 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Anthocyanin accumulation is responsible for the red color of the skin of apple fruits (Malus × domestica Borkh.). Environmental stimuli, such as light, temperature, soil factors, tree factors, and the application of chemicals can alter the synthesis of anthocyanins. Herein, we examined the expression of genes related to anthocyanin biosynthesis in the fruit of apple cultivars, ‘Summer Prince’ and ‘Arisoo’ at the early stage of fruit development 60 DAFB (days after full bloom). The expression of genes, which included structural and regulatory genes involved in anthocyanin biosynthesis, and light-responsive genes was determined in fruits developed under the effect of sunlight exposure for seven days. Apple fruits were divided in to: ‘reflected sunlight’, ‘bagged’, and ‘control’ groups. The expression levels of the anthocyanin synthesis-related genes were significantly different among the groups and between the shaded and reflected sides. We also determined the difference in coloration patterns in the different groups using the colorimetric coordinates method. The shaded side of apple fruits quickly turned more redder after exposure to reflected sunlight and the coloration was almost similar to that on the exposed side after the treatment. Strong correlation coefficients between anthocyanin-related gene expression levels and coloration patterns were obtained. This study shows that sunlight plays an important role in stimulating the coloration of apple fruit by regulating the expression of genes involved in anthocyanin accumulation during the early stage of fruit development. The present study assumes importance because redder fruits, generated through a non-transgenic approach would be more acceptable to consumers.
Introduction
Color development is an important evolutionary trait in fruits and is a major factor contributing to fruit quality that increases the prospects of the fruit in the international market and attracts the attention of consumers. Therefore, researchers have been focusing on understanding the mechanisms underlying the development of color in fruit. Chlorophyll, carotenoids, and anthocyanins are the most important pigments in plant tissues, including fruit (Lancaster et al., Citation1994). Anthocyanin was found to be one of the most important pigments responsible for the color formation in apple fruit. The enzymes operating in this biosynthetic pathway in apple have been well characterized (Honda et al., Citation2002; Kim et al., Citation2003). Anthocyanin biosynthesis in fruits has become an interesting and useful aspect of research considering the need for a better understanding of its mechanism and for the development of fruit cultivars with higher anthocyanin contents.
Higher plants, such as apple trees, operate the well-known basic pathway for anthocyanin biosynthesis. The expression of anthocyanin biosynthesis-related structural genes (PAL, CHS, CHI, F3H, DFR, ANS, UFGluT) have been used as marker genes for study on the anthocyanin accumulation (Honda et al., Citation2002; Ju et al., Citation1997; Kondo et al., Citation2002a, Citation2002b; Liu et al., Citation2013; Zhang et al., Citation2013). MYB transcription factors were also found is involved in anthocyanin accumulation by regulating the expression of anthocyanin biosynthetic genes through interacting of the MYB–bHLH–WD40 protein complex, resulting in the transcription activation of these genes; this interaction is influenced by external environment factors (Allan et al., Citation2008; Koes et al., Citation2005). Transcriptome analysis of the induction of red coloration in apple fruit by light, identified novel regulatory genes, MYBs, that are significantly associated with the red color of the skin of apple fruit (Vimolmangkang et al., Citation2014). The correlation between the expression of MYB10 and anthocyanin accumulation has been verified in the apple (Espley et al., Citation2007; Wang et al., Citation2015). The functional of MYB10 in anthocyanin accumulation has also been studied in other plants. Upon overexpression of FaMYB10 in strawberry Fragaria ananassa, the transgenic strawberry had upregulated anthocyanin synthesis in leaf, root, seed, and fruit with approximately 50% increase in the anthocyanin content (Kui et al., Citation2010). Overexpression of peach MYB10.1/bHLH3 and MYB10.3/bHLH3 activated anthocyanin production by upregulating NtCHS, NtDFR, and NtUFGT (Rahim et al., Citation2014). Overexpression of AcMYB10 elevated the accumulation of anthocyanin in transgenic Actinidia chinensis (Yu et al., Citation2019). These results suggest an essential role of MYB transcription factors in the anthocyanin biosynthesis in plants. Light-responsive genes were also found to plays an important role in the regulatory pathways of anthocyanin synthesis. PIFs were found to interact with phytochromes and participate in anthocyanin biosynthesis by binding to the promoters of anthocyanin biosynthesis-related genes (Liu et al., Citation2015; Shin et al., Citation2007). The functional of COP1 on the expression of anthocyanin biosynthetic genes was recently studied in Arabidopsis (Maier et al., Citation2013), pear (Wu et al., Citation2019), and apple (An et al., Citation2017; Li et al., Citation2012).
The environmental factors involved in the accumulation of anthocyanins have been reviewed (Lancaster, Citation1992; Saure, Citation1990). Light is known to be the most important external factor regulating anthocyanin synthesis. It is a stimulus that activates a broad range of plant photosynthesis-related genes. It regulates photosynthesis in plant tissues leading to the regulation of sugar content, necessary energy, hence, controls the expression of the genes that encode enzymes of the anthocyanin biosynthetic pathway (Kawabata et al., Citation1999; Rachel et al., Citation2001).
