ABSTRACT
Cells control their metabolism through modulating the anabolic and catabolic pathways. TP53INP2/DOR (tumor protein p53 inducible nuclear protein 2), participates in cell catabolism by serving as a promoter of autophagy. Here we uncover a novel function of TP53INP2 in protein synthesis, a major biosynthetic and energy-consuming anabolic process. TP53INP2 localizes to the nucleolus through its nucleolar localization signal (NoLS) located at the C-terminal domain. Chromatin immunoprecipitation (ChIP) assays detected an association of TP53INP2 with the ribosomal DNA (rDNA), when exclusion of TP53INP2 from the nucleolus repressed rDNA promoter activity and the production of ribosomal RNA (rRNA) and proteins. The removal of TP53INP2 also impaired the association of the POLR1/RNA polymerase I preinitiation complex (PIC) with rDNA. Further, TP53INP2 interacts directly with POLR1 PIC, and is required for the assembly of the complex. These data indicate that TP53INP2 promotes ribosome biogenesis through facilitating rRNA synthesis at the nucleolus, suggesting a dual role of TP53INP2 in cell metabolism, assisting anabolism on the nucleolus, and stimulating catabolism off the nucleolus.
Introduction
Precise control of ribosome biogenesis is fundamental for cell growth and proliferation, which are critically dependent on the synthesis of macromolecules including proteins. An initial and key regulating step in ribosome biogenesis for protein production is the synthesis of rRNA precursor from rDNA by POLR1/RNA polymerase I. Transcription of the rDNA starts with the recruitment and assembly of POLR1 and a set of transcription factors into a preinitiation complex (PIC) at the rDNA promoter. In mammalian cells, the basal factors essential for the recruitment of POLR1 to the rDNA promoter for achieving activated transcription, constitute UBTF (upstream binding transcription factor, RNA polymerase I), selectivity factor 1 (SL1), and RRN3 (RRN3 homolog, RNA polymerase I transcription factor).Citation1-3 UBTF binding to the rDNA promoter appears to be the first step in rDNA transcription followed by the recruitment of SL1.Citation4 Through interaction with both POLR1 and SL1, RRN3 guides POLR1 to the rDNA promoter and facilitates multiple rounds of transcription.Citation5-7 The rDNA transcription can be modified in response to cellular environment cues that correlates strongly with cellular demand for protein synthesis, including the availability of growth factors and nutrients.Citation1,8 Nevertheless, the regulation of the recruitment and assembly of POLR1 PIC in rDNA promoter is far from fully understood.
TP53INP2/DOR (tumor protein p53 inducible nuclear protein 2; please note that the mouse nomenclature is TRP53INP2. However, we use TP53INP2 hereafter to refer to both the human and mouse genes/proteins for simplicity), is originally identified as a nuclear protein expressed abundantly in tissues with a high metabolism level.Citation9 By serving as a coactivator of the thyroid hormone receptor, TP53INP2 modulates thyroid hormone function, suggesting a role of TP53INP2 in cell catabolism. Consistently, TP53INP2 is downregulated in the skeletal muscle of Zucker diabetic fatty (ZDF) rats, and exerts a negative impact on myogenesis.Citation9,10 Animals with muscle-specific overexpression of TP53INP2 show reduced muscle mass, while deletion of TP53INP2 leads to muscle hypertrophy.Citation10 In addition, TP53INP2 interacts physically with the autophagy-related protein MAP1LC3A/B (microtubule associated protein 1 light chain 3 α/β), contributing to autophagy, a lysosome-dependent degradation process in eukaryotic cells.Citation11,12 Under nutrient-deprivation conditions, TP53INP2 associates in the nucleus with deacetylated MAP1LC3A/B, and relocates together with MAP1LC3A/B from the nucleus to the cytoplasm, which is essential to the formation of autophagosomes in the cytoplasm.Citation13 TP53INP2 binds to VMP1 (vacuole membrane protein 1) and targets to phagophore membranes,Citation11,12 indicating additional functions of TP53INP2 in the cytoplasm for autophagy. Moreover, it has been recently reported that thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy,Citation14 suggesting that functioning in autophagy may be the major role of TP53INP2 in cell metabolism modulation,Citation10 and that TP53INP2 mediates cell catabolism at both transcriptional and post-transcriptional level either in the nucleus or the cytoplasm.
Interestingly, in addition to the distribution in the nucleoplasm, TP53INP2 is also found at the nucleolus.Citation15 Disruption of the nucleolus impairs neither the nuclear-cytoplasmic shuttling of TP53INP2 nor the function of TP53INP2 in autophagy,Citation15 implying that the nucleolus-localized TP53INP2 may display an effect independent of autophagy. In this study, we have investigated the function of nucleolar TP53INP2 in ribosome biogenesis. Our results suggest an important role of TP53INP2 in rDNA transcription by interacting directly with the components of the POLR1 PIC and facilitating the assembly of the complex at rDNA promoters.
Results
TP53INP2 is localized dynamically to the nucleolus through its C-terminal domain
Before the functional assessment of TP53INP2 in the nucleolus, we began with the verification of the location and property of endogenous TP53INP2 in the nucleolus, because so far the nucleolar localization of TP53INP2 has only been seen in exogenously expressed TP53INP2.Citation15 Using different human cell lines and specific anti-TP53INP2 antibody, we detected by immunostaining the presence of endogenous TP53INP2 at the nucleolus, indicated by a perfect colocalization of TP53INP2 with nucleolar marker proteins POLR1A/RPA194 (polymerase [RNA] I subunit A) and FBL (fibrillarin) (). The nucleolar signals of TP53INP2 but not that of FBL, faded away in cells treated with TP53INP2 siRNAs (Fig. S1), confirming the specificity of the TP53INP2 antibody staining and suggesting that TP53INP2 may not be essential to the assembly of the nucleolus. The distribution of TP53INP2 in the nucleolus was verified by the results from cell fractionation and nucleolus isolation showing that TP53INP2 was enriched in the extracted and purified nucleolus (). We then performed fluorescence recovery after photobleaching in living cells expressing a GFP-tagged TP53INP2. A very fast GFP fluorescence recovery was observed when a selected nucleolar region was photobleached (), indicating a rapid exchange between the nucleoplasmic pool and the nucleolar pool of the GFP-TP53INP2. This exchange mimics very much that of many known nucleolar components involved in ribosome biogenesis.Citation16,17
Figure 1. TP53INP2 is localized dynamically to the nucleolus through its C-terminal domain. (A) Colocalization of TP53INP2 with the nucleolar markers. The cells stained with anti-TP53INP2 and anti-POLR1A or anti-TP53INP2 and anti-FBL antibodies, were visualized by confocal microscopy. (B) Analysis of TP53INP2 distribution in subcellular fractions and purified nucleoli of HeLa cells. TUBB, LMNB1 or FBL was used as indicator of the cytosolic, nuclear or nucleolar fraction respectively. (C) HeLa cells transiently expressing GFP-TP53INP2 were imaged before and after photobleaching the indicated nucleolar region (red circle). (D) MCF-7 cells transiently expressing GFP-TP53INP2, GFP-TP53INP2Δ (191 to 212) or GFP-TP53INP2 (191 to 212) were stained with anti-FBL. Scale bars: 10 μm.
