ABSTRACT
Autophagy is a highly conserved intracellular degradation system, and recently was shown to display circadian rhythms in mice. The mechanisms underlying circadian regulation of autophagy, however, are still unclear. Here, we observed that numbers of autophagosomes and autolysosomes exhibit daily rhythms in the zebrafish liver, and cebpb/(c/ebpβ) and various autophagy genes are rhythmically expressed in zebrafish larvae but significantly upregulated in per1b and TALEN-generated nr1d1/rev-erbα mutant fish, indicating that both Per1b and Nr1d1 play critical roles in autophagy rhythms. Luciferase reporter and ChIP assays show that the circadian clock directly regulates autophagy genes through Nr1d1, and also regulates transcription of cebpb through Per1b. We also found that fasting leads to altered expression of both circadian clock genes and autophagy genes in zebrafish adult peripheral organs. Further, transcriptome analysis reveals multiple functions of Nr1d1 in zebrafish. Taken together, these findings provide evidence for how the circadian clock regulates autophagy, imply that nutritional signaling affects both circadian regulation and autophagy activities in peripheral organs, and shed light on how circadian gene mutations act through autophagy to contribute to common metabolic diseases such as obesity.
Introduction
Virtually all organisms from cyanobacteria to mammals possess the circadian clock that allows for them to anticipate environmental changes and coordinate their behavior and physiology.Citation1,2 The mammalian master clock in the suprachiasmatic nucleus (SCN) coordinates and synchronizes peripheral oscillators in most tissues and organs.Citation3 In both SCN neurons and peripheral tissues and organs, the circadian clock exerts its regulatory role through 2 highly conserved negative feedback loops. The bHLH-PAS transcriptional factors CLOCK and ARNTl1/BMAL1, formed as a heterodimer, bind to the cis element E/E’-box (CACGT[G/T]/CANNTG) in the promoter regions of their target genes including PER/PERIOD and CRY/CRYPTOCHROME and activate their expression.Citation4 PER and CRY, formed as the other heterodimer, translocate to the nucleus and inhibit E/E’-box-mediated transcription by interacting with the CLOCK-ARNTl1/BMAL1 heterodimer, fulfilling the negative feedback loop.Citation5 In addition, 2 subfamilies of nuclear hormone receptors, NR1D/Rev-erbs (NR1D1/Rev-erbα and NR1D2/Rev-erbβ) and NR1F/RORs (NR1F1/RORA/RORα, NR1F2/RORB/RORβ and NR1F3/RORC/RORγ), either repress or activate gene transcription through competing to bind to RORE motifs (RGGTCA/RGGTCACTRGGTCA) in the promoter regions of their downstream genes, constituting the second feedback loop of circadian regulation.Citation6,7
Autophagy is an evolutionarily conserved process in eukaryotes by which the cytoplasmic cargo is sequestered inside double-membrane vesicles and delivered to the lysosome for degradation.Citation8 This process not only removes misfolded proteins and damaged organelles but also is an adaptive response for providing nutrients and energy under various stresses.Citation9 The ATG1/ULK1 complex is an essential positive regulator in autophagy initiation.Citation10,11 In mice, the mechanistic target of rapamycin (serine/threonine kinase) complex 1 (MTORC1) phosphorylates the ULK1 protein and inhibits autophagy initiation when nutrients are abundant.Citation12 Genetic disruption of autophagy genes promotes tumorigenesis and accumulation of ubiquitinated protein aggregates.Citation13
Autophagic vacuoles vary daily in rat tissues,Citation14,15 and transcription factor CEBPB has recently been found to link autophagy to the circadian clock and maintain nutrient homeostasis in mice.Citation16 The nuclear receptor NR1D1/Rev-erbα, highly expressed in the liver and adipose tissue, plays regulatory roles in adipogenesis, macrophage inflammatory response, and lipid, bile acid and glucose metabolism.Citation6,17-19 Heme involved in cellular redox balance and mitochondrial respiration is the endogenous ligand for NR1D1.Citation20,21 NR1D1 deficiency results in reduced mitochondrial content and oxidative function as well as altered autophagy activities in skeletal muscle of mice.Citation22 However, the molecular mechanisms underlying circadian regulation of autophagy rhythms are still not elucidated.
The zebrafish (Danio rerio) is an attractive model for studying the circadian clock and autophagy.Citation23-25 Zebrafish as a diurnal species display robust daily changes in physiology and behavior, for instance, zebrafish locomotor activities peak during the daytime but their melatonin production peaks during the nighttime, reminiscent of humans.Citation25 Using various genetic tools including TALEN (transcription activator-like effector nucleases),Citation26-29 a number of zebrafish circadian mutants have been generated for dissecting molecular genetic pathways underlying their time-keeping mechanisms.Citation30-32 Zebrafish's optical clarity of embryos, external development, and the related versatile transgenic, mutational and imaging tools have made them become important for studying vertebrate autophagy.Citation23,33 Whether zebrafish autophagy displays daily rhythms as well as possible underlying regulatory mechanisms, however, have not been investigated to date.
In this study, we found that autophagy displays robust circadian rhythms in the zebrafish liver and larvae, and autophagy genes and autophagy activities are markedly upregulated and lose rhythmicity in both TALEN-generated nr1d1 mutant fish and viral insertional per1b mutant fish,Citation30 suggesting that both Nr1d1 and Per1b are critical for maintaining autophagy rhythms in zebrafish. We revealed that both E-box-mediated and RORE-mediated transcriptional regulatory mechanisms contribute to circadian regulation of autophagy rhythms. Nr1d1 is also essential for the zebrafish circadian clock, as evidenced by altered expression of core circadian clock genes and disrupted locomotor rhythms in nr1d1 mutant fish. In addition, our transcriptome analysis reveals extensive roles of Nr1d1 in numerous life processes. These findings ascertain circadian roles in autophagy, define Nr1d1's essential functions in zebrafish circadian regulation, and provide insights into how circadian misalignment derived from mutations of circadian clock genes such as nr1d1 likely contributes to common metabolic diseases such as obesity.
