Abstract
Autophagy plays critical roles in plant responses to stress. In contrast to the wealth of information concerning the core process of plant autophagosome assembly, our understanding of the regulation of autophagy is limited. In this study, we demonstrated that transcription factor HsfA1a played a critical role in tomato tolerance to drought stress, in part through its positive role in induction of autophagy under drought stress. HsfA1a expression was induced by drought stress. Virus-induced HsfA1a gene silencing reduced while its overexpression increased plant drought tolerance based on both symptoms and membrane integrity. HsfA1a-silenced plants were more sensitive to endogenous ABA-mediated stomatal closure, while its overexpression lines were resistant under drought stress, indicating that phytohormone ABA did not play a major role in HsfA1a-induced drought tolerance. On the other hand, HsfA1a-silenced plants increased while its overexpression decreased the levels of insoluble proteins which were highly ubiquitinated under drought stress. Furthermore, drought stress induced numerous ATGs expression and autophagosome formation in wild-type plants. The expression of ATG10 and ATG18f, and the formation of autophagosomes were compromised in HsfA1a-silenced plants but were enhanced in HsfA1a-overexpressing plants. Both electrophoretic mobility shift assay and chromatin immunoprecipitation coupled with qPCR analysis revealed that HsfA1a bound to ATG10 and ATG18f gene promoters. Silencing of ATG10 and ATG18f reduced HsfA1a-induced drought tolerance and autophagosome formation in plants overexpressing HsfA1a. These results demonstrate that HsfA1a induces drought tolerance by activating ATG genes and inducing autophagy, which may promote plant survival by degrading ubiquitinated protein aggregates under drought stress.
Abbreviations
| ABA | = | abscisic acid |
| ATG | = | autophagy-related |
| CDS | = | coding DNA sequence |
| ChIP | = | chromatin immunoprecipitation |
| EL | = | electrolyte leakage |
| EMSA | = | electrophoretic mobility shift assay |
| Hsf | = | heat-shock transcription factor |
| HSE | = | heat-shock element |
| Hsp | = | heat-shock protein |
| MDC | = | monodansylcadaverine |
| OE | = | overexpressing |
| PE | = | phosphatidylethanolamine |
| PTM | = | protein post-translational modification |
| RWC | = | relative water content |
| TEM | = | transmission electron microscopy |
| TF | = | transcription factor |
| TOR | = | target of rapamycin |
| TRV | = | tobacco rattle virus |
| Ub | = | ubiquitin |
| UPS | = | ubiquitin-26S proteasome system |
| VIGS | = | virus-induced gene silencing |
| WT | = | wild-type |
Introduction
Plants are often exposed to abiotic and biotic stresses (i.e. extreme temperatures, salt, drought, heavy metals, fungi and bacteria) that adversely affect their physiological traits, metabolic pathways, and protein activity. Repair or degradation of misfolded and damaged proteins, which are induced by stresses and are highly toxic to cell, relies on an elaborately regulated protein quality control system.Citation1 Molecular chaperones such as heat-shock proteins (Hsps) can promote folding and refolding of denatured proteins and the protein degradation systems such as autophagy and the ubiquitin-26S proteasome system (UPS) to remove damaged and misfolded proteins.Citation2,3 These processes are regulated by complex signaling networks including signaling molecules, protein kinases and transcription factors (TFs).Citation4-6
Autophagy is an evolutionarily conserved protein degradation process in eukaryotes. The process is involved in the degradation of unnecessary or dysfunctional cellular components under environmental stress conditions or during certain stages of development.Citation7 During autophagy, cells create double-membrane-bound compartments called phagophores that engulf cytoplasmic components; the phagophore expands and seals to form a double-membrane autophagosome. Then, the outer membrane of the autophagosome fuses with the tonoplast to release the internal vesicle as an autophagic body that will be recycled within the vacuole.Citation8 More than 30 autophagy-related (ATG) genes, which encode core autophagic machineries and are the main genetic regulators of the autophagy process have been identified in plants over the last 20 years.Citation9,10
Although the critical roles of plant autophagy have been well established in a wide spectrum of biological processes, including stress responses, our understanding of the regulation and action mechanisms of plant autophagy is extremely limited. Most research on the mechanisms underlying autophagy regulation has traditionally focused on the TOR (target of rapamycin) kinase.Citation11 Silencing of the TOR gene in Arabidopsis leads to the constitutive formation of autophagosomes, indicating that TOR is a negative regulator of autophagy in plants.Citation12 Furthermore, a NADPH oxidase inhibitor blocks autophagy induction upon nutrient starvation and salt stress but not osmotic stress.Citation13 Thus, ROS may mediate the induction of autophagy during some, but not all, stress conditions. We recently reported that silencing of the tomato WRKY33 transcription factor genes compromise tomato heat tolerance and reduce heat-induced ATG gene expression and autophagosome accumulation, suggesting the possible involvement of tomato WRKY33 in ATG induction.Citation14 However, little information is available concerning the transcriptional regulation of plant autophagy genes under other stress conditions.
Heat-shock transcription factors (Hsfs) are a family of TFs found in all organisms that function as regulators of the genes encoding molecular chaperones and other stress proteins and enable survival following exposure to acute stress.Citation15 Plants possess a uniquely complex Hsf family, with 21 members defined in Arabidopsis thaliana, 25 members in rice (Oryza sativa) and 24 members in tomato (Solanum lycopersicum).Citation16,17 These Hsfs are classified into 3 conserved evolutionary categories (A, B and C) according to the structural features of their oligomerization domains.Citation18 Class A Hsfs contain the transcriptional activation domain that is characterized by aromatic, large hydrophobic, and acidic amino acid residues. Class B and C Hsfs lack this particular amino acid motif and cannot induce transcriptional activation alone.Citation18,19 There are 4 HsfA1 members in the class A1 group (HsfA1a, HsfA1b, HsfA1c and HsfA1e).Citation20,21 HsfA1a is a master regulator that functions both as an activator and as a repressor of its target genes under heat-shock stress.Citation22,23 Exposure to heat stress leads to oligomerization of HsfA1a into active trimers, followed by its translocation into the nucleus and binding to the heat-shock elements (HSEs) located within the promoter regions of target genes (i.e., HsfA2, Hsp70, and Hsp90).Citation24 HsfA1a can also interact with the HsfA2, Hsp70, and Hsp90 proteins to form a heterooligomeric complex that exhibits a versatile ability to regulate the plant stress response.Citation24,25 Transgenic Arabidopsis and tomato plants overexpressing HsfA1a proteins show constitutive heat-shock protein synthesis and exhibit enhanced basic thermotolerance.Citation26,27 In addition to heat stress, HsfA1a also mediates plant tolerance to salt, drought and oxidative stresses by inducing genes encoding Hsp proteins.Citation22,24 However, the functions of HsfA1a in the regulation of the expression of non-heat-shock responsive genes under stress conditions are less well understood.
To better understand the mechanistic basis of autophagy regulation in plant stress responses, we sought to identify and functionally analyze the involvement of HsfA1a in autophagy under drought stress. Gene overexpression and tobacco rattle virus (TRV)-induced gene silencing (VIGS) have been extensively used for functional gene analysis in plants including Arabidopsis,Citation28 Nicotiana speciesCitation29 and tomato,Citation30 despite their potential complications associated with high levels of transgene products and viral infection, which can be largely overcome by using appropriate controls. We showed that HsfA1a, a transcription factor with a broad role in plant abiotic stress responses, was a positive regulator of drought-induced autophagy. Using an electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation coupled with qPCR (ChIP-qPCR) analysis, we revealed that HsfA1a directly bound to the promoters of 2 autophagy genes (ATG10 and ATG18f). Silencing these 2 autophagy genes compromised HsfA1a-mediated plant drought tolerance and autophagosome production. This is the first study to demonstrate that the multifunctional transcription factor HsfA1a is responsive to drought treatment and plays a positive role in induction of the ATG10 and ATG18f genes and the formation of autophagosomes under drought stress. HsfA1a acts as a positive regulator in autophagy-induced drought tolerance in the tomato plants.
Results
Induction of the HsfA1a gene and the phenotypes of plants with silenced or overexpressed HsfA1a under drought stress
To analyze the possible involvement of HsfA1a in plant dehydration tolerance, we first examined the expression pattern of HsfA1a in response to drought stress. As shown in , the transcript level of the HsfA1a gene remained largely constant throughout the 13-day experimental period under normal water supply conditions. However, when water was withheld, the HsfA1a transcript level was increased by as early as the 3rd day and remained elevated up to 13 d under dehydration stress (). Notably, the largest increase in the HsfA1a transcript level was observed after 6 d under drought stress, when the plants began to show symptoms of dehydration ().