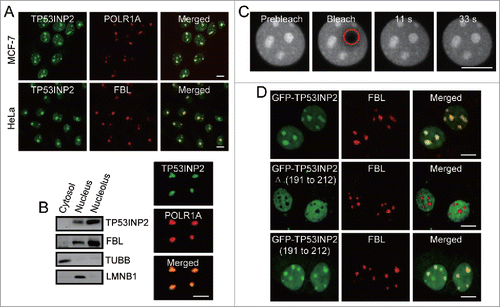
Wild-type full-length TP53INP2 comprises 221 amino acids. To search for the signal sequence in TP53INP2 that is responsible for the localization of TP53INP2 to the nucleolus, we created GFP-tagged truncated TP53INP2 mutants and expressed them in the cells. We found that a truncated TP53INP2 mutant lacking amino acids 191 to 212, failed to locate to the nucleolus, although it was distributed in the nucleoplasm (). Meanwhile, a TP53INP2 mutant that contains merely the 191 to 212 amino acids, was sufficient to associate with the nucleolus (). Together, these data suggest that TP53INP2 is a dynamic nucleolar protein and its nucleolar localization signal (NoLS) is included in its C-terminal domain.
TP53INP2 is required for rDNA transcription
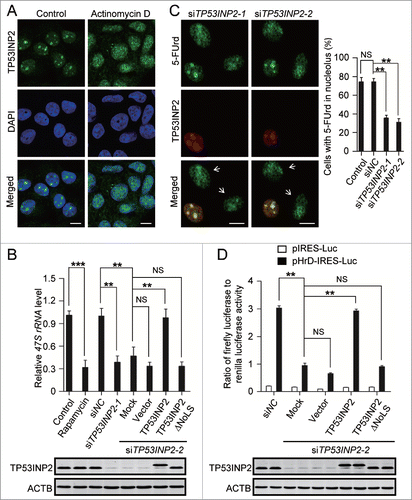
The localization of TP53INP2 in the nucleolus prompted us to investigate a possible role of TP53INP2 in rRNA synthesis. First, we examined the correlation between TP53INP2 nucleolar distribution and rDNA transcription. Treatment of the cells with actinomycin D at low concentrations that specifically inhibit rDNA transcription by POLR1,Citation18,19 abolished TP53INP2 from the nucleolus (), indicating a potential involvement of TP53INP2 in rDNA transcription. We then measured the primary rRNA transcript 47S rRNA production in TP53INP2 knockdown cells. Clearly, treatment with TP53INP2 siRNAs resulted in a significant decrease in 47S rRNA level, which was reversed by expression of a wild-type TP53INP2, but not a TP53INP2 mutant lacking the NoLS (TP53INP2ΔNoLS) (). POLR1 transcription activity was directly assessed by an in situ run-on assay based on the incorporation of 5-fluorouridine (5-FUrd) into nascent RNA.Citation20,21 In TP53INP2 knockdown cells, 5-FUrd incorporation at nucleolar sites detected by an anti-BrdU antibody, was evidently inhibited (). Using the human rDNA promoter luciferase reporter (pHrD-IRES-Luc),Citation22 we found that knockdown of TP53INP2 caused dramatically the inhibition of rDNA promoter activity (). Furthermore, this inhibition could be restored by expression in TP53INP2 knockdown cells of the wild-type TP53INP2 but not the TP53INP2ΔNoLS (). These results therefore suggest that nucleolus-localized TP53INP2 is required for rDNA transcription by preserving rDNA promoter activity.
Figure 2. TP53INP2 is required for rDNA transcription. (A) MCF-7 cells treated with 50 ng/ml of actinomycin D for 2 h, were fixed and stained with anti-TP53INP2 and DAPI. (B) HeLa cells treated by TP53INP2 siRNA2 for 24 h were transfected with pCDNA3.1-myc (vector), TP53INP2-MYC (TP53INP2) or TP53INP2ΔNoLS-MYC (TP53INP2ΔNoLS) respectively. After 24 h, cellular 47S rRNA level was measured by real-time PCR and normalized to ACTB mRNA. Cells treated with rapamycin (80 nM, 4 h) were used as a positive control. TP53INP2 protein levels in the cells were shown by western blot. (C) MCF-7 cells treated with TP53INP2 siRNAs were incubated with 5-FUrd, fixed and stained with anti-TP53INP2 and anti-BrdU. Arrows indicate TP53INP2 knockdown cells. (D) HeLa cells treated with TP53INP2 siRNA2 for 24 h were transfected with the indicated plasmids. After 24 h, luciferase activity was measured. TP53INP2 protein levels in the cells were shown by western blot. Data are presented as mean ± SEM of triplicate experiments. ***, P< 0.001; **, P < 0.01; NS, not significant. Scale bars: 10 μm.
TP53INP2 binds to the rDNA locus
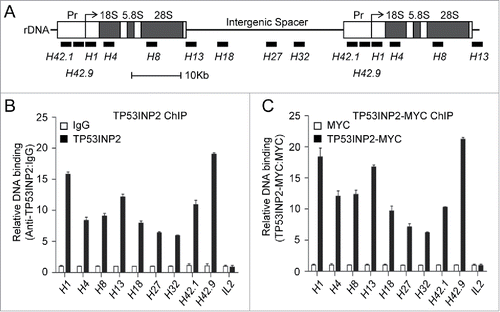
Regulation of rDNA promoter activity by TP53INP2 suggests a potential association of TP53INP2 with rDNA. We therefore performed a chromatin immunoprecipitation (ChIP) assay to check the interaction between TP53INP2 and rDNA. The DNA precipitated by TP53INP2 antibody was amplified by real time PCR using 9 primer sets distributed spanning the entire rDNA repeats (). We found that TP53INP2 is particularly enriched in the promoter regions of rDNA (H42.1, H42.9 and H1), moderately enriched in the coding regions (H4, H8 and H13) and less presented in the untranscribed intergenic spacer (H18, H27 and H32) (). ChIP experiments were also carried out with exogenously expressed TP53INP2-MYC, and a similar distribution of TP53INP2-MYC along the rDNA promoter was detected (). Amplification of the promoter region of IL2 (interleukin 2) where TP53INP2 does not bind,Citation9 was used as a negative control to confirm a specific precipitation of TP53INP2 and TP53INP2-associated DNA by the TP53INP2 antibody ().
