Abstract
The common sea buckthorn or seaberry (Hippophae rhamnoides) is important environmentally, commercially, and as a new berry crop. Some morphological differences among mother plants and lines obtained by cuttings often lead a purchaser to question if the propagated plantlets are true-to-name cultivars. Intersimple sequence repeat (ISSR) markers were employed to assess the genetic stability of lines obtained by cuttings of four H. rhamnoides cultivars and the genetic relationships among them. Fifteen ISSR primers generated 174 bands among the 35 samples of four cultivars, 158 of which (90.8%) were polymorphic. Percentages of polymorphic and unique bands between mother plant and propagated plantlets ranged from 53.21% to 77.17% and from 7.09% to 13.76%, respectively. Cluster analysis based on ISSR data indicated that at a Jaccard coefficient of 0.78, mother plant and lines obtained by cuttings for each cultivar were grouped into different subclusters, respectively, which could be further clustered into different sub-subclusters. Our data first indicated that the genotype was affected by the cutting process to a certain degree, because there were some morphological and low molecular variations between the mother plant and lines obtained by cuttings. Information generated from this study can be used to select parents for hybrid development to maximize desirable agronomic traits in a breeding program aimed at developing segregating populations to map genes controlling special traits in H. rhamnoides.
INTRODUCTION
The genus Hippophae L. is a deciduous, perennial shrub or tree of the Elaeagnaceae. It has six species and 12 subspecies worldwide (CitationBartish et al., 2002; CitationSun et al., 2002; CitationSwenson and Bartish, 2002) that are restricted to the Qinghai-Xizang Plateau and adjacent areas except for Hippophae rhamnoides L., which is naturally distributed from Asia to Europe and has been introduced into Africa, South America, and North America. H. rhamnoides adapts well to extreme conditions, including drought, salinity, and alkalinity. Commercially, it is a multipurpose plant with orange, red, or yellow berries and has substantial agricultural, ecological, nutritional, medical, and ornamental values (CitationCheng et al., 2003; CitationZadernowski et al., 2002).
Hippophae rhamnoides is being planted on a large scale in the world for commercial aims and environmental improvements. This requires a great number of seedlings of specific cultivars. In its natural condition, reproduction of H. rhamnoides occurs through seeds with a low germination rate as well as root suckers. Some previously published results indicated that the application of micropropagation is a valid tool for propagation of this species (CitationLiu et al., 2007; CitationMontpetit and Lalonde, 1988; CitationMou, 1995), but there are low regeneration rates and survival rates after transplantation (CitationRuan et al., 2007). In contrast, multiplication by cuttings that have over an 80% survival rate is still the main protocol used for propagation of H. rhamnoides cultivars (CitationRuan et al., 2007).
Micropropagation protocol is severely hindered due to incidences of somaclonal variations (CitationJoshi and Dhawan, 2007), as in case of oil palm, where aberrant flowering patterns were observed among the regenerated plants (CitationMatthes et al., 2001). Somaclonal variation mostly occurs as a response to the stress imposed on the plant in culture conditions and is manifested in the form of DNA methylations, chromosome rearrangements, and point mutations (CitationKaeppler and Phillips, 1993; CitationPeschke et al., 1987; CitationPhillips et al., 1994; CitationXu et al., 2004). There are many woody species commercially produced by micropropagation (CitationMatthes et al., 2001), and variability induced by cutting multiplication has also been reported in Sansevieria trifasciata (http://www.yhagri.gov.cn/documents/docdetail.asp? documenti =19397&sub_menuid=76) and Vigna radiata (CitationZhang et al., 2002). Hence a stringent quality check in terms of genetic similarity of the lines obtained by cuttings becomes important not only for seedling production but also for lawsuits requiring careful technical investigation (unauthorized commercialization of patented cultivars).
Success of any crop-breeding program is based on the knowledge of and availability of genetic variability for efficient selection (CitationAli et al., 2008). Genetic similarity estimates among genotypes are helpful in selecting parental combinations for creating segregating populations so as to maintain genetic diversity in a breeding program (CitationBecelaere et al., 2005) and the classification of germplasm into heterotic groups for hybrid-crop breeding (CitationMenz et al., 2004). The search for and establishment of heterotic groups can be based on geographical origin, agronomical traits, pedigree data, or molecular marker data (CitationMelchinger, 1999). Neither pedigree- nor morphologically based estimates may reflect the actual genetic difference of the studied populations (CitationAlmanza-Pinzon et al., 2003; CitationFufa et al., 2005; Citationvan Beuningen and Busch, 1997). On the other hand, molecular markers are not influenced by the environment and reflect genetic similarity (and differences) and do not require previous pedigree information (CitationBohn et al., 1999), which is valuable for plants for which pedigree information is lacking.
