ABSTRACT
Autophagy-dependent longevity models in C. elegans display altered lipid storage profiles, but the contribution of lipid distribution to life-span extension is not fully understood. Here we report that lipoprotein production, autophagy and lysosomal lipolysis are linked to modulate life span in a conserved fashion. We find that overexpression of the yolk lipoprotein VIT/vitellogenin reduces the life span of long-lived animals by impairing the induction of autophagy-related and lysosomal genes necessary for longevity. Accordingly, reducing vitellogenesis increases life span via induction of autophagy and lysosomal lipolysis. Life-span extension due to reduced vitellogenesis or enhanced lysosomal lipolysis requires nuclear hormone receptors (NHRs) NHR-49 and NHR-80, highlighting novel roles for these NHRs in lysosomal lipid signaling. In dietary-restricted worms and mice, expression of VIT and hepatic APOB (apolipoprotein B), respectively, are significantly reduced, suggesting a conserved longevity mechanism. Altogether, our study demonstrates that lipoprotein biogenesis is an important mechanism that modulates aging by impairing autophagy and lysosomal lipolysis.
Introduction
Yolk lipoproteins are large lipid transfer modules capable of transporting phospholipids and cholesterol to oocytes, supplying lipids and amino acids for embryonic development. In C. elegans, the synthesis of the yolk lipoprotein, VIT/vitellogenin, is performed in the endoplasmic reticulum of the intestine, and is initiated before the worm enters adulthood. VIT is secreted and transported to the developing oocytes via receptor-mediated endocytosis.Citation1,2 Since vitellogenesis persists throughout and beyond the reproductive stage, lipoproteins accumulate in the passive circulatory system found between tissues (pseudocoelom) as the animal ages.Citation3,4 Proteomic analyses of long-lived insulin/IGF-1 receptor daf-2 mutants have shown that these animals accumulate substantially less VITCitation5 via transcriptional and post-transcriptional mechanisms,Citation6 suggesting that reducing VIT production may be cytoprotective.
Six isoforms encoding VIT (vit-1,-2,-3,-4,-5,-6) are synthesized in the intestine of C. elegans, of which vit-1 to vit-5 display the highest sequence similarities to the N-terminal domain of human APOB (apolipoprotein B), the precursor of hepatic very-low density lipoprotein and intestinal chylomicrons.Citation7 In mammals, lipoprotein production and secretion can be regulated at various levels: (1) transcriptionally via nuclear repressors and transcriptional activators, (2) post-transcriptionally via cotranslational lipidation and degradation mechanisms, and (3) post-translationally via the ER-to-Golgi intermediary compartment, and Golgi checkpoints that govern secretion of lipoprotein-containing particles.Citation8,9 Excess circulating APOB is linked to the development of age-related diseases such as atherosclerosis, which is characterized by a gradual accumulation of lipids in the intima of the arterial walls and subsequently leads to inflammation, fatty streaks, and ultimately the occlusion of blood flow.Citation10 While the overproduction of atherogenic lipoproteins is physiologically and unambiguously detrimental, the impact of lipoprotein biosynthesis on gene regulation and processes within synthesizing tissues (intestine and liver), somatic maintenance, as well as organismal aging is unclear.
A major life-span determinant in several longevity models is the intracellular recycling process of autophagy.Citation11 It encompasses a multistep process that begins with the sequestration of intracellular material into an autophagosome, followed by its fusion to a lysosome filled with proteases and lipases, that ultimately break down the autophagic cargo into reusable metabolites.Citation12,13 Autophagy is negatively regulated by the nutrient sensor TOR (target of rapamycin; LET-363 in C. elegans), an important regulator of life span in several species, including C. elegans.Citation14,15 Under regular nutrient conditions, TOR inhibits autophagy by phosphorylating and inactivating proteins in the autophagy initiation complex.Citation16 LET-363/TOR also regulates autophagy at the transcriptional level by preventing the nuclear translocation of universally conserved transcription factors, including DAF-16/FOXO, PHA-4/FOXA, and HLH-30/TFEB, that increase the expression of autophagy and lysosomal genes necessary for life span extension in C. elegans.Citation17 Interestingly, HLH-30/TFEB also regulates genes involved in lipid metabolism, suggesting that the survival of an animal under nutrient stress relies on the stringent coordination between pathways of lipid metabolism and autophagy.Citation18,19
We have previously proposed that the breakdown of lipids via autophagy and lysosomal degradation, a process called lipophagy,Citation20 represents a novel mechanism by which life span is prolonged.Citation21,22 Subsequent studies have supported the importance of lipophagy, and more specifically lysosomal acid lipases in the extension of life span,Citation19 and lipid signaling.Citation23,24 Interestingly, several nuclear hormone receptors (NHRs) involved in lipid metabolism have been linked to longevity mechanisms in C. elegans.Citation25 Gonadal steroid signals such as dafachronic acid have been implicated in facilitating communication between the reproductive system and the soma, via the NHR DAF-12.Citation26,27 Another NHR, NHR-49, has been associated with the ability of nematodes to activate beta-oxidation genes under conditions of limited nutrients.Citation28 Moreover, in long-lived germline-less nematodes, NHR-80 is activated, resulting in an increased expression of genes involved in lipid desaturation.Citation29 While evidence for complex and conserved relationships between lipid metabolism, reproduction, and life span is accumulating (reviewed in ref. 30), it is still unclear how they are coordinated to maintain optimal cellular conditions that ultimately foster longevity.
Our current study addresses this relationship by investigating the role of VIT production in mediating changes in lipid metabolism, autophagy and longevity. We found that VIT production and accumulation is reduced in various longevity models, and that artificially increasing vitellogenesis leads to a significant reduction in the life span of long-lived animals, by impairing the induction of autophagy-related and lysosomal lipase genes. Along these same lines, we observed that reducing vitellogenesis extends life span by stimulating autophagy and lysosomal lipolysis. We also demonstrate that longevity mediated by reduced VIT, or enhanced lysosomal lipolysis is dependent on NHRs NHR-49 and NHR-80. These observations suggest that lipids produced via lipophagy may act as signals, via these NHRs, to mediate life span extension. Interestingly, we found that dietary restriction in mice decreases the expression of hepatic Apob, the mammalian counterpart of VIT. Our data point to a conserved longevity mechanism whereby a reduction in lipoprotein biogenesis leads to the redirection of lipids previously destined to the periphery toward a cascade of autophagy, lipid remodeling and signaling. Altogether our study demonstrates an important and previously unrecognized role for lipoprotein production in transcriptionally modulating autophagy and determining life span.