Figure 3. TP53INP2 binds to the rDNA locus. (A) Schematic representation of the human rDNA repeats. The positions of primer pairs (in kb relative to the transcription start site) used in ChIP assays are indicated. (B) ChIP assay of the enrichment of rDNA in TP53INP2 immunoprecipitates. (C) ChIP assay of the enrichment of rDNA in TP53INP2-MYC immunoprecipitates from TP53INP2-MYC-expressing HeLa cells. The occupancy of TP53INP2 on the promoter region of interleukin 2 (IL2) was used as a negative control. Data are presented as mean ± SEM of triplicate experiments.
TP53INP2 is required for the recruitment of POLR1 machinery to rDNA
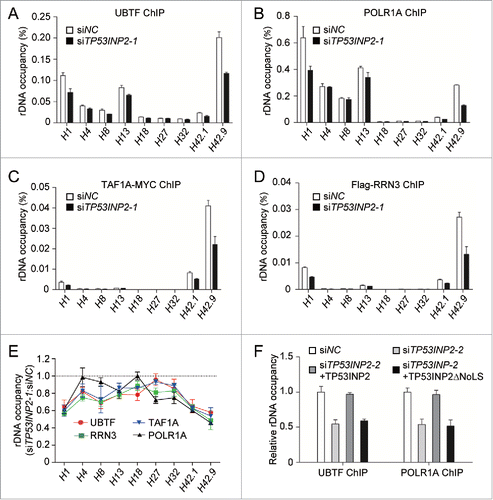
In yeast, rRNA synthesis is regulated by both the proportion of active genes and the transcription from active loci.Citation3,23 However, in mammalian cells, rRNA production is mainly mediated by direct regulation of the activity of the POLR1 transcription machinery in which the formation of POLR1 PIC at rDNA promoters is a primary determiner.Citation24,25 To test whether TP53INP2 promotes rDNA transcription by stimulating the assembly of POLR1 PIC at rDNA promoters, we used a ChIP assay to check the effect of TP53INP2 depletion on rDNA promoter binding of the complex components. Consistent with previous findings,Citation26 UBTF and POLR1A, that function in both transcription initiation and elongation, bound to the rDNA promoter regions (H1, H42.1 and H42.9), as well as the coding regions (H4, H8 and H13) ( and Fig. S2A and B), TAF1A/TAFI48, a subunit of the SL1 complex, and RRN3 were located only in the promoter regions ( and Fig. S2C and D). Significantly, in TP53INP2 knockdown cells, binding of UBTF and POLR1A to the rDNA promoters was impaired, whereas their binding to the coding regions was relatively less affected (, B and E and Fig. S2A and B). The occupancy of TAF1A and RRN3 at the rDNA promoters was also dramatically decreased by TP53INP2 knockdown (, D and E and Fig. S2C and D). Furthermore, the reduced occupancy of UBTF and POLR1A by TP53INP2 RNAi could be restored by expression in TP53INP2 knockdown cells of wild-type TP53INP2, but not TP53INP2ΔNoLS (). These data suggest that nucleolus-localized TP53INP2 contributes to the recruitment of the POLR1 PIC to the rDNA promoters, and imply that TP53INP2 may promote the assembly of the complex. The intracellular protein levels of POLR1A, TAF1A, RRN3 and UBTF were not changed in TP53INP2 siRNAs-treated cells (data not shown), indicating that the effect of TP53INP2 knockdown was not due to a decrease in the expression of the complex components.
Figure 4. TP53INP2 is required for the recruitment of the POLR1 machinery to rDNA. (A to D) Association of rDNA with UBTF (A), POLR1A (B), TAF1A-MYC (C) or Flag-RRN3 (D) was analyzed by ChIP assay in HeLa cells treated with TP53INP2 siRNA1. The percentage of precipitated DNA was calculated relative to the ChIP input DNA. (E) Normalized quantification of the data from (A to D). (F) HeLa cells treated with TP53INP2 siRNA2 for 24 h were transfected with TP53INP2-MYC or TP53INP2ΔNoLS-MYC. After 24 h, the cells were subjected to ChIP assay using an anti-UBTF or anti-POLR1A antibody. The relative enrichment was determined by real time PCR using primer set H42.9. All data are presented as mean ± SEM of triplicate experiments.
TP53INP2 interacts with POLR1 PIC and is required for the assembly of the complex
Given that TP53INP2 associates across the whole rDNA repeats with particular affinity for the promoter region, which is very similar to that of UBTF,Citation26,27 we then tested a potential interaction of TP53INP2 with POLR1 PIC. We found that TP53INP2-MYC expressed in cells was able to coimmunoprecipitate UBTF, TAF1A, RRN3 and POLR1A (). In addition, purified TP53INP2 pulled down each of the proteins from the cell lysates, and DNase I treatment failed to abrogate it (), indicating that DNA did not mediate the observed interactions. Looking for the direct target of TP53INP2 in POLR1 PIC, we carried out an siRNA screening. Knockdown of UBTF dramatically weakened the interaction between TP53INP2 and the other proteins (), while the interaction between TP53INP2 and UBTF was not demonstrably influenced by RNAi of either POLR1A or TAF1A or RRN3 (Fig. S3).
