ABSTRACT
Lysosomes are highly acidic cellular organelles traditionally viewed as sacs of enzymes involved in digesting extracellular or intracellular macromolecules for the regeneration of basic building blocks, cellular housekeeping, or pathogen degradation. Bound by a single lipid bilayer, lysosomes receive their substrates by fusing with endosomes or autophagosomes, or through specialized translocation mechanisms such as chaperone-mediated autophagy or microautophagy. Lysosomes degrade their substrates using up to 60 different soluble hydrolases and release their products either to the cytosol through poorly defined exporting and efflux mechanisms or to the extracellular space by fusing with the plasma membrane. However, it is becoming evident that the role of the lysosome in nutrient homeostasis goes beyond the disposal of waste or the recycling of building blocks. The lysosome is emerging as a signaling hub that can integrate and relay external and internal nutritional information to promote cellular and organismal homeostasis, as well as a major contributor to the processing of energy-dense molecules like glycogen and triglycerides. Here we describe the current knowledge of the nutrient signaling pathways governing lysosomal function, the role of the lysosome in nutrient mobilization, and how lysosomes signal other organelles, distant tissues, and even themselves to ensure energy homeostasis in spite of fluctuations in energy intake. At the same time, we highlight the value of genomics approaches to the past and future discoveries of how the lysosome simultaneously executes and controls cellular homeostasis.
Introduction
The presence of approximately 60 different hydrolases makes the lysosome the primary catabolic center of the cell.Citation1 The products of digestion are ultimately used as building blocks for biosynthetic pathways or to meet energy demands. Lysosomal membrane proteins include exporters of these metabolites allowing their translocation and clearing.Citation2 With the collective action of lysosomal hydrolases that include proteases, glycosidases, lipases, nucleases, phosphatases and sulfatases, macromolecules and even metabolic organelles such as mitochondria and peroxisomes or energy storage compartments such as lipid droplets can be degraded or recycled in the lysosome.Citation3,4 However, because it is the only organelle receiving cargo directly from both the inside and the surroundings of the cell, the lysosome is uniquely positioned to have the additional functions of integrating nutritional information and orchestrating homeostatic responses. In fact, as it gains more attention, the multifaceted and central role in nutrient homeostasis of this formerly neglected organelle becomes increasingly evident. Although there are many essential functions ascribed to the lysosome, here we focus on 3 interrelated but distinct functions relevant to nutrient homeostasis. The first section, “Nutrient sensing at the lysosome,” describes the emerging role of the lysosome in sensing nutrients and locally relaying information to the master nutrient sensors MTOR and AMPK. In section 2, “Nutrient processing by the lysosome,” although we acknowledge the lysosome's role as a processor of damaged organelles, macromolecular complexes, nutrient and growth factor receptors, and proteins, we focus on the role of lysosomal hydrolases in processing energy-dense molecules (glycogen and lipids) to generate energy units that contribute to energy homeostasis. The third section, “Nutrient signaling from the lysosome,” summarizes the role of the lysosome in generating signaling molecules capable of traveling either to the nucleus to activate homeostatic transcriptional programs, or to distant tissues to activate global homeostatic responses. All in all, we describe a picture in which the lysosome plays a central role in providing nutrients and ensuring that organisms invest in growth and reproduction only when the internal and external conditions are favorable to do so.
Nutrient sensing at the lysosome
A key nutrient-sensing node acting in all tested eukaryotes is the kinase complex MTOR complex 1 (MTORC1). This master growth regulator promotes anabolic processes such as protein translation when nutrients are available, and licenses catabolic processes such as macroautophagy when nutrients are scarce.Citation5 Interestingly, nutrients such as amino acidsCitation6 and glucose,Citation7 promote the translocation of MTORC1 to the lysosomal surface. Proteomics approaches revealed that in the presence of nutrients, 2 protein complexes, the Ragulator and the RAG-heterodimer, dock MTORC1 on the surface of the lysosome, and that formation of this multiprotein complex, coined lysosome nutrient sensing machinery or LYNUS,Citation8 is a key event in nutrient signaling through MTORC1.Citation9,10 Ragulator is a multiprotein guanine nucleotide exchange factor (GEF) that acts as a lysosomal anchor for RAG.Citation6,9 RAG is a multiprotein complex comprising the obligate GTPase heterodimers RRAGA or RRAGB in complex with RRAGC or RRAGD. MTORC1 seems to be differentially regulated by specific amino acids.Citation11 The human SLC38A9 (solute carrier family 38 member 9) is part of the Ragulator-GTPase machinery,Citation12 and activates MTOR in the presence of arginine.Citation13 RRAGA and RRAGB are required for MTOR activation by leucine, whereas glutamine does not require RAG GTPases. Instead glutamine-mediated MTORC1 activation occurs via the ARF1 (ADP ribosylation factor 1) GTPase.Citation11
Various proteins interact with Ragulator and RAG GTPases to facilitate and fine-tune MTOR-mediated responses at the lysosome. For example, a Ragulator interacting protein, BORCS6/c17orf59, was recently shown to competitively inhibit RAG binding to the Ragulator, thus preventing RAG GTPase docking to lysosomes, and negatively affecting the amino acid activation of MTORC1.Citation14 However, a loss of BORCS6 in HeLa cells has no effect on the inhibition of MTORC1 signaling during nutrient deprivation suggesting that there could be other roles for the Ragulator-BORCS6 complex independent of MTORC1. BORCS6 may regulate the interaction of Ragulator with BORC, shown to be important for lysosomal positioning.Citation15 Another MTOR modulator is SQSTM1/p62 (sequestosome 1), a multidomain receptor protein involved in intracellular signaling, which interacts with MTORC1 in the presence of amino acids. This interaction is in turn required for the interaction of MTORC1 with RAG GTPases, and thus the translocation of MTORC1 to the lysosomal surface.Citation16 Also, SQSTM1 interacts with TRAF6 (TNF receptor associated factor 6), which is required for the activation of MTOR.Citation17 In addition, the GATOR complex (GTPase activating protein toward Rag GTPases), a multiprotein complex consisting of 2 subcomplexes, GATOR1 (DEPDC5-NPRL2-NPRL3) and GATOR2 (MIOS-SEH1L-WDR24-WDR59-SEC13), regulates the RAG GTPases.Citation18,19 GATOR1 functions as a GTPase activating protein for RRAGA and RRAGB, and GATOR2 acts as a negative regulator of GATOR1.Citation18 Finally, a series of stress responsive growth regulators known as sestrins (SESN1, SESN2, and SESN3) interact with GATOR2 in response to lack of amino acids. Sestrins act as guanine nucleotide dissociation inhibitors for RAG GTPases, thus suppressing the lysosomal localization of MTOR.Citation20-22
A Drosophila cell-based RNA interference screen for genes involved in lysosomal biogenesis or function, unveiled that amino acid signaling to MTORC1 does not begin at the plasma membrane, but begins within the lysosome.Citation23 The vacuolar-type H+ adenosine triphosphatase (V-ATPase), an ATP-dependent proton pump, has a pivotal role in acidifying the lysosomal lumen by pumping protons into the lysosome. However, V-ATPase regulates signaling through MTOR independently of its acidifying capacity.Citation23 Assembly of 2 domains of the V-ATPase, the membrane spanning proton-translocating domain (V0) and the peripheral ATPase domain (V1) is increased upon amino acid starvation, but reversed on re-addition of amino acids. Amino acid-triggered changes in V-ATPase assembly also depend on its catalytic activity as well as the pH of the lysosomal lumen.24 In the presence of amino acids, V-ATPase triggers Ragulator GEF activity for RAG GTPases.Citation9 Interestingly, recruitment of MTOR to the lysosome is dependent on RRAGA/B but seems to be independent of RAG GTP charge.Citation10,25,26