Various types of molecular markers are available for genome analysis in trees and shrubs (CitationTeixeira da Silva et al., 2005) and have been used to assess the genetic uniformity of the micropropagated plantlets (CitationMartins et al., 2004). This is apparent in studies conducted to screen somaclonal variations produced in tissue-cultured plants such as in neem (CitationSingh et al., 2002), tea (CitationSingh et al., 2004), and soybean (CitationHofmann et al., 2004). Intersimple sequence repeat (ISSR) markers in particular have been reported to be very useful to unravel genetic diversity among closely related genotypes, as there are high polymorphic bands (CitationAgostini et al., 2008; CitationBornet et al., 2002; CitationPrevost and Wilkinson, 1999; CitationSreedhar et al., 2007; CitationVijayan et al., 2006) and to detect variations among tissue culture-produced plants (CitationChowdari et al., 1998; CitationLeroy et al., 2000; CitationLeroy et al., 2001; CitationRahman and Rajora, 2001). It has been extensively used in genetic-diversity studies in many plants, including Hagenia abyssinica (CitationFeyissa et al., 2007), fig (CitationIkegami et al., 2009), mulberry (CitationKar et al., 2008) and papayas (CitationCarrasco et al., 2009). CitationJoshi and Dhawan's (2007) results, based on ISSR markers, confirmed the clonal fidelity of the tissue culture-raised Swertia chirayita plantlets and corroborated the fact that axillary multiplication is the safest mode for multiplication of true-to-type plants.
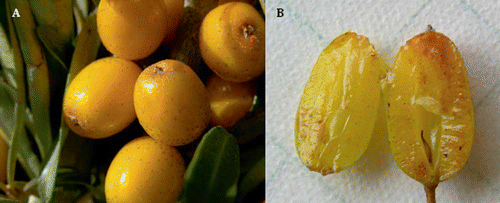
Genetic diversities, variations, originations, and phylogenetic relationships of H. rhamnoides have been studied at a molecular level (isozyme, random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), ISSR, chloroplast DNA (cpDNA) and internal transcribed spacer (ITS); CitationBartish et al., 1999; CitationChen et al., 2008; CitationRuan et al., 2007; CitationTian et al., 2004). Among the six subspecies of H. rhamnoides, ssp. sinensis and ssp. mongolica are mainly and widely distributed in Asia and Europe. Selection and breeding programs for these two subspecies have been running since the 1960s to improve their adaptability, yield, and quality. Some specific cultivars have been successfully selected and bred in many countries (e.g., China, Russia, and Germany) and distributed using plants from multiplication by cuttings. However, some morphological differences among mother plants and lines obtained by cuttings () not only make a purchaser question if the plantlets obtained by cuttings are true-to-name cultivars but has also lead to commercial disputes (unauthorized commercialization of patented cultivars).
TABLE 1 Plant Cultivars Used in this Study that are Planted Widely in China and Other Countries and Potential Materials for Breeding Fine and Resistant Lines
FIGURE 1 Fruits of ‘Yalishandashierhao’ (A) and profile view of ‘Wuheshaji’ fruit without stone (B).
In this study, we first report the use of ISSR markers to assess genetic stability of propagated plantlets of four H. rhamnoides cultivars; then, we analyze genetic relationships among the four cultivars with different characteristics. These data provide a scientific basis for the identification of genetic stability of propagated Hippophae plantlets and the development of crossing strategies in a breeding program.
MATERIALS AND METHODS
Plant Materials and DNA Extraction
Four H. rhamnoides cultivars with different characteristics (), which are being widely planted on a large scale in China and other countries, were used in this study, including ‘Yalishandashierhao’ (‘YLSD’; ssp. mongolica), ‘Hongmaxiongzhu’ (‘HMXZ’; ssp. mongolica), ‘Zhongguowucixiongzhu’ (‘ZGWCXZ’; ssp. sinensis), and ‘Wuheshaji’ (‘WHSJ’; ssp. mongolica; ). In our cultivar-cutting orchard there are just four original genotypes (only one original genotype within a cultivar and the rest of the plants should be clones of these). There are some morphological differences between mother plants and lines obtained by cuttings (). The mother plant and lines obtained by cuttings for each cultivar were used as donor materials for multiplication by cuttings and have some differences in morphological characteristics. They were selected as the samples in this study () to investigate ISSR analyses for assessing the genetic stability of lines obtained by cuttings. In addition, the four cultivars used in this study are potential materials for breeding specific lines with different characters. Genetic relationship estimates among genotypes based on molecular markers is helpful in selecting parental combinations for creating segregating populations so as to maintain genetic diversity in a breeding program (CitationBecelaere et al., 2005) and clone the desired gene. To develop optimal strategies for breeding, we estimated the genetic relationships among the four cultivars using ISSR markers.