Figure 1. Functional analysis of HsfA1a in response to drought stress in tomato leaves. (A) The expression of HsfA1a in the WT plants under drought stress. Six-wk-old tomato WT Ailsa Craig plants were exposed to dehydration by withholding water. Total RNA was isolated from leaf samples of the WT plants at the indicated times. (B and C) Reduced or increased tomato drought tolerance in TRV-HsfA1a or HsfA1aOE plants. Six-wk-old plants in soils were exposed to dehydration by withholding water for 13 d. Bars: 10 cm. (D) Relative water content (RWC) of the terminal leaflets were determined immediately after 13 d of control or drought treatment in TRV and TRV-HsfA1a plants or WT and HsfA1aOE plants. All data are presented as the means of 5 biological replicates (± SE). Means with the same letter did not significantly differ at P< 0.05 according to Duncan multiple range test. Three independent experiments were performed with similar results. 1# and 3#, 2 lines of HsfA1aOE plants; OE, overexpressing; WT, wild-type.
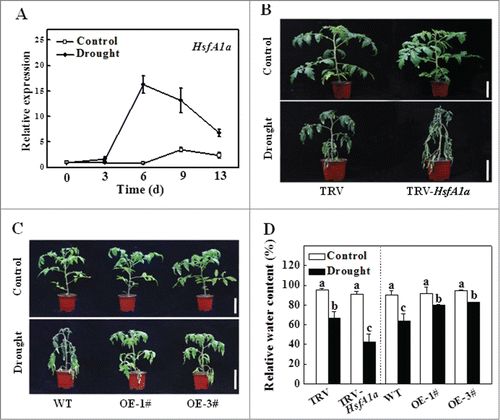
To directly analyze the role of HsfA1a in drought tolerance, we compared WT and F2 progeny of HsfA1a-overexpressing (HsfA1aOE) plants from 2 independent lines (1# and 3#) expressing high HsfA1a protein levels (Fig. S1). We also analyzed HsfA1a-silenced (TRV-HsfA1a) plants, which had only 30 to 40% of HsfA1a transcript levels of the TRV control plants (Fig. S2). The transcript levels of other 3 HsfA1 homologous genes (HsfA1b, HsfA1c and HsfA1e) were not changed in the TRV-HsfA1a plants when compared to those of the TRV control plants (Fig. S2). Only some margins of old leaves displayed symptoms of dehydration after withholding water for 13 d in the drought-treated TRV and WT plants, whereas a majority of the leaves remained green (). In contrast, a majority of the leaves from TRV-HsfA1a plants exhibited extensive wilting after 13 d under drought stress (). The relative water content (RWC) was similar in all the plants under normal water supply (). However, the RWC of TRV-HsfA1a plants was 36.4% less than that of TRV plants under drought stress (). The RWC of HsfA1aOE-1# and HsfA1aOE-3# was 24.7% and 29.4%, respectively, higher than that of WT plants after 13 d under drought stress (). Furthermore, the electrolyte leakage (EL) value of TRV-HsfA1a plants was similar to that of TRV plants when they were grown with a normal water supply. However, the EL value of TRV-HsfA1a plants was 58.7% higher than the TRV plants after 13 d of drought stress (Fig. S3). Drought tolerance was significantly increased in both lines of HsfA1aOE plants, and the EL values of HsfA1aOE-1# and HsfA1aOE-3# were reduced by 47.1% and 53.3%, respectively, when compared with those of the WT plants after 13 d under drought stress ( and Fig. S3). Thus, the membrane integrity was more sensitive in TRV-HsfA1a plants than that of TRV plants. By contrast, the membrane integrity of plants overexpressing HsfA1a was more resistant than that of the WT plants under drought stress.
Association of drought sensitivity or tolerance and the protein–ubiquitin conjugates in HsfA1a-silenced or -overexpressing plants under drought stress
Drought is known to induce plant stomatal closure and abscisic acid (ABA) accumulation. To determine whether HsfA1a was involved in stomatal closure and ABA accumulation under drought stress, we silenced the HsfA1a gene and overexpression of HsfA1a in tomato plants and analyzed the stomatal aperture and ABA content. As shown in , the stomata were open in TRV, TRV-HsfA1a, WT and HsfA1aOE plants, and the ratio of stomatal aperture length over width (which indicates the degree of stomatal closure) and ABA content were similar under normal water conditions. However, the stomata of TRV plants were closed and the ratio of stomatal aperture length over width and ABA accumulation were significantly increased after 6 d under drought stress (84.6% and 137.6%, respectively; ). Interestingly, the stomatal closure was more pronounced in HsfA1a-silenced plants after 6 d under drought stress (). Moreover, the ratio of stomatal aperture length over width and ABA accumulation were more increased in TRV-HsfA1a plants than that of TRV plants after 6 d under drought stress (). Meanwhile, the stomatal aperture in HsfA1aOE plants was higher and ABA contents were significantly decreased when compared with those of WT plants after 6 d under drought stress (). These results indicate that endogenous ABA and stomatal closure are more sensitive in TRV-HsfA1a plants under drought stress and are therefore not likely to account for HsfA1a-mediated tomato drought tolerance.
Figure 2. Stomatal aperture and ABA content in TRV-HsfA1a and HsfA1aOE plants after 6 d under drought stress. (A) Microphotographs of stomata observed in TRV and TRV-HsfA1a plants under drought stress. Bars: 10 µm. (B) Ratio of stomatal aperture length over width were plotted for the TRV and TRV-HsfA1a plants under drought stress. The data are presented as the means of 5 biological replicates (± SE); each replicate represents the average stomatal aperture of 150 randomly selected stomata. (C) Accumulation of ABA in TRV and TRV-HsfA1a plants under drought stress. (D) Microphotographs of stomata observed in WT and HsfA1aOE plants under drought stress. Bars: 10 µm. (E) Ratio of stomatal aperture length over width were plotted for the WT and HsfA1aOE plants under drought stress. The data are presented as the means of 5 biological replicates (± SE); each replicate represents the average stomatal aperture of 150 randomly selected stomata. (F) Accumulation of ABA in WT and HsfA1aOE plants under drought stress. The data are presented as the means of 5 biological replicates (± SE). Means with the same letter did not significantly differ at P< 0.05 according to the Duncan multiple range test. Three independent experiments were performed with similar results. OE, overexpressing; WT, wild-type.
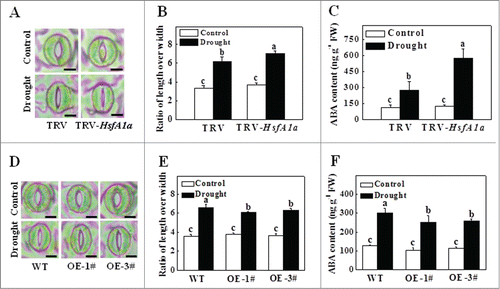
We have previously reported that compromised stress tolerance of mutants for autophagy and WRKY33 is associated with increased accumulation of ubiquitinated protein aggregates under abiotic stress.Citation14,31 Due to the important role of HsfA1a in plant tolerance to abiotic stresses, we examined whether there was an increased accumulation of ubiquitinated drought-induced protein aggregates in HsfA1a-silenced plants. As shown in , the levels of insoluble protein aggregates were similar in TRV and TRV-HsfA1a plants under normal water supply conditions. However, the level of insoluble protein aggregates in TRV-HsfA1a plants was approximately 93.8% higher than in TRV plants after 13 d under drought stress (). We separated the total proteins to soluble and insoluble proteins and stained with Coomassie brilliant blue. As shown in Fig. S4A, the accumulation of insoluble proteins was increased under drought stress, especially in HsfA1a-silenced plants, while, the soluble proteins were decreased which were associated with the increasing of insoluble proteins based on the migration patterns. Thus, the profuse accumulation of insoluble proteins under drought stress was likely to be transferred from the soluble proteins. To determine whether these insoluble proteins were ubiquitinated, we isolated total, soluble and insoluble proteins from TRV and TRV-HsfA1a plants collected under normal water supply conditions and after 13 d of drought stress. The proteins were separated with SDS-PAGE and analyzed for ubiquitination using an anti-ubiquitin monoclonal antibody. As shown in , similar levels of ubiquitinated proteins were observed in the soluble fractions in these plants with or without drought stress. Drought stress induced slight accumulation of ubiquitinated proteins in the insoluble proteins of TRV plants. However, the accumulation of ubiquitinated proteins was drastically increased in the insoluble protein aggregates of TRV-HsfA1a plants compared with the TRV plants after 13 d of drought stress (). Furthermore, the levels of insoluble protein aggregates did not display significant difference between WT and HsfA1aOE plants under normal water supply conditions (). The levels of insoluble protein aggregates in HsfA1aOE-1# and HsfA1aOE-3# were 23.1% and 25.4% lower than those of WT plants after 13 d under drought stress (, Fig. S4B). We also observed similar levels of ubiquitinated proteins in the soluble fractions in WT and HsfA1aOE plants with or without drought stress (). The accumulation of ubiquitinated proteins in the insoluble proteins of HsfA1aOE plants were decreased after 13 d withholding water compared with the WT plants ().