Results
VIT overexpression reduces the life span of long-lived animals
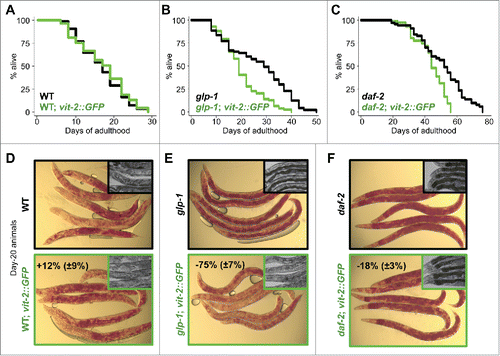
To investigate the role of reduced VIT production in the longevity mechanism of autophagy-dependent, long-lived animals, we assessed the effect of increasing vitellogenesis on their life span. A previously constructed strainCitation2 expressing a highly conserved VIT protein, VIT-2, fused to GFP under its endogenous promoter was utilized to generate elevated levels of VIT (Fig. S1A). Expressed in wild-type animals, VIT-2::GFP localized primarily to the oocytes in early adulthood, as previously reportedCitation2, and accumulated in the pseudocoelomic fluid and gonads after reproduction ended (d 7). A modest decrease in VIT accumulation was also observed when vit-2::GFP was expressed in glp-1(e2141) animals (Fig. S1A). In long-lived insulin/IGF-1 receptor daf-2(e1370) mutants, we found that vit-2::GFP expression was markedly attenuated (Fig. S1A), consistent with previous work showing that animals in which DAF-16/FOXO is active in the intestineCitation31,32 have reduced VIT gene expressionCitation33 and protein accumulation.Citation5 Overexpression of vit-2 had no effect on the life span of wild-type animals (, Table S1). However, it significantly reduced the long life span of glp-1 and daf-2 mutants ( and , and Table S1). Notably, progeny production and pharyngeal pumping were not affected by increased VIT-2 production in wild-type, glp-1 or daf-2 animals (Fig. S1B), suggesting that increasing VIT-2 biogenesis did not reduce the life span of glp-1 and daf-2 animals through alterations in reproduction or food intake, both physiological aspects linked to life span.
Figure 1. VIT overexpression impairs longevity. Life span analyses of wild-type (WT) (A), germline-less glp-1(e2141) (B), insulin/IGF-1 receptor mutant daf-2(e1370) (C) and their corresponding vit-2-overexpressing transgenic counterparts (green) grown on OP50 E. coli. Details on life span analyses are found in Table S1. Micrographs of Oil-Red-O stained day-20 wild-type (WT) (D), germline-less glp-1(e2141) (E), insulin/IGF-1 receptor mutant daf-2(e1370) (F) and their corresponding vit-2-overexpressing transgenic counterparts. Insets in panels (D to F) are corresponding micrographs for each strain (black border, parent strain; green border, transgenic strain) at d 20, displaying the dark intestine phenotype in the proximal intestine associated with intestinal lipid droplet storage.
Since the life span of wild-type animals was not affected by the expression of vit-2::GFP, we explored the effect of further increasing the accumulation of VIT in the pseudocoelom on aging. Silencing rme-2—the gene encoding the VIT receptor, which is expressed in the gonadsCitation2—led to VIT-2::GFP accumulation in the pseudocoelomic fluid (Fig. S2A). As expected, a similar profile of VIT distribution was observed in germline-less glp-1 animals expressing vit-2::GFP (Fig. S1A) due to the lack of rme-2 expression in the absence of a developing germline (Fig. S2B). More importantly, the redistribution of VIT lipoproteins in the pseudocoelom was not detrimental for the life span of wild-type animals, or wild-type animals expressing vit-2::GFP (Fig. S2C and D, and Table S2) in which vit-2 expression was increased by 3 fold (Fig. S2E). Altogether, these observations suggest that the peripheral accumulation of VIT is not likely to precede or initiate the aging process.
VIT overexpression reduces intestinal lipid storage in long-lived animals
Larger stores of neutral lipid droplets as measured by Oil-Red-O staining were maintained throughout the life span of glp-1 and daf-2 mutants, relative to wild-type animals (–, and S1C).Citation34 In contrast, glp-1 and daf-2 mutants expressing vit-2::GFP had significantly lower levels of lipid stores on d 20 (−75% and −18%, respectively), evident from the reduction in Oil-Red-O staining (–, and S1C). The loss of neutral lipid droplets from the intestines of these animals could also be deduced from the lighter and less defined profiles of their mid sections (insets within –). The noticeably smaller effect of VIT overexpression on lipid reduction in daf-2 animals is likely due to the reduced expression of the vit-2::GFP transgene (Fig. S1A). Overall, the failure of long-lived animals to maintain their intestinal lipid stores when vit-2::GFP is expressed may suggest that higher levels of VIT result in the secretion of lipids that would otherwise be stored and/or remodeled.
VIT overexpression impairs the transcriptional induction of lysosomal lipolysis
Longevity in nematodes is generally dependent on the activity of transcription factors including but not limited to, PHA-4/FOXA, DAF-16/FOXO and HLH-30/TFEB.Citation11 The expression of these transcription factors is increased in different longevity models and their levels have been shown to correlate with their activity as they can autoregulate their own expression.Citation11 Hence, to determine the role of lipoprotein biogenesis in the mechanisms promoting longevity, we investigated the effect of increasing VIT levels on the expression and activity of these longevity-associated transcription factors. VIT-2 overproduction led to a significant decrease in pha-4 mRNA levels in glp-1 animals (). The expression of autophagy gene lgg-1 (ortholog of GABARAPCitation35), a PHA-4/FOXA target geneCitation20,36, was decreased in both glp-1 and daf-2 animals, while Igg-2 (ortholog of LC3A and LC3B) was decreased in glp-1 animals (). Overexpressing vit-2 also resulted in decreased daf-16 levels, as well as the DAF-16/FOXO target gene sod-337, in both glp-1 and daf-2 animals (). In contrast, expression of the lysosomal serine carboxypeptidase homolog of cathepsin A, C08H9.1, a DAF-16/FOXO target,Citation38 was unaffected, suggesting that the expression of specific subsets of DAF-16/FOXO targets may be differentially altered by increased vitellogenesis.
Figure 2. VIT overexpression prevents autophagy-associated gene induction. (A) QPCR analysis of pha-4 and pha-4-regulated autophagy targets lgg-1 and lgg-2, (B) daf-16 and daf-16-regulated targets sod-3 and C08H9.1 as well as (C) members of the lysosomal acid lipase gene family (lipl-1 to lipl-8) in d-1 WT, glp-1, daf-2 animals and their transgenic counterparts. N = 3 per condition. Graphs represent mean±SD; *, P<0.05 vs parent strain using the Student t test.