Young leaves were collected from each sample of each cultivar in 2007. Total genomic DNA was extracted from leaf tissue using the protocol of CitationRuan et al. (2004).
ISSR Analysis
Amplification reactions of ISSR analyses were carried out in a total volume of 20 µL in a 0.2 mL microfuge tube: 2µl of the 10X reaction buffer, 6 mM MgCl2, 600 µM dNTPs, 0.5 µM primer, 4% formamide, 20 ng DNA template, and 2U Taq DNA polymerase (TakaRa, China). The thermocycler program for PCR was set at 1.5 minutes at 94°C; followed by 45 cycles of 45 seconds at 94°C, 45 seconds annealing at 53°C, 1.5 minutes extension at 72°C; and a single 7 minutes at 72°C extension cycle. The samples were then placed at 4°C until gel analysis was performed. Primer selection was performed on the samples of four cultivars using 40 primers purchased from the University of British Columbia. Based on the number and quality of polymorphic fragments at 52°C, 52.5°C, 53°C, 53.5°C, and 54°C, fifteen ISSR primers (see ) were selected and amplification was carried out on all samples in a GeneAmp PCR system 9700 (Applied Biosystems). Amplification products were separated on 1.5% agarose gels, and visualized under AutoChemi System TM UVP Bioimaging Systems after ethidium bromide staining.
TABLE 2 Assessment of Genetic Stability of Propagated Plantlets Based on ISSR Markers
Data Analysis
The amplified fragments of each ISSR marker were scored as “1” and “0”, where “1” indicated the present of a band and “0” indicated its absence. The CitationJaccard (1908) coefficient was calculated by using NTSYS2.02 (CitationRohlf, 2000). A similarity tree was produced based on the Jaccard coefficient with the unweighted-combination group method using arithmetic averages (UPGMA) and the sequential agglomerative hierarical nested (SAHN)-clustering program. The goodness of fit of the clustering was tested using the matrix comparison (MXCOMP) program, which directly compares the original similarity matrix and the cophenetic value matrix. Cluster analysis and MXCOMP were conducted by using NTSYS2.02 (CitationRohlf, 2000).
RESULTS
ISSR Analysis
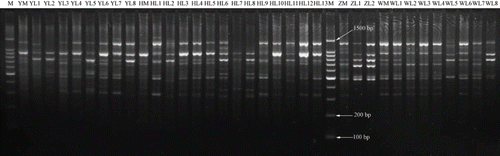
Fifteen ISSR primers generated 174 ISSR bands for all 35 samples, 158 of which (90.8%) were polymorphic. The number of bands over all 35 samples detected by individual primers ranged from six for the UBC primer 827 to 19 for the UBC primer 840, with a mean of 11.6 bands per primer. Among the four cultivars, the number of unique bands detected by an individual primer ranged from one for the UBC primers 809, 810, 825, and 827 to six for the UBC primer 840, with a mean of 3.7. Fifteen primers generated 38 unique bands among four cultivars, two of which, generated from the UBC primer 811, appeared in all lines of ‘HMXZ’ obtained by cuttings, and one generated from the UBC primer 816 appeared in all lines of ‘WHSJ’ obtained by cuttings. In our study all the primers amplified bands between 100 bp to 1,500 bp molecular size range. Amplifications produced by the UBC primer 881 were shown in .
FIGURE 2 Amplifications produced by the UBC primer 881. Lane M represents the 100 bp DNA Ladder Marker and other lanes represent the mother plants or lines obtained by cuttings of four Hippophae cultivars, means of code in .
shows total bands, polymorphic bands, and unique bands generated from each cultivar using 15 ISSR primers. Among mother plants and lines obtained by cuttings, the percentages of polymorphic and unique bands ranged from 53.21% to 77.17% and ranged from 7.09% to 13.76%, respectively. Additionally, 17.1% polymorphic bands for ‘YLSD’ were unique bands, as well as 9.2% for ‘HMXZ’, 25.86% for ‘ZGWCXZ’, and 19.3% for ‘WHSJ’.