Figure 3. Accumulation and ubiquitination of insoluble proteins in TRV, TRV-HsfA1a, WT and HsfA1aOE plants under drought stress. (A) Accumulation of insoluble proteins in TRV and TRV-HsfA1a plants under drought stress. Leaf tissues from TRV and TRV-HsfA1a plants collected at d 13 under drought stress for the preparation of total, soluble and insoluble proteins as described in the Materials and Methods. Total proteins in the starting homogenates and insoluble proteins in the last pellets were determined for the calculation of the percentages of insoluble proteins to total proteins. The data are presented as the means of 5 biological replicates (± SE). Means with the same letter did not significantly differ at P< 0.05 according to the Duncan multiple range test. (B) Ubiquitination of insoluble protein aggregates in TRV and TRV-HsfA1a plants collected at day 13 under drought stress. Proteins from the starting homogenates (T), first supernatant fractions (S) and last pellet fractions (P) were subjected to SDS-PAGE and probed with an anti-ubiquitin monoclonal antibody. Three independent experiments were performed with similar results. (C) Accumulation of insoluble proteins in WT and HsfA1aOE plants under drought stress. Leaf tissues from WT and HsfA1aOE plants collected at d 13 under drought stress for the preparation of total, soluble and insoluble proteins as described in Materials and Methods. Total proteins in the starting homogenates and insoluble proteins in the last pellets were determined for the calculation of the percentages of insoluble proteins to total proteins. The data are presented as the means of 5 biological replicates (± SE). Means with the same letter did not significantly differ at P < 0.05 according to the Duncan multiple range test. (D) Ubiquitination of insoluble protein aggregates in WT and HsfA1aOE plants collected at day 13 under drought stress. Proteins from the starting homogenates (T), first supernatant fractions (S) and last pellet fractions (P) were subjected to SDS-PAGE and probed with an anti-ubiquitin monoclonal antibody. Three independent experiments were performed with similar results.
Expression of autophagy-related (ATG) genes and formation of autophagosomes under drought stress
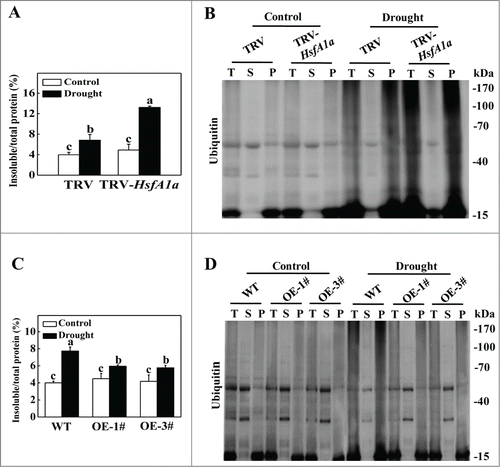
To analyze whether autophagy involved in HsfA1a-induced drought tolerance, we first examined the expression of 26 autophagy genes identified from the Sol Genomics Network (http://solgenomics.net/) in response to drought stress. As shown in Fig. S5, the transcript levels of several ATG genes were slightly increased at the 3rd day and most ATG gene expression was highly elevated at the 6th day under drought stress when the plants started to show symptoms of dehydration. We compared 20 drought-induced ATG genes to further assess the expression patterns of those genes in HsfA1a-silenced and HsfA1aOE plants after 6 d under drought stress. As shown in , the transcript levels of these ATG genes in TRV-HsfA1a or HsfA1aOE plants were similar with the levels in TRV or WT plants when they were grown with a normal water supply. However, the expression levels of ATG10 and ATG18f in TRV-HsfA1a plants were 50.6% and 64.0% lower than those of TRV plants, respectively, under drought stress conditions (). Furthermore, the expression of ATG10 and ATG18f in HsfA1aOE-1# and HsfA1aOE-3# plant was significantly increased compared with WT plants after 6 d of drought stress (). Whereas, other 18 ATG genes, which were induced by drought stress in WT plants, were similar in VIGS and OE plants when compared with their own control plants (). These results indicated that the expression of many ATG genes was induced under drought stress and ATG10 and ATG18f induction under drought stress was HsfA1a-dependent.
Figure 4. Induction of autophagy genes by drought stress in TRV, TRV-HsfA1a, WT and HsfA1aOE plants. Six-wk-old tomato plants were exposed to dehydration by withholding water and total RNA was isolated from leaf samples collected on d 6. Transcript levels were determined using qRT-PCR. All data are presented as the means of 5 biological replicates (± SE). Means with the same letter did not significantly differ at P< 0.05 according to the Duncan multiple range test. Three independent experiments were performed with similar results.
To investigate the effect of HsfA1a on drought-induced autophagy, we used monodansylcadaverine (MDC) as a probe to detect autophagic activity in HsfA1a-silenced and -overexpressing plants. MDC is an autofluorescent dye that stains plant autophagosomes.Citation14,32 Under normal water supply conditions, we observed low numbers of punctate fluorescent signals in TRV, TRV-HsfA1a, WT and HsfA1aOE plants (). After a 6-day drought treatment, the numbers of punctate fluorescent signals increased by approximately 2.0- to 2.5-fold in the TRV and WT plants (). Importantly, the number of punctate fluorescence signals after drought stress was substantially reduced in the TRV-HsfA1a plants compared to the drought-treated TRV plants (). However, the number of punctate fluorescence signals was significantly increased in HsfA1aOE plants compared with the drought-treated WT plants ().
Figure 5. Visualization of the accumulation of autophagosomes in tomato leaves under drought stress. (A and B) MDC-stained autophagosomes in the leaves of TRV and TRV-HsfA1a or WT and HsfA1aOE plants. Six-wk-old plants were exposed to dehydration by withholding water and the leaves were MDC-stained and visualized on day 6 by fluorescence confocal microscopy. MDC-labeled structures are shown as green signals. Bars: 25 μm. (C) Relative autophagic activity normalized to the activity of the TRV or WT control plants in (A and B). The MDC-stained autophagosomes in leaves at each treatment were quantified to calculate the autophagic activity relative to TRV or WT control plants, which was set to 1. More than 300 mesophyll cells for each treatment were used for the quantification. (D and E) Representative transmission electron microscopy (TEM) images of autophagic structures in the mesophyll cells of TRV and TRV-HsfA1a or WT and HsfA1aOE plants. Six-wk-old plants were exposed to dehydration by withholding water and the mesophyll cells were visualized on d 6 by TEM. Autophagic bodies are indicated by red arrows. Bars: 1 µm. (F) Relative autophagic activity normalized to the activity of the TRV or WT control plants in (D and E). More than 20 cells were used to quantify autophagic structures. (G and H) Atg8 protein levels in the leaves of TRV and TRV-HsfA1a plants or WT and HsfA1aOE plants. Atg8 and Atg8–PE are the nonlipidated and lipidated forms of Atg8, respectively. The Rubisco large subunit was used as a loading control for the western blotting analysis. Results represent the means ± SE. Means with the same letter did not significantly differ at P< 0.05 according to the Duncan multiple range test. Three independent experiments were performed with similar results. Cp, chloroplast; V, vacuole; S, starch; OE, overexpressing; WT, wild-type; 1# and 3#, 2 lines of HsfA1aOE plants.
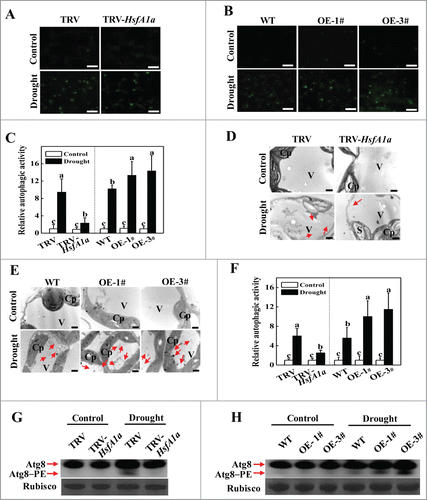
To confirm these results, we used transmission electron microscopy (TEM) to monitor autophagic activity under drought stress. In agreement with the results of MDC staining, TEM showed few autophagosomes and autophagic bodies in all of the plants under normal water supply conditions, whereas the numbers of both classic double-membrane autophagosomes in the cytoplasm and signal-membrane autophagic bodies in the vacuole were increased by 4.0- to 4.5 fold in the TRV and WT plants after 6 d of drought stress (). The number of autophagosomes and autophagic bodies only increased by 1.5-fold in TRV-HsfA1a plants, but increased by 9.0-fold in HsfA1aOE plants after 6 d of drought stress ().