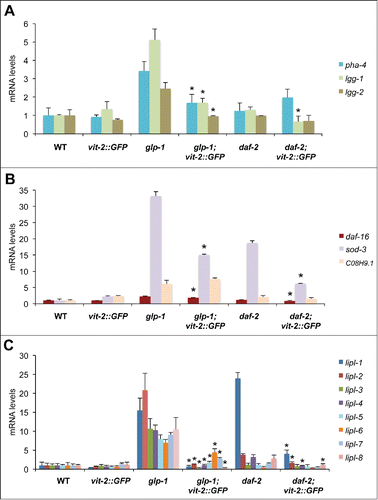
Recent reports suggest that the transcriptional induction of lysosomal lipolysis genes contributes to longevity in glp-1 and daf-2 animals by stimulating lipophagy.Citation21,39 We analyzed the expression of lysosomal lipase genes in glp-1 and daf-2 mutants and found that relative to wild-type animals, these long-lived mutants express significantly higher levels of 4 or more lysosomal acid lipase genes relative to wild-type (), which corresponded to significantly higher lysosomal acid lipase activity (Fig. S1D). Strikingly, vit-2 overexpression markedly inhibited the induction of several lysosomal acid lipase genes of the lipl family, specifically lipl-1 to lipl-8 in glp-1, and lipl-1, −2, −4 and -8 in daf-2 animals (). Accompanying this observation was the significant decrease in lysosomal acid lipase activity in glp-1 mutants, and to a smaller extent in daf-2 animals expressing vit-2::GFP (−85% and −25%, respectively) (Fig. S1D). The effect of expressing vit-2::GFP on lipase expression appeared specific to lysosomal acid lipases, as the expression of cytosolic lipase genes, atgl-1 and hosl-1 had no notable change (Fig. S1E). Therefore, by artificially increasing the production of VITs, we effectively interfered with the expression of pha-4 and daf-16, as well as their targets, which include autophagy and lysosomal lipase genes. This previously unrecognized link between vitellogenesis, autophagy and lysosomal lipolysis, suggests that VIT production may inhibit the ability of organisms to mount and maintain a protective response to age-related metabolic and proteostatic declines.Citation40
Reducing vitellogenesis increases life span and neutral lipid accumulation
To clarify the role of VITs in the life span of C. elegans, we silenced VITs by using E. coli expressing double-stranded RNA (RNAi) against vit genes (vit-1/2, −3, −4 and −5), and studied the effect on life span and lipid accumulation. Vit-1/2 RNAi (hereafter referred as vit RNAi) sufficed in silencing vit-1 to vit-5 ( and S3A and B), largely due to the high level of homology shared by the targeted sequences. The reduction in vit gene expression resulted in a significant (16 to 40%) increase in the life span of wild-type animals ( and Table S3), without affecting progeny production and pumping rates (Fig. S3C). These observations are consistent with a previous report that also demonstrates life span extension when another RNAi, vit-5, was used (Table S3).Citation33 Altogether, these data indicate that reducing the production of VITs has beneficial effects on the life span of C. elegans without relying on a reproductive trade-off.
Figure 3. VIT reduction increases life span, lipid storage, autophagy and lysosomal lipolysis. (A) QPCR analysis of the expression of vit genes in d-1 animals fed control bacteria or bacteria expressing dsRNA against vit-1 from hatching (N = 3 per condition). (B) Life span analysis of animals fed control bacteria or bacteria expressing dsRNA against vit during adulthood. (C) Oil-Red-O staining of d-20 wild-type fed control bacteria or bacteria against vit during adulthood (insets are corresponding micrographs of each treated group). (D) Quantification of GFP::LGG-1 foci in the proximal intestine of d-1 wild-type animals fed control bacteria or bacteria expressing dsRNA against vit from hatching. *, P < 0.001, N = 28 worms per condition, ±SEM, t test. (E) QPCR analysis of the expression of lysosomal acid lipase (lipl-1 to lipl-8) genes in d-3 animals fed control bacteria or bacteria expressing dsRNA against vit from hatching (N = 3 per condition). ±SD; *, P<0.05; N = 3, t test. (F) Measurement of lysosomal acid lipase activity in wild-type animals and animals with reduced vitellogenesis. Fluorometric values normalized to the wild type. ±SD; *, P < 0.05; N = 3 per condition, t test. Life-span analysis of atg-18(gk378) mutants (G), lipl-3(tm4498) (H) and lipl-4(tm4417) mutants (I), daf-12(rh61rh411) mutants (J), nhr-49(nr2014) mutants (K) and glp-1(e2141);nhr-80(tm1011) mutants (raised at the permissive temperature) (L) fed control bacteria or bacteria expressing dsRNA against vit during adulthood. Details on life span analyses are found in Table S3.
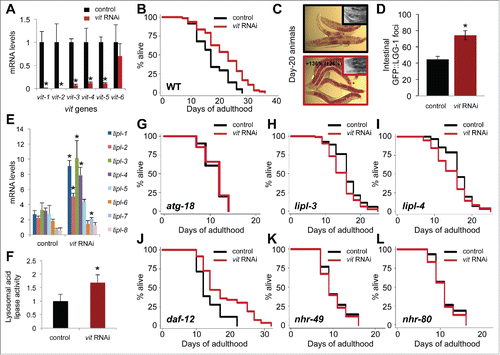
Interestingly, silencing vit genes with RNAi led to a significant increase in neutral lipid accumulation (+136%) in the intestine of wild-type animals, similar to glp-1 and daf-2 animals ( and ), which was evident by Oil-Red-O staining ( and S3D). It is therefore conceivable that the lipids bound for yolk lipoprotein biogenesis and secretion were redirected to the cytosol of intestinal cells in the form of neutral lipid droplets. It is important to note that the life span of wild-type animals was unchanged when intestinal lipid storage was increased by inhibiting the expression of the cytosolic lipase atgl-1 (−67% lipase activity), indicating that solely increasing cytosolic intestinal lipid storage is not sufficient to extend life span (Fig. S4A and B, and Table S4). While intestinal lipid droplet accumulation was also observed in longevity models, including glp-1 and daf-2Citation34,41,42 (–), the origin of these lipids is not clear. Some daf-2 mutant strains, including daf-2(e1370), display an increase in de novo lipogenesis.Citation43 In glp-1 animals, the expression of lipogenic genes, fasn-1 (fatty acid synthase), and pod-2 (acetyl-CoA carboxylase) were elevated (Fig. S4C), suggesting that intestinal lipid accumulation may be driven by increased lipogenesis. Interestingly, these lipogenic genes were also required for the long life span of glp-1 animals (Fig. S4D and E, Table S4), lending further support to the importance of lipogenesis in life span extension. However, expression of fasn-1 and pod-2 was not increased in wild-type animals with reduced vitellogenesis (Fig. S4F). Therefore, we propose that reduced VIT-containing lipoprotein production, along with sustained (or increased) lipogenesis more aptly explain the increase in intestinal neutral lipid accumulation observed in glp-1 and daf-2 mutantsCitation41,43 ( and ), as well as animals with reduced vitellogenesis ( and S3D). Altogether, our data led us to conclude that VIT production play specific roles in intestinal lipid homeostasis and life span extension.
Reducing vitellogenesis increases autophagy and lysosomal lipolysis
To gain insight into the mechanism by which vitellogenesis modulates longevity, we explored the effects of reducing VIT levels on autophagy, lysosomal lipolysis as well as the expression of longevity-related transcription factors and their known targets. Animals subjected to vit RNAi showed an increase in GFP::LGG-1 foci in their proximal intestines (),Citation21,44 which suggests an increase in autophagosome formation. Expression of longevity-related transcription factors pha-4/foxa and daf-16/foxo were also increased in these animals compared to wild type (Fig. S3E), while levels of HLH-30/TFEB were unaffected. Wild-type worms expressing daf-16::GFP displayed increased levels upon vit silencing (Fig. S3F), further corroborating the activity of DAF-16/FOXO.Citation31,45 Loss of VIT production also modified the transcriptional profile of genes required for life span extension. The expression of the autophagy-related targets of PHA-4/FOXA, namely lgg-1 and lgg-2, was increased (Fig. S3E), as was the expression of the DAF-16/FOXO target sod-3. However, the mRNA levels of another DAF-16/FOXO target, CO8H9.1, remained unchanged (Fig. S3E). Expression of HLH-30/TFEB target, atg-18Citation46, was increased, but not that of another target, vha-16 (Fig. S3E). Notably, the expression of lysosomal acid lipase genes (lipl-1, −2, −3, −4 and -7), which are regulated by DAF-16/FOXOCitation39 and HLH-30/TFEB,Citation19 were significantly increased by vit silencing (). Accordingly, lysosomal acid lipase activity was increased significantly (). Notably, the expression of cytosolic lipase genes atgl-1 and hosl-1 was unaffected (Fig. S3E). These analyses highlight specific upregulation of longevity-associated transcription factors, as well as autophagy and lysosomal lipolysis genes that arise from reduced vitellogenesis, reminiscent of the expression profiles of the previously analyzed longevity models (–).
