Abstract
Because of their recognized health benefits, there has been increased demand and consumption of blueberries in recent years. Great strides have been made in blueberry cultivar development since its domestication using traditional breeding approaches. However, genomic tools that could be used to hasten improvement are lacking. The aim of our Specialty Crop Research Initiative project, funded at the end of 2008, is to develop genomic tools for molecular breeding and assessing genetic diversity of blueberry. Marker-assisted breeding would be particularly useful for combining traits for climatic adaptation with those for improved fruit and nutritional quality in highbush blueberry (Vaccinium corymbosum). Genomic resources being developed include expressed sequence tag libraries, expressed sequence tag-based molecular markers, and genetic linkage maps. Transcriptome sequences have been generated from fruit at different stages of development, flower buds at different stages of cold acclimation, and leaves by 454 sequencing. About 600,000 sequences have been assembled into approximately 15,000 contigs. Markers derived from expressed sequence tags (simple sequence repeats and expressed sequence tag-polymerase chain reaction markers) are being used to identify quantitative trait loci associated with cold hardiness, chilling requirement, and fruit quality traits, in studies of genetic diversity, spatial genetic structure, and gene flow in the wild lowbush blueberry (V. angustifolium), and to construct a phylogenetic tree of Vaccinium species in the section Cyanococcus. Availability of these genomic tools will allow future advances, such as the development of a blueberry microarray to facilitate studying gene expression and the use of marker-assisted breeding.
INTRODUCTION
The Ericaceae family includes many commercially important species, such as the berry crops (blueberry, cranberry, and lingonberry) and the floral and nursery crops (rhododendron, azalea, and mountain laurel). The most economically important fruit crop within this family is blueberry.
Blueberry is a high value small fruit crop that is adapted to acidic, imperfectly drained sandy soils that might otherwise be considered worthless for agricultural crop production. North America is the major producer of blueberries. The farm gate value of blueberry in 2007 was estimated to be over $600 million, a three-fold increase over the estimated value in 2000 (USDA Agricultural Statistics). Much of this increased value is due to increased consumption of blueberries because of its many recognized health benefits. Blueberry fruit is very high in anthocyanins, which act as antioxidants. In recent studies, anthocyanins have been linked to many health benefits, such as reducing eyestrain, improving night vision, helping to prevent macular degeneration, and exhibiting anti-cancer activity (CitationCho et al., 2004; CitationKalt et al., 2007). Antioxidants, in general, have been suggested to ameliorate inflammation associated with chronic diseases like cancer and heart disease. The compound resveratrol, found in blueberry and grapes, also has been linked to reducing the risk of heart disease and cancer (CitationRimando et al., 2004). Another compound, pterostilbene, apparently plays a role in lowering cholesterol and other blood fats (CitationRimando et al., 2004).
Great strides have been made in traditional breeding efforts of blueberry in the short time since its domestication in the 20th century. Breeding efforts have focused on development of cultivars with broader climatic adaptation, broader soil adaptation, disease and pest resistance, mechanical handling tolerance, and high fruit quality (CitationGalletta and Ballington, 1996). The availability of genomic tools for molecular breeding could lead to more rapid genetic improvement, particularly when combining traits for climatic adaptation with other important traits like fruit and nutritional quality. Woody perennials, like blueberry, are especially suitable for improvement via marker-assisted breeding because of their long generation times, high heterozygosity, self-incompatibility, inbreeding depression, and polyploidy of commercial types.
Genomic research in blueberry, and in the Ericaceae family in general, is still in its infancy although significant progress has been made in the last few years. The first few thousand expressed sequence tags (ESTs) have been generated and made publicly available for this family, about 5,000 from blueberry and about 1,200 from rhododendron. Both of these sets of ESTs from blueberry and rhododendron were generated as parts of projects focused on cold acclimation research. Thus, ESTs are from non-acclimated and cold acclimated flower bud libraries, in the case of blueberry (CitationDhanaraj et al., 2004, 2007), and from non-acclimated and cold acclimated leaf libraries, in the case of rhododendron (CitationWei et al., 2005). Another ∼16,000 ESTs have been generated from blueberry fruit by the New Zealand Institute for Plant & Food Research Ltd. (formerly HortResearch), but they are not publicly available.