Atg8 has been widely used to monitor autophagosomes. To further examine the induction of autophagy, we used western blotting to analyze the formation of Atg8–phosphatidylethanolamine (PE) conjugates as a marker for autophagic activation using an anti-Atg8a antibody.Citation33 Under normal water supply conditions, we could barely detect the Atg8–PE band in tomato plants (). Faster migration of the Atg8–PE bands was observed in TRV and WT plants after 6 d of drought stress (). In contrast, Atg8–PE was significantly decreased in TRV-HsfA1a plants but was more abundant in HsfA1aOE plants after drought stress (). Taken together, these results suggest that HsfA1a is involved in both drought-induced expression of ATG10 and ATG18f genes and autophagosome formation.
Binding of HsfA1a to the ATG10 and ATG18f promoters
To examine the possible regulation of tomato ATG genes by HsfA1a, we inspected 1.5 kb sequences located upstream of the predicted transcriptional start sites of 26 tomato ATG genes. Promoters of 5 ATG genes (ATG5, ATG8e, ATG10, ATG13b and ATG18f) contain HSE sequence (GAANNTTC) (Fig. S6). We performed an EMSA to analyze whether HsfA1a directly bound in vitro to these promoters. The signaling of probe-protein complex was not detected using ATG5, ATG8e and ATG13b probes (data not shown). However, HsfA1a bound to the ATG10 and ATG18f promoter probes (). When the core sequence of HSE motifCitation26 in the ATG10 probe was mutated to the mutant ATG10−1Δ probe (), the binding to the complexes was reduced (). When additional mutations were introduced to the mutant ATG10–2Δ probe (), the binding by HsfA1a was totally lost (). Similarly, the binding signal to the complex was not detected when the HSE sequence in the ATG18 probe was mutated to the mutant ATG18fΔ probe (). These results suggest that the HsfA1a protein specifically binds to the HSE sequences in the synthesized probes of the ATG10 and ATG18f promoters in vitro.
Figure 6. HsfA1a binds to the promoters of ATG10 and ATG18f in vitro and in vivo. (A) Oligonucleotide used in the electrophoretic mobility shift assays (EMSA). The ATG10 probe contains one direct HSE, whereas in the ATG10−1Δ and ATG10–2Δ probes the HSE core sequence was mutated. The ATG18f probe contains one direct HSE, whereas in the ATG18fΔ probe, the HSE core sequence was mutated. The WT and mutated HSE sequences are underlined. The mutated bases were indicated in red. (B and C) EMSA showing HsfA1a bound to the HSE sequences of the ATG10 or ATG18f promoters. Recombinant HsfA1a was purified from E. coli cells and used for DNA binding assays with ATG10, ATG10−1Δ, ATG10–2Δ, ATG18f, or ATG18fΔ as the probes. His was included as the negative control. (D) Direct binding of HsfA1a to the ATG10 and ATG18f promoters was analyzed using ChIP-qPCR in 35S-HsfA1a-HA-overexpressing (HsfA1aOE) plants. Six-wk-old HsfA1aOE plants were exposed to dehydration by withholding water and input chromatin was isolated from leaf samples on d 6. The epitope-tagged HsfA1a-chromatin complex was immunoprecipitated with an anti-HA antibody. A control reaction was processed side-by-side using mouse IgG. Input- and ChIP-DNA samples were quantified by qRT-PCR using primers specific for the promoters of the ATG genes. The ChIP results are presented as percentage of the input DNA. Means with the same letter did not significantly differ at P< 0.05 according to the Duncan multiple range test. Three independent experiments were performed with similar results. OE, overexpressing; 1#, line of HsfA1aOE plants.
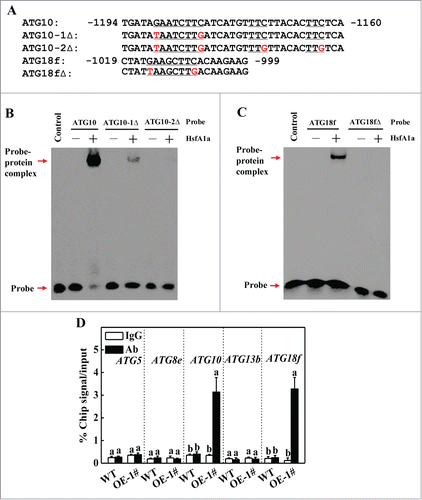
To determine whether tomato HsfA1a directly regulates the expression of these 5 ATG genes in vivo, we performed ChIP-qPCR assays to test HsfA1a protein binding to the promoters of these ATG genes under drought stress. As shown in , only ATG10 and ATG18f promoter sequences were substantially enriched using an anti-HA antibody that immunoprecipitated the 3HA-tagged HsfA1a transgene product in the OE lines and not the WT lines after 6 d of drought stress. In contrast, the IgG control antibody failed to pull down these 2 ATG gene promoter DNA segments (). Thus, HsfA1a binds to the ATG10 and ATG18f gene promoters and may directly regulate their induction under drought stress.
Identification and functional analysis of ATG10 and ATG18f in HsfA1a-induced tolerance and formation of autophagosomes under drought stress
To test the functions of the ATG10 and ATG18f genes in HsfA1a-induced drought tolerance, ATG10 and ATG18f genes were respectively silenced in WT and HsfA1aOE plants, and the expression of their own gene was decreased 60 to 70% in silenced plants compared with their own TRV control plants (Fig. S7A). Silencing of ATG10 or ATG18f gene did not affect HsfA1a gene expression under normal water supply condition or drought stress (Fig. S7B). No significant differences in phenotypes, RWC and EL were observed in the ATG10- and ATG18f-silenced plants compared with TRV control plants under normal water supply condition (Fig. S7C to E). However, most of the leaves from ATG10- and ATG18f-silenced WT and HsfA1aOE plants exhibited serious wilting after 13 d of drought stress (Fig. S7C). The RWC of the TRV-ATG10 and pTRV-ATG18f plants were 30.9% and 15.4% lower than WT TRV plants after 13 d of drought stress, respectively (Fig. S7D). Similarly, the RWC of the HsfA1aOE TRV-ATG10 and TRV-ATG18f plants were decreased by 30.9% and 31.6% compared with the HsfA1aOE TRV plants, respectively (Fig. S7D). Similarly, the EL values of the TRV-ATG10 and TRV-ATG18f plants were increased by 35.4% and 12.1%, respectively, compared with the WT TRV plants (Fig. S7E). The EL values of the HsfA1aOE TRV-ATG10 and TRV-ATG18f plants were 131.2% and 147.2% higher than those of HsfA1aOE TRV plants (Fig. S7E).
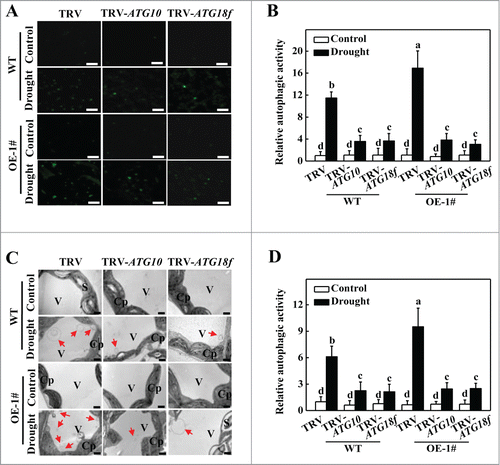
To further investigate the role of the ATG10 and ATG18f genes in the HsfA1a-mediated drought stress response, we used MDC and TEM to detect autophagic activity. We observed low numbers of punctate fluorescent signals in all of the plants under normal water supply conditions (). The numbers of punctate fluorescent signals significantly increased in WT TRV plants after a 6-d drought treatment (). However, the number of punctate fluorescence signals in all TRV-ATG10 and TRV-ATG18f plants was decreased by 66.7 to 73.4%. Furthermore, the TEM results were in agreement with the results MDC staining results. Under normal water supply conditions, few autophagosomes and autophagic bodies were detected in any of the plants. The numbers of autophagosomes and autophagic bodies increased by 6-fold in WT TRV plants. However, the numbers of autophagosomes and autophagic bodies in TRV-ATG10 and TRV-ATG18f plants increased only by 2.1- to 2.5-fold under drought stress (). Thus, HsfA1a-induced drought tolerance and formation of autophagosomes under drought stress were largely Atg10- and Atg18f-dependent.
