abstract
A collaborative consortium, named “TRANSAUTOPHAGY,” has been created among European research groups, comprising more than 150 scientists from 21 countries studying diverse branches of basic and translational autophagy. The consortium was approved in the framework of the Horizon 2020 Program in November 2015 as a COST Action of the European Union (COST means: CO-operation in Science and Technology), and will be sponsored for 4 years. TRANSAUTOPHAGY will form an interdisciplinary platform for basic and translational researchers, enterprises and stakeholders of diverse disciplines (including nanotechnology, bioinformatics, physics, chemistry, biology and various medical disciplines). TRANSAUTOPHAGY will establish 5 different thematic working groups, formulated to cooperate in research projects, share ideas, and results through workshops, meetings and short term exchanges of personnel (among other initiatives). TRANSAUTOPHAGY aims to generate breakthrough multidisciplinary knowledge about autophagy regulation, and to boost translation of this knowledge into biomedical and biotechnological applications.
It is well established that the interest in the process of autophagy has dramatically increased during the past two decades. Indeed, the number of publications found in PubMed with the term “autophagy” appearing in the title or abstract is experiencing an exponential growth. Considering discrete 5-year intervals, the number of papers containing the word “autophagy” published in 1996-2000 was 220, then 694 for the next five years, followed by 4,815, and finally, from 2011 to date, 15,525 (Pubmed search: December 3rd, 2015). The two guidelinesCitation1,2 published in 2008 and 2012 by scientists working in the autophagy field to establish good practices in analysis and interpretation of data, are among the most cited papers in the past years, and a third version was published this year.Citation3 Such interest is not only purely scientific, i.e., aimed at identifìying and characterizing new factors or mechanisms, but also connected to the potential of autophagy modulation for both biomedical applications, including the development or use of drugs, and for addressing environmental and agricultural challenges.
Several national associations for autophagy research such as the Francophone Autophagy Club (CFATG) in France (cftag.org), the Nordic Autophagy Network (nordicautophagy.org) and the Spanish Autophagy Group have risen to celebrate annual meetings that gather most of the national laboratories working on this topic. These associations had their first joint meeting in 2014, and since then more national networks focused on autophagy have been established, including the UK Autophagy Network and in Germany the Autophagy Study Group under the umbrella of the German Society for Biochemistry and Molecular Biology. This underscores the will and necessity for international networking, which has finally found a European dimension under the umbrella of Horizon 2020. More than 150 European scientists (listed in Annex 1) from 21 countries (), along with one COST international partner from the US, have congregated in a consortium named TRANSAUTOPHAGY, and a proposal to the European Union for developing a coordinated action in the fremewok of the Cooperation in Science and Technology (COST) Association has been accepted.
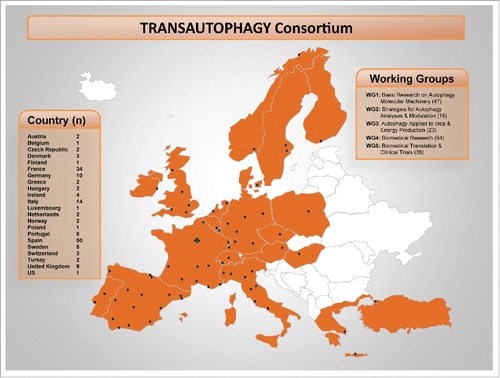
Figure 1. Distribution of European cities where TRANSAUTOPHAGY groups are active and distributed into working groups (WG). Inserts show the number (n) of groups operating in each country and the number of researchers participating in each WG.
Through genetic, molecular and cellular approaches in yeast, animals, and plants, researchers have learned that autophagy plays crucial roles in differentiation and development, cellular and tissue homeostasis, protein and organelle quality control, metabolism, and immunity, but also in protection from aging and diverse diseases (e.g., heart failure, liver inflammation, impaired long-lived humoral immunity, Alzheimer and Parkinson diseases). An increasing number of human diseases are being linked to polymorphisms or mutations in autophagy genes (e.g., Danon cardiomyopathy, Crohn disease, Vici syndrome, asthma, breast and colon cancer), and numerous studies are addressing the clinical effects of deficiencies in autophagy or the altered balance between autophagy and inflammation. There is thus a need for new approaches, diagnosis methods, and therapeutic molecular target identification that requires intensive collaborative effort at the international level.