The accumulation of anthocyanin changes under the effect of many factors and it is complicated. The accumulation of anthocyanin changes with the developmental stage of the fruit, and is increased toward the harvest stage (Honda et al., Citation2002; Ju et al., Citation1997; Marais et al., Citation2001; Reay and Lancaster, Citation2001), together with increased ethylene production (Whale and Singh, Citation2007). Moreover, the temperature is well known to have a major effect on anthocyanin synthesis, and thereby, effects the fruit color (Kui et al., Citation2011; Steyn et al., Citation2009; Ubi et al., Citation2006). Other studies on anthocyanin biosynthesis and the influence of different factors on its biosynthesis have focused on determining the changes in the expression of anthocyanin biosynthetic genes either throughout the different stages of fruit development or at the pre-harvest stage. This makes it complicated to investigate the effect of a given factor on the biosynthesis of anthocyanin in controlling fruit coloration. Therefore, to minimize the interference from the effects of others factors, such as the age of fruit, high temperature, ethylene signaling, and of unknown crosstalk, on the expression of anthocyanin genes, we selected fruit at the young stage just when they start turning red (60 DAFB) for investigating the effect of a single factor is reflected sunlight on the coloration pattern of apple fruit.
To investigate the different coloration patterns of red-skinned apple cultivars, we investigated the fruit under the difference of sunlight exposure after seven days of treatment. Through the detection of differential gene expression analysis, we identified key genes that control the development of coloration in apple fruit. Pearson analysis was also performed to determine the correlation between the coloration of fruits and the expression levels of anthocyanin-related genes. These findings will help to improve our understanding of the molecular mechanism of anthocyanin accumulation in apple under the effect of the environmental factor at the early stage of fruit development.
Materials and Methods
Fruit Materials
The fruit used in this experiment were obtained from 6-year-old trees grown in the orchard of the Apple Research Institute (Gunwi; 36.28 N, 128.47E), Republic of Korea. Two red-skinned apple cultivars, ‘Summer Prince’ and ‘Arisoo’ at the early developmental stage of fruit (60 DAFB) were used to investigate the effect of sunlight exposure. Apple fruits were divided into ‘reflected sunlight’, ‘bagged’, and ‘control’ groups. The treatment of fruits was conducted at the end of June 2019 ().
Figure 1. Orchard-based apple reflected sunlight was conducted to increase light intensity on the fruit of apple ‘Summer Prince’ cultivar at the early developmental stage (60 DAFB). (a) Fruits were sampled for anthocyanin analysis showed the difference red-coloration levels between the three groups: reflected sunlight, bagged, and control. (b) The fruits in the control group showed the significant difference red-coloration level between the exposed and shaded sides in the same fruit. (c). For the reflected sunlight treatment, the fruit was set up with a paper wrapped-aluminum disc (28 cm diameter, V-shaped with angle of 140°) by hanging the disc on below the fruit in order to the shaded side of fruit to be exposed to sunlight (d). The same experimental layout was also applied to apple ‘Arisoo” cultivar (data not show)
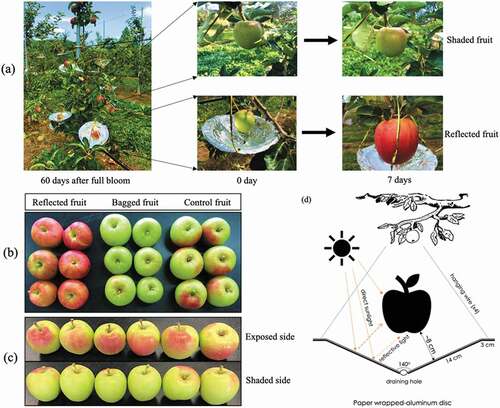
For the reflected sunlight treatment, every single fruit was set up with a paper wrapped-aluminum disc (28 cm diameter, V-shaped with an angle of 140°) by hanging the disc below the fruit such that the shaded side of the fruit can be exposed to sunlight (). For the bagging treatment, each fruit was bagged with double-layered paper bags. The inner layer was made of black paper with a wax coating, and the outer layer was gray. The fruits in the control group were grown under normal sunlight conditions. At the time of sampling, 20 fruits were randomly collected from five trees after 7 days of treatment. Half of the fruits were used to determine the various physiological parameters (soluble solid content (SSC), firmness, acidity, starch, and color indices) (). The other 10 fruits were used for RNA isolation by sampling their peel and flesh; these tissues were immediately frozen in liquid nitrogen and stored at −80°C for further analysis.
Table 1. Fruit characteristics and colorimetric coordinates of two apple ‘Arisoo’ and ‘Summer Prince’ cultivars
Measurements of Fruit Color and Fruit Characteristics
Fruit firmness and color measurements were carried out at three random positions on each side (exposed and shaded side) of fruits. Fruit flesh firmness was determined by penetration at three random positions on two opposite sides of each fruit after removal of the skin (1 mm thick, 1 cm2) using a digital fruit firmness tester (TR Turoni, Italy) with a Ø8 mm plunger. The same fruit was cut in two equatorial parts and used to measure the SSC, expressed as oBrix, with a pocket refractometer (PAL-1, Atago, Japan) as previously described (Minas et al., Citation2016). The colors of the apple skin were measured using a chromameter (CR-400, Konica Minolta, Japan) based on the Hunter L, a, b system. The data are described as L (lightness), a (red-green sensation), b (yellow-blue sensation). The chroma (C) is defined according to the following mathematical function: . The hue angle (h) is defined according to the following mathematical function:
. Color index:
(Goulas et al., Citation2015; Rolle and Guidoni, Citation2007).
RNA Extraction and cDNA Synthesis
Total RNA was isolated from the skin and flesh tissues of ‘Summer Prince’ and ‘Arisoo’ apple cultivars using the CTAB method (Ouyanga et al., Citation2014). To remove genomic DNA contamination, all the RNA samples were treated with DNase (TURBO DNA-free™ Kit, Cat. #AM1907, Invitrogen, Carlsbad, CA, USA). The concentration and quality of RNA were detected by UV spectrophotometry and by electrophoresis on a 1.2% agarose gel, which was visualized by ethidium bromide staining. First-strand cDNA synthesis was carried out using oligo dT according to the manufacturer’s instructions (PrimeScript™ 1st strand cDNA Synthesis Kit, Cat. #6110A, Takara, Japan).