Cluster Analysis
Based on the Jaccard coefficient, a UPGMA dendrogram was constructed (). The measure of goodness of fit of the cluster analysis was given by the “cophenetic correlation” (r) calculated with the MXCOMP program. The dendrogram had a matrix correlation r = 0.893, which was interpreted as a good fit.
FIGURE 3 Dendrogram of 35 samples of four Hippophae cultivars by UPGMA cluster analysis based Jaccard coefficient, using 158 ISSR polymorphic bands obtained by 15 primers, means of code in .
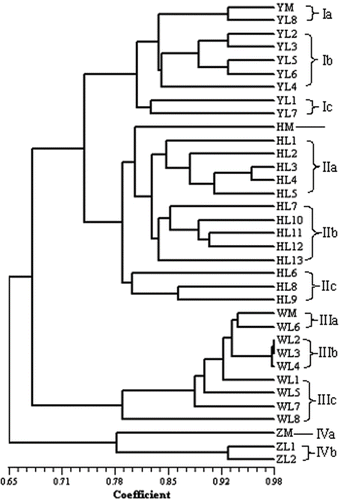
At a Jaccard coefficient of 0.67, the following clusters were formed: cluster I (group IV) included one ssp. sinensis cultivar (‘ZGWCXZ’) from China; cluster II consisted of three ssp. mongolica cultivars from Russia, including ‘YLSD’ (group I), ‘HMXZ’ (group II), and ‘WHSJ’ (group III). At a level of 0.78, mother plant and all lines obtained by cuttings for each cultivar were grouped into different single subcluster (), respectively. Group I could be further clustered three sub-subclusters. Group Ia included YM and YL8, five plantlets (YL2–YL6) were clustered in group Ib, and group Ic consisted of YL1 and YL7. In group II, 13 plantlets of ‘HMXZ’ were clustered in three subgroups, group IIa included five plantlets (HL1–HL5), five plantlets (HL7, HL10–HL13) were clustered in group IIb, and the remaining three (HL6, HL8, and HL9) were consisted of group IIc. For ‘WHSJ’, group IIIa included WM and WL6, three plantlets (WL2–WL4) in group IIIb were not discriminated clearly by the dendrogram, and the remaining four plantlets (WL1, WL5, WL7, and WL8) in group IIIc were separate each other. For ‘ZGWCXZ’, two plantlets in group IVb were discriminated from ZM at a Jaccard coefficient of 0.79.
DISCUSSION
Our results of ISSR bands and cluster analysis indicate that multiplication by cuttings affect the genotype of Hippophae to a certain degree, because there were some morphological and low molecular variations among mother plants and lines obtained by cuttings.
ISSR markers detected the highest levels of polymorphism (CitationJoshi and Dhawan, 2007; CitationSreedhar et al., 2007), that is, they provide the highest level of discrimination between any pair of genotypes (CitationPowell et al., 1996). CitationDevarumath et al. (2002) found that ISSR fingerprinting detected more polymorphic loci (12.8%) than RAPD fingerprinting (4.28%) in micropropagated tea clones. The number of marker bands amplified per cultivar (ranged from 102 to 127) and percentages of polymorphic bands (ranged from 53.21% to 77.17%; ) in our study is sufficient to reveal variations. This is evidenced by the comparable number of bands scored in various plant taxa by employing ISSR-based marker assays. CitationKumar et al. (2009) used 64 bands with 98.14% polymorphism to reveal genetic diversity among 12 Jatropha species, and CitationCarrasco et al. (2009) used 114 bands with 55.3% polymorphism to reveal genetic diversity among 333 samples of Vasconcellea pubescens.
Between mother plant and lines obtained by cuttings of the four Hippophae cultivars, low molecular variations were also detected by ISSR analysis (, ) and there are some differences in morphological characteristics (). These indicate that multiplication by cuttings of H. rhamnoides affect genotypes to some degree. Cluster analysis () provides some evidence for cutting induced variation. In fact when we look at the genetic distance between individuals within a cultivar we see that it is high relative to the total genetic variation (up to half the total Jaccard coefficient value). However, variation induced by cuttings occurs within cultivars. This is because all mean genetic similarities (Jaccard coefficient) among mother plant and lines obtained by cuttings for each cultivar in this study were more than 0.78, which is the upper limit of genetic similarities (ranged from 0.29 to 0.78) among 15 different Hippophae cultivars. At a Jaccard coefficient of 0.78, fifteen Hippophae cultivars were independent of each other and not clustered into any group (CitationRuan and Li, 2005), and in this study, mother plant and lines obtained by cuttings for ‘YLSD’ were group into one cluster, as well as ‘WHSJ’, ‘HMXZ’, and ‘ZGWCXZ’.