Results
Autophagy rhythms in zebrafish
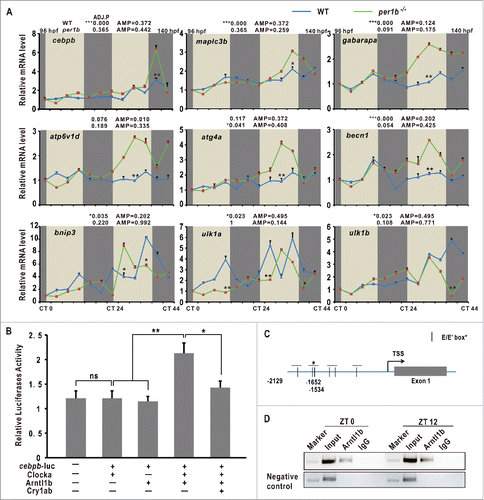
To determine autophagy rhythmicity in zebrafish, we used transmission electron microscopy (TEM) to examine autophagosome and autolysosome numbers in the zebrafish liver at time-of-day-specific points.Citation34 We observed that numbers of autopahgosomes and autolysosomes display robust oscillations, peaking at ZT 0 (Zeitgeber Time), tapering off during the day and rising again during the night, suggesting autophagosome/autolysosome rhythms in zebrafish (). Next, we examined expression patterns of autophagy genes and autophagy-related genes in larvae through quantitative RT-PCR and JTK-CYCLE analyses.Citation2 Results show that atg4a (autophagy related 4A, cysteine peptidase), atp6v1d (ATPase, H+ transporting, lysosomal V1 subunit D), becn1/beclin1 (beclin 1, autophagy related), ulk1a/ulk1b (unc-51 like autophagy activating kinase 1a/b), bnip3 (BCL2/ adenovirus E1B interacting protein 3), gabarapa (GABA(A) receptor-associated protein a), and map1lc3b (microtubule-associated protein 1 light chain 3 β), which are involved in the autophagosome initiation, nucleation, elongation, fusion, induction, substrate targeting, and degradation, are all rhythmically expressed in zebrafish larvae under LD (Light/Dark) condition with statistical significance (9.605 ≤ F11,24 ≤ 521 .855, one-way ANOVA, P ≤ 0 .001) ( and S1A), suggesting that transcription of autophagy and autophagy-related genes is regulated by the circadian clock and development in larvae.Citation35 Like autophagosomes and autolysosomes in the liver, these autophagy or autophagy-related genes peak at ZT 0, trough during the day and rise up in the night (). We also analyzed the expression patterns of these autophagy genes in the adult zebrafish liver. Results show that these genes are downregulated after the morning feeding but upregulated in the night with statistical significance (17.501 ≤ F11,24 ≤ 351 .503, one-way ANOVA, P ≤ 0 .001) ( and S1B). In addition, we observed that autophagy or autophagy-related genes show 2 peaks during daytime compared with nighttime, likely due to scheduled feedings,Citation36 suggesting that the circadian clock and nutritional signals both affect their transcription in the adult zebrafish liver. We also examined expression of several core circadian clock genes including per1b, nr1d1, clockb/clock1b and arntl1b/bmal1b, and found that these 4 circadian clock genes all display rhythmic expression in zebrafish larvae on LD conditions (Fig. S1F), implicating that the circadian clock may play a role in regulating rhythmic expression of these autophagy and autophagy-related genes in zebrafish.
Figure 1. Autophagy rhythms in zebrafish. (A) Transmission electron micrograph of the zebrafish liver sections at ZT 0, ZT 6, ZT 12 and ZT 18. The scale bar in the bottom left corner of ZT 0 panel represents one micron. High magnification in the right low corners highlights a cytosolic region. Note the presence of double-membrane autophagosomes and autolysosomes (red arrowhead). (B) Quantification of autophagosome and autolysosome abundance in (A). Data represent mean ± s.e. **, P ≤ 0.01. The Student t test was conducted. (C) RT-PCR analysis of expression of map1lc3b, bnip3, atp6v1d, becn1, ulk1a and ulk1b in zebrafish larvae from 72 hpf to 96 hpf. Zebrafish were maintained at 14 h light and 10 h dark condition. Approximately 50 zebrafish larvae were pooled for each time point. (D) RT-PCR analysis of expression of map1lc3b, bnip3, atp6v1d, becn1, ulk1a and ulk1b in zebrafish livers. Livers from 5 adult zebrafish were pooled for each time point. The mRNA expression levels were analyzed by the JTK-CYCLE method. ADJ.P for adjusted minimal P values (*, P ≤ 0 .05; ***, P ≤ 0 .001), AMP for amplitude. One-way ANOVA was conducted (***, P ≤ 0 .001). Data represent mean ± s.d. of the 3 independent experiments.
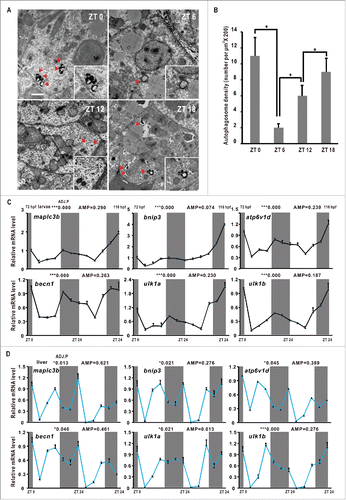
Map1lc3b is a zebrafish ortholog of yeast Atg8 that undergoes conjugation to phosphatidylethanolamine upon autophagy induction.Citation24 The unconjugated LC3 (LC3-I) is localized in the cytosol, whereas the lipid-conjugated form (LC3-II) resides on the phagophore and autophagosome membrane.Citation23,37 SQSTM1/p62 (sequestosome 1) is a protein that interacts with LC3 and delivers autophagy cargos for liposomal degradation.Citation38 Western blotting assays show that Map1lc3b-II displays obvious oscillations in zebrafish larvae under the LD condition, suggesting that autophagy fluxes are regulated by the circadian clock (Fig. S1C, S1D and S1E). However, Sqstm1 does not show daily changes (Fig. S1C, S1D and S1E), likely affected by developmental regulation in zebrafish.Citation39 Together, these results suggest that the autophagy activities, as evidenced by the numbers of autophagosomes and autolysosomes, the expression patterns of autophagy or autophagy-related genes and the conversion ratio of Map1lc3b-I to Map1lc3b-II, display robust daily rhythms in the zebrafish larvae and liver.