Robust molecular markers like simple sequence repeats (SSRs) (CitationBoches et al., 2005) and expressed sequence tag-polymerase chain reaction markers (EST-PCRs) (CitationRowland et al., 2003a, 2003b, 2003c) were recently developed from some of the publicly available ESTs and demonstrated to be useful in genetic diversity and mapping studies in related species. Although it is expected that EST-SSR markers would be highly polymorphic among individuals within a species, the high levels of polymorphisms detected even with the EST-PCR markers are fortuitous. This is not entirely surprising, however, considering that blueberry is a highly heterozygous, primarily outcrossing, polyploid crop. In addition, the SSR and EST-PCR markers derived from highbush blueberry ESTs appear to be useful for all species within the Cyanococcus section and even other sections, including the cranberry species (CitationBassil et al., 2009; CitationBell et al., 2008; CitationBoches et al., 2005, Citation2006a, Citation2006b; CitationRowland et al., 2003a, Citation2003c). Mapping populations and initial genetic linkage maps have been developed (CitationBrevis et al., 2007; CitationRowland et al., 1999, Citation2003b) but inadequate funding has left the maps insufficiently saturated for quantitative trait loci (QTL) mapping. Developing a genetic linkage map for a highly heterozygous tetrasomic species, such as the commercial highbush blueberry, is particularly challenging. Difficulties in developing and maintaining a large enough mapping population and in identifying different allelic states (monoallelic, diallelic, triallelic, and tetra-allelic) at loci must be overcome. The first microarray experiments have been carried out in blueberry and have successfully identified many transcripts whose abundances increase with cold acclimation and identified interesting differences in expression between acclimation under cold room and field conditions and between cold tolerant and cold sensitive genotypes (CitationDhanaraj et al., 2007; CitationRowland et al., 2008). More gene expression studies need to be undertaken using microarrays that are based on a larger collection of gene sequences in order to sort out genes that are expressed in response to various stimuli and during development.
Because of the need to develop more genomic resources for blueberry, the research objectives of our project, “Generating Genomic Tools for Blueberry Improvement” funded through the Specialty Crop Research Initiative, are as follows: (1) to generate more ESTs from different blueberry organs, such as fruit, flower buds, leaves, and stems; (2) to develop EST-PCR and SSR markers based on these ESTs; and (3) to use these markers to map QTL for chilling requirement, cold hardiness, and fruit quality in diploid and tetraploid mapping populations (the tetraploid population is an actual breeding population), in genetic diversity studies on wild populations of the commercial lowbush blueberry, and to examine the evolutionary relationships of the Vaccinium species within section Cyanococcus. Progress on these objectives is presented.
MATERIALS AND METHODS
Transcriptome Sequencing
ESTs were generated and assembled from the highbush cultivar Bluecrop using next generation Roche/454 GS-FLX transcriptome sequencing at the Genomic Sciences Laboratory at North Carolina State University. ESTs were generated from four organs: fruit, flower buds, and a combination of leaves and stems. cDNAs for sequencing were prepared from RNA extracted from fruit collected at different stages of development (green, white, pink, and blue stage), flower buds collected at different stages of cold acclimation (0, 400, 800, and 1300 chill units or number of hours of exposure to temperatures from 0–7°C), and young leaf and stem tissue. All cDNA samples were tagged and multiplexed. The 454 sequences were sent to the Bioinformatics Laboratory at Towson University for annotation. The consensus sequences from the assembly were batch BLASTed to identify the genes expressed in the respective tissues. BLASTX results were passed through a custom pipeline, built with the scripting language PERL, to create annotated, tab-delimited tables, which include information on taxonomy, key words, gene function, tissue specificity, and gene ontology terms (CitationVera et al., 2008).
Marker Development
Publicly available blueberry ESTs were downloaded from GenBank dbEST on February 11, 2009. The ESTs were filtered and assembled using the EST processing pipeline documented in CitationFolta et al. (2005). The unigene sequences were mined for SSRs and used to design new SSR and EST-PCR primers for mapping and genetic diversity studies. SSR primer pairs were designed using the online SSR server freely available through the Genome Database for Rosaceae website (http://www.rosaceae.org/bio/content?title=&url=%2Fcgi-bin%2Fgdr%2Fgdr_ssr&style=height:1024px;width:950px;). EST-PCR primer pairs were designed toward the ends of the ESTs using the P3 website (http://frodo.wi.mit.edu/) as described in CitationRowland et al. (2003c).