It is generally recognized that somaclonal variation mainly occurs in tissue culture (CitationHuang and Liu, 2006; CitationMatthes et al., 2001; CitationPalombi and Damiano, 2002) and that there are typically no problems concerning genetic stability when multiplying cultivars by cuttings. Many woody species are commercially vegetatively reproduced, but some variation is occasionally found in lines obtained by cuttings, such as plantlets of Sansevieria trifasciata.
Transposition in response to environmental stress is a common phenomenon by which plants acquire genetic diversity (CitationFlavell et al., 1994; CitationMcclintock, 1984). Genetic variation in the scion plants of Vigna radiata seems not to be caused simply by DNA transformation, but most likely the nutrient and hormone stress induced mutation (CitationZhang et al., 2002). Some morphological and molecular variations between the mother plant and lines obtained by cuttings for Hippophae used in this study may be related to the cutting process, because the bases of cuttings were often dipped for 1 hour in a mixed 5,000 ppm solution containing indole butyric acid, indoleacetic acid, naphthylacetic acid, and rooting reagent before planting, which may be induce some variation (CitationPhillips et al., 1994).
Information generated from this study can be used to select parents for hybrid development to maximize desirable agronomic traits in a breeding program aimed at developing segregating populations to map genes controlling special traits in sea buckthorn. The use of hybrids between some species in exotic conditions frequently results in significant genetic gains possibly due to either true heterotic effects and/or a combination of traits. The resulting information on genetic relationships can be used as a guideline for the development of crossing strategies in a breeding program (Le Thierry d' Ennequin et al., 2000). Genotype ‘ZGWCXZ’ is adaptable to harsh environments and is fast growing, and ‘YLSD’ has promising characteristics (e.g., large fruits, long fruit-stalks, and high vitamin-C contents in seeds and leaves). Crossing between them (Jaccard coefficient between ‘ZM’ and ‘YM’ = 0.44) may be useful in breeding specific lines with each parent's desirable traits. Similarly, crossing between ‘WHSJ’ and ‘ZGWCXZ’ (Jaccard coefficient between them = 0.50) may be useful in breeding of advanced lines with promising characteristics, such as fruit without stones, fast growth, and strong adaptability to harsh environments. Dried-shrink disease is the main problem restricting the sustainable development of Hippophae, and molecular biotechnology may be able to provide a solution to overcome this problem (CitationRuan et al., 2007). ‘YLSD’ and ‘HMXZ’ can be used in breeding programs aimed at developing cultivars with resistance to dried-shrink disease. This is because ‘YLSD’ is resistant to dried-shrink disease, but ‘HMXZ’ is susceptible to this disease.
ACKNOWLEDGEMENTS
This research was funded by the Keystone Project of the Agriculture Science and Technology Research Item of the Liaoning Scientific and Technological Committee (No. 2007207005) and the preparative fund of Dalian Nationalities University (No. 0212-112012).
LITERATURE CITED
- Agostini , G. , Echeverrigaray , S. and Souza-Chies , T.T. 2008 . Genetic relationships among South American species of Cunila D. Royen ex L. based on ISSR . Plant Syst. Evol. , 274 : 135 – 141 .
- Alamnza-Pinzon , M.I. , Khairullah , M. , Fox , P.N. and Warburton , M.L. 2003 . Comparison of molecular markers and coefficients of parentage for the analysis of genetic diversity among spring bread wheat accessions . Euphytica , 130 : 77 – 86 .
- Ali , M.L. , Rajewski , J.F. , Baenziger , P.S. , Gill , K.S. and Eskridge , K.M. 2008 . Assessment of genetic diversity and relationship among a collection of US sweet sorghum germplasm by SSR markers . Mol. Breeding , 21 : 497 – 509 .
- Bartish , I.V. , Jeppsson , N. and Nybom , H. 1999 . Population genetic structure in the dioecious pioneer plant species Hippophae rhamnoides investigated by random amplified polymorphic DNA (RAPD) markers . Mol. Ecol. , 8 : 791 – 802 .
- Bartish , I.V. , Jeppsson , N. , Swenson , U. and Nybom , H. 2002 . Phylogeny of Hippophae (Elaeagnaceae) inferred from parsimony analysis of chloroplast DNA and morphology . Syst. Bot. , 27 : 41 – 54 .
- Becelaere , G.V. , Edward , L.L. , Paterson , A.H. and Chee , P.W. 2005 . Pedigree-vs. DNA marker-based genetic similarity estimates in cotton . Crop Sci. , 45 : 2281 – 2287 .
