chen Y-P, He Z-Q, Shi Y, Zhou Q, Cai Z-M, Yu B, Wang T. Not all chromosome aberrations can be detected by NIPT in women at advanced maternal age: a multicenter retrospective study. Clin Chim Acta. 2018;486:232–6. doi:10.1016/j.cca.2018.08.018.