Quantification of Gene Expression
Expression analysis was performed using real-time quantitative reverse transcription-PCR (RT-qPCR) on LightCycler 480 II Real-Time PCR System (Roche Diagnostics, Mannheim, Germany). The LightCycler 480 SYBR Green I Master Mix (Roche) was used as described by the manufacturer. The qPCR conditions were as follows: 95°C for 5 min, followed by 40 cycles at 95°C for 10s, 60°C for 15s, and 72°C for 12s, melting curve analysis was performed at 65–95°C. The qPCR efficiency for each gene was obtained by analyzing the standard curve of a serially diluted cDNA of that gene. An apple SGF 29 tudor-like domain-containing protein (MDP0000336547) gene was selected as a reference gene for normalizing the expression (Zhou et al., Citation2017). The raw data were analyzed with Light Cycler® 480 Software release (version 1.5.1, Roche). Specific primers were designed for the region that could easily distinguish genes from one another using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) (Thornton and Basu, Citation2011). The sequences of primers for the 11 candidate genes are listed in . All the assays were carried out in seven technical replicates with those for template-free negative controls performed in parallel.
Table 2. Sequences of primers used in qRT-PCR
Data Analysis
Experimental data of fruit characteristics and colorimetric coordinates for the two apple cultivars, ‘Arisoo’ and ‘Summer Prince’ are the average of three independent biological replicates. Experimental data for the expression levels of anthocyanin-related genes are the average of values for seven independent biological replicates ± S.E.
The Tukey’s test was used to analyze the significance of the differences between three different treatment groups, and the differences in the same treatment group but two different sides (shaded side and exposed side) of the same cultivars using the t-test.
Pearson’s correlation coefficients were determined to obtain an overview of the correlation between the coloration levels and expression levels of the anthocyanin-related genes using the Pearson correlation analysis. The values of p < .05 and <0.01 were considered statistically significant.
Results
Different Coloration Patterns of Apple Fruit
The coloration patterns of apple fruit in the three groups are easily distinguishable (). The fruits in the bagged group were green (; middle panel), those in the control group (; right panel) had red color on the exposed side and green on the opposite side (shaded side) (), and those in the reflected group were completely red (; left panel). The shaded side of fruits in the reflected group, after exposure to reflected sunlight, quickly turned redder and the coloration level was almost similar to that in the exposed part after 7 days of treatment.
The red-coloration level was indicated to be negatively correlated with hue angle values and positively correlated with the chroma value and color index (CI). This correlation is also clear from the data presented in ; the CI for fruits in the bagged group was significantly lower than in other groups. The CI for ‘Summer Prince’, in the bagged group, was not significantly different between the two sides of fruits and was also not different from the CI for the shaded side of fruits in the control group. Significant differences between the exposed and shaded side of fruits were observed in the control group. In the reflected group, CI was not significantly different between the two sides; however, the values were higher than that for the exposed side of fruits in the control group. The coloration pattern of the ‘Arioso’ cultivar, was similar to that of ‘Summer Prince’. However, in the bagged group, the CI for the exposed side was higher than that for the shaded side. The CI for the exposed side of fruit in the control group was also higher than that in the reflected group.
Table 3. Coloration patterns of two apple cultivars with the different light treatments
Expression Profiles of the Anthocyanin Synthesis-Related Structural Genes
The transcript levels of seven synthesis-related structural genes (PAL, CHS, CHI, F3H, DFR, ANS, and UFGluT) in the skin and flesh of apple fruits from the three groups were investigated by qRT-PCR (). In general, in both the apple cultivars, the expression levels of these genes in the three groups of fruits were significantly different. The variation in the expression level of these genes showed a tendency to increase in the following order: bagged group <control group <reflected group. However, the expression of CHS, F3H, ANS, and UFGluT in the flesh of ‘Arisoo’ did not show much variation. Additionally, the expression level in the skin was always higher than in the flesh in both the cultivars.
Figure 2. Expression profiles of anthocyanin synthesis-related structural genes PAL, CHS, CHI, F3H, DFR, ANS, and UFGluT in the skin and flesh of three apple fruit groups: reflected, control, and bagged on the exposed side and shaded side of the apple ‘Arisoo’ (left panel) and ‘Summer Prince’ (right panel). All qRT-PCR reactions were normalized to an apple SGF 29 tudor-like domain-containing protein (MDP0000336547) gene. Data are means ± S.E. of seven replications
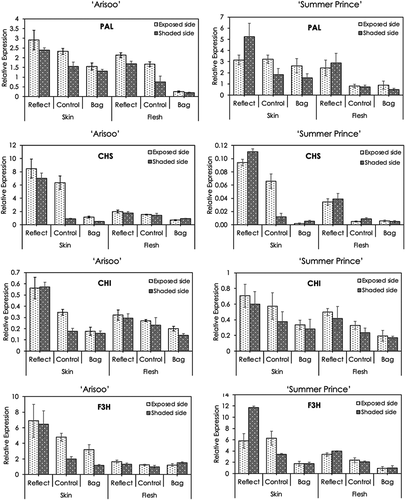
Comparison of the gene expression levels between the exposed and the shaded sides of fruits in the bagged group showed that the expression of anthocyanin synthesis-related structural genes was not altered in the skin and flesh of two cultivars. In the control group, the data showed significant differences in the expression levels between the two sides of the fruit. The exposed side showed a higher expression level than the shaded side, and the expression in the shaded side in the control group was similar to that in the bagged group. Especially, the shaded side of apple fruits after exposure to reflected sunlight (shaded side, reflect group) showed higher expression than the shaded side of the control group (without treatment) and their expression reached the same level as in the exposed side in the control group. Some of these genes, such as PAL, CHS, F3H, DFR, and UFGluT, in the shaded side of the reflected group even showed higher expression levels than the exposed side of ‘Summer Prince’ in the control group. In a comparison of the gene expression level in the exposed side between the reflected and the control groups, we also recognized that the expression of some (CHS, CHI, ANS, and UFGluT) among the seven genes in the reflected group was higher than in the control group.