Although how nutrient sufficiency leads to recruitment of MTORC1 to the lysosomal surface is not fully understood, lysosomal localization and interaction with its activator RHEB are required for full MTOR activation. The small GTPase RHEB (Ras homolog enriched in brain), stimulates the phosphorylation and activation of MTORC1 when bound to GTP in a nutrient-abundant state. Upon amino acid withdrawal or the inhibition of growth factor signaling, RAG GTPases recruit TSC (tuberous sclerosis complex) to the lysosomes.Citation27 TSC, which acts as a GTPase-activating protein for RHEB, is composed of TSC1, TSC2, and the GTPase TBC1D7, which converts GTP-RHEB to GDP-RHEB preventing its stimulatory effect on MTORC1 ().Citation28,29 The GTP/GDP-independent activation of MTORC1 by RHEB may implicate other modulators in RHEB-mediated activation of MTORC1.Citation30
Figure 1. The lysosome is a nutrient-sensing center. When nutrients are sufficient (upper panel), amino acids induce structural changes in the lysosomal vacuolar-type ATPase (V-ATPase), so that it weakens its association with the Ragulator-RAG complex. Thus, Ragulator-RAG can recruit MTOR to the lysosomal membrane.Citation23 The small GTPase RHEB that resides at the lysosomal membrane, can now stimulate the phosphorylation and consequent activation of MTOR.Citation28,29 RRAGA/B facilitates MTOR activation and recruitment of TFEB to the lysosome for its phosphorylation and retention in the cytoplasm by YWHA chaperones.Citation9,26,47 When nutrients are scarce (bottom panel), the RAG GTPases recruit TSC (tuberous sclerosis complex), which converts GTP-RHEB to GDP-RHEB causing inactivation and release of MTOR into the cytosol.Citation27 Fasting stimulates AMPK, which in turn activates the TSC complex. In addition, since MTOR phosphorylates the MiTs transcription factors, upon MTOR inhibition, these transcriptional regulators are not phosphorylated and are free to translocate to the nucleus and activate genes involved in lysosomal biogenesis and function.Citation46,48,50,51
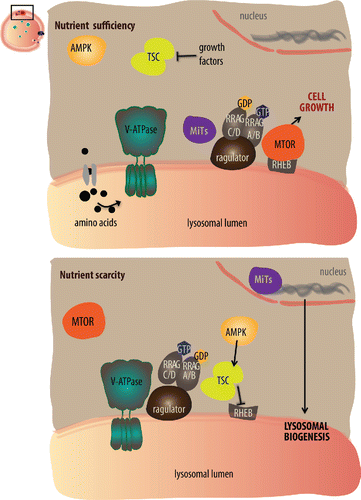
In addition to nutrients, metazoans couple growth rates to growth factors. Notably, insulin-mediated activation of MTORC1 requires amino acids and Ragulator present on the lysosomal surface.Citation6,25 Insulin and growth factors stimulate the class I phosphoinositide 3-kinase (PI3K), which phosphorylates AKT. In turn, AKT phosphorylates TSC2 of the TSC complex.Citation31-33 In the absence of growth factors, the TSC complex is localized to the lysosome in a RHEB-dependent manner. In response to insulin and AKT-mediated phosphorylation, the TSC complex gets acutely released from lysosomes eventually leading to MTORC1 activation.Citation34 Thus, the lysosome is a nexus between nutrients, growth factors, and MTORC1-mediated regulation of cellular and organismal growth.
Glucose regulates MTORC1 activity through its regulation of RHEB. The enzyme GAPDH (glyceraldehyde-3-phosphate dehydrogenase), directly interacts with RHEB independent of GDP/GTP-RHEB binding when glucose levels are low, thereby preventing RHEB from activating MTORC1.Citation35 Knocking down or perturbing the interaction between GAPDH and RHEB renders MTORC1 unable to sense changes in glucose levels. Interestingly, this GAPDH-RHEB interaction is observed even under high levels of glucose suggesting that GAPDH shuttles between glycolysis and the MTOR pathways acting as a direct mediator of MTORC1 signaling in response to glucose levels. Low glucose levels also affect MTORC1 signaling indirectly through decreasing ATP levels, which leads to activation of AMP-activated protein kinase (AMPK). AMPK inhibits MTORC1 activity by phosphorylating TSC2, which inhibits RHEB-mediated MTORC1 activation.Citation36,37 The RHEB-mediated signaling of glucose to MTORC1 suggests that lysosomal localization would also be an important component of glucose sensing; however, this has so far not been directly tested. Whether or not MTORC1 relocates to the cytoplasm when inactivated by GAPDH-RHEB signaling remains to be determined.
Lipids also interact with and regulate MTOR activity. The saturated free fatty acid palmitate induces MTORC1 activation by increasing its translocation onto the lysosomal surface.Citation38 Palmitate supplementation also decreases AMPK phosphorylation leading to hypophosphorylation of RPTOR and the activation of MTORC1; this is reversed upon addition of the mono-unsaturated fatty acid oleate.Citation39 Oleate and the polyunsaturated fatty acid eicosapentanoic acid, inhibit MTORC1 activation.Citation38 Thus, saturated and unsaturated FFAs could have opposing effects on MTORC1 regulation.
AMPK is another major intracellular energy sensor that can inhibit MTORC1 by direct phosphorylation of RPTORCitation40 or by activating TSC2.Citation41 The V-ATPase-Ragulator complex also plays a role in sensing low energy levels by forming a complex with the AMPK regulators AXIN and STK11/LKB1 at the lysosome, and subsequent activation of AMPK. In addition, the GEF activity of Ragulator for RAG is inhibited by AXIN, leading to inactivation of MTOR and thus activation of the catabolic activities of the cell.Citation42 Therefore, the lysosome is not a passive loading dock for important nutrient sensors; instead, the lysosome is sensing and relaying information to warrant that the cell will fully commit to growth only when long-range growth factor signals, local building blocks, and energy are present.
The location of the lysosome in the cell is also found to be important in coordinating catabolic and anabolic processes that respond to nutrients. When nutrients are not limiting, lysosomes are found at the periphery of the cell associated with an activated MTORC1. By contrast, starvation causes perinuclear clustering of lysosomes facilitating autophagosome-lysosome fusion and the consequent release of nutrients during starvation.Citation43
An emerging line of investigation in lysosomal biology is how lysosomal biogenesis, function, and turnover are regulated through the lysosome. Integrated transcriptomics analysis revealed that several genes encoding lysosomal proteins are co-expressed after genetic, chemical, or environmental perturbations.Citation44 Promoter analysis of these lysosomal genes revealed a common regulatory sequence known as an E-box.Citation45 Together these data led to the identification of the coordinated lysosomal enhancement and regulation (CLEAR) gene network, which controls lysosomal biogenesis, and lysosome-related functions such as autophagy, exocytosis, endocytosis, and phagocytosis. The related E-box transcription factors MITF (microphthalmia-associated transcription factor), TFE3 (transcription factor binding to IGHM enhancer 3), TFEC (transcription factor EC), and TFEB (transcription factor EB) can bind to CLEAR sites. MITF, TFE3 and TFEB, hereafter MiTs, respond to starvation promoting autophagosome formation and lysosomal biogenesis.Citation46-48 When MTORC1 is active, it phosphorylates MiTs. Given that active MTOR is on the lysosomal membrane, the MiT transcription factors must be recruited to the lysosome to be phosphorylated. MiTs transiently localize to the lysosomal membrane through binding to the same RAG GTPases that recruit MTORC1 to the lysosome. Phosphorylation causes their binding to the cytosolic chaperone YWHA/14-3-3 and sequestration in the cytoplasm.Citation46-50 The current model suggests that in fed conditions the MiT transcription factors continuously cycle between the lysosome and the cytosol. When MTORC1 is inhibited, unphosphorylated MiTs are released from the YWHA chaperones and are free to enter the nucleus to transcriptionally regulate lysosomal homeostasis and autophagy.Citation46,48,51 Additionally, the class III phosphatidylinositol 3-kinase PIK3C3/VPS34 (a pro-autophagic lipid kinase) controls lysosomal tubulation downstream of MTOR. Upon phosphorylation by MTORC1, UVRAG activates PIK3C3; mutant versions of UVRAG result in reduced PIK3C3 activity, which in turn reduces the lysosomal pool of phosphatidylinositol 3-phopshate (PtdIns3P) causing increased lysosomal tubulation and failure to generate normal lysosomes during starvation.Citation52 Therefore, through the control of MTORC1, lysosomes control their own biogenesis and function.