Mapping Studies
The interspecific diploid mapping population that segregates for mid-winter cold tolerance and chilling requirement has been described in detail elsewhere (CitationRowland et al., 1999). Briefly, the population was generated by crossing a V. darrowii selection Fla4B (low chilling, cold sensitive, evergreen) x diploid V. corymbosum also known as V. caesariense selection W85-20 (high chilling, cold hardy, deciduous) hybrid (Fl #10) to another diploid V. corymbosum W85-23. The tetraploid mapping population that segregates for cold tolerance, chilling requirement, and fruit quality traits was generated by crossing the northern highbush cultivar Draper (high chilling, cold hardy) and the southern highbush cultivar Jewel (low chilling, cold sensitive). SSR and EST-PCR primers were tested on parents of mapping populations to identify polymorphic markers suitable for mapping. Markers that detected differences were followed in the appropriate mapping populations and added to the current genetic linkage maps using MAPMAKER, JoinMap, or Tetraploid Map. The method for amplifying EST-PCR products, separating the products on agarose gels, and scoring gels was essentially that described by CitationRowland et al. (2003c). For SSR analysis, standard PCR protocols were used and PCR products were pooled into multiplexes and separated by capillary electrophoresis using the CEQ 8000 (Beckman Coulter Inc., Fullerton, CA, USA) or similar equipment. Allele sizing and visualization was performed using the fragment analysis module of the CEQ 8000 software by calling internal size standards included with each sample.
Cold hardiness evaluations were performed on the diploid mapping population in two separate years (2009 and 2010) using the freeze-thaw protocol of CitationArora et al. (2000). The tetraploid mapping population was asexually propagated and planted at five locations with varying winter temperatures and chilling hours: Gainesville, FL; Waycross, GA; Invergowrie, Scotland; Corvallis, OR; and Benton Harbor, MI.
Other Genetic Relationship Studies
EST-PCR primers are being tested on DNA from different collections of lowbush blueberry genotypes to investigate spatial genetic structure of wild populations on a local (touching individuals within the same field) and a wide range of distribution. EST-PCR primers were also tested on DNA from a collection of three or more representatives of each of the blueberry species at each ploidy level within the section Cyanococcus (including the diploids V. boreale, V. corymbosum, V. darrowii, V. elliottii, V. myrtilloides, V. pallidum, V. tenellum; the tetraploids V. angustifolium, V. corymbosum, V. hirsutum, V. myrsinites, V. pallidum, V. simulatum; and the hexaploids V. constablaei and V. virgatum), for a total of more than 50 genotypes. Length polymorphisms of EST-PCR markers were scored and analyzed using The Numerical Taxonomy and Multivariate Analysis System (NTSYS-PC version 2.2, Exeter Software, Setauket, NY, USA). For dendrogram construction, the unweighted pair-group method (UPGMA) and Neighbor Joining (NJ) cluster analyses were performed from a DICE similarity matrix to examine evolutionary relationships of the species.
RESULTS AND DISCUSSION
Transcriptome Sequencing
A high-throughput pyrosequencing technology (454 EST sequencing) was used for transcriptome profiling of fruit during different stages of development, of flower buds during different stages of cold acclimation, and of leaves and stems, from ‘Bluecrop’. About 1.35 million reads were obtained altogether to yield approximately 390 Mb of data. Of these, about 600,000 sequences were assembled. A cluster analysis of all the assembled sequences resulted in approximately 15,000 contigs (). The types of genes and their abundances in the different libraries are currently being compared to identify potential differentially expressed genes. For example, the five most highly abundant genes in the ripe fruit library encode aspartic proteinase, burp domain-containing protein, flavonoid 3-hydroxylase, ethylene-forming enzyme, and aminocyclopropane-1-carboxylate oxidase. In comparison, the five most abundant genes in the green fruit library encode metallothionein-like protein, burp domain-containing protein, lipid transfer protein, dehydrin protein, and 2s albumin. Differential expression of these and other interesting genes will be confirmed by quantitative RT-PCR.