Generation of zebrafish nr1d1 mutants
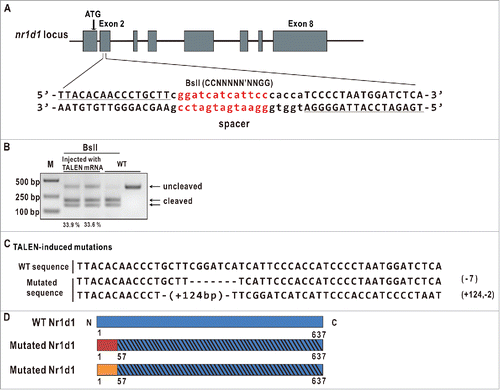
We used an on-line TALEN design web site (https://tale-nt.cac.cornell.edu/) to design a pair of TALEN arms targeting the second exon of zebrafish nr1d1, and the targeted fragment harbors a BslI enzyme site for evaluating mutagenesis efficiency and subsequent mutant identification ().Citation40 We employed the “unit assembly” method to construct the 2 nr1d1 TALEN arms.Citation41 Capped mRNAs of these 2 TALEN arms were comicroinjected into one-cell stage zebrafish embryos at a concentration of 250 pg each. To evaluate the mutagenesis efficiency, a 430-bp (base pair) fragment containing the TALEN-targeted site was PCR amplified with DNAs extracted from injected and uninjected control embryos (10 each). Enzymatic digestion of the PCR-amplified fragment with BslI estimated approximately 33% of mutagenesis frequency (). Siblings of TALEN-injected F0 embryos were raised to adulthood, and then outcrossed with wild type to produce F1s. Enzymatic and DNA sequencing analyses of the targeted fragment PCR-amplified from F1 fin-clipped DNAs identified 2 types of heritable mutations: one has a 7-bp deletion and the other a 122 bp (+124, −2) insertion; both induce frame shift mutations resulting in a truncated peptide of 57 AA (). The homozygous mutant fish with the 7-bp deletion were used for most of the subsequent experiments.
Figure 2. Generation of zebrafish nr1d1 mutants. (A) Diagram of the TALEN target fragment in zebrafish nr1d1. The left and right TALEN arms are underlined; the TALEN spacer containing a BslI restriction enzyme site (in red) is in small case. (B) Examine TALEN efficiencies by gel analysis. The nr1d1 target fragment was amplified by PCR from genomic DNAs of approximately 10 embryos microinjected with capped mRNAs of nr1d1 TALEN left and right arms at a concentration of 300 pg each, and digested with BslI. Mutagenesis efficiencies were estimated by the ratios of the uncleaved bands and the sum of the cleaved bands and the uncleaved bands quantified with software ImageJ. WT, wild type; M, marker. (C) Two mutated fish lines were identified with DNA sequencing. One has a 7-bp deletion, the other has a 124-bp insertion and a 2-bp deletion, and both lead to frame shift mutations resulting in truncated proteins. (D) The predicted truncated Nr1d1 proteins induced by TALEN. The 2 truncated Nr1d1 proteins each have only 57 amino acids.
Altered expression of key circadian clock genes and disrupted locomotor activities in nr1d1 mutant zebrafish
Locomotor activities of zebrafish larvae exhibit robust circadian rhythmicity and peak during the subjective day.Citation42 We performed locomotor experiments for the nr1d1 mutant and wild-type larvae from 5 d postfertilization (dpf) to 7 dpf. Under LD conditions, nr1d1 larvae display significantly increased locomotor activities compared with wild type (). Indeed, the amplitude of nr1d1 mutant larvae increases more than 40% during the whole duration of a 24-h day and night (). Under the DD (dark/dark) condition, nr1d1 larvae also display increased amplitude, though much dampened, approximately half-an-hour phase delay and 0.25-h shortened period compared with wild types (). Altered behaviors of nr1d1 mutant fish are consistent with nr1d1 knockout mice that display mania-like behavior.Citation43
Figure 3. Disrupted rhythms of locomotor activities and altered expression of key circadian clock genes in homozygous nr1d1 mutant zebrafish. (A, C) The locomotor assays of nr1d1 mutant and wild-type larvae from 4 dpf to 7 dpf under LD (A) and DD (C) conditions. nr1d1 mutants, n = 48; wild types, n = 48. (B, D) Heat maps represent the total swimming or resting area of a wild-type or nr1d1 mutant larva in each well under LD (B) and DD (D). The spatial bin size is 10X10 mm (see scale on the right). The red colors represent more resting activities of the larvae, and the blue color more locomotor activities of the larvae. nr1d1 mutant larvae (bottom) swim more of the perimeter in the well compared with wild types (top). (E) The average swimming distances of single wild-type and nr1d1 larva during one day. The upper dot box indicates the swimming distance of single larva in the day, and the bottom box the swimming distance of single larva in the night. Data represent mean ± s.d. (***, P ≤ 0 .001). (F) Histogram of the average period of wild-type and nr1d1 larvae. Data represent mean±s.d. (*, P ≤ 0 .05). (G) Histogram of the average swimming distances of wild-type and nr1d1 larva during the 3 d. Data represent mean ± s.d. (*, P ≤ 0 .05). (H) Histogram of the average phase during the 3 d. Data represent mean±s.d. (**, P ≤ 0 .01) (I) Altered expression of key circadian clock genes arntlb/bmal1b, arntl2/bmal2, cry1aa, cry1ab, per1b, per2, and per3 in nr1d1 mutant fish under DD condition shown by qRT-PCR analyses. The mRNA expression levels were analyzed by the JTK-CYCLE method. ADJ.P for adjusted minimal P-values (***, P ≤ 0 .001), AMP for amplitude. Two-way ANOVA with the Tukey post hoc test was conducted (***, P ≤ 0 .001). Approximately 50 zebrafish larvae were pooled for each time point. Data represent mean ± s.d. of the 3 independent experiments.
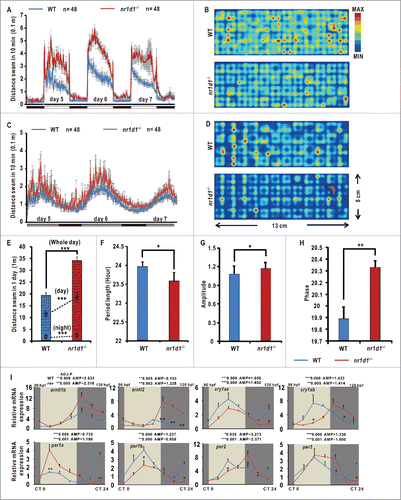
We also examined expression patterns of core circadian clock genes in nr1d1 mutant larvae under DD condition. Both arntl1b/bmal1b and arntl2/bmal2 are significantly upregulated in nr1d1 mutant larvae (1845.768 ≤ F6,28 ≤ 3435 .709, 2-way ANOVA, P ≤ 0 .001), consistent with our previous study exhibiting that Nr1d1 represses arntl1b/bmal1b transcription through RORE ().Citation32 Moreover, per1a is also significantly upregulated (F6,28= 2006.444, 2-way ANOVA, P ≤ 0 .001), cry1aa, and per1b are significantly downregulated (106.233 ≤ F6,28 ≤ 206 .737, 2-way ANOVA, P ≤ 0 .001), and cry1ab, per2, and per3 are phase-delayed (146.821 ≤ F6,28 ≤ 1619 .126, 2-way ANOVA, P ≤ 0 .001), in nr1d1 mutant larvae (), even though they still maintain rhythmicity (). Together, these results suggest that Nr1d1 plays an essential role in regulating circadian clock genes and behavior in zebrafish.