Longevity associated with vit silencing requires autophagy, lysosomal lipases, DAF-16/FOXO and HLH-30/TFEB
As the longevity-associated processes of autophagy and lysosomal lipolysis were induced when animals were subjected to vit RNAi (–), we sought to confirm the role of autophagy and lysosomal lipases in the long life span mediated by reduced vitellogenesis. ATG-18 (ortholog of mammalian WIPI1/2) is a fundamental protein involved in the initiation of autophagy.Citation47 We found that loss of atg-18 prevented the life-span extension conferred by vit RNAi (). Silencing VIT in animals bearing mutations in lysosomal lipases lipl-3 or lipl-4 did not increase life span (). Collectively, these results indicate that both autophagy and lysosomal lipolysis contribute to the longevity mechanism associated with reduced vitellogenesis.
The long life span of glp-1 animals requires the LET-363/TOR-regulated transcription factors DAF-16/FOXO, HLH-30/TFEB, as well as the NHR DAF-12, in order to activate the expression of longevity-associated genes.Citation11 Since glp-1 animals display reduced accumulation of VIT-2::GFP (Fig. S1A), we aimed to determine whether these longevity modulators also mediate the life-span extension achieved by reducing vitellogenesis. We found that vit silencing failed to extend the life span of daf-16 and hlh-30 mutants (Fig. S3H and I and Table S3), indicating a role for these transcription factors in this longevity regiment, and highlighting potential functional overlap between HLH-30 and DAF-16.Citation19,39,48 Interestingly, loss of daf-12 did not prevent vit silencing from extending the life span of wild-type animals ( and Table S3) and vit silencing had no effect on progeny production and pharyngeal pumping (Fig. S3C), suggesting that germline signaling does not contribute to the longevity mechanism associated with reduced vitellogenesis.
Lipophagy-mediated longevity requires NHR-49 and NHR-80
NHRs NHR-49 and NHR-80 are required for the longevity of various longevity models,Citation28,29 including the long-lived glp-1 animals (Table S4).Citation27,49 NHR-49 regulates the expression of several genes involved in beta-oxidation,Citation28 while NHR-80 regulates the expression of lipid remodeling genes, including fatty acid desaturases.Citation29 We assessed the role of these NHRs in longevity and lipid remodeling, specifically in the context of how vitellogenesis and lysosomal lipolysis modulate lipid homeostasis and lipid signaling.Citation23 In animals subjected to vit silencing, major targets of NHR-49, acs-2 (acyl-CoA synthetase), and lbp-8 (lipid binding protein), showed elevated levels of expression while ech-1 remained unaffected (Fig. S5A). The expression levels of delta-9 fatty acid desaturases fat-5, fat-6 and fat-7, which are key targets of NHR-80, were also significantly elevated (Fig. S5B). It is particularly noteworthy that vit silencing failed to extend the life span of nhr-49 and nhr-80 mutants ( and ) as this demonstrates the importance of NHR-49 and NHR-80 in the longevity associated with reduced vitellogenesis. Additionally, these mutants were unable to increase lipid storage under conditions of reduced vitellogenesis (Fig. S5C and D), demonstrating the function of these NHRs extends beyond lipid remodeling and into lipid storage. These findings support our previous work on the interdependent relationship between autophagy, lipid remodeling and lipid storage.Citation34 Altogether, these observations suggest that in order to promote longevity, reduced vitellogenesis needs to be coordinated with transcriptional changes in genes involved in lipid remodeling, transport and breakdown.
As reduced vitellogenesis increases lysosomal lipolysis gene expression, we utilized animals overexpressing lipl-4Citation39—a gene previously shown to be elevated when vitellogenesis is reduced ()—to verify the role of both NHR-49 and NHR-80 in longevity, and establish the potential of lysosomal lipolytic productsCitation23 as signaling molecules for NHR signaling. In wild-type animals, silencing of either nhr-49 or nhr-80 resulted in modest reductions in life span (Fig. S5E and Table S5). In contrast, life span was markedly reduced when nhr-49 or nhr-80 was silenced in lipl-4-overexpressing animals (Fig. S5F and Table S5), suggesting that products of lysosomal lipolysis likely contribute to the activation of NHR signaling necessary for maintaining autophagy and promoting longevity, as recently suggested.Citation24,50,51 In addition to our initial findings that increased VIT production impairs lysosomal lipase gene expression (), these observations reveal VIT as a central player in the modulation of autophagy and lysosomal lipolysis, which are key mechanisms that promote longevity in several models.Citation39,52 Our data also suggest that long-lived animals coordinate lipid metabolism with their transcriptional output to enhance autophagy and lysosomal lipolysis.
Dietary-restricted animals display reduced vitellogenesis and elevated lysosomal lipase gene expression
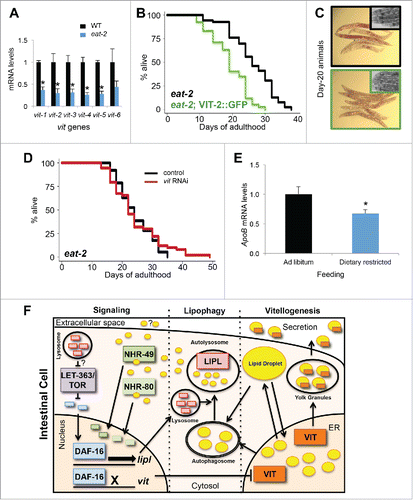
Having established the importance of vitellogenesis in autophagy and lysosomal lipolysis, we sought to determine whether VIT production also plays a role in the ability of an animal to survive nutrient stress, especially since enhanced autophagy mediates life-span extension by dietary restriction.Citation53 The expression of vit genes was significantly decreased in pumping-defective, dietary-restricted, eat-2(ad1116) mutants (), which mirrors the markedly decreased levels of VIT protein observed in glp-1 and daf-2 animals (Fig. S1A).Citation5 This suggests that both animal models may have a similar requirement of reduced vitellogenesis for longevity. Concomitantly, the expression of lysosomal acid lipases (lipl-1, −2, −3, −4 and -5) in eat-2 animals was significantly increased (Fig. S6A). Similarly, silencing the nutrient sensor let-363/tor also led to an increase in lysosomal lipase expression (lipl-1, −2 and −4) (Fig. S6B). These observations demonstrated that multiple, mechanistically distinct longevity models display increased lysosomal lipolysis. As seen in other longevity models (Fig. S1A), we observed that vitellogenesis is suppressed in eat-2 animals (Fig. S6C). Increasing vitellogenesis by expressing vit-2::GFP in eat-2 animals reduced their life span significantly (, Table S1), in addition to producing animals devoid of neutral lipid stores (). Silencing let-363/tor in vit-2::GFP-expressing animals failed to extend their life span (Fig. S6D, Table S2), suggesting that the life-span extension associated with reduced nutrient sensing is sensitive to VIT production. Likewise, further reduction of vitellogenesis by subjecting eat-2 animals to vit RNAi did not lead to additional extension in life span, as seen in glp-1 animals (, Table S3). Altogether, these observations indicate that the reduction in vitellogenesis is an important mechanism in distinct longevity models.