Various ion channels present in the lysosomal membrane also help sense the presence of nutrients. MTORC1 associates with an ATP-sensitive sodium channel, a complex of TPCN1 and TPCN2 (2 pore segment channels) on the lysosomal membrane. Upon nutrient depletion and reduced ATP, MTORC1 is translocated away and the channel is open. This channel controls membrane potential, pH stability, and amino acid homeostasis.Citation53 The activity of a lysosomal Ca2+ channel, MCOLN1 (mucolipin 1) is also increased during starvation. Increased Ca2+ release promotes autophagosome-lysosome fusion as well as lysosome reformation from autolysosomes.Citation54 A lysosome specific phosphoinositide PtdIns(3,5)P2, activates both ion channels, TPCN and MCOLN1.Citation55,56 Thus, regulation of lysosomal cation channels is another mechanism by which lysosomes control their own health and abundance.
Nutrient processing by the lysosome
In this section we focus on the role of the lysosome in digesting energy-dense substrates like glycogen and lipids, and make a brief reference to the processing of proteins, growth factors and their receptors, micronutrients, and metabolic organelles. For a comprehensive description of lysosomal storage disorders, refer to recent reviews on the subject.Citation57,58
In addition to its role in nutrient sensing, the lysosome contributes to energy homeostasis through its direct role in the mobilization of energy stores. Specialized lysosomal hydrolases process energy-rich molecules such as lipids and glycogen to generate energy units and building blocks. Lysosomal hydrolases digest and mobilize nutrients in growth-promoting conditions (fed state), as made evident by the hyper-accumulation of undigested lipids or glycogen when the function of the lysosomal hydrolases is impaired, or of digested products when these cannot be cleared via lysosomal membrane transporters or other unknown exporting mechanisms. Undigested lipids or glycogen accumulate inside the lysosome and become toxic, leading to pathological states ranging from mild disease to death.Citation59,60 The essential role of the lysosome in nutrient homeostasis is illustrated by the compromised survival observed in organisms with impaired lysosomal hydrolase activity; this selective pressure has led to a high level of conservation of the hydrolases and their modulators across eukaryotes ().
Table 1. The lysosome is an essential energy generator.
The enzyme GAA (glucosidase, α; acid) is responsible for breaking down glycogen into glucose within the lysosome, and mutations in this gene lead to Pompe disease.Citation59 Pompe disease is characterized by the accumulation of glycogen within and beyond the lysosome, most prominently in glycogen-storing tissues like skeletal and cardiac muscle.Citation61 Humans with penetrant mutations in GAA experience severe muscle weakness in skeletal and respiratory muscles, and many die as infants.Citation62 Additionally, the disrupted mobilization of sugars from the lysosome can also lead to disease, as is the case in Salla disease, a sialic acid storage disease, where export of the monosaccharide sialic acid is defective due to mutations in its transporter, SLC17A5/sialin.Citation63 It is unclear how glucose is transported from the lysosome to the cytoplasm to be processed through the glycolytic pathway. Of the 3 sugar transporters that have been described to reside in the lysosome, only one has been shown to transport glucose out of rat liver lysosomes.Citation64,65 More recently, SLC2A8/GLUT8 (solute carrier family 2 [facilitated glucose transporter], member 8) was found to contain a highly conserved late endosomal/lysosomal motif. SLC2A8 was observed within endosomal/lysosomal membranes, and it does not translocate to the plasma membrane like the better-known transporter SLC2A4/GLUT4; however, the functional relevance of SLC2A8 remains to be determined.Citation66
Unlike glucose, which is soluble in the bloodstream and internalized into the cytoplasm through dedicated transporters, lipids circulate as parts of various lipoproteins and are taken up from the extracellular environment through specialized internalization mechanisms.Citation67 In one of the best-known examples, receptor-mediated endocytosis mediates the uptake of low-density lipoprotein (LDL) particles and directs them to the lysosome.Citation68 First, LDL binds the LDL receptor, then the plasma membrane invaginates forming a clathrin-coated vesicle containing LDL bound to its receptor, and these vesicles eventually fuse with the lysosome.Citation67 Within the lysosome, LIPA (lipase A, lysosomal acid, cholesterol esterase) is responsible for hydrolyzing triglycerides and cholesteryl esters contained in the LDL particle, converting them into free fatty acids and cholesterol.Citation69,70 The lysosome membrane protein NPC1 (Niemann-Pick disease, type C1), deletion of the gene that causes Niemann-Pick disease, type C, facilitates the efflux of cholesterol out of the lysosome.Citation71,72 It is unclear how free fatty acids are transported from the lysosome to the mitochondria, or the mitochondria and peroxisomes in the case of lower eukaryotes for their processing through β-oxidation.
In addition to processing bloodstream-circulating lipids, the lysosome digests lipids stored in cytoplasmic lipid droplets. Lipid droplet hydrolysis in fasted hepatocytes occurs mainly in autolysosomes, a process termed lipophagy.Citation73 When cells are under nutritional stress, the small GTPase RAB7A is activated and promotes trafficking of lipid droplets to multivesicular bodies and lysosomes for lipophagy.Citation74 Once in the lysosome, lipids are broken down by specialized lipases. Lysosomal acid lipasesCitation75 and Atg15,Citation76 an autophagy-related protein with predicted triglyceride-lipase activity, are proposed to mediate lipophagy. Epistatic analyses were used to establish that lysosomal lipases are responsible for breaking down fats through lipophagy in C. elegans. C. elegans mutants for the lysosomal lipase genes lipl-1 and lipl-3 accumulate 2-fold more fat than wild-type animals, and this obesity phenotype is not additive with the genetic inactivation of autophagy.Citation77 High-content in vivo RNA interference screening in C. elegans revealed that the MAX-like transcription factor MXL-3 represses lysosomal lipolysis in the presence of nutrients. MXL-3 shares its target sequence with HLH-30 (the C. elegans TFEB ortholog).Citation78 Opposite of MXL-3, HLH-30 induces the expression of lysosomal lipase genes upon fasting and this response is conserved in mouse and human cells in culture.Citation77 Interestingly, whereas mammals have only one lysosomal acid lipase, LIPA, yeast defective in either ATG15 or TGL1 (the homolog of human LIPA) accumulate more lipid droplets and mobilize lipids at a slower rate.Citation79 C. elegans has at least 3 lysosomal acid lipases.Citation77,80 This higher functional divergence in the lysosome of lower organisms suggests a need for more specialized processing, possibly to distinguish nutrients from lipid signals and biotoxins abundant in the complex habitats of these organisms. Mice deficient in lysosomal acid lipase show massive storage of triglycerides and cholesteryl esters in adult liver, adrenal glands, and small intestine, and die at 7 to 8 mo of age.Citation81 In humans, lysosomal acid lipase deficiency causes cholesteryl ester storage disease and the more severe Wolman disease, which is characterized by infant mortality accompanied by increased fat stores.Citation60
Two additional roles for the lysosome in lipid homeostasis were recently reported. Mice with impaired chaperone-mediated autophagy (CMA), a chaperone-dependent targeting of soluble cytosolic proteins to the lysosome, show liver steatosis in the presence of functional macroautophagy, suggesting that CMA is involved in lipid droplet breakdown.Citation82 Like macroautophagy, CMA is activated during prolonged starvation.Citation83 However, as expected from the consensus that the main substrates of CMA are proteins and not lipids, the role of CMA in lipid homeostasis is mediated by the proteolytic and not the lipolytic function of the lysosome. PLIN2 (perilipin 2) and PLIN3 are lipid droplet-coating proteins that protect lipid droplets from cytosolic lipases such as LIPE (lipase, hormone-sensitive). In basal conditions, and more so upon fasting, CMA translocates PLIN2 and PLIN3 to the lysosome for their degradation. Therefore, the lysosomal degradation of perilipins promotes fat breakdown by licensing cytosolic lipases access to their substrates contained in lipid droplets.Citation84 Second, recent data suggest that the lysosome could also be involved in scavenging of lipids for long-term provision of energy during prolonged starvation.Citation85
The lysosome also stores and provides nutrients, generates building blocks (i.e., amino acids), recycles nutrient and growth factor receptors, and participates in the quality control for important metabolic organelles. Proteins delivered to the yeast equivalent of the lysosome, the vacuole, are degraded by proteases and other vacuolar hydrolases.Citation86 Newly recycled amino acids such as leucine are effluxed to the cytosol via various vacuolar effluxers including Atg22, enabling protein synthesis.Citation87 Lysosomal proteases can nonselectively digest endogenous or exogenous proteins. Lysosomal proteases are generally termed cathepsins, which are mainly cysteine and aspartic proteases. Cathepsins are differentially active in various cell types and tissues, providing some level of substrate specificity.Citation88-90 Additionally, mammals break down cytosolic proteins selectively delivered to the lysosomes through CMA. The cytosolic HSPA8/HSC70 chaperone (heat shock protein family A [Hsp70] member 8) recognizes proteins containing a sequence similar to KFERQ.Citation91 These complexes formed of CMA substrate bound to HSPA8 bind to LAMP2A (lysosomal-associated membrane protein 2A) promoting its multimerization. CMA substrate proteins undergo unfolding and pass through the LAMP2A multimers helped by intralysosomal HSPA8. Lysosomal proteases then degrade the protein substrates after translocation.Citation92 Disease can arise from the improper breakdown or efflux of protein-derived amino acids from the lysosome. For example, an amino acid transporter called CYNS/cystinosin helps the translocation of cysteine across the lysosomal membrane; deletion of the corresponding gene leads to cystinosis, a lysosomal storage disease.Citation93 In C. elegans, loss of the lysosomal lysine/arginine transporter LAAT-1 causes accumulation of lysine and arginine and results in enlarged and defective lysosomes that compromise embryonic development.Citation94 Lysosomes are not only involved in digesting nutrients but also in temporarily storing essential elements such as zinc or iron.Citation95,96
In addition to the mobilization of macro- and micronutrients, the lysosome processes critical growth factors, and growth factor-receptor complexes. GHR (growth hormone receptor) bound to GH1 (growth hormone 1) is degraded in the lysosome after selective delivery of the receptors that will be recycled back to the plasma membrane.Citation97 Inhibition of the sorting of EGFR (epidermal growth factor receptor) into multivesicular bodies and its subsequent degradation in the lysosome leads to tumorigenesis in mice, developmental defects in Drosophila, as well as vulval abnormalities in C. elegans.Citation98-100 In addition, secretory granules containing growth factors such as insulin are also processed in the lysosome.Citation101 Therefore, lysosomal degradation of growth factors and their receptors contributes to fine tuning cellular responses to growth signals.