Loss of rhythmic expression of cebpb, autophagy genes and autophagy-related genes in nr1d1 and per1b mutant fish
We used an mCherry-map1lc3b vector to monitor the acidic autolysosomes in nr1d1 mutants.Citation44 Results show that the numbers of autolysosomes are higher in nr1d1 mutants than in the wild-type control (). Transmission electron microscopy analysis of the adult zebrafish livers shows the increased numbers of autophagosomes and autolysosomes in nr1d1 mutants at ZT 0 (). Thus, it appears that Nr1d1 plays an essential role in autophagy activities in zebrafish.
Figure 4. Disrupted autophagy activities and altered expression of autophagy genes in nr1d1 mutant zebrafish. (A) Zebrafish larval autolysosomes were estimated by expression of mCherry-Map1lc3b at 56 hpf. The scale bar in the lower left of the wild-type panel represents 100 microns. The right upper panels show high magnification highlighting a hindbrain region in both wild-type and nr1d1 mutant larva labeled with mCherry-Map1lc3b. (B) Transmission electron micrograph of wild-type and nr1d1 mutant liver sections at ZT 0. The scale bar in the lower left of the wild-type panel represents one micron. (C) Quantification of autolysosomes in (A), calculated manually. (D) Quantification of autophagosome abundance in (B) with software ImageJ. The figure represents one of the 3 independent sets of samples. Data represent mean ± s.d.(**, P≤0 .01). The Student t test was conducted, (**, P ≤ 0 .01). (E) RT-PCR analysis of autophagy genes in nr1d1 (−7bp or +124,-2bp) mutant zebrafish at different time points under DD condition. Zebrafish larvae were raised under light/dark (14 h/10 h) condition for the first 3 dpf, and then placed under constant darkness condition. Approximately 50 zebrafish larvae were pooled for each time point. The mRNA expression levels were analyzed by the JTK-CYCLE method. ADJ.P for adjusted minimal p-values (*, P ≤ 0 .05; ***, P ≤ 0 .001), AMP for amplitude. Two-way ANOVA with the Tukey post hoc test was conducted (*, P ≤ 0 .05; **, P ≤ 0 .01).
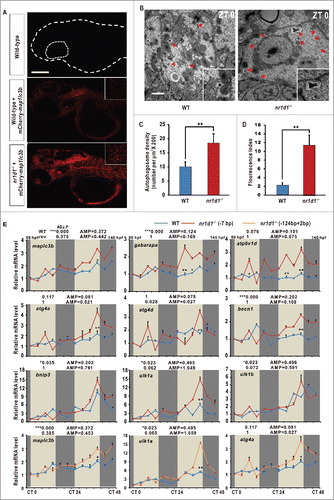
Rhythmic expression of autophagy genes suggests that transcriptional regulation plays an important role in driving cyclic autophagy. The molecular components of autophagy have been extensively investigated,Citation37,45 and previous studies show that clock-controlled transcription factors CEBPB and FOXO3 play important roles in regulating transcription of autophagy genes in mice.Citation16,46 However, how the circadian clock regulates cebpb and whether the circadian clock directly regulates autophagy genes are still not clear. To underscore circadian regulation, we examined expression of autophagy and autophagy-related genes in nr1d1 mutant and wild-type larvae under DD condition.Citation35,47,48 Here, qRT-PCR analyses show that map1lc3b and gabarapa are significantly upregulated in nr1d1 mutants (0.964 ≤ F11,48 ≤ 58 .517, 2-way ANOVA, P ≤ 0 .001) (). Meanwhile, significantly upregulated atp6v1d mRNA level (F11,48= 138.475, 2-way ANOVA, P ≤ 0 .001) suggests that more autophagosomes fuse with lysosomes, forming autolysosomes in nr1d1 mutants (). Likewise, other autophagy genes or autophagy-related genes including atg4a, atg4d, becn1, and bnip3 are also significantly upregulated (38.545 ≤ F11,48 ≤ 162 .689, 2-way ANOVA, P ≤ 0 .001); in particular, JTK-CYCLE analysis showed all these genes lose rhythmicity in nr1d1 mutants compared with the wild-type zebrafish (). As a second internal nonrhythmic gene, tuba1b (tubulin, α1b) expression is not rhythmic in wild-type and nr1d1 mutant larvae (ADJ.P (P > 0 .05), WT = 1.356, nr1d1 = 1.384), and has no significant changes between wild-type and nr1d1 mutant larvae (P = 0.972) (Fig. S3D). Moreover, similar to what observed in the nr1d1 (−7bp) mutant line, map1lc3b, ulk1a and atg4a are also significantly upregulated in the other nr1d1 (+124,-2 bp) mutant line (35.367 ≤ F11,48 ≤ 141 .612, 2-way ANOVA, P ≤ 0 .001) (), further corroborating that altered expression of these autophagy genes and autophagy-related genes is resulted from loss of Nr1d1 function.
qRT-PCR and JTK-CYCLE analyses also show that cebpb is rhythmically expressed in zebrafish with statistical significance (F11,48 = 153.434, 2-way ANOVA, P ≤ 0 .001) (), and its expression of autophagy genes and autophagy-related genes map1lc3b, gabarapa, atp6v1d, atg4a, atg4d, becn1, bnip3, ulk1a and ulk1b are all significantly upregulated without rhythmicity in per1b mutant zebrafish (36.824 ≤ F11,48 ≤ 570 .718, 2-way ANOVA, P ≤ 0 .001) (),Citation30 suggesting that Per1b also plays a regulatory role in expression of cebpb, autophagy genes and autophagy-related genes.
Figure 5. Upregulation of autophagy genes in per1b mutants and cebpb is controlled by the circadian clock in zberafish. (A) Upregulation of autophagy genes in zebrafish per1b mutant larvae under DD, shown by qRT-PCR analysis. The mRNA expression levels were analyzed by the JTK-CYCLE method. ADJ.P for adjusted minimal p-values (*, P ≤ 0 .05; ***, P ≤ 0 .001), AMP for amplitude. Two-way ANOVA with the Tukey post hoc test was conducted (*, P ≤ 0 .05; **, P ≤ 0 .01). (B) Luciferase reporter assays. Activities of the cebpb promoter are enhanced by cotransfection with Clocka/Clock1a and arntl1b but repressed by Cry1ab. The cebpb-pGL4, Clocka/Clock1a-pcDNA3.1, Arntl1b/Bmal1b-pcDNA3.1 and Cry1ab-pcDNA3.1 plasmids were transfected at the concentration of 100 ng/ul each. Data represent mean ± s.d. (**, P ≤ 0 .01). The Student t test was applied. (C) Diagram of the cebpb promoter containing multiple E-boxes and E’-boxes. (D) ChIP assays. Arntl1b/Bmal1b binds to the E-boxes in the cebpb promoter.