Figure 4. Dietary-restricted animals have reduced lipoprotein expression. (A) QPCR analysis of vit genes in d-1 adult wild-type and eat-2(ad1116) animals. ±SD; *, P <0.05, N = 3 per condition, t test. (B) Life-span analysis of eat-2(ad1116) animals and eat-2(ad1116) animals expressing vit-2::GFP. Details on life-span analyses are found in Table S1. (C) Oil-Red-O staining of day-20 eat-2(ad1116) animals and eat-2(ad1116) animals expressing vit-2::GFP (insets are corresponding micrographs of each strain). (D) Life-span analysis of eat-2(ad1116) animals fed control bacteria or bacteria expressing dsRNA against vit. Details on life-span analyses are found in Table S3. (E) QPCR analysis of hepatic Apob expression of mice fed ad libitum or dietary restricted at 3 mo of age for 5.5 wk. ±SEM; *, P<0.05; N = 8 to 10 per condition, t test. (F) Model depicting the connections associated with vitellogenesis, lipophagy and lipid signaling. During vitellogenesis, ER lipids are packaged into secretable yolk lipoproteins. Reduction in vitellogenesis results in the redistribution of lipids into neutral lipid droplets and leads to an increase in the expression of autophagy genes and lipl genes involved in lipophagy. Lipid signaling products of lysosomal lipolysis can serve as activators of nuclear hormone receptors NHR-49 and NHR-80, as well as potential inter-tissue signaling molecules. Their translocation to the nucleus stimulates lipid metabolic gene expression in conjunction with DAF-16 (which is negatively regulated by LET-363/TOR) activating lipl gene expression and repressing vit gene expression, thereby maintaining elevated autophagy and lysosomal lipolysis.
Reduced lipoprotein production is conserved in dietary-restricted animals
To evaluate whether the reduction in lipoprotein biogenesis in dietary-restricted animals is conserved, we measured the expression of the murine VIT ortholog, apolipoprotein B (Apob). We found that hepatic levels of Apob expression were significantly decreased in dietary-restricted mice (), suggesting that lowering lipoprotein biogenesis is conserved and integrated in longevity models. Concomitantly, we found that the hepatic expression of Foxa3 (ortholog of pha-4) is modestly increased. Moreover, the targets Lc3a and Lc3b (orthologs of lgg-235) were significantly increased in dietary-restricted mice, compared to ad libitum-fed mice (Fig. S6E), suggesting the potential conservation of transcriptional induction of autophagy induction mediated by reduced lipoprotein biogenesis across phyla. Overall, these observations highlight a shared mechanism in long-lived and nutrient-restricted animals by which lipoprotein biogenesis is reduced to enable long-term survival. Altogether, our data supports a model in which lipoprotein biogenesis prevents life span extension by distributing lipids away from the intestine, and by negatively regulating the induction of autophagy-related and lysosomal lipase genes, thereby challenging the animal’s ability to maintain lipid homeostasis and somatic maintenance ().
Discussion
Lipoprotein biogenesis is an important process that facilitates the transport of lipids between tissues, but its role in somatic maintenance has not been clearly established. Here, we provide evidence of an important role for lipoprotein production in the transcriptional induction of autophagy, a recently-uncovered longevity mechanism.Citation17 We observed that lipoprotein biogenesis governs the ability of animals to induce autophagy- and lysosomal lipolysis-related genes involved in lipid remodeling and signaling associated with long-term survival.
VIT production modulates the transcriptional regulation of autophagy and lysosomal lipase genes
Our study shows that regulating yolk lipoprotein biogenesis in C. elegans can modulate aging by preventing the transcriptional activation of autophagic and lysosomal lipolytic genes. Increasing vitellogenesis can impair the activities of PHA-4/FOXA and DAF-16/FOXO, while reducing vitellogenesis enhances their expression, as well as that of some of their targets, which includes autophagy and lysosomal acid lipase genes. The life-span extension mediated by VIT silencing requires the activity of both DAF-16/FOXO and HLH-30/TFEB, pointing to potential overlapping roles of these transcription factors in the induction of lysosomal acid lipase expression ().Citation19,39 Since LET-363/TOR regulates both DAF-16/FOXO and HLH-30/TFEB, VIT production may be integrated with LET-363/TOR signaling to modulate life span (). Indeed, we found that life-span extension by TOR silencing was reliant on modest level of vitellogenesis (Fig. S6D). As LET-363/TOR inhibition leads to the DAF-16-mediated induction of lipl-452 and since VIT expression was markedly increased in glp-1 animals in which daf-16 was mutated (Fig. S3G), we conclude that DAF-16 plays a central role in the balance between vitellogenesis and lysosomal lipolysis (). Autophagy enhancements from reduced lipoprotein production may also involve the activation of the lysosomal and autophagy regulator HLH-30/TFEB.Citation54 Interestingly, cessation of vitellogenesis in the fat body of mosquitoes increases lysosomal activity, suggesting that the relationship between VIT and lysosomal activity may be conserved across phyla.Citation55 Indeed, nematodes and mice both display reduced lipoprotein gene expression and elevated expression of autophagy proteins, LC3A and LC3B, key proteins involved in autophagosome formation. We conclude that lipoprotein biogenesis is coordinated with the transcriptional activation of genes in the autophagy-lysosomal pathway.
Lipophagy-mediated lipid signaling, a novel mechanism for life-span extension
Redistribution of lipids by reducing vitellogenesis concomitant with changes in lipogenesis, fatty acid chain length and saturation degree,Citation43,56 appears to contribute to lipid metabolic changes necessary for life-span extension. To mediate these changes, lipid remodeling and desaturation are driven by the activity of NHRs, including NHR-49 and NHR-80. Here, we find that these NHRs play a crucial role in life-span extension associated with reduced vitellogenesis or enhanced lysosomal acid lipase overexpression. Signaling to these NHRs appears to be mediated by products of lysosomal lipolysis, including the polyunsaturated fatty acid eicosanopentanoic acid (EPA)Citation23 and in a recent report, by the fatty acid oleoylethanolamide.Citation24 As VITs are also major transporters of cholesterolCitation57 and C. elegans obtain cholesterol from their diet,Citation58 it is possible that maintenance of cholesterol stores in the intestine also improves survival. Recent work in yeast has shown that the maintenance of vacuolar microdomains involved in lipophagy is dependent on sterols.Citation59 Notably, artificial, cholesterol-producing C. elegans can live longerCitation60 and a cholesterol-binding protein modulates DAF-16 activity.Citation61 Additionally, cholesterol-mediated signaling via lipophagy may be enhanced when vitellogenesis is reduced as lysosomal lipases also possess physiologically important cholesterol esterase activity.Citation62 Interestingly, 2 recent studies have uncovered roles for NHRs NR1H4/FXR and PPARA/PPARα (NHR-49 ortholog) in modulating TFEB-mediated transcriptional regulation of autophagy.Citation50,51 Therefore, the action of lipophagy on NHR-49 and NHR-80 may extend beyond lipid remodeling to maintain beneficial levels of autophagic flux, potentially by interacting with longevity-associated transcription factors.