Figure 5. TP53INP2 interacts with POLR1 PIC and is required for the assembly of the complex. (A) Coimmunoprecipitation of UBTF, POLR1A, Flag-RRN3, TAF1A-flag with TP53INP2-MYC in HeLa cells. TP53INP2-MYC was immunoprecipitated with anti-MYC. (B) Cell lysates from HeLa cells expressing Flag-RRN3 and TAF1A-MYC were treated with DNase I (100 units/ml, 10 min), and incubated with purified GST-TP53INP2. POLR1A, Flag-RRN3, TAF1A-MYC or UBTF bound with affinity isolated GST-TP53INP2 was detected by western blot. (C) GST-TP53INP2 pulldown assay in HeLa cells treated with UBTF siRNA for 24 h followed by Flag-RRN3 and TAF1A-MYC transfection. The affinity isolated GST-TP53INP2 is shown. (D) HeLa cells expressing CFP-UBTF and TP53INP2-YFP were imaged before and after photobleaching with a high-intensity 514 nm laser. Note the increased CFP-UBTF signals at the nucleolar region after photobleaching. Changes of CFP and YFP signals within the nucleolus were shown. (E) In vitro binding of TP53INP2 with UBTF. Purified TP53INP2 was incubated with purified GST-UBTF or GST-TAF1A immobilized on glutathione-sepharose beads, and bound TP53INP2 was analyzed by western blot. (F) Gel filtration analysis of the nuclear extract of HeLa cells treated with TP53INP2 siRNAs. The fractions were analyzed by western blot using anti-POLR1A antibody. (G) TP53INP2 siRNA2-treated cells were transfected with Flag-RRN3 and TAF1A-MYC, together with TP53INP2-MYC or TP53INP2ΔNoLS-MYC. After 24 h, the cells were subjected to immunoprecipitation by anti-UBTF, followed by western blot to detect each of the indicated proteins.
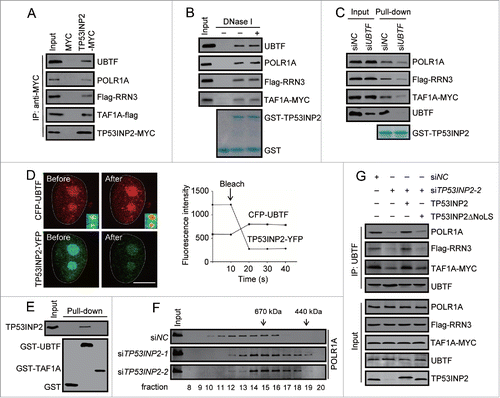
Further, we performed fluorescence resonance energy transfer assay, which detects the proximity of interacting proteins. In living HeLa cells expressing CFP-UBTF and TP53INP2-YFP, photobleaching TP53INP2-YFP, the energy acceptor, significantly enhanced the fluorescence intensity of CFP-UBTF, the energy donor, at the nucleolus (). Quantification of the CFP-UBTF fluorescence at the nucleolus showed a more than 1.35-fold increase after TP53INP2-YFP photobleaching, indicating a strong energy transfer between the 2 proteins. Furthermore, recombinant GST-UBTF but not GST-TAF1A, purified from Escherichia coli, pulled down purified TP53INP2 from the coculture of the proteins (), confirmed a direct interaction of TP53INP2 with UBTF.
To investigate a potential role of TP53INP2 in the assembly of POLR1 PIC, we first carried out gel filtration experiments. We found that POLR1 was present in a broad peak with different molecular mass as previously reported ().Citation28 Knockdown of TP53INP2 evidently shifted POLR1 sedimentation from larger complexes to smaller ones (), suggesting a function of TP53INP2 in POLR1 PIC formation. Further, we performed immunoprecipitation to test a possible need of TP53INP2 for the association of UBTF with the other components of POLR1 PIC. In HeLa cells, when endogenous UBTF coimmunoprecipitated endogenous POLR1A and exogenously expressed Flag-RRN3 and TAF1A-MYC, demonstrating the formation of POLR1 PIC in the cells, knockdown of TP53INP2 evidently dampened the coimmunoprecipitation (). Expressing wild-type TP53INP2, but not TP53INP2ΔNoLS, reversed the effect of TP53INP2 knockdown ().
Together, these results suggest that TP53INP2 regulates the assembly of POLR1 PIC at the rDNA promoter by interacting with UBTF.
TP53INP2 is required for protein synthesis and cell proliferation
Ribosome biogenesis is a prerequisite for protein synthesis and cell proliferation. To verify the effect of TP53INP2 in rRNA production, we examined cell protein synthesis through the surface sensing of translation (SUnSET),Citation29,30 detecting the amount of puromycin incorporation into nascent proteins. Clearly, mimicking that of serum depletion, TP53INP2 knockdown led to a decrease in protein synthesis rate (). Expression of wild-type TP53INP2, but not TP53INP2ΔNoLS, cancelled the effect of TP53INP2 knockdown (), indicating a function of TP53INP2 at the nucleolus. Consistently, knockdown of TP53INP2 inhibited cell proliferation indicated by reduced BrdU incorporation, and the inhibition was reversed by wild-type TP53INP2 but not TP53INP2ΔNoLS mutant expression ().
Figure 6. TP53INP2 is required for protein synthesis and cell proliferation. (A) Global protein synthesis detected by SUnSET in HeLa cells. The cells were either cultured with serum-free medium for 12 h or treated with TP53INP2 siRNA1. The specificity of the anti-puromycin antibody was demonstrated by a sample without puromycin incubation. Coomassie blue staining was used as loading control. (B and C) Puromycin incorporation in TP53INP2 siRNA1-treated cells detected by anti-puromycin antibody. Arrows indicate TP53INP2 knockdown cells. Scale bar: 10 μm. (D) HeLa cells treated with TP53INP2 siRNA2 for 24 h were transfected with TP53INP2-MYC or TP53INP2ΔNoLS-MYC. After 24 h, the nascent proteins were analyzed by SUnSET. (E and F) HeLa cells treated with TP53INP2 siRNA2 for 24 h were transfected with TP53INP2-MYC or TP53INP2ΔNoLS-MYC. After 24 h, cells were subjected to BrdU incorporation assay. Scale bar: 50 μm. (G) Intracellular distribution of TP53INP2 in HeLa cells. The cells were either cultured in amino acid-free medium for 3 h with or without replacement to normal medium for 3 h, or treated with 250 nM of Torin1 for 3 h with or without replacement to Torin1-free medium for 3 h. Scale bars: 10 μm. All the quantitative data are presented as mean ± SEM of triplicate experiments. **, P< 0.01; NS, not significant.