TABLE 1 Transcriptome Sequencing of Blueberry—Overall Results (The Number of Reads, Number of Reads Assembled, Number of Contigs, and Average Contig Length Are Shown for Each of the Blueberry Samples)
Marker Development and Mapping Studies
ESTs, beginning with those previously generated from flower bud libraries by Sanger sequencing (CitationDhanaraj et al., 2004, 2007), were mined for SSRs and used to design new SSR and EST-PCR primers for mapping and genetic diversity studies. (parts A and B) shows how the primers were designed, either flanking SSRs within ESTs, in the case of SSR primers, or toward the ends of the ESTs, in the case of EST-PCR primers, to amplify as much of each gene as possible from the available sequences. For mapping purposes, parents of the diploid and tetraploid mapping populations were screened for polymorphic EST-PCR and SSR markers. shows EST-PCR markers being screened against the parent plants of the diploid population. Polymorphic markers identified in this way are then followed in the appropriate mapping populations and added to the current genetic linkage maps. Although it was expected that the SSR markers would be highly polymorphic among the parent plants, the EST-PCR markers have also proved to be quite polymorphic, as we have shown previously on a collection of highbush (CitationRowland et al., 2003c) and lowbush genotypes (CitationBell et al., 2008). Forty of 120 (33.3%) EST-PCR primer pairs screened so far this year on the diploid parents detected polymorphisms useful for mapping. Currently, 122 markers have been followed in the diploid mapping population, and 110 have been mapped to 16 linkage groups.
FIGURE 1 Design of (A) SSR and (B) EST-PCR primers using the P3 website (http://frodo.wi.mit.edu/), and (C) screening of the diploid Vaccinium parent plants for polymorphic EST-PCR markers. In the SSR example shown (A), primers were designed flanking (GA)14(CGA)5 repeats to give a product in the 250 bp range. For EST-PCR markers (B), primers were designed near the ends of the ESTs. In the gel picture shown (C), an EST-PCR marker (lower band) is present in the original parent Fla4B, absent in the other parent W85-20, present in the F1#10, and absent in the testcross parent W85-23, making it “mappable” in the diploid population. PCRs of each of the parent plants were run in duplicate. In the ‘M’ lane are molecular weight standards.

Potted plants of the diploid mapping population, held in an unheated greenhouse, were evaluated for cold hardiness in February of 2009 and 2010, after receiving approximately 500 chilling hours. Plants will be evaluated for chilling requirement over the next 2 years. The tetraploid mapping population, propagated and planted at various field locations, will be evaluated for chilling requirement, cold hardiness, and fruit quality traits in the coming few years. The tetraploid mapping population has also been propagated and placed in a greenhouse to evaluate chilling requirement over the next 2 years.
Other Genetic Relationship Studies
EST-PCR primers are also being used to investigate gene flow and spatial genetic structure of wild lowbush blueberry populations on a local (touching individuals within the same field) and a wide range of distribution (individuals collected at multiple sites throughout Maine and other regions). Details of this work are described in presentations by CitationBell et al. (2009, Citation2010, CitationIn press) and CitationBeers et al. (2010).
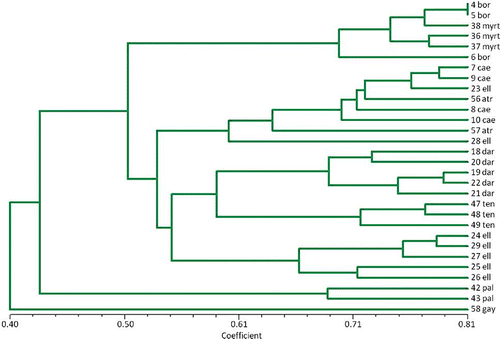
EST-PCR markers were also used to examine evolutionary relationships among the different blueberry species within the section Cyanococcus of the Vaccinium genus. At least three wild representatives of each of the blueberry species at each ploidy level were included in the study. shows a UPGMA dendrogram of only the diploid species (the NJ tree was nearly identical) based on 249 polymorphic EST-PCR markers. Interestingly, almost all the V. elliottii representatives grouped together and separate from the V. caesariense (or diploid V. corymbosum) and V. atrococcum (also known as V. fuscatum Ait.) representatives, providing support for considering V. elliottii a separate species from V. corymbosum. The V. boreale and V. myrtilloides representatives clustered together, and the V. darrowi and V. tenellum groups were closely related. Indeed, this tree was very similar to the isozyme dendrogram reported previously by CitationBruederle and Vorsa (1994). Sequences from two DNA regions, the nuclear granule bound starch synthase (waxy) gene and the nuclear internal transcribed spacer (nrITS) region, are also being used to examine evolutionary relationships between the species. Details of this work are described in a presentation by CitationDurchholz et al. (2010).