Additionally, in the comparison of gene expression between ‘Arisoo’ (, left panel) and ‘Summer Prince’ (, right panel), we found that all the seven genes showed similar expression patterns. However, by comparing the expression levels of the same genes, we found that the levels of these genes between ‘Arisoo’ and ‘Summer Prince’ were also significantly different. The expression level of CHS in ‘Arisoo’ (, left panel) was about 10-fold higher than in ‘Summer Prince’ (, right panel). Moreover, the expression levels of DFR, ANS, and UFGluT in ‘Arisoo’ were lower than in ‘Summer Prince’ about 10-, 3-, and 300-fold, respectively.
Expression Profiles of Anthocyanin Synthesis-Related Transcription Factors
The transcript levels of two synthesis-related transcription factors genes (MYB10 and MYB110a) in the skin and flesh of apple fruits in the three groups were examined by qRT-PCR (). In general, in both the cultivars, the expression levels of MYB10 and MYB110a in the three groups were significantly different, especially in the skin. The expression level of these genes was the highest in the reflected group and lowest in the bagged group. However, the expression of MYB10 in the flesh of ‘Arisoo’ did not show much significant difference.
Figure 3. Expression profiles of anthocyanin synthesis-related transcription factors MYB1 and MYB110a in the skin and flesh of three apple fruit groups: reflected, control, and bagged on the exposed side and shaded side of the apple ‘Arisoo’ (left panel) and ‘Summer Prince’ (right panel). All qRT-PCR reactions were normalized to an apple SGF 29 tudor-like domain-containing protein (MDP0000336547) gene. Data are means ± S.E. of seven replications
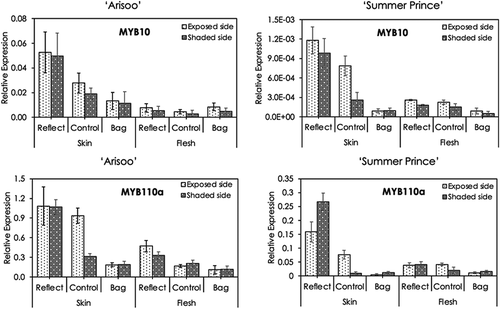
Based on the data analysis of the transcript levels of the two regulatory genes, we recognized that there was a consistent pattern of variation in their expression with respect to the expression of most of the synthesis-related structural genes (). The results of the comparison of expression of the two regulatory genes between the exposed and shaded sides in the three groups showed there was not much significant difference in the bagged group. However, there was a significant difference in the expression in both the sides of the control group fruits, with the levels in the exposed side being higher than the respective levels in the shaded side. Following the consistent pattern for most of the synthesis-related structural genes, the expression levels in the shaded side of fruits in the reflected group were much higher than in the shaded side of fruits in the control group. The expression of MYB110a in ‘Summer Prince’ was boosted and reached the highest in the shaded side in the reflected group relative to that of the shaded side of fruits in the control group; it was also higher than in the exposed side of fruits in the same group.
A comparison of the expression levels of the same genes, we found that the levels of anthocyanin biosynthesis genes in ‘Arisoo’ and ‘Summer Prince’ were also significantly different. Considering the shaded side of the control group as standard, the expression level of both the regulatory genes, MYB10 and MYB110a, in ‘Arisoo’ was 115- and 16-fold higher than in ‘Summer Prince’, respectively.
Expression Profiles of Light-Responsive Genes
The transcript levels of the light-responsive genes in the skin and flesh of fruits in the three groups were investigated by qRT-PCR (). The expression pattern of the PIF3 gene in the skin was lower than that in the flesh in both the cultivars. In ‘Arisoo’, the expression pattern of the COP1 gene in the skin was lower than that in the flesh. However, in ‘Summer Prince, the expression pattern of COP1 was not much significant difference in the skin and flesh.
Figure 4. Expression profiles of anthocyanin synthesis-related light-responsive genes PIF3 and COP1 in the skin and flesh of three apple fruit groups: reflected, control, and bagged on the exposed side and shaded side of the apple ‘Arisoo’ (left panel) and ‘Summer Prince’ (right panel). All qRT-PCR reactions were normalized to an apple SGF 29 tudor-like domain-containing protein (MDP0000336547) gene. Data are means ± S.E. of seven replications
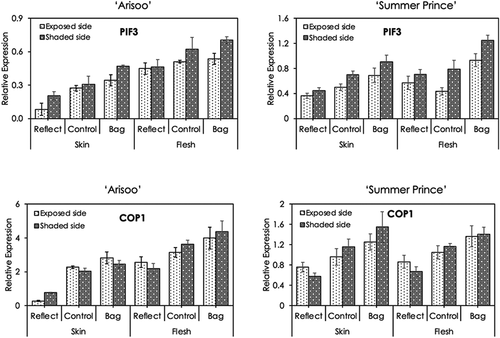
In contrast to the results for structural and regulatory genes, the expression of PIF3 and COP1 genes reached the highest levels in the bagged group and lowest levels in the reflected group. The expression levels of PIF3 in the shaded side were higher than in the exposed side. Especially, the expression level of PIF3 was significantly different between the exposed and the shaded sides in the skin of ‘Arisoo’ fruits in the reflected group. The expression levels of COP1 in the two side did not show much difference, except in the skin of ‘Arisoo’ fruits in the reflected group.