Regulation of cebpb, autophagy genes and autophagy-related genes by the circadian clock
We observed that the zebrafish cebpb promoter region harbors several E-boxes and E’-boxes (). Luciferase reporter assays show that activities of the zebrafish cebpb promoter are enhanced by Clocka/Clock1a and Arntl1b/Bmal1b but repressed by Cry1ab (). ChIP assays show that Arntl1b/Bmal1b binds to the second E’-box in the zebrafish cebpb promoter region (). Together, these results indicate that cebpb is regulated directly by the circadian clock via E’-box in zebrafish. Thus, it is likely that the circadian clock regulates autophagy rhythms via direct control of transcription factor cebpb that in turn regulates autophagy genes and autophagy-related genes.Citation16
We also observed motifs including E-box, RORE, RevDR2, CCAAT (cebpb binding) motif in approximately 2, 500-bp promoter regions of these autophagy and autophagy-related genes in zebrafish,Citation7,49,50 suggesting transcription of these autophagy genes and autophagy-related genes may be regulated directly by the circadian clock components (Fig. S2).
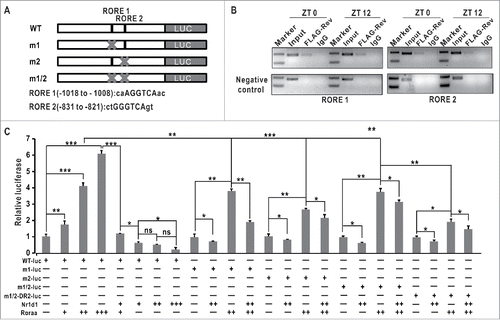
Altered patterns of ulk1a and ulk1b in nr1d1 or per1b mutants led us to hypothesize that ulk1 transcription might be regulated directly by the circadian clock. As shown by luciferase assays, while Clocka/Clock1a and Arntl1b/Bmal1b have no effects on ulk1a promoter activities (Fig. S6F), Nr1d1 or Roraa can repress or activate ulk1a in a dosage-dependent manner (). We observed 2 canonical RORE motifs in the ulk1a promoter (). Luciferase assays show that deleting either RORE1 or RORE2 or both significantly reduces the ulk1a promoter activities, indicating that ulk1a is regulated directly by Nr1d1 through RORE in vitro (). To further investigate the possible role of Rev-DR2, we generated a construct wherein the Rev-DR2 motif and the 2 RORE motifs were all deleted. Results show that ulk1a promoter activities are significantly reduced but still not completely abolished compared with both RORE site deleted ulk1a promoter (), suggesting that an unknown motif or motifs play certain regulatory roles in ulk1a transcription. ChIP assays show that Nr1d1 binds directly to the RORE sites in the ulk1a promoter in vivo (), indicating that Nr1d1 directly regulates ulk1a in zebrafish. In addition, we also found that another autophagy-related gene atp6v1d is a direct target of Nr1d1, as evidenced by luciferase reporter and ChIP assays (Fig. S3B and S3C). Together, these results demonstrate that Nr1d1 directly regulates autophagy genes and autophagy-related genes in zebrafish.
Figure 6. ulk1a is regulated directly by Nr1d1 (A) Schematic of the wild-type and site-directed mutagenized 2,000-bp zebrafish ulk1a promoter. (B) ChIP assays. Nr1d1 binds to the RORE in the ulk1a promoter. (C) ulk1a is regulated directly by Nr1d1 through RORE, as shown by luciferase reporter assays. The wild-type ulk1a promoter (WT-luc) and the mutagenized ulk1a promoter with site-specific deletions of RORE1 (m1-luc), RORE2 (m2-luc), both (m1/2-luc) and (m1/2) plus RevDR2 (m1/2-DR2-luc) were cotransfected with Nr1d1 and/or Roraa, respectively. Luciferase activities were normalized by cotransfected pcDNA3.1; mean values of wild-type promoter activities in empty vector-transfected cells were set to 1. For the dose-dependent assays, the concentration of nr1d1 -pcDNA3.1 and roraa-pcDNA3.1 were 100 ng/μl, 200 ng/ul, 300 ng/ul, respectively. Data represent mean ± s.d. of the 3 independent experiments (**, P ≤ 0 .01). The Student t test was applied.
Pharmacological activation of Nr1d1 functions in zebrafish
A previous study showed that GSK4112, a Nr1d1 agonist, can increase the repressive efficiency of Nr1d1 through competing with the ligand heme.Citation51 We treated zebrafish larvae with 20 μM GSK4112 and observed that arntl1a/bmal1a and arntl1b/bmal1b are both downregulated in GSK4112-treated fish, indicating that GSK4112 can enhance the repressive efficiency of Nr1d1 in zebrafish larvae (Fig. S6A). Moreover, to evaluate effects of pharmacological treatment on expression of autophagy or autophagy-related genes, we performed qRT-PCR experiment and found that autophagy genes ulk1a, ulk1b, and atg4d, and autophagy-related genes bnip3 and atp6v1d are all downregulated in GSK4112-treated fish (Fig. S6B). In addition, injecting capped nr1d1 mRNAs could rescue expression of these autophagy genes in nr1d1 mutants (Fig. S6B). We also examined protein levels of autophagy markers, Map1lc3b and Sqstm1, in nr1d1 mutant and pharmacologically treated wild-type zebrafish larvae at different time points. Results show that Map1lc3b and Sqstm1 are significantly upregulated in nr1d1 mutants but no significant changes of Map1lc3b and Sqstm1 in GSK4112-treated wild-type fish (Fig. S6C, 6D and 6E), even though Map1lc3b and Sqstm1 are antiphasic in zebrafish larvae (Fig. S1C, D and E). Thus, GSK4112 treatment can affect transcription but not translation of autophagy and autophagy genes in zebrafish. These results further support that rhythmic expression of autophagy genes are under control of Nr1d1 in zebrafish.