FIGURE 2 A UPGMA dendrogram of diploid Vaccinium species in the section Cyanococcus constructed from length polymorphisms of EST-PCR markers. In the tree, bor, myrt, cae, eli, atr, dar, ten, pal, and gay stand for V. boreale, V. myrtilloides, V. caesariense, V. elliottii, V. atrococcum, V. darrowii, V. tenellum, V. pallidum, and Gaylussacia brachycera (which served as an outlier), respectively. Most of the V. elliottii representatives formed a distinct group, suggesting V. elliottii should probably not be lumped with V. caesariense and V. atrococcum as diploid V. corymbosum (color figure available online).
CONCLUSIONS
Genomic tools are being developed to aid in blueberry improvement. New transcriptome sequences are being used to develop molecular markers, and markers are being used to identify QTL associated with cold hardiness, chilling requirement, and fruit quality traits for marker-assisted breeding in highbush blueberry, and in studies of genetic diversity, spatial genetic structure, and gene flow in lowbush blueberry, as well as to construct a phylogenetic tree of Vaccinium species in the section Cyanococcus. Transcriptome sequences will also be useful for developing a blueberry microarray to allow study of expression of thousands of genes in response to biotic and abiotic stresses and during development.
ACKNOWLEDGMENTS
We wish to acknowledge the highbush (Berry Blue LLC and Driscoll Strawberry Associates, Inc.) and the lowbush blueberry (Wild Blueberry Commission of Maine) industries for providing matching funds in support of this project.
LITERATURE CITED
- Arora , R. , Rowland , L.J. , Lehman , J.S. , Lim , C.C. , Panta , G.R. and Vorsa , N. 2000 . Genetic analysis of freezing tolerance in blueberry (Vaccinium section Cyanococcus) . Theor. Appl. Genet. , 100 : 690 – 696 .
- Bassil , N.V. , Oda , A. and Hummer , K. 2009 . Blueberry microsatellite markers identify cranberry cultivars . Acta Hort , 810 : 181 – 186 .
- Beers , L. , Rowland , L.J. and Drummond , F.A. Preliminary analysis of genetic diversity in lowbush blueberry (Vaccinium angustifolium) across its native range . 11th North American Blueberry Research and Extension Workers’ Conference . July 25–28 2010 , Kalamazoo , MI . pp. 55
- Bell , D.J. , Rowland , L.J. , Polashock , J.J. and Drummond , F.A. 2008 . Suitability of EST-PCR markers developed in highbush blueberry for genetic fingerprinting and relationship studies in lowbush blueberry and related species . J. Amer. Soc. Hort. Sci. , 133 : 701 – 707 .
- Bell , D.J. , Rowland , L.J. , Zhang , D. and Drummond , F.A. 2009 . Spatial genetic structure of lowbush blueberry, Vaccinium angustifolium, in four fields in Maine . Botany , 87 : 932 – 946 .
- Bell , D.J. , Rowland , L.J. , Stommel , J. and Drummond , F.A. 2010 . Yield variation among clones of lowbush blueberry as a function of genetic similarity and self-compatibility . J. Amer. Soc. Hort. Sci. , 135 : 259 – 270 .
- Bell , D.J. , Rowland , L.J. and Drummond , F.A. 2012 . Does pollen ‘neighborhood’ affect yield in lowbush blueberry (Vaccinium angustifolium Ait.)? Intl . J. Fruit Sci , 12 : 65 – 74 .
- Boches , P.S. , Bassil , N.V. and Rowland , L.J. 2005 . Microsatellite markers for Vaccinium from EST and genomic libraries . Mol. Ecol. Notes , 5 : 657 – 660 .
- Boches , P. , Bassil , N.V. and Rowland , L.J. 2006a . Genetic diversity in the highbush blueberry Vaccinium corymbosum L. evaluated with microsatellite markers . J. Amer. Soc. Hort. Sci. , 131 : 674 – 686 .
- Boches , P. , Rowland , L.J. , Hummer , K. and Bassil , N.V. 2006b . Cross-species amplification of SSR loci in the genus Vaccinium . Acta Hort. , 715 : 119 – 128 .
- Brevis , P. , Hancock , J. and Rowland , L.J. 2007 . Development of a genetic linkage map for tetraploid highbush blueberry using SSR and EST-PCR markers . HortScience , 42 : 963
- Bruederle , L.P. and Vorsa , N. 1994 . Genetic differentiation of diploid blueberry, Vaccinium section Cyanococcus (Ericaceae). System. Botany . 19 : 337 – 349 .