Correlation Analysis of the Expression Levels of Anthocyanin-Related Genes and Coloration Patterns
Pearson analysis was performed to determine the correlation coefficients between the coloration of fruits and the expression levels of anthocyanin-related genes (). In the ‘Arisoo” cultivar, the CI was significantly positively correlated with the expression levels of PAL, CHS, F3H, DFR, ANS, and MYB110a. The CI of fruits of ‘Summer Prince’ was significantly positively correlated with the expression levels of CHS, CHI, F3H, DFR, ANS, UFGluT, MYB10, and MYB110a. The expression of the light-responsive genes, COP1 and PIF3, in ‘Summer Prince’, showed a very strong negative correlation with the CI but not in ‘Arisoo’. However, in general, the correlation coefficients, indicating that had strong to the extremely strong correlation between the expression levels of anthocyanin-related genes and the CI, excepted for COP1 in ‘Arisoo’ for which the correlation was intermediate.
Table 4. Correlation analysis of anthocyanin-related gene expression levels and coloration patterns
Discussion
Sunlight Stimulates Coloration in Apple Fruit
In this study, the different treatments of sunlight resulted in different coloration patterns in apple fruit (). In the dark condition, the fruits were red-colorless (bagged group). In the same fruit, the side of fruits obscured by tree canopy (shaded side) showed less color than the opposite side, which was directly exposed to sunlight (exposed side). Upon enhanced sunlight irradiation, the shaded side of fruits quickly changed to red. Liu et al. (Citation2013) found that in the bagged non-red apple cultivars (‘Granny Smith’ and ‘Golden Delicious’), after 15 days of bag removal, the skin of exposed fruit rapidly turned red, with the accumulation of anthocyanins. The effect of sunlight on the expression levels of anthocyanin synthesis genes was investigated at the juvenile stage of non-red ‘Mutsu’ apple fruit. Anthocyanin was produced in the exposed side but not in the shaded side (Kondo et al., Citation2002b).
In ‘Summer Prince’, the CI in the reflected group was higher than that of the exposed side in the control group. This may be since fruits in the reflected group received more irradiation, which was reflected from the aluminum-wrapped disc. Similarly, previous reports have shown a linear relationship between anthocyanin accumulation and light intensity (Lancaster, Citation1992; Saure, Citation1990).
Sunlight Stimulates the Coloration of Apple Fruit by Enhancing the Expression of Anthocyanin Synthesis-Related Structural Genes
The level of anthocyanin pigment depends on the expression level of the genes involved in their synthesis-related genes pathway. In this study, we analyzed the expression of seven synthesis-related structural genes and compared their expression levels. The data showed significant differences in the expression levels in the three groups and also between the two sides of the fruit. The shaded side of fruits after exposure to reflected sunlight showed an expression level higher than in the shaded side of the fruit in the control group. Kondo et al. (Citation2002a) found that CHS, ANS, and UFGluT in the non-bagged fruits were higher than in the skin of bagged fruits of ‘Tsugaru’ at 98 DAFB. This suggests the fruits receiving more irradiation showed higher expression of genes
The expression level of CHS, CHI, ANS, and UFGluT in the exposed side of the reflected group was higher than in the control group. This can be explained by the fact that the exposed side of fruit in the reflected group, besides receiving direct light, also received additional from reflected light. Our data show that the lower expression levels of DFR, ANS, and UFGluT in ‘Arisoo’ than ‘Summer Prince’ could be the reason for lower coloration values in ‘Arisoo’ (). This shows the role of genetic factors in the differential expression of these genes.
Among the seven genes selected for expression analysis, CHS, ANS, and UFGluT showed greater fluctuation in the expression levels in the three groups for both the apple cultivars. Similar results were obtained in other studies on apple (Kondo et al., Citation2002b; Liu et al., Citation2013; Zhang et al., Citation2013) and pears (Yang et al., Citation2015, Citation2013; Zhang et al., Citation2011). This shows that CHS, ANS, and UFGluT are candidate marker genes that should be chosen for analysis of differences in the coloration of fruits.
Sunlight Stimulates the Coloration of Apple Fruit by Modulating the Expression of Transcription Factors
In our study, two transcription factors, MYB10 and MYB110a, were found to be involved in the anthocyanin biosynthesis in both the apple cultivars. The transcription of MYB10 and MYB110a genes was lowest in the bagged group, increased in the control group, and was the highest in the reflected group of fruits exposed to sunlight (). Many studies have confirmed that transcription factor genes can positively regulate anthocyanin biosynthesis, and light is required for their function in anthocyanin accumulation (Cominelli et al., Citation2008; Feng et al., Citation2013; Takos et al., Citation2006; Yu et al., Citation2019). It was reported that UV-B irradiation enhanced the anthocyanin accumulation by upregulated MYBA in the fruit skin of ‘Tsugaru’ cultivar (Ban et al., Citation2007). Sato et al. (Citation2019) determined the correlation between anthocyanin accumulation and MYB110a expression in ‘Nakano Shinku’ apple; the expression level of MYB110a in the red part was significantly higher than in the white part. When dark-grown fruit was exposed to sunlight, the MYB1 transcript levels increased over several days, correlating with anthocyanin synthesis in the skin (Takos et al., Citation2006). In another study, anthocyanin content and the transcript levels of MYB1 and four late genes in the anthocyanin pathway were dramatically elevated in the re-exposed fruits compared to that in the fruits of the control group (Zhang et al., Citation2013). Taken together, sunlight is one of the factors that trigger the expression of transcription factors, thereby, enhancing the accumulation of anthocyanin.