Figure 7. Visualization of the accumulation of autophagosomes by confocal microscopy and transmission electron microscopy (TEM) in ATG10 and ATG18f-silenced plants under drought stress. (A) MDC-stained autophagosomes in the leaves of WT TRV, TRV-ATG10 and TRV-ATG18f plants or HsfA1aOE TRV, TRV-ATG10 and TRV-ATG18f plants. Six-wk-old plants were exposed to dehydration by withholding water and the leaves were MDC-stained and visualized on day 6 by fluorescence confocal microscopy. MDC-labeled structures are visible as green signals. Bars: 25 μm. (B) Relative autophagic activity normalized to the activity of WT TRV control plants in (A). Quantification of the MDC-stained autophagosomes in the leaves at each treatment was performed to calculate the autophagic activity relative to WT TRV control plants, which was set to 1. More than 300 mesophyll cells for each treatment were used for the quantification. (C) TEM images of autophagic structures in the mesophyll cells of WT TRV, TRV-ATG10 and TRV-ATG18f plants or HsfA1aOE TRV, TRV-ATG10 and TRV-ATG18f plants. Six-wk-old plants were exposed to dehydration by withholding water and the mesophyll cells were visualized on d 6 by TEM. Autophagic bodies are indicated by red arrows. Bars: 1 µm. (D) Relative autophagic activity normalized to the activity in WT TRV control plants in (C). More than 20 cells were used to quantify autophagic structures. Results represent the means ± SE. Means with the same letter did not significantly differ at P< 0.05 according to the Duncan multiple range test. Three independent experiments were performed with similar results. Cp, chloroplast; V, vacuole; S, starch; WT, wild-type; OE, overexpressing; 1#, line of HsfA1aOE plants.
Discussion
Studies over the past 20 years have provided a large body of evidence that autophagy plays a critical role in a wide range of important biological processes in plants.Citation9,13 In contrast, our understanding of the regulation of autophagy in plants is extremely limited. In the present study, we provided genetic evidence that HsfA1a, a transcription factor involved in plant stress responses, contributed to the upregulation of autophagy genes and the formation of autophagosomes. Furthermore, autophagy was involved in HsfA1a-mediated tomato drought tolerance. These results provide important new insights into the regulation of autophagy in plant stress responses.
Water scarcity has a dramatic impact on crop growth and productivity.Citation34 The phytohormone ABA is produced under water-deficit conditions and plays an important role in the adaptation of vegetative tissues to drought.Citation35 ABA promotes stomatal closure in guard cells to reduce transpiration and conserve water, and regulates the expression of many genes that may function in plant dehydration tolerance.Citation36 Additionally, various transcription factors play critical roles in plant signaling networks to regulate plant drought tolerance and activate gene transcription and the downstream metabolic system.Citation37 The present study demonstrated that drought stress induced the expression of transcription factor HsfA1a and silencing of the HsfA1a gene made tomato plants hypersensitive to drought stress, while overexpression of the HsfA1a gene increased plant drought tolerance (). Collectively, these results indicate that transcription factor HsfA1a plays a critical role in plant drought tolerance. Silencing of the HsfA1a gene resulted in an increased reduction of the stomatal aperture compared to the TRV control under drought stress, whereas drought stress induced increased levels of ABA accumulation in TRV-HsfA1a plants compared to the TRV control plants (). These results argued against a critical role of ABA-dependent pathways in HsfA1a-mediated plant drought tolerance.
Drought stress is an important threat to plant protein activities through diminution in cellular volume, macromolecular crowding and oxidative damage.Citation38 Abiotic stresses, such as drought, cause damage to a variety of cellular structures and macromolecules, including protein denaturation and aggregation.Citation39 Molecular chaperones, such as Hsp70, and the LEA proteins (which bind with high affinity to solvent-exposed, nonstructural and hydrophobic regions of denatured proteins), can maintain targeted proteins in their functional conformations and prevent the aggregation of denatured proteins, thereby contributing to cellular homeostasis under mild to moderate stress conditions.Citation40,41 Numerous studies have shown that stress-generated misfolded proteins that cannot reestablish their normal conformation are recognized, ubiquitinated and targeted for degradation by the cellular protein quality control machinery.Citation31,42,43 We have previously observed that denatured or otherwise damaged cellular proteins are ubiquitinated under heat and oxidative stress conditions in Arabidopsis plants.Citation43 Ubiquitination is an enzymatic protein post-translational modification (PTM) process in which ubiquitin (Ub, a highly conserved 8-kD protein) is attached to substrate proteins, such as denatured or damaged cellular proteins, in 3 consecutive steps catalyzed by the E1, E2, and E3 enzymes.Citation44,45 Here, we observed a significant increase in the level of insoluble proteins in TRV-HsfA1a plants compared with the TRV control plants (). Under drought stress, the TRV-HsfA1a plants but not the TRV control plants displayed a major elevation in the levels of ubiquitinated proteins (), indicating that drought-induced ubiquitinated proteins are aggregated and accumulated in HsfA1a-silenced plants due to reduced degradation.
For targeted degradation, aggregated or damaged proteins formed under stress conditions must be specifically recognized and transferred to the protein degradation systems. When native structures cannot be achieved, molecular chaperones can facilitate their degradation through association with chaperone-dependent ubiquitin E3 ligases that can specifically catalyze the ubiquitination of denatured or misfolded but not native proteins in a chaperone-dependent manner.Citation46 Ubiquitinated proteins can be degraded by the UPS and autophagic pathway.Citation31,47, 48 However, the UPS is limited in its capacity to degrade oligomeric and aggregated proteins that are not sufficiently small to fit through the narrow proteasome entrance channel.Citation49 The increased accumulation of ubiquitinated proteins in drought- and heat-treated autophagy mutants indicates that autophagy is also a major mechanism underlying the degradation of ubiquitinated proteins under stress conditions in plants.Citation31 Indeed, drought stress induced an increase in the transcript levels of ATG genes (Fig. S5) and the formation of autophagosomes in WT tomato plants (). Moreover, the accumulation of ubiquitinated aggregates in HsfA1a-silenced plants was associated with compromised autophagosome formation (). The expression of most ATGs was still induced under drought stress; however, induction of ATG10 and ATG18f gene expression was compromised in TRV-HsfA1a plants under drought stress (), indicating that HsfA1a is necessary for the sustained induction of autophagy in the plant response to drought stress.
The oligomerization domain of HsfA1a is required for the formation of functional trimers under stress conditions,Citation50 which can then bind to the HSEs of target genes and regulate their transcription.Citation51 The expression of ATG10 and ATG18f was compromised in TRV-HsfA1a plants under drought stress, and, interestingly, the promoters of these genes contained HSE boxes (). HsfA1a directly bound the HSE boxes in the promoters of these 2 autophagy genes based on results from in vitro EMSA and in vivo ChIP-qPCR assays (), indicating that ATG10 and ATG18f are direct target genes of the HsfA1a transcription factor. In Arabidopsis, Atg10 has been identified as the Atg12-conjugating enzyme that was essential for autophagic vesicle formation.Citation52 atg10 mutant plants are hypersensitive to nitrogen and carbon starvation and fail to accumulate autophagic bodies inside the vacuole.Citation52 In the present study, genomic database searches with BLASTP identified a single tomato ATG10 gene that contained several regions with strong amino acid conservation, including a presumed active-site cysteine (Cys176) that forms the thioester intermediate with Atg12 prior to its transfer to Atg5.Citation52 Silencing of the tomato ATG10 gene made the plants hypersensitive to drought stress (Fig. S7) and blocked autophagy formation under drought stress (). The plant ATG18f gene is a member of the ATG18 gene family and possesses a sequence similar to the yeast autophagy gene ATG18.Citation53 In Arabidopsis, the transcript level of ATG18f was upregulated during nutrient starvation and a combination of drought and heat stress,Citation53,54 indicating that Atg18f potentially played a role in the response to these stress conditions. Indeed, silencing of the tomato ATG18f gene compromised plant tolerance against drought stress (Fig. S7) and inhibited autophagosome formation (). Thus, the Atg18f protein is likely to be required for autophagosome formation in tomato plants. Importantly, ATG10- or ATG18f silencing in HsfA1a-overexpressing plants resulted in hypersensitivity to drought stress and completely blocked HsfA1a-mediated plant drought tolerance (Fig. S7). Likewise, induced autophagy formation in HsfA1a-overexpressing plants was significantly reduced by silencing ATG10 or ATG18f (), supporting that the ATG10 and ATG18f were involved in HsfA1a-mediated formation of autophagosomes and drought tolerance. In human breast adenocarcinoma cells, HSF1 (heat-shock factor 1), a master regulator of the heat-shock responses in mammal, was identified as the regulator of cytoprotective autophagy induced by the chemotherapeutic agent carboplatin through an increase in LC3 punctate structures and LC3 and Atg7 protein expression.Citation55 Thus, the transcriptional upregulation of autophagy genes does not appear to be as independent in plants as the process observed in mammalian systems.