Effects of fasting and loss of Nr1d1 on expression of circadian clock genes, and autophagy and autophagy-related genes in peripheral organs
Previous studies showed that nr1d1 is expressed extensively in zebrafish tissues/organs and nutritional signals can affect zebrafish circadian behavior.Citation35,52,53 However, whether nutritional signaling or loss of Nr1d1 affects the circadian clock or autophagy in peripheral organs has not yet been investigated in zebrafish. We first examined expression patterns of core circadian genes in the liver of fed nr1d1 mutants, fasted nr1d1 mutants, fed wild-type and fasted wild-type fish. Results show that arntl1b/bmal1b, clocka/clock1a, cry1ab and per1b are all significantly upregulated in the nr1d1 mutant group in comparison with the wild-type control (57.125 ≤ F11,48 ≤4270 .745, 2-way ANOVA, P ≤ 0 .001); and expression of both clocka/clock1a is significantly higher in fasted wild types than the fed wild-type control (F11,48 = 224.366, 2-way ANOVA, P ≤ 0 .001), and even higher in fasted nr1d1 mutant group than the fed nr1d1 mutant group (F11,48 = 157.281, 2-way ANOVA, P ≤ 0 .001) (); suggesting that both nutritional signals and nr1d1 contribute to the zebrafish liver clock.
Figure 7. Effects of fasting and loss of Nr1d1 on expression of autophagy genes and circadian clock genes. (A) qRT-PCR analysis of core circadian clock genes in the liver of nr1d1 mutant (fed), fasted nr1d1 mutant, wild-type (fed) and fasted wild-type fish. (B) qRT-PCR analysis of autophagy genes map1lc3b, atp6v1d, ulk1a and atg4a in the adult peripheral organs of nr1d1 mutant, fasted nr1d1 mutant, fasted wild-type and wild-type fish under LD. Following 2-wk fasting, 4-mo-old wild-type and nr1d1 mutant fish, and their controls were sacrificed, and the brains, the livers, the hearts, the eyes, and the intestines were dissected out and collected for RNA extraction at 12 time points with 4-h interval under LD for a total of consecutive 2 d. Each sample contains organs from at least 5 male fishes. Three independent sets of samples were used. The mRNA expression pattern was analysis by JTK-CYCLE method. ADJ.P for adjusted minimal P values (*, P ≤ 0 .05; **, P ≤ 0 .01; ***, P ≤ 0 .001), AMP for amplitude. Two-way ANOVA with the Tukey post hoc test was conducted (*, P≤0 .05; **, P ≤ 0 .01; ***, P ≤ 0 .001). (C) Heatmap comparing ADJ.P values of autophagy and autophagy-related genes in different zebrafish organs of wild-type (fed), wild-type-fasted, nr1d1 (fed) and nr1d1 -fasted groups. (D) Heatmap showing correlation coefficiencies (R) of P values from 2-way ANOVA of autophagy and autophagy-related genes in different zebrafish organs in terms of wild-type (fed) vs wild-type-fasted, wild-type (fed) vs nr1d1 (fed), wild-type-fasted vs nr1d1-fasted, and nr1d1 (fed) vs nr1d1-fasted. WT-f, WT-fasted.
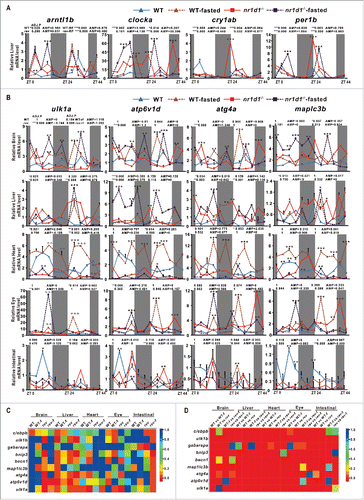
We then examined several key autophagy and autophagy-related genes in the brain, the liver, the heart, the eye and the intestine of fed nr1d1 mutant, fasted nr1d1 mutant, fed wild-type and fasted wild-type fish ( and Fig. S3A). Consistent with what observed in the wild-type liver, these autophagy or autophagy-related genes still display 2 peaks in one day's cycling in the zebrafish heart, eye, brain and intestinal organs, suggesting synergistic regulation of their transcription by the nutrition status and the circadian clock. Autophagy and autophagy-related genes ulk1a, atp6v1d, atg4a, map1lc3b, bnip3 and ulk1b were significantly upregulated in the brain of the fasted wild-type group in comparison with those of fed wild types (23.193 ≤ F11,48 ≤ 99 .003, 2-way ANOVA, P ≤ 0 .001), and also in those of the fasted nr1d1 mutant group in comparison with those of the fed nr1d1 mutant group (30.418 ≤ F11,48 ≤ 41025 .151, 2-way ANOVA, P ≤ 0 .001) ( and Fig. S3A), indicating fasting can upregulate these genes in the brain. Meanwhile, cebpb has only a modest increase in the brain of the fasted wild-type group compared with fed wild types (F11,48 = 15.588, 2-way ANOVA, P ≤ 0 .001) ( and Fig. S3A). In the liver, ulk1a, atp6v1d, atg4a, becn1, bnip3 and ulk1b still maintain rhythmicity in fed nr1d1 mutant and fasted nr1d1 mutant groups (659.009 ≤ F11,48 ≤ 1468 .932, 2-way ANOVA, P ≤ 0 .001), indicating that robustness of autophagy rhythms in the zebrafish liver ( and Fig. S3A). Interestingly, we found that in the heart, loss of Nr1d1 results in a relatively low levels of expression changes of most autophagy and autophagy-related genes (23.348≤F11,48 ≤ 929 .824, 2-way ANOVA, P ≤ 0 .001), but fasting wild-type fish leads to significant upregulation of the majority of these genes (22.885 ≤ F11,48 ≤ 560 .429, 2-way ANOVA, P ≤ 0 .001) ( and Fig. S3A), suggesting that autophagy activities in the heart are contingent more upon nutritional status. However, fasting nr1d1 mutant fish leads to significant downregulation of most autophagy and autophagy-related genes (15.386 ≤ F11,48 ≤ 1080 .438, 2-way ANOVA, P ≤ 0 .001) ( and S3A). In the zebrafish eye, fasted wild-type (117.605 ≤ F11,48 ≤ 1179 .692, 2-way ANOVA, P ≤ 0 .001) and fed nr1d1 mutant fish (95.442 ≤ F11,48 ≤ 318 .904, 2-way ANOVA, P ≤ 0 .001) and loss of Nr1d1 (25.283 ≤ F11,48 ≤ 230 .818, 2-way ANOVA, P ≤ 0 .001) all significantly upregulate cebpb (F11,48 = 29.459, 17.588, 18.038, 2-way ANOVA, P ≤ 0 .001) and most autophagy and autophagy-related genes ( and Fig. S3A), implicating that nutritional signal, Cebpb and Nr1d1 may act together to regulate autophagy and autophagy-related genes in the zebrafish eye. For the intestine, we observed significant downregulation of autophagy and autophagy-related genes in fasted wild-type (30.852 ≤ F11,48 ≤ 230 .350, 2-way ANOVA, P ≤ 0 .001), fed nr1d1 mutant (48.069 ≤ F11,48 ≤ 308 .333, 2-way ANOVA, P ≤ 0 .001) and fasted nr1d1 mutant groups (36.827 ≤ F11,48 ≤ 310 .199, 2-way ANOVA, P ≤ 0 .001) compared with the fed wild-type group; surprisingly, autophagy and autophagy-related genes still maintain rhythmicity in the fasted nr1d1 mutant group ( and Fig. S3A). As shown in a heatmap comparing ADJ.P (JTK-CYCLE) values of these autophagy and autophagy-related genes in wild-type-fed, wild-type-fasted, nr1d1-fed and nr1d1-fasted groups (), ulk1a is rhythmically expressed in most cases but gabarap and map1lc3b show rhythmic expression only in a few cases. We also compared correlation coeffiencies (R) of P values derived from 2-way ANOVA of these autophagy and autophagy-related genes in different organs in terms of wild-type-fed vs wild-type-fasted, wild-type-fed vs nr1d1-fed, wild-type-fasted vs nr1d1-fasted, and nr1d1-fed vs nr1d1- fasted, and found that more than 92% of them show statistical differences (). These data indicated that autophagy and autophagy-related genes, and circadian clock genes display distinct rhythmic expression patterns in different organs, which are all significantly disrupted in fasted wild-type, fasted nr1d1 mutant and fed nr1d1 mutant fishes ( and Fig. S3A).