44The difference in the expression of different MYBs are specific and depend on the cultivar, tissue type, tissue age, and interaction with bHLH and WD40 domains that are regulated by MYB–bHLH–WD40 complex (Espley et al., Citation2007). In the expression analysis of MYBA in the different tissues (skin and flesh) of the three apple cultivars, expression was only detected in the skin of ‘Jonathan’ and ‘Maypole’ but not in the ‘Niedzwetzkyana’. The expression level of MYBA is positively correlated with the accumulation of anthocyanins in apple skin after a certain age but not in young fruit skin (Ban et al., Citation2007). Moreover, our data showed that MYB10 and MYB110a were expressed in both the apple cultivars, with the expression levels in the skin being much higher than in the flesh. Similar results were also found in the cultivar Cripps’ Red; the transcript level of MYB1 in the skin was up-regulated, whereas it was in the flesh was down-regulated or even showed no expression in the flesh (Takos et al., Citation2006).
Sunlight Stimulates Coloration of Apple Fruit by Down-Regulating the Expression of Light-Responsive Genes
Our results show that the expression level of PIF3 and COP1 genes in the bagged group (darkness condition) was up-regulated and reached the highest level. Moreover, their expression decreased in the control and reflected groups. This can be explained, based on the fact that when the fruits were exposed to the sunlight, PIF3 was degraded and COP1 was inhibited by photomorphogenesis. The expression pattern of PIF3 in the skin was lower than in the flesh in both the cultivars (), probably because the flesh of fruits absorb less sunlight than the skin.
Similar phenomena were also found in other studies. It has been reported that PIF3 degrades rapidly after irradiation of dark-grown seedlings in a process controlled by phytochromes (Bauer et al., Citation2004). Transgenic pears overexpressing of PbCOP1.1 showed decreased expression levels of anthocyanin-related synthetic genes and negatively regulated the coloration of fruits (Wu et al., Citation2019). Through ubiquitination, COP1 degrades HY5, a promoter of anthocyanin biosynthesis, and thereby, reduces the accumulation of anthocyanin in apple (An et al., Citation2017). The E3 ubiquitin ligase COP1/SPA, together with the light control of the protein stability of the MYB transcription factors, PAP1 and PAP2, was reported to be involved in anthocyanin accumulation in Arabidopsis (Maier et al., Citation2013). COP1 degrades the MYB protein, which inhibits fruit coloration was observed in apples (Li et al., Citation2012).
Conclusion
To investigate the role of sunlight in the different coloration patterns of apple fruit, we analyzed the differential expression of anthocyanin synthesis-related genes and identified the key genes involved in controlling the development of coloration in the skin and flesh of the fruit of two red-skinned apple cultivars, ‘Arisoo’ and ‘Summer Prince’. These findings will help in improving our understanding of the molecular mechanisms underlying anthocyanin accumulation in apple, and the important role of light, an environmental stimulus, in regulating the expression of genes involved in anthocyanin accumulation in apple fruit at the early stages of fruit development. Based on a better understanding of the genetic regulation of anthocyanin biosynthesis and the effect of environmental stimuli, we can intervene in the anthocyanin synthesis pathway to boost the anthocyanin content of fruits.
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Research involving human participants and/or animals
This article does not contain any studies with human participants performed by any of the authors. Informed consent Informed consent was obtained from all individual participants included in the study.
Additional information
Funding
Literature cited
- Allan, A.C., R.P. Hellens, and W.A. Laing. 2008. MYB transcription factors that color our fruit. Trends Plant Sci. 13:99–102. doi: https://doi.org/10.1016/j.tplants.2007.11.012.
- An, J.P., F.J. Qu, J.F. Yao, X.N. Wang, C.X. You, X.F. Wang, and Y.J. Hao. 2017. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4:17023. doi: https://doi.org/10.1038/hortres.2017.23.
- Ban, Y., C. Honda, Y. Hatsuyama, M. Igarashi, H. Bessho, and T. Moriguchi. 2007. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48:958–970. doi: https://doi.org/10.1093/pcp/pcm066.
- Bauer, D., A. Viczián, S. Kircher, T. Nobis, R. Nitschke, T. Kunkel, K.C. Panigrahi, E. Adám, E. Fejes, E. Schäfer, et al. 2004. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 16:1433–1445. doi: https://doi.org/10.1105/tpc.021568.
- Cominelli, E., G. Gusmaroli, D. Allegra, M. Galbiati, H.K. Wade, G.I. Jenkins, and C. Tonelli. 2008. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 165:886–894. doi: https://doi.org/10.1016/j.jplph.2007.06.010.
- Espley, R.V., R.P. Hellens, J. Putterill, D.E. Stevenson, S. Kutty-Amma, and A.C. Allan. 2007. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49:414–427. doi: https://doi.org/10.1111/j.1365-313X.2006.02964.x.
- Feng, F., M. Li, F. Ma, and L. Cheng. 2013. Phenylpropanoid metabolites and expression of key genes involved in anthocyanin biosynthesis in the shaded peel of apple fruit in response to sun exposure. Plant Physiol. Biochem. 69:54–61. doi: https://doi.org/10.1016/j.plaphy.2013.04.020.