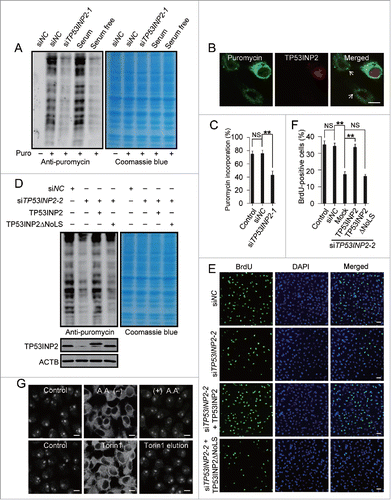
As a key regulator of autophagy, TP53INP2 relocates from the nucleus to the cytoplasm during cell starvation or rapamycin treatment which involves an inhibition of MTOR (mechanistic target of rapamycin [serine/threonine kinase]) to shut down protein synthesis.Citation11,12 To test whether MTOR inhibition leads to a dissociation of TP53INP2 from the nucleolus, the cells were either cultured with amino acid-free medium or treated with a specific MTOR inhibitor Torin1.Citation31 Clearly, amino acid starvation or Torin1 treatment caused release of TP53INP2 from both the nucleoplasm and nucleoli to the cytoplasm in the cells (). Intriguingly, placing the cells back to amino acid-rich or Torin1-free medium, TP53INP2 re-emerged not only in the nucleoplasm but also the nucleoli (), suggesting a strong correlation between MTOR activity and TP53INP2 nucleolar localization. Finally, we tested whether the nucleolus-untargeted TP53INP2 could support autophagy. We found, overexpression of TP53INP2ΔNoLS not only elevated the basal level but also enhanced amino acid starvation-induced autophagosome formation (Fig. S4), which mimicked highly that of TP53INP2 overexpression,Citation12,13 supporting a stimulating effect of nucleolus-disassociated TP53INP2 on autophagy.
Discussion
A major conclusion of this study is that TP53INP2 protein, a key player in autophagy initiation, is required for ribosome biogenesis, by serving as a promoter of rDNA transcription under adequate nutrition condition. This role of TP53INP2 is conferred by its localization at the nucleolus and interaction with POLR1 PIC.
Identification of the nucleolus-targeting domain in TP53INP2 allowed not only the ascertainment of TP53INP2 location at the nucleolus, but also the dissection of nucleolar function of the TP53INP2 protein. It has been shown that TP53INP2 shuttles between the nuclear pool and the cytoplasmic poor under fed conditions, and during the shuttling, TP53INP2 passes through the nucleolus.Citation15 However, our data from the GFP-TP53INP2 photobleaching experiments suggest that TP53INP2 is constitutively localized to the nucleolus and cycles between the nucleolus and the nucleoplasm. This is verified by the association of TP53INP2 with the rDNA, suggesting that the nucleolar localization of TP53INP2 is organized and significative, although disruption of the nucleolus shows no influence to the function of TP53INP2 in autophagy.Citation15 We show that the nucleolus-localized TP53INP2 contributes to rDNA transcription, and the mechanism for this function of TP53INP2 involves an interaction of TP53INP2 with the POLR1 PIC. In addition, we identified that UBTF is seemingly the direct target of TP53INP2. A plethora of cellular pathways control rRNA synthesis by directly regulating the activity of the POLR1 transcription machinery. When in many cases it relates to the post-translational modifications of the components of the PIC including UBTF and subunits of the SL1 complex,Citation32-36 our data suggest that the TP53INP2-UBTF interaction mediates the recruitment and assembly of the complex at rDNA promoter. Currently, it remains unclear if TP53INP2 is the driving factor for the rDNA association of UBTF or the reverse, because both proteins present similar rDNA binding pattern, and when TP53INP2 knockdown disrupts the association of UBTF, knockdown of UBTF also affects the association of TP53INP2 (data not shown). Possibly, an interdependence between TP53INP2 and UBTF applies in the rDNA binding of the 2 proteins.
Stringent regulatory mechanisms operate to ensure a precise balance between cell anabolism and catabolism based the requirement for and availability of cell nutrition. Finding the role of TP53INP2 in ribosome biogenesis overthrows our previous acquaintance with the function of TP53INP2, which is that it participates mainly in cell catabolism by mediating autophagy, indicating a novel function of TP53INP2 in cell anabolism. The accomplishment of these opposite dual functions of TP53INP2 is dependent on the subcellular distributions of the protein. Under nutrient-rich conditions, it locates to the nucleolus, promoting protein synthesis. Meanwhile, by a slow shuttling between the nucleus and cytoplasm,Citation15 it contributes to the sustainment of basal autophagy. Under nutrient deficiency, TP53INP2 dissociates from the nucleolus, which, on the one hand, shuts down protein synthesis, the most energy consuming process, and, on the other hand, initiates autophagosome formation for autophagy.
Our results suggest that the targeting of TP53INP2 to the nucleolus is regulated by MTOR activity, although this may be due to the rapid cycling between the nucleolus and the nucleoplasm. So far, the potential mechanism by which MTOR regulates intracellular distribution of TP53INP2 is unclear. However, it seems like the phosphorylation of TP53INP2 is not responsible for the distribution, because we have found that the C-terminal domain of TP53INP2 determines its nuclear localization, and mutation of each of the 5 serines and/or threonines in the region showed little influence on the localization (data not shown). In addition, mutation of each of the lysine residues in the region changes neither the distribution of TP53INP2 at basal conditions nor starvation-induced nucleus-to-cytoplasm translocation of TP53INP2 (data not shown), excluding preliminarily an involvement of acetylation-deacetylation changes in the process. Nevertheless, our data indicating a dissociation of TP53INP2 from the nucleolus by inactivation of MTOR suggest that TP53INP2 is a novel mediator of MTOR activity for the control of ribosome biogenesis, in addition to the known RPS6KB1 (ribosomal protein S6 kinase B1) which governs both the biogenesis and translational efficiency of ribosomes.
Materials and methods
Cell cultures, reagents, and antibodies
HeLa and MCF-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C under an atmosphere of 5% CO2. Amino acid-free medium (24020–117) was from Invitrogen. Puromycin (A610593) was from Sangon Biotech; 5-fluorouridine (F5130), Brdu (B5002), actinomycin D (A1410), and rapamycin (R8781) were from Sigma-Aldrich; Torin1 (4247) was from Tocris Bioscience. The following antibodies were used: anti-TP53INP2 (LifeSpan Biosciences, C119137), anti-FBL (Abcam, ab4566), anti-puromycin (EMD Millipore, MABE343), anti-ACTB (Sigma-Aldrich, A5316), anti-TUBB (Sigma-Aldrich, T5293), anti-BrdU (Sigma-Aldrich, B8434), anti-UBTF (Santa Cruz Biotechnology, sc-13125), anti-POLR1A (Santa Cruz Biotechnology, sc-48385), anti-LMNB1 (Santa Cruz Biotechnology, sc-20682), anti-Flag (Santa Cruz Biotechnology, sc-807), anti-MYC (Santa Cruz Biotechnology, sc-40), Alexa Fluor 488 (A-11008 and A-11001) and 546 (A-11035 and A-11003) tagged secondary antibodies for immunostaining were from Molecular Probes. The secondary antibodies for western blot were donkey anti-rabbit IRDye800CW (LI-COR Biosciences, 926–32213) and donkey anti-mouse IRDye680 (LI-COR Biosciences, 926–32222).