Nr1d1 also contributes to stress response, metabolism, signal transduction and post-translational modification
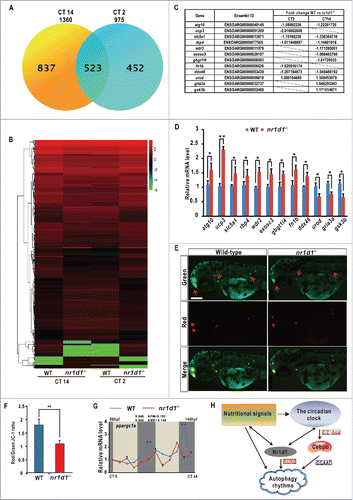
To examine possible effects of Nr1d1 on other life processes, we conducted transcriptome sequencing (RNA-seq) analysis of nr1d1 mutant and wild-type fish, and uncovered 975 differentially expressed genes (DEGs) at CT2 and 1, 360 DEGs at CT14 (2 ≤ P ≤ 5) (). Clustering analysis shows more upregulated genes than downregulated genes in nr1d1 mutant fish at the indicated time points (), likely because Nr1d1 acts as a transcription repressor in zebrafish.Citation32 Through gene Ontology (GO) classification, we found that DEGs are enriched for the binding, catalytic activity in molecular functions, and metabolic process, biological regulation, developmental process, immune system and rhythmic process in biological processes (Figs. S4A and S5A). For the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, we found that the MAPK signaling pathway shows the highest numbers of DEGs (13 at CT2 and 29 at CT12). While at CT2, we found a large number of DEGs involved in metabolism pathways like purine, pyrimidine, steroid and cytochrome P450; at CT12, DEGs trend to distribute in ribosome processing, the calcium signaling pathway, the insulin signaling pathway and oxidative phosphorylation (Figs. S4B and S5B). Further, we reconfirmed transcriptome analysis results by qRT-PCR and found upregulation of atg10 (autophagy), ucp3 (brown adipose tissue metabolism), rbp4 (adipose tissue metabolism), slc5a1 and wdr3 (signal transduction), exosc3 (RNA processing), gbgt1l4 (immune response), gria3a (neurotransmission) and ddx46 (posttranslational modification); and downregulation of urod (heme biosynthesis), fn1b (cell function) and gsk3b (energy metabolism) in nr1d1 mutants (). In addition, we performed the JC-1 staining experiment,Citation54 and found that the ratio of Red/Green is significantly decreased in nr1d1 mutants, implying the low mitochondrial content in nr1d1 mutants (). We also observed that ppargc1a is significantly downregulated in nr1d1 mutants (F11,48 = 244.142, 2-way ANOVA, P ≤ 0 .001) (). Together, our transcriptome and qRT-PCR analyses suggest that Nr1d1 also contributes to numerous life processes including stress response, metabolism, signal transduction and post-translational modification in zebrafish.
Figure 8. Transcriptome analysis and a model for circadian regulation of autophagy. (A) Numbers of differentially expressed genes (DEGs) in nr1d1 mutant fish at CT2 and CT12, revealed by transcriptome analysis. (B) Histogram of 523 DEGs in nr1d1 mutant fish shared between CT12 and CT2. Red colors represent upregulation, green colors downregulation. (C) List of genes with significant fold-changes, revealed by transcriptome analysis. The fold-changes were calculated by values of WT vs. nr1d1/rev-erbα. Genes with positive values were downregulated in the nr1d1 mutant, and those with negative values upregulated. (D) Upregulated genes atg10, ucp3, slc5a1, rbp4, wdr3, exosc3, gbgt1l4, fn1b and ddx46 and downregulated genes urod, gria3a and gsk3b, revealed by transcriptome analysis, are reconfirmed by independent qRT-PCR analysis. Three independent experiments were performed. Data represent mean ± s.d. (E) JC-1 staining assays. Mitochondria with low membrane potential are in green and mitochondria with high membrane potential in red. (F) Relative Red/Green ratio was quantified by ImageJ. Data represent mean±s.d. of the 3 independent experiments. **, P ≤ 0 .01. The Student t test was applied. (G) qRT-PCR analysis of ppargc1a in nr1d1 mutants under DD condition. The mRNA expression pattern was analysis by JTK-CYCLE method. ADJ.P for adjusted minimal P-values (*, P ≤ 0 .05), AMP for amplitude. Two-way ANOVA with the Tukey post hoc test was conducted (***, P ≤ 0 .001). (H) A model for circadian regulation of autophagy in zebrafish. Transcription factor cebpb is regulated by the Clock-Arntl/Bmal heterodimer and induces autophagy rhythms in zebrafish and mice, while Nr1d1 directly controls autophagy genes through RORE. Further, nutritional signals can affect both autophagy and circadian rhythms in zebrafish.