The lysosome is also required for the rejuvenation of metabolic organelles. In yeast, selective digestion of the endoplasmic reticulum happens in the vacuole during excessive ER stress. Termed reticulophagy/ER-phagy, this mechanism is distinct in that it does not require autophagosomes or proteins implicated in autophagy.Citation102 Additionally, the lysosome degrades mitochondria brought to it through a specialized form of macroautophagy termed mitophagy.Citation103 Lysosomes also recycle ribosomes,Citation104 peroxisomes,Citation105 and even other impaired lysosomes.Citation106
Long-range nutrient signaling from the lysosome
Significant progress has been made in our understanding of the role of the lysosome in providing a platform and local signaling to MTORC1 and AMPK. By contrast, little is known about the contribution of the lysosome to distal signaling. However the few known examples, described below, suggest the lysosome generates short- and long-range signals with important roles in cellular and organismal homeostasis.
As described in section 2, cholesteryl esters are taken up by receptor-mediated endocytosis, and degraded through the action of LIPA to release cholesterol through specialized transporters. In addition to being a precursor of many metabolites and a structural component of membranes, cholesterol released from the lysosomes also functions as a signaling molecule. The SREBF (sterol regulatory element binding transcription factor) proteins control the expression of genes involved in lipid uptake and biosynthesis.Citation107 When lysosome-derived cholesterol levels are high, SREBF resides in the ER, bound to SCAP (SREBF chaperone) and INSIG1 ().Citation108-110 Low cholesterol levels lead to dissociation of INSIG1, freeing the SREBF-SCAP complex to traffic to the Golgi where it is cleaved by the proteases MBTPS1/S1P and MBTPS2/S2P. Free SREBF translocates to the nucleus to activate the transcription of genes involved in lipid uptake and biosynthesis.Citation110 Conversely, binding of cholesterol to SCAP inhibits cleavage of the SREBF-SCAP complex. In this way, lysosomal cholesterol represses its own synthesis. Additionally, an excess of lysosome-derived cholesterol causes activation of the transcription factor NR1H/LXR (nuclear receptor subfamily 1 group H), which transcribes genes involved in the removal of cholesterol from cells.Citation111 Thus, sterol signals originated in the lysosome are an integral part of cholesterol homeostasis.
Figure 2. Long-range signals from the lysosome coordinate nutrient homeostasis. The lysosome generates signals that travel to activate cell autonomous or systemic responses that promote nutrient homeostasis. Some of these signaling pathways are depicted here: I. Cholesterol uptake and synthesis is controlled from the lysosome. Cholesterol is taken up and processed by the lysosomal system. When the lysosome releases enough cholesterol, the transcription factor SREBF/SREBP is in the ER. By contrast, low cholesterol promotes SREBF trafficking from the ER to the Golgi (not shown), and then to the nucleus where it transcribes genes involved in lipid uptake and biosynthesis.Citation110 II. Lysosome fatty-acid derivatives distally control autophagy and the transcription of β-oxidation genes. In C. elegans, fasting leads to increased lysosomal lipase activity (LIPL-4).Citation112 Increased LIPL-4 activity is capable of: 1) generating lipid signals including ω-3 and ω-6 polyunsaturated fatty acids (ω-FA) and oleoylethanolamide (OEA),Citation80,112 2) inhibiting LET-363/MTOR,Citation113 3) activating autophagy,Citation112,113 and 4) inducing β-oxidation and other metabolic genes through NHR-49 and NHR-80.Citation80 ω-3 and ω-6 polyunsaturated fatty acids are transported to distant tissues by LBP-3 and LBP-5, and OEA is transported to the nucleus by LBP-8. Green arrows indicate unconfirmed activation during fasting conditions. Dotted lines illustrate likely pathways that have not been directly tested (intermediate steps are likely). III. Lysosomal calcium activates lysosomal biogenesis and autophagy. Starvation triggers calcium release from the lysosome through the MCOLN1 channel. Calcium then activates the phosphatase PPP3/calcineurin, which dephosphorylates TFEB promoting its translocation to the nucleus where it transcribes genes involved in lysosomal biogenesis and autophagy.Citation116 LAL, lysosomal acid lipases.
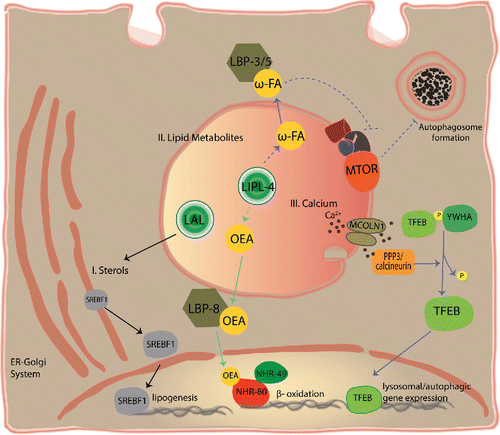
In fed as well as in starvation conditions, lipids stored in lipid droplets and the membranes of organelles are processed in the lysosome. Lysosomal acid lipases then break down these lipids into fatty acids. Mammals have a single lysosomal lipase, LIPA, whereas other animals like C. elegans have several lipases within their lysosomes. In C. elegans, the lysosomal acid lipase gene lipl-4 is upregulated upon starvation.Citation112 Increased expression of LIPL-4 leads to an enrichment in ω-3 and ω-6 polyunsaturated fatty acids during starvation,Citation112 and oleoylethanolamide (OEA) in nonphysiological conditions.Citation80 Both ω-3 and ω-6 polyunsaturated fatty acids (PUFAs) and OEA act as lipid signals. The lipid binding proteins LBP-3 and LBP-5, whose encoding genes are also upregulated upon fasting, transport ω-3 and ω-6 PUFAs to distant tissues where they activate autophagy in response to nutrient deprivation.Citation112 Lapierre et al. reported that upregulation of LIPL-4 leads to the inactivation of LET-363/MTOR;Citation113 it would be interesting to test if supplementation of the diet with ω-3 and ω-6 PUFAs is sufficient to reduce MTOR activity and thus explain the beneficial effects of fish oils on health span. Folick et al. reported a more intriguing lipid signaling mechanism by which the lipid binding protein LBP-8, whose encoding gene is paradoxically downregulated upon fasting,Citation112 translocates OEA into the nucleus.Citation80 OEA then binds and activates the nuclear hormone receptors NHR-49 and NHR-80; that among others, regulate the expression of genes involved in fatty acid β-oxidation (). Interestingly, in MCF-7 breast cancer cells, ω-3 PUFA-derived ethanolamines stimulate the mammalian homolog of NHR-49, PPARG/PPARγ (peroxisome proliferator-activated receptor gamma), inhibit the AKT-MTOR pathway, and induce phosphorylation of BCL2, thereby promoting its dissociation from BECN1/Beclin 1 which results in the activation of autophagy.Citation114 These observations, in addition to ω-3 and ω-6 PUFAs activating autophagy in human cells in culture,Citation112 suggest that the role of lysosome-derived lipid metabolites in organismal homeostasis may be conserved all the way up to humans.