Moving forward, identifying key lysosomal lipolysis-mediated lipid signals will be critical for gaining a deeper understanding of the mechanism by which lipophagy increases longevity. It will also be interesting to determine the role of other recently uncovered longevity-associated nuclear hormone receptors (e.g., NHR-8 and NHR-62)Citation63-65 in mediating transcriptional and lipid metabolic changes in VIT-depleted animals. Taken together, our study presents the interesting possibility whereby intracellular lipid distribution is integrated with the induction of autophagy, a quintessential process in somatic maintenance ().
Dietary-restriction effects on lipoprotein biogenesis are conserved
Because of their role in reproduction and their impact on intestinal lipid metabolism, yolk lipoproteins are relevant in the discussion of potential molecular trade-offs between reproduction and somatic maintenance.Citation66 Our data from nematodes demonstrate that increasing vitellogenesis has no effect on the life span of wild-type animals, but is detrimental for the long-term survival of dietary-restricted (eat-2), germline-less (glp-1), LET-363/TOR-inhibited animals, and mutants of the insulin/IGF-1 receptor (daf-2). The life span of animals with reduced expression of the VIT receptor, rme-2, remains unaffected, despite increased levels of VIT, suggesting that the accumulation of VIT in the pseudocoelemic fluid is not detrimental to the animal. Our data suggest that the accumulation of lipids outside of the intestine, at least via vitellogenesis, may not be a major contributor to nematode aging, as recently speculated.Citation67 Indeed, increasing VIT expression in wild-type flies has recently been found to have no effect on life span.Citation68 A previous study suggests that reduction of VIT production in dietary-restricted flies does not contribute to life span extension.Citation69 Unexpectedly, we find that dietary-restricted nematodes require low VIT levels to live long. In dietary-restricted worms and mice, the transcription of vit genes and hepatic Apob is significantly reduced, suggesting that the promoter regions of these homologous lipoprotein genes may contain conserved elements responsive to the nutrient status of the animal. Further studies of the promoter region of these lipoprotein genes may reveal novel factors and epigenetic changes that contribute to longevity. Altogether, our study of lipoprotein metabolism in the context of aging supports a specific molecular trade-off in which lipids transported away from the soma via lipoprotein secretion prevent the initiation of the key protective cellular processes of autophagy.
Implications in human diseases
Recently, rare functional variants of the APOB gene have been found to correlate with centenarians, suggesting a role for the regulation APOB protein production in extreme longevity.Citation70 Indeed, the production of APOB is a critical marker for the development of atherosclerosis and remains an attractive target for the prevention of cardiovascular diseases.Citation71 Notably, use of specific RNAi to silence APOB gene expression in primates results in significant reduction in serum APOB.Citation72 As lipid metabolic pathways are conserved between C. elegans and mammalsCitation73, it will be interesting to determine the effect of reduced APOB production on autophagy activation and nuclear hormone signaling. Our data provides evidence on the potential benefits of reducing APOB-containing lipoprotein production, not only with respect to cardiovascular risks, but also for preventing age-related decline in liver or intestine, by enhancing autophagy, lysosomal lipolysis and lipid-mediated signaling, which improves somatic maintenance. Therefore, silencing APOB could provide an alternative strategy to enhance autophagy in order to combat age-related metabolic and proteostatic decline.
Overall, this study uncovers a physiological balance between lipoprotein production and autophagy, providing a molecular mechanism that links lipid metabolism and somatic maintenance in the context of aging. Our data suggest a conserved mechanism between dietary-restricted worms and mice leading to reduced lipoprotein expression and increased induction of autophagy-related genes. Therefore, we propose a model () in which long-term survival is initiated by reduced lipoprotein biogenesis, resulting in the activation of longevity transcription factors, autophagy and lysosomal lipolysis, and ultimately nuclear hormone receptor signaling to promote and maintain the transcriptional activation of genes that extend life span.
Methods
Strains
Nematodes were maintained at 20°C on OP50 E. Coli as previously describe.Citation74 A strain list is available in Table S6.
Life-span analysis
Synchronized animals were prepared with hypochlorous acid and transferred onto bacteria-seeded plates. All life-span analyses were carried out at 20°C during adulthood. Viability was scored every 2 or 3 d, as previously described.Citation75 RNAi clones from the Ahringer library were usedCitation76 and the plasmid expressing dsRNA against let-363/tor was previously published.Citation77
Gene expression analysis
Each sample contained approximately 500 worms, RNA was extracted and cDNA was prepared as described.Citation46 Biological triplicates were analyzed using SYBR green (Roche, 04887352001) and a Roche LightCycler 480 (Indianapolis, IN). Expression was normalized using 3 to 6 housekeeping genes, including act-1, cyn-1, cdc-42, pmp-3, ama-1 and nhr-23. For the mice study, dietary restriction protocol, sample preparation and analysis were carried out as described.Citation46 A primer list is available upon request.
Oil-Red-O staining
Synchronized worms were aged to d 7 or d 20 and fixed in a 2% paraformaldehyde solution and stained with Oil-Red-O (Sigma, O0625) as described in.Citation41
Fluorescence microscopy
Transgenic worms were mounted and aligned on a 2% agarose pad using a M9 solution (42 mM Na2HPO4 22 mM KH2PO4 86 mM NaCl, 1 mM MgSO4-7H2O) containing 0.1% sodium azide. Oil-Red-O stains and expression of GFP fusion proteins was observed using a fluorescence dissecting scope (Leica DFC310 FX, Buffalo Grove, IL) and images were taken with a Leica DFC310 FX camera.
Lipase assays
Lysosomal lipase activity was assayed using a fluorimetric assay as previously described.Citation19 Neutral lipase activity was measured using a colorimetric assay as previously described.Citation52 Fluorometric and colorimetric values were normalized to the wild type. For each sample, ∼300 worms were homogenized.
Statistical analysis
Statistical significance was determined by the Student t test or one-way ANOVA (Dennett test) using the Graph Pad Prism 6 Software. Kaplan-Meier survival curves and P values for life span assays were obtained by plotting data and using Log Rank (Mantel-Cox) test with the software Stata 13.
Abbreviations
| APOB | = | apolipoprotein B |
| ER | = | endoplasmic reticulum |
| FOXA | = | forkhead protein box A |
| FOXO | = | forkhead protein box O |
| NHR | = | nuclear hormone receptor |
| TFEB | = | transcription factor EB |
| TOR | = | target of rapamycin |
| VIT | = | vitellogenin |
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
KAUP_A_1127464_Supplemental_Info.zip
Download Zip (16.5 MB)Acknowledgments
The authors would like to thank Jane Park and Andrew Davis for technical assistance. We would also like to thank Drs. Ryan Birse, Carissa Perez Olsen and Marc Tatar for comments. We are grateful to Dr. Shohei Mitani and the National BioResource Project for providing lysosomal lipase mutant strains.