The molecular mechanisms and the physiology of autophagy in plant cells are less clear than in animals. The details of how autophagy functions in pathogen-triggered programmed cell death in plants, seed protein accumulation, senescence, tolerance to nutrient deprivation, abiotic stresses, and redox homeostasis are not well understood. Basic questions such as why and how mechanistically reduced autophagy renders plant more sensitive to environmental cues are still unanswered. Moreover, it is unknown how many types of autophagy pathways are active in plant cells, which types of autophagy are involved in pathogen-triggered programmed cell death, or how some pathogens use the autophagy pathway to manipulate the plant immune system. Due to the presence of specific autophagy cargo receptors, autophagy can also be a selective process that targets specific proteins, protein aggregates and organelles for degradation. Not only the “core” autophagy machinery, but also the receptors and adapters of selective autophagy, are evolutionarily conserved. This fact justifies the broad transkingdom approach in this action.
To cover, at least partially, some of these gaps in our knowledge, TRANSAUTOPHAGY plans to reach several challenging objectives in a 4-year program, which include:
increasing knowledge and technological breakthroughs in autophagy;
standardizing technological procedures through multidisciplinary joined guidelines;
reporting white-papers for policy makers;
bridging separate disciplines, such as nanotechnology, bioinformatics, physics, chemistry, biology and medicine;
increasing common projects on healthy aging, prevention, diagnosis or treatment of diseases;
finding new methods to improve crop production, generate alternative energy, and reduce CO2 emissions;
promoting and integrating new discoveries into systems-biology-based databases;
increasing autophagy modulators for translational uses; boosting the number of biomedical products related to autophagy with patent licensing for biomedical or biotechnological purposes;
popularizing the scientific theme of autophagy within the society and especially for youth;
supervising and increasing the inclusion of female cohorts in pre-clinical and clinical studies.
In order to reach the aforementioned goals, researchers of the TRANSAUTOPHAGY network have been organized into 5 working groups (WG) (), which will be highly collaborative within them, and will spend strong interactive efforts among disciplines and specific areas; half of the WG leaders are women. Several activities will start, and new instruments will be created, which include: “think-tank” committees for biotechnology and biomedical translation (composed of experts); annual and periodic foresight workshops to promote synergies, open innovation, collaboration in common projects, specific translational research actions; deal-making between basic researchers and technological and pharmaceutical companies; participation of younger researchers at informal meetings for brainstorming at the annual conferences; annual conferences that will promote breakthrough visibility to non-expert audiences; and training activities for fostering scientific and technical exchanges. Early career investigators will benefit from opportunities for partnering. A dissemination committee will maintain an active website that incorporates links to companies and patients' associations. Moreover, researchers from small and medium-sized enterprises will be encouraged to participate in all activities, and working groups with a multidisciplinary composition of stakeholders, including small and medium-sized enterprises and big companies, will be created.
In conclusion, the TRANSAUTOPHAGY network will facilitate an easy and fast exchange of scientific discoveries, techniques and resources. For the new generation of researchers, this action will favor co-mentoring and increase opportunities to perform high-quality science. New discoveries will enhance creation of high-quality job opportunities, ideas for spin-off companies, and market opportunities. In addition, gender balance in biomedical research will benefit due to persistent alert on this issue within the consortium. Finally, the network, open to European scientists in the field, will help to bridge and circumvent traditional gaps and obstacles in knowledge translation by sharing experiences and offering direct links that could open business opportunities. It will also serve as the catalyst for implementation of scientific discoveries to the benefit of the society.
Disclosure of potential conflicts of interest
Authors declare that there is no conflict of interest regarding the publication of this article.
References
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4:151-75; PMID:18188003; http://dx.doi.org/10.4161/auto.5338
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/10.4161/auto.19496
- Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2016; 12:1–222; http://dx.doi.org/10.1080/15548627.2015.1100356
Annex 1.
List of participating groups and cities
Austria: C. Kraft (Vienna), S. Martens (Vienna).
Belgium: H. Batoko (Louvain).
Czech Republic: M. Cahova (Prague), V. Zarsky (Prague).
Denmark: F. Cecconi (Copenhagen), M. Jäättelä (Copenhagen), M. Petersen (Copenhagen).
Finland: E.-L. Eskelinen (Helsinki).
France: S. Lanone (Creteil), A. Moris (Paris), A. Esclatine (Paris), A. Rozieres (Lyon), B. Dehay (Bordeaux), V. Cecile (Toulouse), C. Masciaux-Daubresse (Versailles), C. Boulanger (Paris), C. Viret (Bron), C. Berlioz-Torrent (Paris), F. Mechta-Grigoriou (Paris), F. Pinet (Lille), F. Lafont (Lille), F. Lezoualc'h (Toulouse), G. Kroemer (Paris), H. Nguyen (Clermont-Ferrand), J. Iovanna (Marseille), I. Vergne (Toulouse), C. Marelli (Montpellier), M. Faure (Lyon), M. Bianchi (Gif sur Yvette), M. Djavaheri-Mergny (Bordeaux), N. Pallet (Paris), P. Codogno (Paris), P. Auberger (Nice), S. Pattingre (Montpellier), P.E. Rautou (Paris), M. Priault (Bordeaux), R. Legouis (Gif sur Yvette), S. Lotersztajn (Paris), S. Giuriato (Toulouse), S. Muller (Strasbourg), D. Tosi (Montpellier), F. Ichas (Pessac).