During the plant stress response, HsfA1a interacts with HsfA2 to form hetero-oligomeric complexes that can synergistically regulate the expression of some Hsfs and molecular chaperone Hsps, including a number of small Hsps, DnaJ/Hsp40, and Hsp70.Citation22,25,56,57 These chaperones assist with a wide range of protein-folding processes to prevent protein aggregation in plant cells under normal and stress conditions.Citation41 The transcript levels of several Hsp-encoding genes are significantly reduced in Arabidopsis hsfA1a and hsfA1b double-knockout mutants,Citation51 indicating that HsfA1s play a positive role in protein folding and in preventing aggregation through the regulation of molecular chaperone Hsps. Indeed, silencing of HsfA1a induced the accumulation of insoluble protein aggregates in tomato plants under drought stress, and these proteins were highly ubiquitinated (). We have previously observed that silencing of the tomato autophagy genes ATG5 and ATG7 or tomato autophagy receptors NBR1a and NBR1b significantly reduces the heat-induced transcript levels of some Hsp-encoding genes,Citation14 indicating that the formation of autophagosomes has a positive feedback effect on the upregulation of Hsfs-activated Hsps. Thus, HsfA1a has dual functions in the regulation of protein aggregates and proteotoxicity in the plant stress response, including the stabilization of unfolded proteins, the prevention of unwanted protein aggregation through the transcriptional upregulation of molecular chaperones Hsps, and the degradation of stress-induced protein aggregation through induction of some autophagy genes and the formation of autophagosomes.
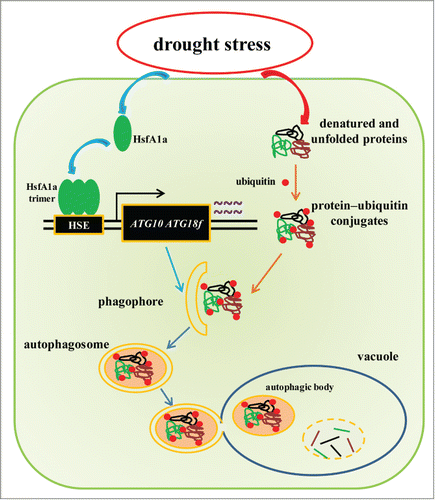
In conclusion, we have provided comprehensive genetic and molecular analyses of tomato HsfA1a in induced autophagy and plant drought tolerance (). We demonstrated that HsfA1a acts as a positive regulator of the expression of the autophagy genes ATG10 and ATG18f and the formation of autophagosomes during induction of autophagy under stress conditions, such as drought. Stress-damaged proteins or protein aggregates are recognized and polyubiquitinated by specific E3 ubiquitin ligases and delivered to the expanding phagophores. Expansion and closure of the phagophores result in the formation of autophagosomes, which then can fuse with vacuoles to form autophagic vacuoles in which ubiquitinated proteins or protein aggregates are degraded (). Processing and degradation of stress-induced protein aggregates would increase stress tolerance, leading to increased survival of plants under stress conditions.
Figure 8. A proposed model for the induction of autophagy by HsfA1a in tomato plants under drought stress. HsfA1a is first activated under drought stress by increasing transcription and trimer formation. Then, activated HsfA1a binds to and upregulates the expression of ATG10 and ATG18f leading to an increase in autophagy. The increase in autophagy leads to an increase in the degradation of insoluble ubiquitinated protein aggregates and cell survival under drought stress.
Materials and Methods
Plant materials and experimental design
The tomato Solanum lycopersicum cv. Ailsa Craig genotype was used in all experiments. Seeds were germinated and grown in 250 cm3 plastic pots filled with a mixture of peat and vermiculite (2:1, v:v). The plants were watered daily with Hoagland nutrition solution in the chamber. The growth conditions were as follows: a 14/10 light/dark cycle, 23/21°C day/night temperature, and 600 μmol m−2 s−1 photosynthetic photon flux density (PPFD).
To induce drought stress, plants in soil were watered to saturation and then exposed to dehydration by withholding water for 13 d. During drought periods, the relative soil water content was measured using a ZigWSN recorder (Ziteng, China) daily to ensure that every pot maintained the same level of water content.
Generation and selection of transgenic plants
To obtain the tomato HsfA1a overexpression construct, the 1581 bp full-length coding DNA sequence (CDS) was amplified with the primers HsfA1aOE-F (5′-TTGGCGCGCCATGGAGCCGAATTCTTAT-3′) and HsfA1aOE-R (5′-GGGGTACCGATCATATGTTTTTGTTG-3′) using tomato cDNA as the template. The PCR product was digested with AscI and KpnI and inserted behind the CaMV 35S promoter in the plant transformation vector pFGC1008-HA. The resulting HsfA1aOE-HA plasmid was transformed into Agrobacterium tumefaciens strain EHA105. Tomato seeds of Ailsa Craig were used for transformation, as described by Fillatti et al.Citation58 Transgenic plants overexpressing the HsfA1a transgene were identified by western blotting using an anti-HA (Pierce, 26183) monoclonal antibody (Fig. S1). Two independent homozygous lines of the F2 progeny were used in the study.
VIGS constructs and Agrobacterium-mediated virus infection
TRV VIGS constructs were used to silence the tomato HsfA1a, ATG10, and ATG18f genes. These genes were generated by PCR amplification using gene-specific primers (Table S1), digested with the appropriate restriction enzymes and ligated into the same sites in TRV2. The resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101. A. tumefaciens-mediated virus infection was performed as previously described.Citation59 The plants were kept at 22°C and used for experiments 3 wk after A. tumefaciens infiltration. Leaflets in the middle of the fifth fully expanded leaves, which showed about 30 to 40% transcript levels of control plants, were used.
Stomatal conductance and relative water content determination
Stomatal conductance was detected according to Xia et al.Citation60 After 6 d drought, tomato leaves abaxial epidermises were peeled with forceps and floated on buffer (30 mM KCl, 10 mM 2-[N-morpholino]-ethanesulfonic acid [MES], pH 6.15). The stomatal apertures were measured using a light microscope equipped with a digital camera (Leica Microsystems, Germany).
The leaves of relative water content was measured according to Zhou et al.Citation61 The fresh weights of the terminal leaflets were measured as Fw. Then, the leaves were immersed in distilled water in darkness for 24 h to obtain the fully turgid weight (Tw). Leaves were dried in an oven at 70°C for 48 h and weight the dry weight (Dw). The relative water content was calculated as follows: RWC (%) = (Fw-Dw)/(Tw-Dw) ×100.
ABA measurements
ABA contents were measured according to Welsch et al.Citation62 with modification. Briefly, 0.3 g of frozen leaf tissues was ground and homogenized in 3 ml of ethyl acetate containing 100 ng ml−1 [2H6](+)-cis,trans-ABA (OlChemlm Ltd., 35671–08–0) as an internal standards. The homogenate was shaken for 12 h in the dark at 4°C and then centrifuged at 15,000 × g for 10 min. The supernatant fraction was transferred to a new tube; then, the pellet was re-extracted with 3 ml of ethyl acetate and combined with the supernatant fraction. The supernatant fraction was eluted through a Sep-Pak C18 cartridge (Waters, WAT020805) to remove the polar compounds and dried under nitrogen. Dried samples were resuspended in 0.5 ml of 70% methanol (v/v). Then, the samples (20 µL) were analyzed by HPLC electrospray ionization/MS-MS using an Agilent 1290 HPLC (Agilent Technologies, Germany) coupled to an Agilent 6460 triple Quad LC/MS (Agilent Technologies, Germany). Chromatographic separation was performed on an Agilent XDB 3.5 µm C18 150 mm × 2.1 mm column at 40°C. The solvent gradient consisted of solvent A (0.1% formic acid aqueous water) and solvent B (100% methanol) at a flow rate of 0.3 ml/min over 20 min with the following gradient setup: 0 to 1.5 min, A: B at 60: 40; a 6.5 min; switch to A: B of 0: 100; and the return of solvent A: B to 60: 40 for 5 min until the end of the run. [2H6](+)-cis,trans-ABA was used to estimate the recovery rate of the ABA content, which was quantified using the external standard.
Total RNA extraction and gene expression analysis
Total RNA was isolated from tomato leaves using RNAsimple Total RNA Kit (Tiangen, DP419) according to the manufacturer's instructions. Total RNA (1 μg) was used to reverse transcribed for cDNA template using the ReverTra Ace qPCR RT Kit (Toyobo, FSQ-301).
