Abstract
Although autophagy is a highly conserved mechanism among species and cell types, few are the molecules involved with the autophagic process that display cell- or tissue- specific expression. We have unraveled the positive regulatory role on autophagy of RUFY4 (RUN and FYVE domain containing 4), which is expressed in subsets of immune cells, including dendritic cells (DCs). DCs orchestrate the eradication of pathogens by coordinating the action of the different cell types involved in microbe recognition and destruction during the immune response. To fulfill this function, DC display particular regulation of their endocytic and autophagy pathways in response to the immune environment. Autophagy flux is downmodulated in DCs upon microbe sensing, but is remarkably augmented, when cells are differentiated in the presence of the pleiotropic cytokine IL4 (interleukin 4). From gene expression studies aimed at comparing the impact of IL4 on DC differentiation, we identified RUFY4, as a novel regulator that augments autophagy flux and, when overexpressed, induces drastic membrane redistribution and strongly tethers lysosomes. RUFY4 is therefore one of the few known positive regulators of autophagy that is expressed in a cell-specific manner or under specific immunological conditions associated with IL4 expression such as allergic asthma.
In addition to removing defective proteins or deteriorated subcellular organelles, autophagy is key to eliminate parasitic microbes in response to a variety of stress and metabolic changes. Most of our current knowledge about autophagy regulation was obtained through the study of starvation induced-autophagy in yeast. Autophagy being conserved among species, this pioneering work has served as a blueprint for chartering the molecular basis of autophagic processes in higher eukaryotes; however, little is known about the existence of cell- or tissue-specific factors controlling autophagy in multicellular organisms and in other circumstances than amino acid starvation.
The evolution of immune systems has increased the complexity of protein degradation pathways, including specific lysosomal protease expression or the appearance of MHC-encoded proteasome subunits to favor exogenous and endogenous antigen processing and presentation. Autophagy is used by MHC II-expressing professional antigen-presenting cells such as DCs, as a means to capture and degrade intracellular antigens for T helper cell-mediated immunosurveillance, and is therefore expected to obey unique regulatory mechanisms dependent on immune conditions.
Toll-like receptor activation by microbial components (e.g., LPS) activates MTORC1, which results in a reduction of autophagy in DCs. This reduction causes accumulation of unprocessed ubiquitinated defective neo-synthetized proteins, together with LC3/ATG8 and the autophagy receptor proteins SQSTM1/p62 and NBR1 in dendritic cell aggresome-like inducible structures (DALIS; ). DALIS represent depots of neosynthetized self-antigens that are slowly processed and therefore poorly presented by the MHC at a key period for the acquisition by DC of their immunostimulatory function. TLR-driven inhibition of autophagy also causes reactive oxygen species-producing mitochondria accumulation. DALIS formation in activated DC is therefore the result of a concomitant MTORC1-dependent increase in protein synthesis and reduction in autophagy, together with an increased production of mitochondrial reactive oxygen species, that augment damage and denaturation of neosynthesized proteins and furthe promotes their aggregation with SQSTM1.
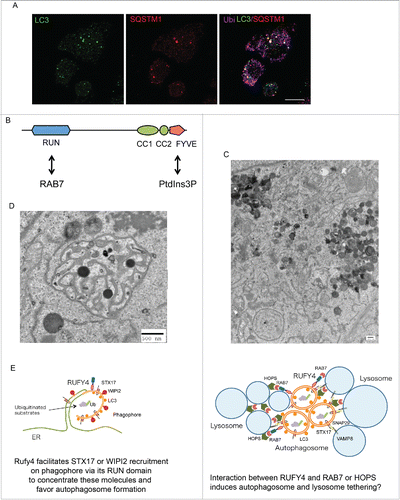
Figure 1. Autophagy inhibition in activated dendritic cells, and RUFY4 molecular structure and function. (A) Detection of DALIS by confocal microscopy in LPS-stimulated DC using staining for LC3 (green), SQSTM1 (red) and polyubiquitinated proteins (Ub, purple), Scale bar 10 µm. (B) Schematic representation of RUFY4 domain organization. (C and D) Electron microscopy micrographs of RUFY4-overexpressing HeLa cells showing lysosomes clustering (C) and abnormal autophagosome-like structures (D). Scale bars 500 nm. (E) Modeling of the different sites of RUFY4 activity with key molecules and organelles.