- Bohn , M. , Utz , H.F. and Melchinger , A.E. 1999 . Genetic similarities among winter wheat cultivars determined on the basis of RFLPs and SSRs and their use for predicting progeny variance . Crop Sci. , 39 : 228 – 237 .
- Bornet , B. , Goraguer , F. , Joly , G. and Branchard , G. 2002 . Genetic diversity in European and Argentinean cultivated potatoes (Solanum tuberosum subsp. tuberosum) detected by inter-simple sequence repeats (ISSRs) . Genome , 45 : 481 – 484 .
- Carrasco , B. , A´vila , P. , Perez-Diaz , J. , Munoz , P. , García , R. , Lavandero , B. , Zurita-Silva , A. , Retamales , J.B. and Caligari , P.D.S. 2009 . Genetic structure of highland papayas (Vasconcellea pubescens (Lenné et C. Koch) Badillo) cultivated along a geographic gradient in Chile as revealed by Inter Simple Sequence Repeats (ISSR) . Genet. Resour. Crop Evol , 56 : 331 – 337 .
- Chen , G. , Wang , Y. , Zhao , C. , Korpelainen , H. and Li , C. 2008 . Genetic diversity of Hippophae rhamnoides populations at varying altitudes in the Wolong natural reserve of China as revealed by ISSR . Silvae Genet. , 57 : 29 – 36 .
- Cheng , J. , Kondo , K. , Suzuki , Y. , Ikeda , Y. , Meng , X. and Umemura , K. 2003 . Inhibitory effects of total flavones of Hippophae rhamnoides L. on thrombosis in mouse femoral artery and in vitro platelet aggregation . Life Sci. , 72 : 2263 – 2271 .
- Chowdari , K.V. , Ramakrishna , W. , Tamhankar , S.A. , Hendre , R.R. , Gupta , V.S. , Sahasrabudhe , N.A. and Ranjekar , P.K. 1998 . Identification of minor DNA variations in rice somaclonal variants . Plant Cell Rpt. , 18 : 55 – 58 .
- Devarumath , R. , Nandy , S. , Rani , V. , Marimuthu , S. , Muraleedharan , N. and Raina , S. 2002 . RAPD, ISSR and RFLP fingerprints as useful markers to evaluate genetic integrity of micropropagated plants of three diploid and triploid elite tea clones representing Camellia sinensis (China type) and C. assamica ssp. assamica (Assam-India type) . Plant Cell Rpt. , 21 : 166 – 173 .
- Feyissa , T. , Nybom , H. , Bartish , I.V. and Welander , M. 2007 . Analysis of genetic diversity in the endangered tropical tree species Hagenia abyssinica using ISSR markers . Genet. Resource Crop Evolution , 54 : 947 – 958 .
- Flavell , A.J. , Pearce , S.R. and Kumar , A. 1994 . Plant transposable elements and the genome . Curr. Opin. Genet. Dev. , 4 : 838 – 844 .
- Fufa , H. , Baenziger , P.S. , Beecher , B.S. , Dweikat , I. , Graybosch , R.A. and Eskridge , K.M. 2005 . Comparison of phenotypic and molecular marker-based classifications of hard red winter wheat cultivars . Euphytica , 145 : 133 – 146 .
- Hofmann , N.E. , Raja , A. , Nelson , R.L. and Korban , S.S. 2004 . Mutagenesis of embryogeneic cultures of soybean and detecting polymorphism using RAPD markers . Biol. Plant. , 48 : 173 – 177 .
- Huang , C.Y. and Liu , Q.L. 2006 . Detection of somaclonal variation by RAPD in Robinia pseudoacacia . Mol. Plant Breeding , 4 : 251 – 254 . in Chinese with an English abstract
- Ikegami , H. , Nogata , H. , Hirashima , K. , Awamura , M. and Nakahara , T. 2009 . Analysis of genetic diversity among European and Asian fig varieties (Ficus carica L.) using ISSR, RAPD, and SSR markers . Genet. Resources Crop Evolution. , 56 : 331 – 337 .
- Jaccard , P. 1908 . Nouvelles rescherches sur la distribution florale . Bul. Soc.Vaud. Sci. Nat. , 44 : 223 – 270 .
- Joshi , P. and Dhawan , V. 2007 . Assessment of genetic fidelity of micropropagated Swertia chirayita plantlets by ISSR marker assay . Biol. Plant. , 51 : 22 – 26 .