The qRT-PCR assays were performed using an LightCycler® 480 II Real-Time PCR detection system (Roche, Swiss). PCRs were performed using the SYBR Green PCR Master Mix (Takara, RR420A). The PCR conditions consisted of denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 58°C for 15 s and extension at 72°C for 30 s. The tomato Ubi3 gene was used as an internal control. Gene-specific primers were designed according to cDNA sequences as described in supplemental Table S2. Relative gene expression was calculated as described as Livak and Schmittgen.Citation63
Protein extraction and western blotting
For protein extraction, tomato leaves were ground in liquid nitrogen and homogenized in extraction buffer (100 mM Tris-HCl, pH 8.0, 10 mM NaCl, 1 mM EDTA, 1% Triton X-100 [Aladdin, T109026], 1 mM phenylmethylsulphonyl fluoride and 0.2% β-mercaptoethanol). The soluble, insoluble and ubiquitinated proteins were detected as described as our previous study.Citation31 The homogenates were filtered through a 300-μm and then 100-μm nylon mesh. Half of the filtered samples were kept to detect total proteins. Other samples were clarified by centrifugation at 2,200 × g for 5 min. Supernatant fractions were kept to detect soluble proteins. The pellet fractions were resuspended in the same buffer and subjected to the low speed centrifugation. The process was repeated twice and after the last centrifugation the pellet fractions were resuspended to detect insoluble protein. The concentrations of proteins in the homogenates (total proteins), the first supernatant fractions (soluble proteins) and last pellet fractions (insoluble proteins) were determined using Bio-Rad protein assay kit (500–0001). The denatured protein extracts were separated using 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). For Atg8a detection, the denatured protein was separated on a 13.5% SDS–PAGE gel in the presence of 6 M urea. For western blotting, the proteins on the SDS–PAGE gel were transferred to a nitrocellulose membrane. The membrane was blocked for 1 h in TBS buffer (20 mM Tris, pH 7.5, 150 mM NaCl, and 0.1% Tween 20 [Amresco, 0777]) with 5% skim milk powder at room temperature, and then incubated for 1 h in TBS buffer with 1% BSA (Amresco,0332) containing a mouse anti-ubiquitin monoclonal antibody (Sigma-Aldrich, U0508), or rabbit anti-Atg8a polyclonal antibody (Abcam, ab77003). After incubation with a goat anti-mouse HRP-linked antibody (Millipore, AP124P) or goat anti-rabbit HRP-linked antibody (Cell Signaling Technology, 7074), the complexes on the blot were visualized using the SuperSignal™ West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, 34080) following the manufacturer's instructions.
MDC staining
To visualize the accumulation of autophagosomes, tomato leaves were excised and then immediately vacuum infiltrated with 100 μM MDC (Sigma-Aldrich, 30432) for 30 min, followed by 2 washes with phosphate-buffered saline (PBS; Solarbio, P1020) buffer. MDC-incorporated structures were monitored under a LSM 780 confocal microscope (Zeiss, Germany), excited by a wavelength of 405 nm and detected at 400 to 580 nm according to the method of Zhou et al.Citation14
TEM analysis
For transmission electron microscopy, tomato leaves were excised and immediately cut into small pieces (∼1 mm × 4 mm), then fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate (PBS) buffer (pH 7.0) for 12 h in the dark. After washes with PBS buffer, the samples were fixed for 2 h in 1% (v/v) osmium tetroxide at room temperature; then, the samples were dehydrated in a graded ethanol series (30 to 100%; v/v) and embedded in Epon 812. Ultrathin sections (70 nm) were prepared on an ultramicrotome (Leica EM UC7, Germany) with a diamond knife and collected on Formvar-coated grids. The sections were examined using a H7650 transmission electron microscope (Hitachi, Japan) at an accelerating voltage of 75 kV to observe autophagosomes and autophagic bodies.Citation32,64
Recombinant protein and EMSA analysis
The tomato HsfA1a recombinant protein was prepared as previously described.Citation65 Briefly, the full-length HsfA1a CDS was amplified with the gene-specific primers: forward primer, 5′-AAGGAAAAAAGCGGCCGCATGGAGCCGAATTCTTAT-3′ and reverse primer, 5′-CCGCTCGAGTTAGATCATATGTTTTTG-3′. The PCR product was digested with NotI and XhoI and ligated into the same sites of pET-32a vector. The recombinant vector was transformed into E.coli strain BL21 (DE3). Expression of the recombinant proteins was induced by isopropyl β-D-1-thiogalactopyranoside and purified according to the instructions of the Novagen pET purification system.
The probes were biotin end-labeled according to the instructions of the Biotin 3′ End DNA Labeling Kit (Pierce, 89818) and annealed to double-stranded probe DNA. EMSA of the HsfA1a–DNA complexes was performed according to the instructions of the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific, 20148).
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed according to the instructions of the EpiQuik™ Plant ChIP Kit (Epigentek, P-2014). Approximately 1 g of leaf tissue was harvested from drought stress 35S-HsfA1a-HA and wild-type plants. Chromatin was immunoprecipitated with an HA antibody (Pierce, 26183); goat anti-mouse IgG (Millipore, AP124P) was used as the negative control. ChIP-qPCR was performed with primers specific (Table S3) for the ATG5, ATG8e, ATG10, ATG13b and ATG18f promoters.
Statistical analysis
At least 5 independent replicates were used for each determination. Statistical analysis of the bioassays was performed using the SPSS18 statistical package. Experimental data were analyzed with Duncan multiple range test at P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1098798_Supplemental_Material.zip
Download Zip (581.1 KB)Acknowledgment
We thank Dr. Zhixiang Chen for his careful reading of the manuscript.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by the National Natural Science Foundation of China (31430076, 31401877) and the Geological Exploration Foundation of Zhejiang Province, China (2014002–02).
References
- Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ 2013; 20:21-30; PMID:22722335; http://dx.doi.org/10.1038/cdd.2012.72
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature 2011; 475:324-32; PMID:21776078; http://dx.doi.org/10.1038/nature10317
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728-41; PMID:22078875; http://dx.doi.org/10.1016/j.cell.2011.10.026
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 2005; 6:973-9; PMID:16177805; http://dx.doi.org/10.1038/ni1253
- Suzuki N, Koussevitzky S, Mittler R, Miller G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 2012; 35:259-70; PMID:21486305; http://dx.doi.org/10.1111/j.1365-3040.2011.02336.x
- Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 2011; 14:290-5; PMID:21377404; http://dx.doi.org/10.1016/j.pbi.2011.02.001
- Bassham DC. Plant autophagy-more than a starvation response. Curr Opin Plant Biol 2007; 10:587-93; PMID:17702643; http://dx.doi.org/10.1016/j.pbi.2007.06.006
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717-21; PMID:11099404; http://dx.doi.org/10.1126/science.290.5497.1717
- Il Kwon S, Park OK. Autophagy in Plants. J Plant Biol 2008; 51:313-20; http://dx.doi.org/10.1007/BF03036132
- Kim SH, Kwon C, Lee JH, Chung T. Genes for plant autophagy: Functions and interactions. Mol Cells 2012; 34:413-23; PMID:22772908; http://dx.doi.org/10.1007/s10059-012-0098-y
- Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 2008; 4:851-65; PMID:18670193; http://dx.doi.org/10.4161/auto.6555
- Liu YM, Bassham DC. TOR is a negative regulator of autophagy in Arabidopsis thaliana. Plos One 2010; 5(7): e11883; http://dx.doi.org/10.1371/journal.pone.0011883
- Liu YM, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009; 5:954-63; PMID:19587533; http://dx.doi.org/10.4161/auto.5.7.9290
- Zhou J, Wang J, Yu JQ, Chen Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci 2014; 5:174; PMID:24817875
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Bio 2010; 11:545-55; http://dx.doi.org/10.1038/nrm2938
- von Koskull-Doring P, Scharf KD, Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci 2007; 12:452-7; PMID:17826296; http://dx.doi.org/10.1016/j.tplants.2007.08.014
- Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Bba-Gene Regul Mech 2012; 1819:104-19.