Plasmid constructs and transfection
The TP53INP2 cDNA was kindly provided by Antonio Zorzano (Institute for Research in Biomedicine, Spain) and cloned into a pEGFP-C1 vector using EcoRI and BamHI to generate GFP-TP53INP2. GFP-tagged, TP53INP2-truncated mutants were constructed by inserting the corresponding DNA domains into a pEGFP-C1 vector using the HindIII and KpnI restriction sites. TP53INP2-MYC and TP53INP2ΔNoLS-MYC were constructed by inserting the DNA domains into a pCDNA3.1-myc vector using the HindIII and KpnI restriction sites. Flag-RRN3 was a gift from Ingrid Grummt (German Cancer Research Center, Germany). The TAF1A cDNA was kindly provided by Jiahuai Han (Xiamen University, China) and cloned into a pCDNA3.1-myc vector using KpnI and BamHI to generate TAF1A-MYC. TAF1A-flag was made by changing the MYC in TAF1A-MYC to Flag. The rDNA promoter reporter (pHrD-IRES-Luc) and the corresponding empty vector (pIRES-luc) were gifts from Ghoshal Kalpana (Ohio State University, Columbus). The pRLTK plasmid was provided by Ximei Wu (Zhejiang University, China). Transient transfections were performed using Lipofectamine 2000 (Invitrogen, 11668–019) according to the manufacturer's instructions. The following siRNA sequences (GenePharma) were used: TP53INP2 siRNA1, GACGAGAGCUGGUUUGUUATT; TP53INP2 siRNA2, CCUUACAUGUCUCACACUATT; UBTF siRNA, GAAGUUCCGUACAUUGACATT; RRN3 siRNA, AAAUAUGCGUGCAUUAGAGAATT; TAF1A siRNA, AAGAGGUACUCACCAAUUAUGTT; POLR1A siRNA, CAACUACGAGGUGAUAAUGAATT; nontargeting siRNA, UUCUCCGAACGUGUCACGUTT. In all the experiments, the cells were treated with the indicated siRNA for 48 h, unless otherwise specified. For all rescue experiments, TP53INP2 siRNA2 targeting the untranslated region of TP53INP2 was used.
Cell fractionation
Nuclear and cytoplasmic fractions were purified as described previously.Citation13 Nucleoli were prepared from HeLa cells using the method developed by Andersen et al.Citation37
Western blot and immunoprecipitation
Western blot was performed as described previously.Citation38 Briefly, the proteins from lysed cells were denatured and loaded on sodium dodecyl sulfate polyacrylamide gels. Then the proteins were transferred to PVDF membranes, blocked in TBST (150 mM NaCl, 10 mM TRIS-HCl, pH 7.5, and 0.1% Tween 20 [Sangon Biotech, B413BA0004]) containing 5% (w/v) bovine serum albumin (Amersco, 0332), and incubated with the corresponding primary and secondary antibodies. The specific bands were analyzed by the western blot infrared imaging system (LI-COR Biosciences, Nebraska, USA). For immunoprecipitation, cells were lysed with Triton X-100 (Beyotime, ST795) lysis buffer (50 mM TRIS-HCl, pH 7.5, 50 mM NaCl, 0.1% Triton X-100, 10% glycerol, 1 mM PMSF [Sigma-Aldrich, P7626], 1 mM DTT [Beyotime, ST040]) containing protease inhibitors (Roche, 04693116001). After centrifugation, the supernatant fractions were incubated with the indicated antibodies overnight and then Protein A/G agarose (Pierce, 20333 and 20398) for 2 h at 4°C. Immunocomplexes were washed and analyzed by western blot.
Immunostaining and confocal microscopy
For immunostaining, HeLa or MCF-7 cells were fixed in 4% formaldehyde followed by permeabilization with 0.1% Triton X-100. After washing twice in phosphate-buffered saline (Beyotime, C0221A), cells were incubated in phosphate-buffered saline with fetal calf serum (10%) to block nonspecific sites of antibody adsorption. The cells were then incubated with appropriate primary and secondary antibodies. Cell images were acquired on a laser scanning confocal microscope (LSM 510; Carl Zeiss, Oberkochen, Germany) and analyzed with Zeiss LSM Image Examiner Software. For fluorescence recovery after photobleaching analysis, a selected nucleolar region of a cell was photobleached with 488 nm laser at full power and the fluorescence recovery of the region was monitored at low-intensity illumination.
RNA purification and real-time PCR
Total cellular RNA was isolated using Trizol reagent (Invitrogen, 15596026) and reverse transcribed using random hexamers, dNTPs mixture and M-MLV reverse transcriptase (Promega Corporation, M1701) according to the manufacturer's protocol. Real-time PCR analysis was performed in a 10-µl reaction mixture using the SYBR GREEN PCR Master Mix (Takara, DRR041A) and the ABI7500 real-time PCR system (Applied Biosystems, Massachusetts, USA). The primers used are listed in Table S1.
ChIP assay
ChIP assay was performed as described previously.Citation39 Briefly, HeLa cells were cross-linked using formaldehyde, and lysed with SDS lysis buffer (50 mM TRIS-HCl, pH 8.1, 10 mM EDTA, pH 8.0, 1% SDS) containing protease inhibitors, then followed by sonication. The cross-linked, sonicated chromatin was precleared before being incubated with 5 μg of the indicated antibodies and rotated at 4°C overnight. After extensive washes, immunocomplexes were treated with proteinase K (Beyotime, ST532) and decrosslinked. Bound DNA in the precipitates, as well as input DNA, was extracted, purified, and subjected to real-time PCR analysis using primers corresponding to different regions of the rDNA repeats. The primers used are listed in Table S2.