The lysosome is the second largest store of calcium in the cell. The presence of calcium microdomains on the surface of the lysosome suggests a role for calcium in relaying messages from this organelle.Citation115 During starvation, TFEB becomes dephosphorylated enabling it to enter the nucleus and transcribe its target genes (). High-content short interfering RNA screening based on cytoplasm-to-nucleus shuttling of TFEB during starvation revealed that the calcium/calmodulin-dependent phosphatase PPP3/calcineurin is responsible for dephosphorylating TFEB, an essential requirement for its translocation to the nucleus.Citation116 Starvation of HeLa cells induces the release of calcium from the lysosome through the MCOLN1 channel without affecting endoplasmic reticulum calcium levels. Inhibition of MCOLN1 impairs the nuclear translocation of TFEB, and the induction of autophagy.Citation116 Thus, lysosomal calcium signaling controls autophagy through PPP3/calcineurin-mediated activation of TFEB.
Beyond its classic role in protein quality control and selective clearance lies a regulatory role for CMA where the upregulation of this autophagy mechanism allows for adaptation to stress and the activation of homeostatic transcriptional programs.Citation117 Cuervo et al. showed that during nutrient deprivation, the increase in transcription factor NFKB (nuclear factor of kappa light polypeptide gene enhancer in B-cells) activity is dependent on lysosomal degradation of NFKBIA/IκB (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, α).Citation118 Thus, lysosomal proteolysis is required for the regulation of genes involved in inflammation, cell proliferation and cell death in conditions of nutritional stress.
Future directions
We have described the role of the lysosome in: 1) nutrient sensing, 2) processing of energy-dense nutrients, and 3) the emission of signals that distally control or modulate energy homeostasis (). The critical role of the lysosome in ensuring organismal homeostasis is made evident by the striking conservation of its regulation and function. Comprehensive understanding of the role of the lysosome in essential biological functions, such as growth and reproduction, and in human disease requires broad approaches. Genomics, transcriptomics, proteomics, and lipidomics of patient-derived samples may reveal variations in sequence, expression, and activity of lysosomal proteins as well as its derived metabolites. These variants may either have predisposing, protective, or no effect on disease onset or progression. Therefore, functional genomics approaches in simpler model systems may help elucidate how these variants, and others too disruptive to be found in human populations, affect how the lysosome senses, processes, and relays information, and ultimately defines organismal homeostasis.
Figure 3. The lysosome is a nutrient sensing, processing and signaling center. The lysosome is the only organelle that receives nutrients and nutritional information from the cell (autophagy) and from the environment (endocytosis). Some of the roles of the lysosome in nutrient homeostasis include: I) sensing of nutrients and growth factors by the Lysosome Nutrient Sensing (LYNUS) machinery, II) digestion and recycling of circulating nutrients (i.e., cholesterol), growth factors (i.e., GH1 [growth hormone 1]), and nutrient regulators (i.e., perilipins); III) digestion and recycling of intracellular macromolecules and organelles; IV) recycling of growth factors, growth factor receptors (i.e., for GH1 and insulin), and nutrient receptors (i.e., LDLR/LDL receptor), V) coordination of responses to fluctuations in nutrient availability by releasing signaling molecules that activate homeostatic responses (i.e., cholesterol biosynthesis or activation of autophagy) locally and in distant cells and tissues; VI) controlling its own biogenesis, and VII) storage. All together, these functions provide building blocks and energy units to promote growth and reproduction, but most importantly the lysosome integrates nutritional information from the cell and the environment so that growth and reproduction are only promoted when conditions are favorable to do so.
![Figure 3. The lysosome is a nutrient sensing, processing and signaling center. The lysosome is the only organelle that receives nutrients and nutritional information from the cell (autophagy) and from the environment (endocytosis). Some of the roles of the lysosome in nutrient homeostasis include: I) sensing of nutrients and growth factors by the Lysosome Nutrient Sensing (LYNUS) machinery, II) digestion and recycling of circulating nutrients (i.e., cholesterol), growth factors (i.e., GH1 [growth hormone 1]), and nutrient regulators (i.e., perilipins); III) digestion and recycling of intracellular macromolecules and organelles; IV) recycling of growth factors, growth factor receptors (i.e., for GH1 and insulin), and nutrient receptors (i.e., LDLR/LDL receptor), V) coordination of responses to fluctuations in nutrient availability by releasing signaling molecules that activate homeostatic responses (i.e., cholesterol biosynthesis or activation of autophagy) locally and in distant cells and tissues; VI) controlling its own biogenesis, and VII) storage. All together, these functions provide building blocks and energy units to promote growth and reproduction, but most importantly the lysosome integrates nutritional information from the cell and the environment so that growth and reproduction are only promoted when conditions are favorable to do so.](/cms/asset/8e5b5c79-fca0-4abf-9e09-3d0200edef40/kaup_a_1147671_f0003_c.gif)
A few examples of important advances would be: 1) the discovery of protein complexes specialized in sensing other amino acids, nutrients other than amino acids, growth factors, or stress signals. The existence of complexes sensing specific amino acids, and the fact that these complexes share most but not all of their components supports the hypothesis that different lysosomal sensors could assess nutrients like fats or carbohydrates, as well as growth factors, or stress signals, so as to coordinate growth with the physiological status of the organism and the environment; 2) determining how lysosome-generated signals influence the function of other nutrient sensors, organelles, or distant cells; 3) defining the role of the lysosome in the flow of energy/nutrients to growth or reproduction. Lysosomes are directly involved in integrating nutrients and nutritional information to decide when to promote growth. Lysosomes also control reproduction through the mobilization of yolk particles. Thus, lysosomes are uniquely positioned to play a role in deciding soma vs. germline nutrient allocation; 4) assessing to which extent and how lysosomal signaling and function contribute to health span. We predict the lysosome will have roles in health span beyond being the digestive companion of autophagy.
We expect the body of knowledge on lysosomal structure, function, and regulation generated by the combination of ‘omics’ with traditional and emerging genetics and cell biological approaches will provide us with the ability to target lysosomal function to improve human health.
Abbreviations
| AMPK | = | AMP-activated protein kinase |
| BORCS6 | = | BLOC-1 related complex subunit 6 |
| CMA | = | chaperone-mediated autophagy |
| GAPDH | = | glyceraldehyde 3-phosphate dehydrogenase |
| GATOR | = | GTPase-activating protein toward Rag GTPases |
| GEF | = | guanine nucleotide exchange factor |
| LDL | = | low density lipoprotein |
| MCOLN | = | mucolipin 1 |
| MITF | = | microphthalmia-associated transcription factor |
| MiTs | = | MITF, TFE3 and TFEB transcription factors |
| MTOR | = | mechanistic target of rapamycin (serine/threonine kinase) |
| NHR | = | nuclear hormone receptor |
| NPC1 | = | Niemann-Pick disease, type C1 |
| OEA | = | oleoylethanolamide |
| PUFA | = | polyunsaturated fatty acid |
| RHEB | = | Ras homolog enriched in brain |
| SCAP | = | SREBP chaperone |
| SREBF | = | sterol regulatory element binding transcription factor |
| TFEB | = | transcription factor EB |
| TPCN | = | two pore segment channel |
| TSC | = | tuberous sclerosis complex |
| V-ATPase | = | vacuolar-type H+ adenosine triphosphatase |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Funding for this work was provided by the National Institutes of Health to E.O'R (DK087928) and SB (T32GM008136-30).