- Kaeppler , S.M. and Phillips , R.L. 1993 . Tissue culture-induced DNA methylation variation in maize . Proc. Nat. Acad. Sci. USA , 90 : 8773 – 8776 .
- Kar , P.K. , Srivastava , P.P. , Awasthi , A.K. and Urs , S.R. 2008 . Genetic variability and association of ISSR markers with some biochemical traits in mulberry (Morus spp.) genetic resources available in India . Tree Genet. Genomes , 4 : 75 – 83 .
- Kumar , R.S. , Parthiban , K.T. and Rao , M.G. 2009 . Molecular characterization of Jatropha genetic resources through inter-simple sequence repeat (ISSR) markers . Mol. Biol. Rep. , 56 : 201 – 209 .
- Leroy , X.J. , Leon , K. , Charles , G. and Branchard , M. 2000 . Cauliflower somatic embryogenesis and analysis of regenerant stability by ISSRs . Plant Cell Rep. , 19 : 1102 – 1107 .
- Leroy , X.J. , Leon , K. , Hily , J.M. , Chaumeil , P. and Branchard , M. 2001 . Detection of in vitro culture-induced instability through inter-simple sequence repeat analysis . Theor. Appl. Genet. , 102 : 885 – 891 .
- Le Thierry D'Ennequin , M. , Panay , O. and Toupance , B. 2000 . Assessment of genetic relationships between Setaria italica and its wild relatives S. viridis using AFLP markers . Theor. Appl. Genet. , 100 : 1061 – 1066 .
- Liu , C.Q. , Xia , X.L. , Yin , W.L. , Zhou , J.H. and Tang , H.R. 2007 . Direct somatic embryogenesis from leaves, cotyledons and hypocotyls of Hippophae rhamnoides . Biol. Plant. , 51 : 635 – 640 .
- Martins , M. , Sarmento , D. and Oliveira , M.M. 2004 . Genetic stability of micropropagated almond plantlets, as assessed by RAPD and ISSR markers . Plant Cell Rpt. , 23 : 492 – 496 .
- Matthes , M. , Singh , R. , Cheah , S.C. and Karp , A. 2001 . Variation in oil palm Elaeis guineensis (Jacq.) tissue culture-derived regenerants revealed by AFLPs with methylation-sensitive enzymes . Theor. Appl. Genet. , 102 : 971 – 979 .
- Mcclintock , B. 1984 . The significance of responses of the genome to challenges . Science , 226 : 792 – 801 .
- Melchinger , A.E. 1999 . “ Genetic diversity and heterosis ” . In The genetics and exploitation of heterosis in crops. Amer. Soc. Agron , Edited by: Coors , G. and Pandey , S. 99 – 118 . Wisc : Madison .
- Menz , M.A. , Klein , R.R. , Unruh , N.C. , Rooney , W.L. , Klein , P.E. and Mullet , J.E. 2004 . Genetic diversity of public inbreds of sorghum determined by mapped AFLP and SSR markers . Crop Sci. , 44 : 1236 – 1244 .
- Montpetit , D. and Lalonde , M. 1988 . In vitro and subsequent nodulation of the actinorhizal Hippophae rhamnoides L . Plant Cell Tiss. Org. , 15 : 189 – 199 .
- Mou , Y.Y. 1995 . Micropropagation of seabuckthorn (Hippophae rhamnoides L.) . Agr. Sci. Finland , 4 : 503 – 512 .
- Palombi , M. and Damiano , V. 2002 . Comparison between RAPD and SSR molecular markers in detecting genetic variation in kiwifruit (Actinidia deliciosa A. Chev.) . Plant Cell Rpt. , 20 : 1061 – 1066 .
- Peschke , V.M. , Phllips , R.L. and Gengenbach , B.G. 1987 . Discovery of transposable element activity among progeny of tissue-cultured-derived maize plants . Science , 238 : 804 – 807 .
- Phillips , R.L. , Kaeppler , S.M. and Olhoft , P. 1994 . Genetic instability of plant tissue cultures: Breakdown of normal controls . Proc. Nat. Acad. Sci. USA , 91 : 5222 – 5226 .
- Powell , W. , Morgante , R. , Andre , C. , Hanafey , M. , Vogel , J. , Tingey , S. and Rafalsky , A. 1996 . The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis . Mol. Breeding , 2 : 225 – 238 .
- Prevost , A. and Wilkinson , M. J. 1999 . A new system of comparing PCR primers applied to ISSR fingerprinting of potato accessions . Theoretical Appl. Genet. , 98 : 107 – 112 .