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperon 2001; 6:177-89; http://dx.doi.org/10.1379/1466-1268(2001)006%3c0177:AATHST%3e2.0.CO;2
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Doring P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 2004; 39:98-112; PMID:15200645; http://dx.doi.org/10.1111/j.1365-313X.2004.02111.x
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim JM, Seki M, Todaka D, et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 2011; 286:321-32; PMID:21931939; http://dx.doi.org/10.1007/s00438-011-0647-7
- Nishizawa-Yokoi A, Nosaka R, Hayashi H, Tainaka H, Maruta T, Tamoi M, Ikeda M, Ohme-Takagi M, Yoshimura K, Yabuta Y, et al. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol 2011; 52:933-45; PMID:21471117; http://dx.doi.org/10.1093/pcp/pcr045
- Liu HC, Liao HT, Charng YY. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 2011; 34:738-51; PMID:21241330; http://dx.doi.org/10.1111/j.1365-3040.2011.02278.x
- Scharf KD, Heider H, Hohfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol 1998; 18:2240-51; PMID:9528795
- Liu Y, Zhang C, Chen J, Guo L, Li X, Li W, Yu Z, Deng J, Zhang P, Zhang K, et al. Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol Biochem 2013; 64:92-8; PMID:23399534; http://dx.doi.org/10.1016/j.plaphy.2012.12.013
- Hahn A, Bublak D, Schleiff E, Scharf KD. Crosstalk between Hsp90 and Hsp70 Chaperones and Heat Stress Transcription Factors in Tomato. Plant Cell 2011; 23:741-55; PMID:21307284; http://dx.doi.org/10.1105/tpc.110.076018
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. In the complex family of heat stress transcription factors, HSfA1 has a unique role as master regulator of thermotolerance in tomato. Gene Dev 2002; 16:1555-67; PMID:12080093; http://dx.doi.org/10.1101/gad.228802
- Lee JH, Hubel A, Schoffl F. Derepression of the Activity of Genetically-Engineered Heat-Shock Factor Causes Constitutive Synthesis of Heat-Shock Proteins and Increased Thermotolerance in Transgenic Arabidopsis. Plant J 1995; 8:603-12; PMID:7496404; http://dx.doi.org/10.1046/j.1365-313X.1995.8040603.x
- Turnage MA, Muangsan N, Peele CG, Robertson D. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J 2002; 30:107-14; PMID:11967097; http://dx.doi.org/10.1046/j.1365-313X.2002.01261.x
- Brigneti G, Martin-Hernandez AM, Jin HL, Chen J, Baulcombe DC, Baker B, Jones JD. Virus-induced gene silencing in Solanum species. Plant J 2004; 39:264-72; PMID:15225290; http://dx.doi.org/10.1111/j.1365-313X.2004.02122.x
- Liu YL, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J 2002; 31:777-86; PMID:12220268; http://dx.doi.org/10.1046/j.1365-313X.2002.01394.x
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 2013; 9:e1003196; PMID:23341779; http://dx.doi.org/10.1371/journal.pgen.1003196
- Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, et al. Autophagy contributes to leaf starch degradation. Plant Cell 2013; 25:1383-99; PMID:23564204; http://dx.doi.org/10.1105/tpc.112.108993
- Kwon SI, Cho HJ, Kim SR, Park OK. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol 2013; 161:1722-36; PMID:23404918; http://dx.doi.org/10.1104/pp.112.208108
- Lobell DB, Burke MB, Tebaldi C, Mastrandrea MD, Falcon WP, Naylor RL. Prioritizing climate change adaptation needs for food security in 2030. Science 2008; 319:607-10; PMID:18239122; http://dx.doi.org/10.1126/science.1152339
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 2002; 53:247-73; PMID:12221975; http://dx.doi.org/10.1146/annurev.arplant.53.091401.143329
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 2006; 57:781-803; PMID:16669782; http://dx.doi.org/10.1146/annurev.arplant.57.032905.105444
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot 2007; 58:221-7; PMID:17075077; http://dx.doi.org/10.1093/jxb/erl164
- Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends Plant Sci 2001; 6:431-8; PMID:11544133; http://dx.doi.org/10.1016/S1360-1385(01)02052-0
- Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotech 2005; 16:123-32; PMID:15831376; http://dx.doi.org/10.1016/j.copbio.2005.02.001
- Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J 2005; 388:151-7; PMID:15631617; http://dx.doi.org/10.1042/BJ20041931
- Wang WX, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 2004; 9:244-52; PMID:15130550; http://dx.doi.org/10.1016/j.tplants.2004.03.006
- Liu LJ, Cui F, Li QL, Yin BJ, Zhang HW, Lin BY, Wu Y, Xia R, Tang S, Xie Q. The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res 2011; 21:957-69; PMID:21187857; http://dx.doi.org/10.1038/cr.2010.181
- Zhou J, Zhang Y, Qi JX, Chi YJ, Fan BF, Yu JQ, et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet 2014; 10(1): e1004116; http://dx.doi.org/10.1371/journal.pgen.1004116
- Kraft E, Stone SL, Ma LG, Su N, Gao Y, Lau OS, Deng XW, Callis J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 2005; 139:1597-611; PMID:16339806; http://dx.doi.org/10.1104/pp.105.067983
- Walsh CK, Sadanandom A. Ubiquitin chain topology in plant cell signaling: a new facet to an evergreen story. Front Plant Sci 2014; 5:122; http://dx.doi.org/10.3389/fpls.2014.00122
- Gamerdinger M, Carra S, Behl C. Emerging roles of molecular chaperones and co-chaperones in selective autophagy: focus on BAG proteins. J Mol Med 2011; 89:1175-82; PMID:21818581; http://dx.doi.org/10.1007/s00109-011-0795-6
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu Rev Biochem 2013; 82:323-55; PMID:23746257; http://dx.doi.org/10.1146/annurev-biochem-060208-092442
- Dokladny K, Myers OB, Moseley PL. Heat shock response and autophagy—cooperation and control. Autophagy 2015; 11:200-13; PMID:25714619; http://dx.doi.org/10.1080/15548627.2015.1009776
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2001; 292:1552-5; PMID:11375494; http://dx.doi.org/10.1126/science.292.5521.1552
- Peteranderl R, Rabenstein M, Shin YK, Liu CW, Wemmer DE, King DS, Nelson HC. Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochem 1999; 38:3559-69; http://dx.doi.org/10.1021/bi981774j
- Busch W, Wunderlich M, Schoffl F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 2005; 41:1-14; PMID:15610345; http://dx.doi.org/10.1111/j.1365-313X.2004.02272.x
- Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008; 178:1339-53; PMID:18245858; http://dx.doi.org/10.1534/genetics.107.086199
- Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 2005; 42:535-46; PMID:15860012; http://dx.doi.org/10.1111/j.1365-313X.2005.02397.x
- Rizhsky L, Liang HJ, Shuman J, Shulaev V, Davletova S, Mittler R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 2004; 134:1683-96; PMID:15047901; http://dx.doi.org/10.1104/pp.103.033431
- Desai S, Liu ZX, Yao J, Patel N, Chen JQ, Wu Y, Ahn EE, Fodstad O, Tan M. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7). J Biol Chem 2013; 288:9165-76; PMID:23386620; http://dx.doi.org/10.1074/jbc.M112.422071
- Chan-Schaminet KY, Baniwal SK, Bublak D, Nover L, Scharf KD. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J Biol Chem 2009; 284:20848-57; PMID:19491106; http://dx.doi.org/10.1074/jbc.M109.007336
- Li SX, Liu JX, Liu ZY, Li XR, Wu FJ, He YK. HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in Arabidopsis. Plant Cell 2014; 26:1764-80; PMID:24728648; http://dx.doi.org/10.1105/tpc.114.124883
- Fillatti JJ, Kiser J, Rose R, Comai L. Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Bio-Technol 1987; 5:726-30; http://dx.doi.org/10.1038/nbt0787-726
- Ekengren SK, Liu YL, Schiff M, Dinesh-Kumar SP, Martin GB. Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 2003; 36:905-17; PMID:14675454; http://dx.doi.org/10.1046/j.1365-313X.2003.01944.x
- Xia XJ, Gao CJ, Song LX, Zhou YH, Shi K, Yu JQ. Role of H2O2 dynamics in brassinosteroid-induced stomatal closure and opening in Solanum lycopersicum. Plant Cell Environ 2014; 37:2036-50; PMID:24428600; http://dx.doi.org/10.1111/pce.12275
- Zhou YH, Lam HM, Zhang JH. Inhibition of photosynthesis and energy dissipation induced by water and high light stresses in rice. J Exp Bot 2011; 58:1207-17; http://dx.doi.org/10.1093/jxb/erl291
- Welsch R, Wust F, Bar C, Al-Babili S, Beyer P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol 2008; 147:367-80; PMID:18326788; http://dx.doi.org/10.1104/pp.108.117028
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/10.1006/meth.2001.1262
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004; 16:2967-83; PMID:15494556; http://dx.doi.org/10.1105/tpc.104.025395
- Lai ZB, Li Y, Wang F, Cheng Y, Fan BF, Yu JQ, Chen Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 2011; 23:3824-41; PMID:21990940; http://dx.doi.org/10.1105/tpc.111.090571