References
- Sleat DE, Della Valle MC, Zheng H, Moore DF, Lobel P. The mannose 6-phosphate glycoprotein proteome. J Proteome Res 2008; 7:3010-21; PMID:18507433; http://dx.doi.org/10.1021/pr800135v
- Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol 2003; 13:137-45; PMID:12628346; http://dx.doi.org/10.1016/S0962-8924(03)00005-9
- Bainton DF. The Discovery of Lysosomes. J Cell Biol 1981; 91:S66-S76; http://dx.doi.org/10.1083/jcb.91.3.66s
- Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J Cell Biol 2014; 205:435-45; PMID:24862571; http://dx.doi.org/10.1083/jcb.201402054
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12:21-35; PMID:21157483; http://dx.doi.org/10.1038/nrm3025
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010; 141:290-303; PMID:20381137; http://dx.doi.org/10.1016/j.cell.2010.02.024
- Efeyan A, Sabatini DM. Nutrients and growth factors in mTORC1 activation. Biochem Society Transactions 2013; 41:902-5; http://dx.doi.org/10.1042/BST20130063
- Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 2013; 14:283-96; PMID:23609508; http://dx.doi.org/10.1038/nrm3565
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012; 150:1196-208; PMID:22980980; http://dx.doi.org/10.1016/j.cell.2012.07.032
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008; 10:935-45; PMID:18604198; http://dx.doi.org/10.1038/ncb1753
- Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, Tagliabracci VS, Guan KL. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 2015; 347:194-8; PMID:25567907; http://dx.doi.org/10.1126/science.1259472
- Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015; 519:477-81; PMID:25561175; http://dx.doi.org/10.1038/nature14107
- Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015; 347:188-94; PMID:25567906; http://dx.doi.org/10.1126/science.1257132
- Schweitzer LD, Comb WC, Bar-Peled L, Sabatini DM. Disruption of the Rag-Ragulator Complex by c17orf59 Inhibits mTORC1. Cell Reports 2015; 12:1445-55; PMID:26299971; http://dx.doi.org/10.1016/j.celrep.2015.07.052
- Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a Multisubunit Complex that Regulates Lysosome Positioning. Developmental Cell 2015; 33:176-88; PMID:25898167; http://dx.doi.org/10.1016/j.devcel.2015.02.011
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 Is a Key Regulator of Nutrient Sensing in the mTORC1 Pathway. Mol Cell 2011; 44:134-46; PMID:21981924; http://dx.doi.org/10.1016/j.molcel.2011.06.038
- Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 Polyubiquitination and Activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell 2013; 51:283-96; PMID:23911927; http://dx.doi.org/10.1016/j.molcel.2013.06.020
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013; 340:1100-6; PMID:23723238; http://dx.doi.org/10.1126/science.1232044
- Panchaud N, Peli-Gulli MP, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal 2013; 6:ra42; PMID:23716719; http://dx.doi.org/10.1126/scisignal.2004112
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Reports 2014; 9:1-8; PMID:25263562; http://dx.doi.org/10.1016/j.celrep.2014.09.014
- Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan KL, Karin M, Budanov AV. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Reports 2014; 9:1281-91; PMID:25457612; http://dx.doi.org/10.1016/j.celrep.2014.10.019
- Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell 2014; 159:122-33; PMID:25259925; http://dx.doi.org/10.1016/j.cell.2014.08.038
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334:678-83; PMID:22053050; http://dx.doi.org/10.1126/science.1207056
- Stransky LA, Forgac M. Amino Acid Availability Modulates Vacuolar H+-ATPase assembly. J Biol Chem 2015; 290:27360-9; PMID:26378229; http://dx.doi.org/10.1074/jbc.M115.659128
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496-501; PMID:18497260; http://dx.doi.org/10.1126/science.1157535
- Oshiro N, Rapley J, Avruch J. Amino Acids Activate Mammalian Target of Rapamycin (mTOR) Complex 1 without Changing Rag GTPase guanyl nucleotide charging. J Biol Chem 2014; 289:2658-74; PMID:24337580; http://dx.doi.org/10.1074/jbc.M113.528505
- Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 2014; 156:786-99; PMID:24529380; http://dx.doi.org/10.1016/j.cell.2014.01.024
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 2003; 13:1259-68; PMID:12906785; http://dx.doi.org/10.1016/S0960-9822(03)00506-2
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003; 17:1829-34; PMID:12869586; http://dx.doi.org/10.1101/gad.1110003
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol 2005; 15:702-13; PMID:15854902; http://dx.doi.org/10.1016/j.cub.2005.02.053
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 2002; 10:151-62; PMID:12150915; http://dx.doi.org/10.1016/S1097-2765(02)00568-3
- Inoki K, Li Y, Zhu TQ, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 2002; 4:648-57; PMID:12172553; http://dx.doi.org/10.1038/ncb839
- Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol 2002; 4:658-65; PMID:12172554; http://dx.doi.org/10.1038/ncb840
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 2014; 156:771-85; PMID:24529379; http://dx.doi.org/10.1016/j.cell.2013.11.049
- Lee MN, Ha SH, Kim J, Koh A, Lee CS, Kim JH, Jeon H, Kim DH, Suh PG, Ryu SH. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol Cell Biol 2009; 29:3991-4001; PMID:19451232; http://dx.doi.org/10.1128/MCB.00165-09
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003; 115:577-90; PMID:14651849; http://dx.doi.org/10.1016/S0092-8674(03)00929-2
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http://dx.doi.org/10.1038/ncb2152
- Yasuda M, Tanaka Y, Kume S, Morita Y, Chin-Kanasaki M, Araki H, Isshiki K, Araki S, Koya D, Haneda M, et al. Fatty acids are novel nutrient factors to regulate mTORC1 lysosomal localization and apoptosis in podocytes. Biochimica Et Biophysica Acta 2014; 1842:1097-108; PMID:24726883; http://dx.doi.org/10.1016/j.bbadis.2014.04.001
- Kwon B, Querfurth HW. Palmitate activates mTOR/p70S6K through AMPK inhibition and hypophosphorylation of raptor in skeletal muscle cells: Reversal by oleate is similar to metformin. Biochimie 2015; 118:141-50; PMID:26344902; http://dx.doi.org/10.1016/j.biochi.2015.09.006
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008; 30:214-26; PMID:18439900; http://dx.doi.org/10.1016/j.molcel.2008.03.003
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 2006; 126:955-68; PMID:16959574; http://dx.doi.org/10.1016/j.cell.2006.06.055
- Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, et al. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab 2014; 20:526-40; PMID:25002183; http://dx.doi.org/10.1016/j.cmet.2014.06.014
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 2011; 13:453-60; PMID:21394080; http://dx.doi.org/10.1038/ncb2204
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science 2009; 325:473-7; PMID:19556463
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994; 8:2770-80; PMID:7958932; http://dx.doi.org/10.1101/gad.8.22.2770
- Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014; 7:ra9; PMID:24448649; http://dx.doi.org/10.1126/scisignal.2004754
- Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol 2013; 200:475-91; PMID:23401004; http://dx.doi.org/10.1083/jcb.201209135
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423; http://dx.doi.org/10.1126/scisignal.2002790
- Bronisz A, Sharma SM, Hu R, Godlewski J, Tzivion G, Mansky KC, Ostrowski MC. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol Biol Cell 2006; 17:3897-906; PMID:16822840; http://dx.doi.org/10.1091/mbc.E06-05-0470