Funding
This work was primarily funded by a K99 award (K99 AG042494) and an R00 grant (R00 AG042494) to LRL from the NIH/NIA. This work was also funded by an AFAR Junior Faculty Research Grant to LRL, and R01 AG038664/R01 AG039756 to MH and R01 AG027463 to AD from the NIA/NIH.
References
- Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev Biol 1983; 96:189–96; PMID:6825952; http://dx.doi.org/10.1016/0012-1606(83)90322-6
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell 1999; 10:4311–26; PMID:10588660; http://dx.doi.org/10.1091/mbc.10.12.4311
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 2002; 419:808–14; PMID:12397350; http://dx.doi.org/10.1038/nature01135
- Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, Morimoto RI, Dobson CM, Vendruscolo M, Mann M, et al. Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell 2015; 161:919–32; PMID:25957690; http://dx.doi.org/10.1016/j.cell.2015.03.032
- Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR, 3rd. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 2007; 317:660–3; PMID:17673661; http://dx.doi.org/10.1126/science.1139952
- DePina AS, Iser WB, Park SS, Maudsley S, Wilson MA, Wolkow CA. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol 2011; 11:11; PMID:21749693; http://dx.doi.org/10.1186/1472-6793-11-11
- Babin PJ, Bogerd J, Kooiman FP, Van Marrewijk WJ, Van der Horst DJ. Apolipophorin II/I, apolipoprotein B, vitellogenin, and microsomal triglyceride transfer protein genes are derived from a common ancestor. Journal of molecular evolution 1999; 49:150–60; PMID:10368443; http://dx.doi.org/10.1007/PL00006528
- Fisher E, Lake E, McLeod RS. Apolipoprotein B100 quality control and the regulation of hepatic very low density lipoprotein secretion. Journal of biomedical research 2014; 28:178–93; PMID:25013401
- Brodsky JL, Fisher EA. The many intersecting pathways underlying apolipoprotein B secretion and degradation. Trends Endocrinol Metab 2008; 19:254–9; PMID:18691900; http://dx.doi.org/10.1016/j.tem.2008.07.002
- Olofsson SO, Boren J. Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J Intern Med 2005; 258:395–410; PMID:16238675; http://dx.doi.org/10.1111/j.1365-2796.2005.01556.x
- Lapierre LR, Hansen M. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol Metab 2012; 23:637–44; PMID:22939742; http://dx.doi.org/10.1016/j.tem.2012.07.007
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67–93; PMID:19653858; http://dx.doi.org/10.1146/annurev-genet-102808-114910
- Gelino S, Hansen M. Autophagy - An Emerging Anti-Aging Mechanism. J Clin Exp Pathol 2012; Suppl 4, 1-12.
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab 2010; 11:453–65; PMID:20519118; http://dx.doi.org/10.1016/j.cmet.2010.05.001
- Hansen M, Kapahi P. TOR Signaling and Aging. In: Hall MN, Tamanoi F, eds. The Enzymes. Burlington: Academic Press, 2010:279–99.
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol 2011; 32:2–11; PMID:22025673; http://dx.doi.org/10.1128/MCB.06159-11
- Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 2015; 11:867–80; PMID:25836756; http://dx.doi.org/10.1080/15548627.2015.1034410
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Jurgen Klisch T, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 2013; 15:647–58; PMID:23604321; http://dx.doi.org/10.1038/ncb2718
- O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 2013; 15:668–76; PMID:23604316; http://dx.doi.org/10.1038/ncb2741
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009; 458:1131–5; PMID:19339967; http://dx.doi.org/10.1038/nature07976
- Lapierre LR, Gelino SR, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate aging in germline-less C. elegans. Curr Biol 2011; 21:1507–14; PMID:21906946; http://dx.doi.org/10.1016/j.cub.2011.07.042
- Lapierre LR, Melendez A, Hansen M. Autophagy links lipid metabolism to longevity in C. elegans. Autophagy 2012; 8:144–6; PMID:22186228; http://dx.doi.org/10.4161/auto.8.1.18722
- O'Rourke EJ, Kuballa P, Xavier R, Ruvkun G. omega-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev 2013; 27:429–40; PMID:23392608; http://dx.doi.org/10.1101/gad.205294.112
- Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, Wang MC. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 2015; 347:83–6; PMID:25554789; http://dx.doi.org/10.1126/science.1258857
- Wollam J, Antebi A. Sterol regulation of metabolism, homeostasis, and development. Annu Rev Biochem 2011; 80:885–916; PMID:21495846; http://dx.doi.org/10.1146/annurev-biochem-081308-165917
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, Antebi A. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci U S A 2007; 104:5014–9; PMID:17360327; http://dx.doi.org/10.1073/pnas.0700847104
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 2006; 124:1209–23; PMID:16529801; http://dx.doi.org/10.1016/j.cell.2006.01.037
- Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 2005; 3:e53; PMID:15719061; http://dx.doi.org/10.1371/journal.pbio.0030053
- Goudeau J, Bellemin S, Toselli-Mollereau E, Shamalnasab M, Chen Y, Aguilaniu H. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol 2011; 9:e1000599; PMID:21423649; http://dx.doi.org/10.1371/journal.pbio.1000599
- Hansen M, Flatt T, Aguilaniu H. Reproduction, fat metabolism, and life span: what is the connection? Cell Metab 2013; 17:10–9; PMID:23312280; http://dx.doi.org/10.1016/j.cmet.2012.12.003
- Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 2006; 124:1055–68; PMID:16530050; http://dx.doi.org/10.1016/j.cell.2006.01.039
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 2003; 115:489–502; PMID:14622602; http://dx.doi.org/10.1016/S0092-8674(03)00889-4
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003; 424:277–83; PMID:12845331; http://dx.doi.org/10.1038/nature01789
- Lapierre LR, Silvestrini MJ, Nunez L, Ames K, Wong S, Le TT, Hansen M, Melendez A. Autophagy genes are required for normal lipid levels in C. elegans. Autophagy 2013; 9:278–86; PMID:23321914; http://dx.doi.org/10.4161/auto.22930
- Alberti A, Michelet X, Djeddi A, Legouis R. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 2010; 6:622–33; PMID:20523114; http://dx.doi.org/10.4161/auto.6.5.12252
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet 2010; 6:e1000848; PMID:20174564; http://dx.doi.org/10.1371/journal.pgen.1000848
- Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 2000; 349:629–34; PMID:10880363; http://dx.doi.org/10.1042/bj3490629
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab 2010; 12:260–72; PMID:20816092; http://dx.doi.org/10.1016/j.cmet.2010.08.004
- Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science 2008; 322:957–60; PMID:18988854; http://dx.doi.org/10.1126/science.1162011