Luciferase reporter assay
HeLa cells were treated with TP53INP2 siRNA2 for 24 h. Then pIRES-Luc or pHrD-IRES-Luc (expressing firefly luciferase) along with the internal control pRLTK (expressing Renilla luciferase, Promega Corporation, E2241) and the corresponding plasmids were cotransfected into HeLa cells. Cells were lysed in passive lysis buffer (from the luciferase assay kit) 24 h after transfection, and assayed for luciferase activity using a luciferase assay kit (Promega Corporation, E1910) according to the manufacturer's protocol.
Recombinant protein purification
Full-length TP53INP2, TAF1A or UBTF was cloned into pGEX-4T-1 and expressed as the GST-tagged form in Escherichia coli BL21 by induction with 0.1 mM isopropyl β-D-thiogalactopyranoside (Beyotime, ST097) for 12 h at 28°C. The recombinant proteins were purified using glutathione sepharose 4B beads (GE Healthcare Life Sciences, 17-0756-01), eluted with glutathione (Beyotime, S0073), or incubated with thrombin (GE Healthcare Life Sciences, 27-0846-01) at 4°C for 4 h to release the proteins from the GST. Then the eluates were appropriately concentrated with Amicon Ultra-4 filter (EMD Millipore, UFC801024) and glycerol was added to a final concentration of 25% for storage at −80°C.
In vitro affinity isolation assay
Recombinant proteins that were purified from Escherichia coli BL21 were incubated with another purified protein or whole cell protein extract at 4°C for 4 h. Glutathione sepharose 4B beads were added to the mixture, followed by further incubation at 4°C for 2 h. Immunocomplexes were washed 4 times with wash buffer (50 mM TRIS-HCl, pH 7.5, 50 mM NaCl, 0.1% Triton X-100, 10% glycerol, protease inhibitors, 1 mM PMSF, 1 mM DTT) and subjected to western blot.
Gel filtration analysis
Nuclear extract was resuspended in lysis buffer (50 mM TRIS-HCl, pH 7.5, 150 mM NaCl, 0.5% CHAPS [Sigma-Aldrich, C3023], 10% glycerol, 1 mM DTT and 1 mM PMSF), and centrifuged at 100,000 × g for 1 h. The resulting supernatant fraction was loaded onto a Superose 6 column (GE Healthcare Life Sciences, 17-5172-01) and eluted at a flow rate of 0.4 ml/min with 20 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM DTT. Aliquots from each flow through fraction were analyzed by western blot.
Protein synthesis and cell proliferation assays
For puromycin incorporation assay, cells were either treated with 1 μM puromycin for 30 min (for western blot), or 100 μM puromycin for 5 min (for immunostaining) followed by extraction with 0.015% digitonin (Sigma-Aldrich, D141)-supplemented permeabilization buffer (50 mM TRIS-HCl, pH 7.5, 5 mM MgCl2, 25 mM KCl, protease inhibitors) to release free puromycin (for immunostaining). Nascent protein was detected with anti-puromycin antibody using western blot or immunostaining. For 5-FUrd incorporation assay, cells were incubated with 2 mM 5-FUrd for 15 min and fixed with 4% paraformaldehyde. Nascent RNA was labeled with anti-BrdU antibody. For BrdU cell proliferation assay, cells were incubated with 20 μM BrdU for 1 h at 37°C. After fixation with 4% paraformaldehyde, the cells were treated with 2 M HCl, neutralized by boric acid (pH 8.4), and stained with anti-BrdU antibody and DAPI.
Statistical analysis
All the statistical data are presented as mean ± SEM. Statistical significance of the differences was determined using the Student t test. P < 0.05 was considered statistically significant.
Abbreviations
| 5-Furd | = | 5-fluorouridine |
| ACTB | = | actin, β |
| CFP | = | cyan fluorescent protein |
| ChIP | = | chromatin immunoprecipitation |
| FBL | = | fibrillarin |
| GFP | = | green fluorescent protein |
| GST | = | glutathione S-transferase |
| LMNB1 | = | lamin B1 |
| MAP1LC3A/B | = | microtubule associated protein 1 light chain 3 α/β |
| MTOR | = | mechanistic target of rapamycin (serine/threonine kinase) |
| NoLS | = | nucleolar localization signal |
| PIC | = | preinitiation complex |
| POLRl | = | RNA polymerase I |
| POLR1A/RPA194 | = | polymerase (RNA) I subunit A |
| rDNA | = | ribosomal DNA |
| RRN3/TIFIA | = | RRN3 homolog:RNA polymerase I transcription factor |
| rRNA | = | ribosomal RNA |
| SL1 | = | selectivity factor 1 |
| SUnSET | = | surface sensing of translation |
| TAF1A/TAFI48 | = | TATA-box binding protein associated factor, RNA polymerase I subunit A |
| TP53INP2/DOR | = | tumor protein p53 inducible nuclear protein 2 |
| TUBB | = | tubulin β class I |
| UBTF/UBF | = | upstream binding transcription factor, RNA polymerase I |
| YFP | = | yellow fluorescent protein |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KAUP_A_1175693_Supplementary_material.zip
Download Zip (1.8 MB)Acknowledgments
We are grateful to the Imaging Center of Zhejiang University School of Medicine for their assistance in confocal microscopy. We thank Jinghao Sheng and Chong Wang for their help with ChIP assay and gel filtration.
Funding
This study was supported by the National Natural Science Foundation of China (31530040, 31271431 and 31171288) and the National Basic Research Program of China (2011CB910100 and 2013CB910200).