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012; 8:903-14; PMID:22576015; http://dx.doi.org/10.4161/auto.19653
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095-108; PMID:22343943; http://dx.doi.org/10.1038/emboj.2012.32
- Munson MJ, Allen GFG, Toth R, Campbell DG, Lucocq JM, Ganley IG. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. Embo J 2015; 34:2272-90; PMID:26139536; http://dx.doi.org/10.15252/embj.201590992
- Cang C, Zhou Y, Navarro B, Seo YJ, Aranda K, Shi L, Battaglia-Hsu S, Nissim I, Clapham DE, Ren D. mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell 2013; 152:778-90; PMID:23394946; http://dx.doi.org/10.1016/j.cell.2013.01.023
- Wang W, Gao Q, Yang M, Zhang X, Yu L, Lawas M, Li X, Bryant-Genevier M, Southall NT, Marugan J, et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc Natl Acad Sci U S A 2015; 112:E1373-81; PMID:25733853; http://dx.doi.org/10.1073/pnas.1419669112
- Wang X, Zhang XL, Dong XP, Samie M, Li XR, Cheng XP, Goschka A, Shen DB, Zhou YD, Harlow J, et al. TPC Proteins Are Phosphoinositide-Activated Sodium-Selective Ion Channels in Endosomes and Lysosomes. Cell 2012; 151:372-83; PMID:23063126; http://dx.doi.org/10.1016/j.cell.2012.08.036
- Dong XP, Shen DBA, Wang X, Dawson T, Li XR, Zhang Q, Cheng XP, Zhang YL, Weisman LS, Delling M, et al. PI(3,5)P-2 controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat Commun 2010; 1:38; http://dx.doi.org/10.1038/ncomms1037
- Boustany RMN. Lysosomal storage diseases-the horizon expands. Nat Rev Neurol 2013; 9:583-98; PMID:23938739; http://dx.doi.org/10.1038/nrneurol.2013.163
- Filocamo M, Morrone A. Lysosomal storage disorders: molecular basis and laboratory testing. Human Genomics 2011; 5:156-69; PMID:21504867; http://dx.doi.org/10.1186/1479-7364-5-3-156
- van der Ploeg AT, Reuser AJ. Pompe's disease. Lancet 2008; 372:1342-53; PMID:18929906; http://dx.doi.org/10.1016/S0140-6736(08)61555-X
- Anderson RA, Rao N, Byrum RS, Rothschild CB, Bowden DW, Hayworth R, Pettenati M. In situ localization of the genetic locus encoding the lysosomal acid lipase/cholesteryl esterase (LIPA) deficient in Wolman disease to chromosome 10q23.2-q23.3. Genomics 1993; 15:245-7; PMID:8432549; http://dx.doi.org/10.1006/geno.1993.1052
- Hesselink RP, Wagenmakers AJ, Drost MR, Van der Vusse GJ. Lysosomal dysfunction in muscle with special reference to glycogen storage disease type II. Biochimica Et Biophysica Acta 2003; 1637:164-70; PMID:12633905; http://dx.doi.org/10.1016/S0925-4439(02)00229-6
- Manganelli F, Ruggiero L. Clinical features of Pompe disease. Acta Myol 2013; 32:82-4; PMID:24399863
- Verheijen FW, Verbeek E, Aula N, Beerens CE, Havelaar AC, Joosse M, Peltonen L, Aula P, Galjaard H, van der Spek PJ, et al. A new gene, encoding an anion transporter, is mutated in sialic acid storage diseases. Nat Genetics 1999; 23:462-5; PMID:10581036; http://dx.doi.org/10.1038/70585
- Mancini GMS, Beerens CEMT, Verheijen FW. Glucose-Transport in Lysosomal Membrane-Vesicles - Kinetic Demonstration of a Carrier for Neutral Hexoses. J Biol Chem 1990; 265:12380-7; PMID:2373697
- Maguire GA, Docherty K, Hales CN. Sugar-Transport in Rat-Liver Lysosomes - Direct Demonstration by Using Labeled Sugars. Biochem J 1983; 212:211-8; PMID:6409099; http://dx.doi.org/10.1042/bj2120211
- Augustin R, Riley J, Moley KH. GLUT8 contains a [DE]XXXL[LI] sorting motif and localizes to a late endosomal/lysosomal compartment. Traffic 2005; 6:1196-212; PMID:16262729; http://dx.doi.org/10.1111/j.1600-0854.2005.00354.x
- Goldstein JL, Brown MS, Anderson RG, Russell DW, Schneider WJ. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annual Rev Cell Biol 1985; 1:1-39; http://dx.doi.org/10.1146/annurev.cb.01.110185.000245
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science 1986; 232:34-47; PMID:3513311; http://dx.doi.org/10.1126/science.3513311
- Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J Biol Chem 1975; 250:8487-95; PMID:172501
- Bona G, Bracco G, Gallina MR, Iavarone A, Artesani L, Perona A, Zaffaroni M. A case of acid lipase deficiency: Wolman's disease. Panminerva Medica 1989; 31:49-53; PMID:2726290
- Sokol J, Blanchette-Mackie J, Kruth HS, Dwyer NK, Amende LM, Butler JD, Robinson E, Patel S, Brady RO, Comly ME, et al. Type C Niemann-Pick disease. Lysosomal accumulation and defective intracellular mobilization of low density lipoprotein cholesterol. J Biol Chem 1988; 263:3411-7; PMID:3277970
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 1997; 277:228-31; PMID:9211849; http://dx.doi.org/10.1126/science.277.5323.228
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009; 458:1131-5; PMID:19339967; http://dx.doi.org/10.1038/nature07976
- Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, McNiven MA. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatol 2015; 61:1896-907; http://dx.doi.org/10.1002/hep.27667
- Czaja MJ, Cuervo AM. Lipases in lysosomes, what for? Autophagy 2009; 5:866-7; PMID:19502773; http://dx.doi.org/10.4161/auto.9040
- Kovsan J, Bashan N, Greenberg AS, Rudich A. Potential role of autophagy in modulation of lipid metabolism. Am J Physiol Endocrinol Metab 2010; 298:E1-7; PMID:19887596; http://dx.doi.org/10.1152/ajpendo.00562.2009
- O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 2013; 15:668-76; PMID:23604316; http://dx.doi.org/10.1038/ncb2741
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 2009; 138:314-27; PMID:19632181; http://dx.doi.org/10.1016/j.cell.2009.04.058
- Jandrositz A, Petschnigg J, Zimmermann R, Natter K, Scholze H, Hermetter A, Kohlwein SD, Leber R. The lipid droplet enzyme Tgl1p hydrolyzes both steryl esters and triglycerides in the yeast, Saccharomyces cerevisiae. Biochim Et Biophysica Acta 2005; 1735:50-8; http://dx.doi.org/10.1016/j.bbalip.2005.04.005
- Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, Wang MC. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 2015; 347:83-6; PMID:25554789; http://dx.doi.org/10.1126/science.1258857
- Du H, Heur M, Duanmu M, Grabowski GA, Hui DY, Witte DP, Mishra JY. Lysosomal acid lipase-deficient mice: depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. J Lipid Res 2001; 42:489-500; PMID:11290820
- Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab 2014; 20:417-32; PMID:25043815; http://dx.doi.org/10.1016/j.cmet.2014.06.009
- Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol 1995; 269:C1200-8; PMID:7491910
- Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol 2015; 17:759-70; PMID:25961502; http://dx.doi.org/10.1038/ncb3166
- Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty Acid Trafficking in Starved Cells: Regulation by Lipid Droplet Lipolysis, Autophagy, and Mitochondrial Fusion Dynamics. Dev Cell 2015; 32:678-92; PMID:25752962; http://dx.doi.org/10.1016/j.devcel.2015.01.029
- Dunn WA, Jr. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol 1990; 110:1923-33; PMID:2351689; http://dx.doi.org/10.1083/jcb.110.6.1923
- Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell 2006; 17:5094-104; PMID:17021250; http://dx.doi.org/10.1091/mbc.E06-06-0479
- Kominami E, Tsukahara T, Bando Y, Katunuma N. Distribution of cathepsins B and H in rat tissues and peripheral blood cells. J Biochem 1985; 98:87-93; PMID:3900059
- Ii K, Hizawa K, Kominami E, Bando Y, Katunuma N. Different immunolocalizations of cathepsins B, H, and L in the liver. J Histochem Cytochem 1985; 33:1173-5; PMID:4056381; http://dx.doi.org/10.1177/33.11.4056381