- Goulas, V., I.S. Minas, P.M. Kourdoulas, A. Lazaridou, A.N. Molassiotis, I.P. Gerothanassis, and G.A. Manganaris. 2015. (1)H NMR metabolic fingerprinting to probe temporal postharvest changes on qualitative attributes and phytochemical profile of sweet cherry fruit. Front Plant Sci. 6:959. doi: https://doi.org/10.3389/fpls.2015.00959.
- Honda, C., N. Kotoda, M. Wada, S. Kondo, S. Kobayashi, J. Soejima, Z. Zhang, T. Tsuda, and T. Moriguchi. 2002. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol. Biochem. 40:955–962. doi: https://doi.org/10.1016/S0981-9428(02)01454-7.
- Ju, Z., Y. Yuan, C. Liu, Y. Wang, and X. Tian. 1997. Dihydroflavonol reductase activity and anthocyanin accumulation in ‘Delicious’, ‘Golden Delicious’ and ‘Indo’ apples. Sci. Hortic. 70:31–43. doi: https://doi.org/10.1016/S0304-4238(97)00040-X.
- Kawabata, S., Y. Kusuhara, Y. Li, and R. Sakiyama. 1999. The regulation of anthocyanin biosynthesis in Eustoma grandiflorum under low light conditions. J. Jpn. Soc. Hortic. Sci. 68:519–526. doi: https://doi.org/10.2503/jjshs.68.519.
- Kim, S. H., J. R. Lee, S. T. Hong, Y. K. Yoo, G. An and S. R. Kim. 2003. Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci 165:403–13. doi:https://doi.org/10.1016/S0168-9452(03)00201-2.
- Koes, R., W. Verweij, and F. Quattrocchio. 2005. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10:236–242. doi: https://doi.org/10.1016/j.tplants.2005.03.002.
- Kondo, K., K. Hiraoka, S. Kobayashi, C. Honda, and N. Terahara. 2002a. Changes in the expression of anthocyanin biosynthetic genes during apple development. J. Am. Soc. Hortic. Sci. 127:971–976. doi: https://doi.org/10.21273/JASHS.127.6.971.
- Kondo, K., M. Maeda, S. Kobayashi, and C. Honda. 2002b. Expression of anthocyanin biosynthetic genes in Malus sylvestris L. ‘Mutsu’ non-red apples. J. Hortic. Sci. Biotech. 77:718–723. doi: https://doi.org/10.1080/14620316.2002.11511562.
- Kui, L.W., B. Karen, G. Karryn, K. Anne, K. Sakuntala, K.M. Tony, V.E. Richard, P.H. Roger, and C.A. Andrew. 2010. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50. doi: https://doi.org/10.1186/1471-2229-10-50.
- Kui, L.W., M. Diego, P. John, V. Richard, L. Lidia, E. Richard, P.H. Roger, C. David, D.R. Daryl, T. Michela, et al. 2011. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 34:1176–1190. doi: https://doi.org/10.1111/j.1365-3040.2011.02316.x.
- Lancaster, J.E. 1992. Regulation of skin colour in apples: A review. Crit. Rev. Plant Sci. 10:487–502. doi: https://doi.org/10.1080/07352689209382324.
- Lancaster, J.E., J.E. Grant, C.E. Lister, and M.C. Taylor. 1994. Skin color in apples-influence of copigmentation and plastid pigments on shade and darkness of red color in five genotypes. J. Am. Soc. Hortic. Sci. 119:63–66. doi: https://doi.org/10.21273/JASHS.119.1.63.
- Li, Y.Y., K. Mao, C. Zhao, X.Y. Zhao, H.L. Zhang, H.R. Shu, and Y.J. Hao. 2012. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160:1011–1022. doi: https://doi.org/10.1104/pp.112.199703.
- Liu, Y., F. Che, L. Wang, R. Meng, X. Zhang, and Z. Zhao. 2013. Fruit coloration and anthocyanin biosynthesis after bag removal in non-red and red apples (Malus × domestica Borkh.). Molecules. 18:1549–1563. doi: https://doi.org/10.3390/molecules18021549.
- Liu, Z., Y. Zhang, J. Wang, P. Li, C. Zhao, Y. Chen, and Y. Bi. 2015. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 238:64–72. doi: https://doi.org/10.1016/j.plantsci.2015.06.001.
- Maier, A., A. Schrader, L. Kokkelink, C. Falke, B. Welter, E. Iniesto, V. Rubio, J.F. Uhrig, M. Hülskamp, and U. Hoecker. 2013. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74:638–651. doi: https://doi.org/10.1111/tpj.12153.
- Marais, E., G. Jacobs, and D.M. Holcroft. 2001. Colour response of ‘Cripps’ Pink’ apples to postharvest irradiation is influenced by maturity and temperature. Sci Hortic. 90:31–41. doi: https://doi.org/10.1016/S0304-4238(00)00256-9.
- Minas, I.S., G. Tanou, E. Karagiannis, M. Belghazi, and A. Molassiotis. 2016. Coupling of physiological and proteomic analysis to understand the ethylene- and chilling-induced kiwifruit ripening syndrome. Front Plant Sci. 7:120. doi: https://doi.org/10.3389/fpls.2016.00120.
- Ouyanga, K., J. Li, H. Huang, Q. Que, P. Li, and X. Chen. 2014. A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. Biotechnol. Biotechnol. Equip. 28:1008–1013. doi: https://doi.org/10.1080/13102818.2014.981086.
- Rachel, G., F. Sharon, and P. Avihai. 2001. Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Sci. 161:579–588. doi: https://doi.org/10.1016/S0168-9452(01)00445-9.