References
- Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 2003; 17:1691-702; PMID:12865296; http://dx.doi.org/10.1101/gad.1098503R
- Moss T, Stefanovsky VY. At the center of eukaryotic life. Cell 2002; 109:545-8; PMID:12062097; http://dx.doi.org/10.1016/S0092-8674(02)00761-4
- Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci 2005; 30:87-96; PMID:15691654; http://dx.doi.org/10.1016/j.tibs.2004.12.008
- Bell SP, Learned RM, Jantzen HM, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 1988; 241:1192-7; PMID:3413483; http://dx.doi.org/10.1126/science.3413483
- Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JC. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J 2001; 20:1373-82; PMID:11250903; http://dx.doi.org/10.1093/emboj/20.6.1373
- Beckmann H, Chen JL, O'Brien T, Tjian R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science 1995; 270:1506-9; PMID:7491500; http://dx.doi.org/10.1126/science.270.5241.1506
- Tuan JC, Zhai W, Comai L. Recruitment of TATA-binding protein-TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol Cell Biol 1999; 19:2872-9; PMID:10082553; http://dx.doi.org/10.1128/MCB.19.4.2872
- Kusnadi EP, Hannan KM, Hicks RJ, Hannan RD, Pearson RB, Kang J. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene 2015; 556:27-34; PMID:25447905; http://dx.doi.org/10.1016/j.gene.2014.11.010
- Baumgartner BG, Orpinell M, Duran J, Ribas V, Burghardt HE, Bach D, Villar AV, Paz JC, González M, Camps M, et al. Identification of a novel modulator of thyroid hormone receptor-mediated action. PLoS One 2007; 2:e1183; PMID:18030323; http://dx.doi.org/10.1371/journal.pone.0001183
- Sala D, Ivanova S, Plana N, Ribas V, Duran J, Bach D, Turkseven S, Laville M, Vidal H, Karczewska-Kupczewska M, et al. Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes. J Clin Invest 2014; 124:1914-27; PMID:24713655; http://dx.doi.org/10.1172/JCI72327
- Nowak J, Archange C, Tardivel-Lacombe J, Pontarotti P, Pebusque MJ, Vaccaro MI, Velasco G, Dagorn JC, Iovanna JL. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell 2009; 20:870-81; PMID:19056683; http://dx.doi.org/10.1091/mbc.E08-07-0671
- Mauvezin C, Orpinell M, Francis VA, Mansilla F, Duran J, Ribas V, Palacín M, Boya P, Teleman AA, Zorzano A. The nuclear cofactor DOR regulates autophagy in mammalian and Drosophila cells. EMBO Rep 2010; 11:37-44; PMID:20010805; http://dx.doi.org/10.1038/embor.2009.242
- Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell 2015; 57:456-66; PMID:25601754; http://dx.doi.org/10.1016/j.molcel.2014.12.013
- Sinha RA, You SH, Zhou J, Siddique MM, Bay BH, Zhu X, Privalsky ML, Cheng SY, Stevens RD, Summers SA, et al. Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. J Clin Invest 2012; 122:2428-38; PMID:22684107; http://dx.doi.org/10.1172/JCI60580
- Mauvezin C, Sancho A, Ivanova S, Palacin M, Zorzano A. DOR undergoes nucleo-cytoplasmic shuttling, which involves passage through the nucleolus. FEBS Lett 2012; 586:3179-86; PMID:22750142; http://dx.doi.org/10.1016/j.febslet.2012.06.032
- Chen D, Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol 2001; 153:169-76; PMID:11285283; http://dx.doi.org/10.1083/jcb.153.1.169
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science 2002; 298:1623-6; PMID:12446911; http://dx.doi.org/10.1126/science.1076164
- Perry RP, Kelley DE. Persistent synthesis of 5S RNA when production of 28S and 18S ribosomal RNA is inhibited by low doses of actinomycin D. J Cell Physiol 1968; 72:235-46; PMID:5724574; http://dx.doi.org/10.1002/jcp.1040720311
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 2006; 20:1075-80; PMID:16618798; http://dx.doi.org/10.1101/gad.1399706
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 2007; 447:730-4; PMID:17554310; http://dx.doi.org/10.1038/nature05842
- Torrano V, Navascues J, Docquier F, Zhang R, Burke LJ, Chernukhin I, Farrar D, León J, Berciano MT, Renkawitz R, et al. Targeting of CTCF to the nucleolus inhibits nucleolar transcription through a poly(ADP-ribosyl)ation-dependent mechanism. J Cell Sci 2006; 119:1746-59; PMID:16595548; http://dx.doi.org/10.1242/jcs.02890
- Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem 2004; 279:6783-93; PMID:14610093; http://dx.doi.org/10.1074/jbc.M309393200
- Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 2003; 4:641-9; PMID:12923526; http://dx.doi.org/10.1038/nrm1171
- Panov KI, Friedrich JK, Zomerdijk JC. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol Cell Biol 2001; 21:2641-9; PMID:11283244; http://dx.doi.org/10.1128/MCB.21.8.2641-2649.2001
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol 2010; 50:131-56; PMID:20055700; http://dx.doi.org/10.1146/annurev.pharmtox.010909.105844
- O'Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 2002; 22:657-68; PMID:11756560; http://dx.doi.org/10.1128/MCB.22.2.657-668.2002
- Copenhaver GP, Putnam CD, Denton ML, Pikaard CS. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res 1994; 22:2651-7; PMID:8041627; http://dx.doi.org/10.1093/nar/22.13.2651
- Seither P, Iben S, Grummt I. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J Mol Biol 1998; 275:43-53; PMID:9451438; http://dx.doi.org/10.1006/jmbi.1997.1434
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 2009; 6:275-7; PMID:19305406; http://dx.doi.org/10.1038/nmeth.1314
- Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J 2011; 25:1028-39; PMID:21148113; http://dx.doi.org/10.1096/fj.10-168799
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009; 284:8023-32; PMID:19150980; http://dx.doi.org/10.1074/jbc.M900301200
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, et al. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 2003; 23:8862-77; PMID:14612424; http://dx.doi.org/10.1128/MCB.23.23.8862-8877.2003
- Panova TB, Panov KI, Russell J, Zomerdijk JC. Casein kinase 2 associates with initiation-competent RNA polymerase I and has multiple roles in ribosomal DNA transcription. Mol Cell Biol 2006; 26:5957-68; PMID:16880508; http://dx.doi.org/10.1128/MCB.00673-06
- Zhao J, Yuan X, Frodin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell 2003; 11:405-13; PMID:12620228; http://dx.doi.org/10.1016/S1097-2765(03)00036-4
- Halkidou K, Logan IR, Cook S, Neal DE, Robson CN. Putative involvement of the histone acetyltransferase Tip60 in ribosomal gene transcription. Nucleic Acids Res 2004; 32:1654-65; PMID:15016909; http://dx.doi.org/10.1093/nar/gkh296
- Pelletier G, Stefanovsky VY, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum LI, Côté J, Moss T. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell 2000; 6:1059-66; PMID:11106745; http://dx.doi.org/10.1016/S1097-2765(00)00104-0
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol 2002; 12:1-11; PMID:11790298; http://dx.doi.org/10.1016/S0960-9822(01)00650-9
- Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, et al. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy 2014; 10:416-30; PMID:24401568; http://dx.doi.org/10.4161/auto.27286
- Sheng J, Yu W, Gao X, Xu Z, Hu GF. Angiogenin stimulates ribosomal RNA transcription by epigenetic activation of the ribosomal DNA promoter. J Cell Physiol 2014; 229:521-9; PMID:24122807; http://dx.doi.org/10.1002/jcp.24477