- Cho , E. , Seddon , J.M. , Rosner , B. , Willett , W.C. and Hankinson , S.E. 2004 . Prospective study of intake of fruits, vegetables, vitamins, and carotenoids and risk of age-related maculopathy . Arch. Ophthalmol. , 122 : 883 – 892 .
- Dhanaraj , A.L. , Slovin , J.P. and Rowland , L.J. 2004 . Analysis of gene expression associated with cold acclimation in blueberry floral buds using expressed sequence tags . Plant Sci. , 166 : 863 – 872 .
- Dhanaraj , A.L. , Alkharouf , N.W. , Beard , H.S. , Chouikha , I.B. , Matthews , B.F. , Wei , H. , Arora , R. and Rowland , L.J. 2007 . Major differences observed in transcript profiles of blueberry during cold acclimation under field and cold room conditions . Planta , 225 : 735 – 751 .
- Durchholz , E.J. , Williams , M.A. , Polashock , J.J. , Rowland , L.J. and Powell , E.A. Evolutionary relationships in Vaccinium section Cyanococcus . 11th North American Blueberry Research and Extension Workers’ Conference . July 25–28 2010 , Kalamazoo , MI . pp. 56
- Folta , K.M. , Staton , M. , Stewart , P.J. , Jung , S. , Bies , D.H. , Jesudurai , C. and Main , D. 2005 . Expressed sequence tags (ESTs) and simple sequence repeat (SSR) markers from octoploid strawberry (Fragaria x ananassa) . BMC Plant Biol. , 5 : 12
- Galletta , G.J. and Ballington , J.R. 1996 . “ Blueberries, cranberries and lingonberries ” . In Fruit breeding, volume II. Vine and small fruit crops , Edited by: Janick , J. and Moore , J.N. 1 – 107 . New York , NY : John Wiley and Sons, Inc .
- Kalt , W. , Joseph , J.A. and Shukitt-Hale , B. 2007 . Blueberries and human health: A review of the current research . J. Amer. Pomol. Soc. , 61 : 151 – 160 .
- Rimando , A.M. , Kalt , W. , Magee , J.B. , Dewey , J. and Ballington , J.R. 2004 . Resveratrol, pterostilbene, and piceatannol in Vaccinium berries . J. Agric. Food Chem. , 52 : 4713 – 4719 .
- Rowland , L.J. , Ogden , E.L. , Arora , R. , Lim , C-C. , Lehman , J.S. , Levi , A. and Panta , G.R. 1999 . Use of blueberry to study genetic control of chilling requirement and cold hardiness in woody perennials . HortScience , 34 : 1185 – 1191 .
- Rowland , L.J. , Dhanaraj , A.L. , Polashock , J.J. and Arora , R. 2003a . Utility of blueberry-derived EST-PCR primers in related Ericaceae species . HortScience , 38 : 1428 – 1432 .
- Rowland , L.J. , Mehra , S. , Dhanaraj , A. , Ogden , E.L. and Arora , R. 2003b . Identification of molecular markers associated with cold tolerance in blueberry . Acta Hort. , 625 : 59 – 69 .
- Rowland , L.J. , Mehra , S. , Dhanaraj , A.L. , Ogden , E.L. , Slovin , J.P. and Ehlenfeldt , M.K. 2003c . Development of EST-PCR markers for DNA fingerprinting and genetic relationship studies in blueberry (Vaccinium, section Cyanococcus). J. Amer. Soc. Hort. Sci . 128 : 682 – 690 .
- Rowland , L.J. , Dhanaraj , A.L. , Naik , D. , Alkharouf , N. , Matthews , B. and Arora , R. 2008 . Study of cold tolerance in blueberry using EST libraries, cDNA microarrays, and subtractive hybridization . HortScience , 43 : 1975 – 1981 .
- Vera , J.C. , Wheat , C.W. , Fescemyer , H.W. , Frilander , M.J. , Crawford , D.L. , Hanski , I. and Marden , J.H. 2008 . Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing . Mol. Ecol. , 17 : 1636 – 1647 .
- Wei , H. , Dhanaraj , A.L. , Rowland , L.J. , Fu , Y. , Krebs , S.L. and Arora , R. 2005 . Comparative analysis of expressed sequence tags from cold-acclimated and non-acclimated leaves of Rhododendron catawbiense Michx. Planta . 221 : 406 – 416 .