Independently of MTORC1 signaling, we found that the presence of the cytokine IL4 augments autophagy flux in DCs, and prevents DALIS formation. By comparing differently expressed genes in response to IL4, we identified RUFY4, as a novel positive autophagy regulator, which influences autophagosome dynamics and can prevent infection by the intracellular pathogen Brucella abortus.
The RUFY family consists of 5 proteins, RUFY1 to RUFY4, and the closest RUFY4 paralog, FYCO1, all bearing a small RAS-like GTPase interacting domain (RUN) and a PtdIns3P interacting site (FYVE) (). Several RUN and FYVE domain-containing proteins such as FYCO1, RUBCN/RUBICON and PLEKHM1 are involved in endosome or autophagosome trafficking and are effectors of the small RAS-like GTPase RAB7. Similar to these molecules, the RUN and FYVE domains of RUFY4 interact respectively with RAB7 and PtdIns3P, pointing toward a function of RUFY4 in autophagosome generation and/or fusion with the endocytic pathway. Indeed RUFY4 overexpression facilitates degradation of LC3/ATG8, while deletion of its RUN domain prevents RUFY4 interaction with RAB7 and abolishes its positive effect on autophagy, further suggesting that RUFY4 functions as a RAB7 effector. RUFY4 expression organizes the LAMP1-positive late endosomal compartments in clusters (), which remain distinct from induced large abnormal autophagosome-like structures positive for STX17 (syntaxin 17), a Qa SNAREs involved in autophagosome formation and fusion (). RUFY4 colocalizes alternatively with RAB7 and STX17 or with LAMP1-positive vesicles, but not with EEA1-positive early endosomes. Interestingly, RUFY4 expression also promotes the formation of WIPI2 puncta, indicative of increased PtdIns3P accumulation as well as omegasome and early autophagosome formation. Together, these results suggest that RUFY4 harnesses the classical autophagy machinery to facilitate autophagosome formation and increase flux by acting at different biochemical steps of the processes involving PtdIns3P generation, RAB7 or STX17 recruitment. In cells stably expressing low levels of RUFY4, enhanced autophagy was only observed upon starvation, suggesting that RUFY4 is a downstream effector of the MTORC1 signaling cascade.
By optimizing effector protein activity and organelle distribution, RUFY4 expression and IL4-induced differentiation facilitate damaged mitochondria or intracellular bacteria elimination, but also promote endogenous antigen presentation to CD4+ T cells. RUFY4 expression in HeLa cells directly prevents Brucella abortus replication, suggesting that the immune system capitalizes on autophagy to mount both innate and adaptive immune responses through long-term IL4 signaling and expression of specific RAB7 effectors. RUFY4 is therefore an important factor defining the nature of the cellular response to its immediate immunological environment, as shown by its induction in a specific DC subset exposed to asthma-like conditions. Mechanistically RUFY4 seems to act both at the level of autophagosome formation and tethering/fusion to lysosomes (). We still need to address how RUFY4 can have such a broad regulatory function and whether it could assemble in different molecular complexes, each specific for a different facet of its activity, like PLEKHM1 that controls endosome and autophagosome dynamics by interacting with the homotypic fusion and protein sorting (HOPS)–tethering complex and RAB7. RUFY4 is therefore the first example of a positive regulator of autophagy, whose activity is restricted to specific immune conditions, suggesting that other factors are likely to exist and regulate autophagy in different physiological contexts.
Disclosure of potential conflicts of interest
The authors declare no further competing financial interests.
Acknowledgments
We thank Christiane Rondeau and Michel Desjardins from the Université de Montréal for electron microscopy analysis.
Funding
The laboratory is funded by Agence Nationale de la Recherche grants: “ANR-12-BSV2-0025-01," “ANR-FCT 12-ISV3-0002-01." DCBiol Labex ANR-11-LABEX-0043- grant ANR-10-IDEX-0001-02 PSL* and A*MIDEX project ANR-11-IDEX-0001-02 funded by the Investissements d'Avenir French government program. The laboratory is funded by a grant from the Fondation pour la Recherche Médicale “Equipe FRM DEQ20140329536. S.T. was a Uehara Memorial Foundation and a Fondation pour la Recherche Médicale (FRM) fellow.