- Rahman , M. and Rajora , O. 2001 . Microsatellite DNA somaclonal variation in micropropagated trembling aspen (Populus tremuloides) . Plant Cell Rpt. , 20 : 531 – 536 .
- Rohlf , F. J. 2000 . NTSYS-pc: numerical taxonomy and multivariate analysis system. Version 2.1 Manual , New York : Applied Biostatistics .
- Ruan , C.J. and Li , D.Q. 2005 . AFLP Fingerprinting analysis of some cultivated varieties of sea buckthorn (Hippophae rhamnoides) . J. Genet. , 83 : 311 – 316 .
- Ruan , C.J. , Qin , P. , Zheng , J.W. and He , Z.X. 2004 . Genetic relationships among some cultivars of sea buckthorn from China, Russia and Mongolia based on RAPD analysis . Sci. Horti. , 101 : 417 – 426 .
- Ruan , C.J. , Teixeira da Silva , J.A. , Jin , H. , Li , H. and Li , D.Q. 2007 . Research and biotechnology in sea buckthorn (Hippophae spp.) . Med. Aromat. Plant Sci. Biotechnol. , 1 : 47 – 60 .
- Singh , A. , Negi , M.S. , Moses , V.K. , Venkateswarlu , B. , Srivastava , P.S. and Lakshmikumaram , M. 2002 . Molecular analysis of micropropagated neem plants using AFLP markers for ascertaining clonal fidelity . In Vitro Cell. Dev. Biol. Plant. , 38 : 519 – 521 .
- Singh , M. , Saroop , J. and Dhiman , B. 2004 . Detection of intra-clonal genetic variability in vegetatively propagated tea using RAPD markers . Biol. Plant. , 48 : 113 – 115 .
- Sreedhar , R.V. , Venkatachalam , L. and Bhagyalakshmi , N. 2007 . Genetic fidelity of long-term micropropagated shoot cultures of vanilla (Vanilla planifolia Andrews) as assessed by molecular markers . Biotechnol. J. , 2 : 1007 – 1013 .
- Sun , K. , Chen , X.L. , Ma , R.J. , Li , C.B. , Wang , Q. and Ge , S. 2002 . Molecular phylogenetics of Hippophae L. (Elaeagnaceae) based on the internal transcribed spacer (ITS) sequences of nrDNA . Plant Syst. Evol. , 235 : 121 – 134 .
- Swenson , U. and Bartish , I.V. 2002 . Taxonomic synopsis of Hippophae (Elaeagnaceae) . Nord. J. Bot. , 22 : 369 – 374 .
- Teixeira da Silva , J.A , Nhut , D.T. , Giang , D.T.T. and Rashid , S.Z. 2005 . “ The phylogeny, breeding and ecology of trees using molecular markers ” . In Genetic resources and biotechnology (Vol III) , Edited by: Thangadurai , D. , Pullaiah , T. and Tripathy , L. 279 – 308 . New Delhi : Regency Publications .
- Tian , C.J. , Lei , Y.D. , Shi , S.H. , Nan , P. , Chen , J.K. and Zhong , Y. 2004 . Genetic diversity of sea buckthorn (Hippophae rhamnoides) populations in northeastern and northwestern China as revealed by ISSR markers . New For. , 27 : 229 – 237 .
- van Beuningen , L.T. and Busch , R.H. 1997 . Genetic diversity among North American spring wheat cultivars: III. Cluster analysis based on quantitative morphological traits . Crop Sci. , 37 : 981 – 988 .
- Vijayan , K. , Srivatsava , P.P. , Nair , C.V. , Awasthi , A.K. , Tikader , A. , Sreenivasa , B. and Urs , S.R. 2006 . Molecular characterization and identification of markers associated with yield traits in mulberry using ISSR markers . Plant Breeding , 125 : 298 – 301 .
- Xu , M. , Li , X. and Korban , S.S. 2004 . DNA-methylation alterations and exchanges in vitro cellular differentiation in rose (Rosa hybrida L.) . Theor. Appl. Genet. , 109 : 899 – 910 .
- Zadernowski , R. , Naczk , M. , Nowak-Polakowsk , A.H. and Nesterowicz , J. 2002 . Effect of sea buckthorn (Hippophae rhamnoides L.) berry extracts on the activity of lipase and lipoxygenase . J. Food Lipids , 9 : 249 – 258 .
- Zhang , D.H. , Meng , Z.H. , Xiao , W.M. and Wang , X.C. 2002 . Graft-induced inheritable variation in Mungbean and its application in Mungbean breeding . Acta Bot. Sinica , 44 : 832 – 837 . Sodmergon