- Rahim, M.A., N. Busatto, and L. Trainotti. 2014. Regulation of anthocyanin biosynthesis in peach fruits. Planta. 240:913–929. doi: https://doi.org/10.1007/s00425-014-2078-2.
- Reay, P.F., and J.E. Lancaster. 2001. Accumulation of anthocyanins and quercetin glycosides in ‘Gala’ and ‘Royal Gala’ apple fruit skin with UV-B–visible irradiation: Modifying effects of fruit maturity, fruit side, and temperature. Sci Hortic. 90:57–68. doi: https://doi.org/10.1016/S0304-4238(00)00247-8.
- Rolle, L., and S. Guidoni. 2007. Color and anthocyanin evaluation of red winegrapes by CIE L*, a*, b* parameters. OENO One. 41:193–201. doi: https://doi.org/10.20870/oeno-one.2007.41.4.838.
- Sato, H., S. Otagaki, Y. Ono, K. Shiratake, and S. Matsumoto. 2019. Upregulation of MdMYB110a is responsible for ABA-mediated colouration of type 2 red-fleshed apples. J. Hortic. Sci. Biotech. 94:33–40. doi: https://doi.org/10.1080/14620316.2018.1452638.
- Saure, M.C. 1990. External control of anthocyanin formation in apple. Sci Hortic. 42:181–218. doi: https://doi.org/10.1016/0304-4238(90)90082-P.
- Shin, J., E. Park, and G. Choi. 2007. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49:981–994. doi: https://doi.org/10.1111/j.1365-313X.2006.03021.x.
- Steyn, W.J., S.J. Wand, G. Jacobs, R.C. Rosecrance, and S.C. Roberts. 2009. Evidence for a photoprotective function of low‐temperature‐induced anthocyanin accumulation in apple and pear peel. Physiol. Plant. 136:461–472. doi: https://doi.org/10.1111/j.1399-3054.2009.01246.x.
- Takos, A.M., F.W. Jaffé, S.R. Jacob, J. Bogs, S.P. Robinson, and A.R. Walker. 2006. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142:1216–1232. doi: https://doi.org/10.1104/pp.106.088104.
- Thornton, B., and C. Basu. 2011. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 39:145–154. doi: https://doi.org/10.1002/bmb.20461.
- Ubi, B.E., C. Honda, H. Bessho, S. Kondo, M. Wada, S. Kobayashi, and T. Moriguchi. 2006. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV‐B and temperature. Plant Sci. 170:571–578. doi: https://doi.org/10.1016/j.plantsci.2005.10.009.
- Vimolmangkang, S., D. Zheng, Y. Han, M.A. Khan, R.E. Soria-Guerra, and S.S. Korban. 2014. Transcriptome analysis of the exocarp of apple fruit identifies light-induced genes involved in red color pigmentation. Gene. 534:78–87. doi: https://doi.org/10.1016/j.gene.2013.10.007.
- Wang, X., Z. Wei, and F. Ma. 2015. The effects of fruit bagging on levels of phenolic compounds and expression by anthocyanin biosynthetic and regulatory genes in red-fleshed apples. Process Biochem. 50:1774–1782. doi: https://doi.org/10.1016/j.procbio.2015.06.024.
- Whale, S.K., and Z. Singh. 2007. Endogenous ethylene and color development in the skin of ‘Pink Lady’ apple. J. Am. Soc. Hortic. Sci. 132:20–28. doi: https://doi.org/10.21273/JASHS.132.1.20.
- Wu, M., M. Si, X. Li, L. Song, J. Liu, R. Zhai, L. Cong, R. Yue, C. Yang, F. Ma, et al. 2019. PbCOP1.1 contributes to the negative regulation of anthocyanin biosynthesis in pear. Plants. 8:39. doi: https://doi.org/10.3390/plants8020039.
- Yang, Y.N., G. Zhao, W.Q. Yue, S.L. Zhang, C. Gu, and J. Wu. 2013. Molecular cloning and gene expression differences of the anthocyanin biosynthesis-related genes in the red/green skin color mutant of pear (Pyrus communis L.). Tree Genet. Genomes. 9:1351–1360. doi: https://doi.org/10.1007/s11295-013-0644-6.
- Yang, Y.N., G.F. Yao, D. Zheng, S.L. Zhang, C. Wang, M.Y. Zhang, and J. Wu. 2015. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 34:189–198. doi: https://doi.org/10.1007/s00299-014-1698-0.
- Yu, M., Y. Man, and Y. Wang. 2019. Light- and temperature-induced expression of an R2R3-MYB gene regulates anthocyanin biosynthesis in red-fleshed kiwifruit. Int. J. Mol. Sci. 20:e5228. doi: https://doi.org/10.3390/ijms20205228.
- Zhang, X., A.C. Allan, Q. Yi, L. Chen, K. Li, Q. Shu, and J. Su. 2011. Differential gene expression analysis of Yunnan red pear, Pyrus pyrifolia, during fruit skin coloration. Plant Mol. Biol. Rep. 29:305–314. doi: https://doi.org/10.1016/j.sajb.2013.07.009.
- Zhang, X.J., L.X. Wang, Y.L. Liu, X.X. Chen, Y.Z. Yang, and Z.Y. Zhao. 2013. Differential gene expression analysis of ‘Granny Smith’ apple (Malus domestica Borkh.) during fruit skin coloration. S. Afr. J. Bot. 88:125–131. doi: https://doi.org/10.1007/s11105-010-0231-z.
- Zhou, Z., P. Cong, Y. Tian, and Y. Zhu. 2017. Using RNA-seq data to select reference genes for normalizing gene expression in apple roots. PLoS One. 12:e0185288. doi: https://doi.org/10.1371/journal.pone.0185288.