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A 2009; 106:14914–9; PMID:19706382; http://dx.doi.org/10.1073/pnas.0902882106
- O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab 2009; 10:430–5; PMID:19883620; http://dx.doi.org/10.1016/j.cmet.2009.10.002
- Yen K, Le TT, Bansal A, Narasimhan SD, Cheng JX, Tissenbaum HA. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS One 2010; 5:pii: e12810; http://dx.doi.org/10.1371/journal.pone.0012810
- Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab 2008; 8:266–74; PMID:18762027; http://dx.doi.org/10.1016/j.cmet.2008.08.007
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003; 301:1387–91; PMID:12958363; http://dx.doi.org/10.1126/science.1087782
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 2001; 28:139–45; PMID:11381260; http://dx.doi.org/10.1038/88850
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 2013; 4:2267; PMID:23925298
- Grimmel M, Backhaus C, Proikas-Cezanne T. WIPI-Mediated Autophagy and Longevity. Cells 2015; 4:202–17; PMID:26010754; http://dx.doi.org/10.3390/cells4020202
- Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol 2013; 15:491–501; PMID:23604319; http://dx.doi.org/10.1038/ncb2720
- Ratnappan R, Amrit FR, Chen SW, Gill H, Holden K, Ward J, Yamamoto KR, Olsen CP, Ghazi A. Germline Signals Deploy NHR-49 to Modulate Fatty-Acid beta-Oxidation and Desaturation in Somatic Tissues of C. elegans. PLoS Genet 2014; 10:e1004829; PMID:25474470; http://dx.doi.org/10.1371/journal.pgen.1004829
- Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 2014; 516(7529):108–11; PMID:25383523; http://dx.doi.org/10.1038/nature13949
- Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 2014; 516(7529):112–5; http://dx.doi.org/10.1038/nature13961
- Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol 2011; 21:1507–14; PMID:21906946; http://dx.doi.org/10.1016/j.cub.2011.07.042
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 2008; 4:e24; PMID:18282106; http://dx.doi.org/10.1371/journal.pgen.0040024
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science 2009; 325:473–7; PMID:19556463
- Raikhel AS. Lysosomes in the cessation of vitellogenin secretion by the mosquito fat body; selective degradation of Golgi complexes and secretory granules. Tissue & Cell 1986; 18:125–42; PMID:3961792; http://dx.doi.org/10.1016/0040-8166(86)90012-1
- Shmookler Reis RJ, Xu L, Lee H, Chae M, Thaden JJ, Bharill P, Tazearslan C, Siegel E, Alla R, Zimniak P, et al. Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany NY) 2011; 3:125–47; PMID:21386131
- Matyash V, Geier C, Henske A, Mukherjee S, Hirsh D, Thiele C, Grant B, Maxfield FR, Kurzchalia TV. Distribution and transport of cholesterol in Caenorhabditis elegans. Mol Biol Cell 2001; 12:1725–36; PMID:11408580; http://dx.doi.org/10.1091/mbc.12.6.1725
- Rauthan M, Pilon M. The mevalonate pathway in C. elegans. Lipids Health Dis 2011; 10:243; PMID:22204706; http://dx.doi.org/10.1186/1476-511X-10-243
- Wang CW, Miao YH, Chang YS. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol 2014; 206:357–66; PMID:25070953; http://dx.doi.org/10.1083/jcb.201404115
- Lee EY, Shim YH, Chitwood DJ, Hwang SB, Lee J, Paik YK. Cholesterol-producing transgenic Caenorhabditis elegans lives longer due to newly acquired enhanced stress resistance. Biochem Biophys Res Commun 2005; 328:929–36; PMID:15707967; http://dx.doi.org/10.1016/j.bbrc.2005.01.050
- Cheong MC, Lee HJ, Na K, Joo HJ, Avery L, You YJ, Paik YK. NSBP-1 mediates the effects of cholesterol on insulin/IGF-1 signaling in Caenorhabditis elegans. Cell Mol Life Sci 2013; 70:1623–36; PMID:23255046; http://dx.doi.org/10.1007/s00018-012-1221-0
- Ouimet M, Marcel YL. Regulation of lipid droplet cholesterol efflux from macrophage foam cells. Arterioscler Thromb Vasc Biol 2012; 32:575–81; PMID:22207731; http://dx.doi.org/10.1161/ATVBAHA.111.240705
- Heestand BN, Shen Y, Liu W, Magner DB, Storm N, Meharg C, Habermann B, Antebi A. Dietary restriction induced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans. PLoS Genet 2013; 9:e1003651; PMID:23935515; http://dx.doi.org/10.1371/journal.pgen.1003651
- Magner DB, Wollam J, Shen Y, Hoppe C, Li D, Latza C, Rottiers V, Hutter H, Antebi A. The NHR-8 nuclear receptor regulates cholesterol and bile acid homeostasis in C. elegans. Cell Metab 2013; 18:212–24; PMID:23931753; http://dx.doi.org/10.1016/j.cmet.2013.07.007
- Thondamal M, Witting M, Schmitt-Kopplin P, Aguilaniu H. Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nat Commun 2014; 5:4879; PMID:25209682; http://dx.doi.org/10.1038/ncomms5879
- Kirkwood TB. Understanding the odd science of aging. Cell 2005; 120:437–47; PMID:15734677; http://dx.doi.org/10.1016/j.cell.2005.01.027
- Ackerman D, Gems D. The mystery of C. elegans aging: An emerging role for fat: Distant parallels between C. elegans aging and metabolic syndrome? Bioessays 2012; 34(6):466–71; PMID:22371137
- Ren Y, Hughes KA. Vitellogenin family gene expression does not increase Drosophila lifespan or fecundity. F1000Research 2014; 3:125; PMID:25110583
- Mair W, Sgro CM, Johnson AP, Chapman T, Partridge L. Lifespan extension by dietary restriction in female Drosophila melanogaster is not caused by a reduction in vitellogenesis or ovarian activity. Exp Gerontol 2004; 39:1011–9; PMID:15236760; http://dx.doi.org/10.1016/j.exger.2004.03.018
- Cash TP, Pita G, Dominguez O, Alonso MR, Moreno LT, Borras C, Rodriguez-Manas L, Santiago C, Garatachea N, Lucia A, et al. Exome sequencing of three cases of familial exceptional longevity. Aging Cell 2014; 13(6):1087–90; PMID:25116423
- Brown WV, Myers GL, Sniderman AD, Stein E. Should we use apoB for risk assessment and as a target for treatment? Journal of clinical lipidology 2010; 4:144–51; PMID:21122646; http://dx.doi.org/10.1016/j.jacl.2010.03.004
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. RNAi-mediated gene silencing in non-human primates. Nature 2006; 441:111–4; PMID:16565705; http://dx.doi.org/10.1038/nature04688
- Watts JL. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol Metab 2009; 20:58–65; PMID:19181539; http://dx.doi.org/10.1016/j.tem.2008.11.002
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974; 77:71–94; PMID:4366476
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet 2005; 1:119–28; PMID:16103914; http://dx.doi.org/10.1371/journal.pgen.0010017
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 2003; 421:231–7; PMID:12529635; http://dx.doi.org/10.1038/nature01278
- Long X, Spycher C, Han ZS, Rose AM, Muller F, Avruch J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr Biol 2002; 12:1448–61; PMID:12225660; http://dx.doi.org/10.1016/S0960-9822(02)01091-6