Germany: A. Fernie (Potsdam), C. Behrends (Frankfurt), E. Isono (Munchen), F. Baluska (Bonn), I. Dikic (Frankfurt), J. Dengjel (Freiburg), M. Thumm (Goettingen), S. Fulda (Frankfurt), T. Avin-Wittenberg (Potsdam), T. Proikas-Cezanne (Tuebingen).
Greece: L. Stefanis (Athens), N. Tavernarakis (Heraklion).
Hungary: G. Juhasz (Budapest), Z. Gaspari (Budapest).
Ireland: F. Bertoli (Dublin), J. Murray (Dublin), K. Dawson (Dublin), M. Rehm (Dublin).
Italy: A. Ballabio (Pozzuoli), A. Cossarizza (Modena), D. Goletti (Rome), E. Emanuele (Robbio), F. Penna (Turin), G.M. Fimia (Rome), G. Buonocore (Siena), M. Pinti (Modena), M. Sandri (Padua), M.C. Albertini (Urbino), M. Chiariello (Siena), M. Mori (Siena), S. Carloni (Urbino), W. Balduini (Urbino).
Luxembourg: B. Janji (Luxembourg City).
Netherlands: A. Meijer (Leiden), F. Reggiori (Groningen)
Norway: A. Simonsen (Oslo), T. Johansen (Tromso).
Poland: A. Sirko (Warsaw).
Portugal: H. Vieira (Lisbon), H. Girão (Coimbra), M. Ricardo Pereira (Coimbra), R. Baptista (Coimbra), R. Matthiesen (Lisbon).
Spain: C. Casas (Barcelona), A. Alonso (Leioa), A. Zorzano (Barcelona), A. Serrano (Sevilla), C. Guillen (Madrid), C. Gotor (Sevilla), M. Gonzalo Claros (Malaga), C. Alonso (Madrid), D. Ruiz-Molina (Madrid), D. Ruano Caballero (Sevilla), F.X. Pimentel-Muinos (Salamanca), F. Goni (Leioa), F. Suarez (Malaga), F. Novio (Barcelona), F. Sevilla (Murcia), F. Wandosell (Madrid), G. Velasco (Madrid), J. Diaz-Nido (Madrid), J.M. de la Fuente (Zaragoza), J. Ruberte (Barcelona), J.A. Sanchez Alcazar (Sevilla), J.M. Fuentes (Caceres), J.M. Garcia Fernandez (Sevilla), J.L. Crespo (Sevilla), J.M. Jerez (Malaga), J. Casas (Barcelona), J.A.G. Ranea (Malaga), J.M. Sepulveda (Madrid), J. Lorenzo (Barcelona), L.M. Sandalio (Granada), I.M. Ricardo (Zaragoza), M.L. Martinez Chantar (Derio), M. Moran (Madrid), M. Soengas (Madrid), M.D. Cordero (Sevilla), M. Martinez-Vicente (Barcelona), M.A. Martin (Madrid), M. Lopez (Santiago de Compostela), N. Apostolova (Castellon), N.S. Coll (Barcelona), P. Boya (Madrid), P.S. Testillano (Madrid), P. Munoz-Canoves (Barcelona), R. Delgado (Madrid), R. Escalante (Madrid), R. Sanchez-Prieto (Albacete), J. Farres (Barcelona), D. Prous (Barcelona), J. Ballesteros (Madrid), A. Lucia (Madrid).
Sweden: A. De Milito (Stockholm), D. Grandér (Stockholm), D. Hofius (Uppsala), J. Martinsson (Stockholm), P. Bozhkov (Uppsala).
Switzerland: H-U. Simon (Bern), J. Puyal (Lausanne), M.P. Tschan (Bern)
Turkey: A. Kosar (Istanbul), D. Gozuacik (Istanbul).
United Kingdom: D. Rubinsztein (Cambridge), J. King (Sheffield), J. Lane (Bristol), H. Jungbluth (London), K. Ryan (Glasgow), M. Gegg (London), N. Ktistakis (Cambridge), P. Lovat (Newcastle), S. Tooze (London).
European Commission: B. Mograbi (Nice).