- Baricos WH, Zhou YW, Fuerst RS, Barrett AJ, Shah SV. The role of aspartic and cysteine proteinases in albumin degradation by rat kidney cortical lysosomes. Arch Biochem Biophy 1987; 256:687-91; http://dx.doi.org/10.1016/0003-9861(87)90625-4
- Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Topics Dev Biol 2006; 73:205-35; http://dx.doi.org/10.1016/S0070-2153(05)73007-6
- Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J 2006; 25:3921-33; PMID:16917501; http://dx.doi.org/10.1038/sj.emboj.7601283
- Town M, Jean G, Cherqui S, Attard M, Forestier L, Whitmore SA, Callen DF, Gribouval O, Broyer M, Bates GP, et al. A novel gene encoding an integral membrane protein is mutated in nephropathic cystinosis. Nat Genetics 1998; 18:319-24; PMID:9537412; http://dx.doi.org/10.1038/ng0498-319
- Liu B, Du H, Rutkowski R, Gartner A, Wang X. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science 2012; 337:351-4; PMID:22822152; http://dx.doi.org/10.1126/science.1220281
- Blaby-Haas CE, Merchant SS. Lysosome-related organelles as mediators of metal homeostasis. J Biol Chem 2014; 289:28129-36; PMID:25160625; http://dx.doi.org/10.1074/jbc.R114.592618
- Roh HC, Collier S, Guthrie J, Robertson JD, Kornfeld K. Lysosome-related organelles in intestinal cells are a zinc storage site in C. elegans. Cell Metab 2012; 15:88-99; PMID:22225878; http://dx.doi.org/10.1016/j.cmet.2011.12.003
- Beguinot L, Lyall RM, Willingham MC, Pastan I. Down-regulation of the epidermal growth factor receptor in KB cells is due to receptor internalization and subsequent degradation in lysosomes. Proc Natl Acad Sci U S A 1984; 81:2384-8; PMID:6326124; http://dx.doi.org/10.1073/pnas.81.8.2384
- Aroian RV, Koga M, Mendel JE, Ohshima Y, Sternberg PW. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 1990; 348:693-9; PMID:1979659; http://dx.doi.org/10.1038/348693a0
- Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, Bellen HJ. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 2002; 108:261-9; PMID:11832215; http://dx.doi.org/10.1016/S0092-8674(02)00611-6
- Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem 1997; 272:2927-35; PMID:9006938; http://dx.doi.org/10.1074/jbc.272.5.2927
- Hoboth P, Muller A, Ivanova A, Mziaut H, Dehghany J, Sonmez A, Lachnit M, Meyer-Hermann M, Kalaidzidis Y, Solimena M. Aged insulin granules display reduced microtubule-dependent mobility and are disposed within actin-positive multigranular bodies. Proc Natl Acad Sci U S A 2015; 112:E667-76; PMID:25646459; http://dx.doi.org/10.1073/pnas.1409542112
- Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci 2014; 127:4078-88; PMID:25052096; http://dx.doi.org/10.1242/jcs.154716
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 2007; 462:245-53; PMID:17475204; http://dx.doi.org/10.1016/j.abb.2007.03.034
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 2008; 10:602-10; PMID:18391941; http://dx.doi.org/10.1038/ncb1723
- Tuttle DL, Lewin AS, Dunn WA, Jr. Selective autophagy of peroxisomes in methylotrophic yeasts. Eur J Cell Biol 1993; 60:283-90; PMID:8330626
- Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 2013; 4:2111; PMID:23817530; http://dx.doi.org/10.1038/ncomms3111
- Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev 1996; 10:1096-107; PMID:8654925; http://dx.doi.org/10.1101/gad.10.9.1096
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 2002; 110:489-500; PMID:12202038; http://dx.doi.org/10.1016/S0092-8674(02)00872-3
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997; 89:331-40; PMID:9150132; http://dx.doi.org/10.1016/S0092-8674(00)80213-5
- Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A 1999; 96:11041-8; PMID:10500120; http://dx.doi.org/10.1073/pnas.96.20.11041
- Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, Tontonoz P. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR α. Proc Natl Acad Sci U S A 2000; 97:12097-102; PMID:11035776; http://dx.doi.org/10.1073/pnas.200367697
- O'Rourke EJ, Kuballa P, Xavier R, Ruvkun G. omega-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev 2013; 27:429-40; PMID:23392608; http://dx.doi.org/10.1101/gad.205294.112
- Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol 2011; 21:1507-14; PMID:21906946; http://dx.doi.org/10.1016/j.cub.2011.07.042
- Rovito D, Giordano C, Vizza D, Plastina P, Barone I, Casaburi I, Lanzino M, De Amicis F, Sisci D, Mauro L, et al. Omega-3 PUFA ethanolamides DHEA and EPEA induce autophagy through PPARgamma activation in MCF-7 breast cancer cells. J Cell Physiol 2013; 228:1314-22; PMID:23168911; http://dx.doi.org/10.1002/jcp.24288
- Rizzuto R, Pozzan T. Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol Rev 2006; 86:369-408; PMID:16371601; http://dx.doi.org/10.1152/physrev.00004.2005
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 2015; 17:288-99; PMID:25720963; http://dx.doi.org/10.1038/ncb3114
- Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res 2014; 24:92-104; PMID:24281265; http://dx.doi.org/10.1038/cr.2013.153
- Cuervo AM, Hu W, Lim B, Dice JF. IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell 1998; 9:1995-2010; PMID:9693362; http://dx.doi.org/10.1091/mbc.9.8.1995
- Teste MA, Enjalbert B, Parrou JL, Francois JM. The Saccharomyces cerevisiae YPR184w gene encodes the glycogen debranching enzyme. FEMS Microbiol Lett 2000; 193:105-10; PMID:11094287; http://dx.doi.org/10.1111/j.1574-6968.2000.tb09410.x
- Sikora J, Urinovska J, Majer F, Poupetova H, Hlavata J, Kostrouchova M, Ledvinova J, Hrebicek M. Bioinformatic and biochemical studies point to AAGR-1 as the ortholog of human acid α-glucosidase in Caenorhabditis elegans. Mol Cell Biochem 2010; 341:51-63; PMID:20349118; http://dx.doi.org/10.1007/s11010-010-0436-3
- Khanna R, Flanagan JJ, Feng J, Soska R, Frascella M, Pellegrino LJ, Lun Y, Guillen D, Lockhart DJ, Valenzano KJ. The pharmacological chaperone AT2220 increases recombinant human acid α-glucosidase uptake and glycogen reduction in a mouse model of Pompe disease. PloS One 2012; 7:e40776; PMID:22815812; http://dx.doi.org/10.1371/journal.pone.0040776
- van der Beek NA, van Capelle CI, van der Velden-van Etten KI, Hop WC, van den Berg B, Reuser AJ, van Doorn PA, van der Ploeg AT, Stam H. Rate of progression and predictive factors for pulmonary outcome in children and adults with Pompe disease. Mol Genetics Metab 2011; 104:129-36; http://dx.doi.org/10.1016/j.ymgme.2011.06.012
- Seifert BL, Snyder MS, Klein AA, O'Loughlin JE, Magid MS, Engle MA. Development of obstruction to ventricular outflow and impairment of inflow in glycogen storage disease of the heart: serial echocardiographic studies from birth to death at 6 months. Am Heart J 1992; 123:239-42; PMID:1729839; http://dx.doi.org/10.1016/0002-8703(92)90779-U
- Kostera-Pruszczyk A, Opuchlik A, Lugowska A, Nadaj A, Bojakowski J, Tylki-Szymanska A, Kaminska A. Juvenile onset acid maltase deficiency presenting as a rigid spine syndrome. Neuromuscular Disorders 2006; 16:282-5; PMID:16531044; http://dx.doi.org/10.1016/j.nmd.2006.02.001
- Du H, Heur M, Duanmu M, Grabowski GA, Hui DY, Witte DP, Mishra J. Lysosomal acid lipase-deficient mice: depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. J Lipid Res 2001; 42:489-500; PMID:11290820
- Hoffman EP, Barr ML, Giovanni MA, Murray MF. Lysosomal Acid Lipase Deficiency. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Smith RJH, Stephens K, eds. GeneReviews(R) Seattle (WA), 1993