Discussion
In this study, we demonstrate that zebrafish autophagy activities including autophagosome and autolysosome numbers, expression of autophagy or autophagy-related genes and Map1lc3b protein all display robust daily rhythms in the larvae and liver ( and S1). We employed TALEN, a versatile genome-editing tool, to generate mutant lines for nuclear hormone receptor gene nr1d1 (). We found that autophagy activities including autophagosome and autolysosome numbers, expression of autophagy or autophagy-related genes and Map1lc3b protein are all upregulated in nr1d1 mutant zebrafish ( and S6C), suggesting that Nr1d1 plays a role in regulating zebrafish autophagy rhythms. Consistent with a previous mouse study,Citation16 we also show that transcription factor cebpb is rhythmically expressed in zebrafish but its expression and expression of autophagy or autophagy-related genes are all upregulated in a circadian mutant per1b,Citation30 suggesting that Perb1 also acts to regulate zebrafish autophagy rhythms.
The circadian clock modulates autophagy rhythms in zebrafish
We found that the circadian clock exerts its regulatory role in autophagy rhythms in 2 ways: one is that the Clock-Arntll/Bmal heterodimer binds to the E-Boxes to regulate transcription of cebpb that in turn controls transcription of autophagy and autophagy-related genes, the other is that Nr1d1 and Roraa binds to the RORE motifs to directly regulate transcription of autophagy and autophagy-related genes ().
To support this notion, we found several Cebpb-binding motifs in promoter regions of a number of autophagy or autophagy-related genes (Fig. S2), and luciferase reporter and ChIP assays indicate that cebpb is a circadian clock-controlled gene in zebrafish (), providing clear evidence for how the circadian clock regulates cebpb.Citation16 We also observed RORE motifs in promoter regions of a number of autophagy and autophagy-related genes (Fig. S2). In particular, luciferase reporter assays show that ulk1a is activated by Roraa but repressed by Nr1d1 via competing to bind to 2 RORE motifs in the ulk1a promoter in vitro (); ChIP assays show that Nr1d1 binds to 2 RORE motifs in the ulk1a and atp6v1d promoters in vivo ( Fig. S3B and S3C); thus indicating a direct role of Nr1d1 in regulation autophagy rhythms. Further evidence shows that Nr1d1 acts to regulate rhythmic expression of autophagy and autophagy-related genes in zebrafish peripheral organs such as the liver, the brain and the heart in a tissue and organ-specific manner (). We found that most autophagy and autophagy-related genes are upregulated and maintain rhythmicity in the brain of the nr1d1 mutant ( and Fig. S3A). However, cebpb is barely altered, implicating that upregulation of autophagy activities might be resulted from loss of Nr1d1 rather than Cebpb ( and Fig. S3A). For the heart, fasting, rather than loss of Nr1d1, significantly upregulates expression of autophagy and autophagy genes ( and Fig. S3A). Unsurprisingly, nutritional signaling affects both the zebrafish circadian clock and autophagy rhythms in the liver and cebpb is also upregulated in the nr1d1 mutant ( and Fig. S3A), indicating that the zebrafish liver and the mouse liver display similar autophagy regulatory mechanisms via the circadian clock and Cebpb.Citation16,46 Intriguingly, most of these autophagy and autophagy-related genes maintain strong rhythmicity in the liver regardless of nutritional stress and circadian dysregulation (), implicating that autophagy rhythms are robust in the zebrafish liver. In contrast, most of these autophagy and autophagy-related genes lose their rhythmicity in the eye and heart in nr1d1-fed and nr1d1-fasted groups (), suggesting that both the circadian clock and normal nutritional status are required for cycling autophagy activities. Even though how the E-Box-mediated and RORE-mediated transcription processes may coordinate to regulate autophagy rhythms needs to be investigated in the future, these 2 circadian regulatory mechanisms unambiguously play important roles in generation and maintenance of autophagy rhythms in zebrafish ().
Nr1d1 is essential for the zebrafish circadian clock
We found that under LD condition, nr1d1 mutant fish display hyperactivity, similar to the phenotypes of zebrafish per1b null mutant but completely different from the zebrafish per2 mutant ().Citation30,32 Under DD conditions, nr1d1 mutant fish show half-an-hour phase delay and 0.25-h shortened period, the per1b mutant fish show 2-h phase advance and half-hour shortened period,Citation30 and per2 mutant fish show 2-h phase delay and 1.5-h lengthened period.Citation32 However, nr1d1 knockout mice also display a 0.5-h shortened period under DD condition and mania-like behaviors.Citation43 Thus, Nr1d1 plays a conserved but distinct role in maintaining zebrafish locomotor rhythms.
We also found that arntll/bmal genes are significantly upregulated in nr1d1 mutant fish, suggesting a key regulatory role of Nr1d1 in arntl/bmal transcription, consistent with previous results ().Citation32,55 Both cry1aa and cry1ab are downregulated in nr1d1 mutant zebrafish larvae (), which is at odds with a previous report of upregulation of Cry1 and Cry2 in the nr1d1 knockout mouse liver,Citation55 suggesting different roles of Nr1d1 in regulating Cry genes in zebrafish and mice. Further, we found that in the zebrafish liver, fasting upregulates arntl1b/bmal1b and clocka/clock1a, while per1b, cry1ab and nr1d1 are phase-shifted (). Thus, the disrupted locomotor rhythms and the altered expression of circadian clock genes we observed in the nr1d1 mutant fish indicate that Nr1d1 is an indispensable component in the zebrafish circadian clock.
Extensive roles of Nr1d1 by transcriptome analysis
Previous studies have revealed multiple functions of NR1D1 in mice.Citation22,43,56,57 Here, our transcriptome analysis shows that DEGs of nr1d1 mutant zebrafish are involved in numerous life processes including autophagy, stress response, metabolism, and signal transduction (Figs. S4, S5 and Table S3). For instance, atg10 (autophagy-related 10 homolog) and exoc3 (exocyst complex component 3) (), involved in autophagy signaling pathway or endoplasmic reticulum stress-induced autophagy,Citation58 are significantly upregulated in nr1d1 mutant fish. For stress response, we observed significant upregulation of DNA repair genes including 6-4 photolyase, ddb2 (damage-specific DNA binding protein 2) and rbp4 (retinol binding protein 4, plasma) ( and Table S3), and heat shock gene serpinb1 (serine protease inhibitor, clade b (ovalbumin), member 1), implicating the pathway for DNA repair of UV-induced damages or entrainment of the circadian clock by temperature are both disrupted in the nr1d1 mutant fish.Citation59-64 A large number of DEGs such as ucp3 (uncoupling protein 3), gsk3b (glycogen synthase kinase 3 β), urod (uroporphyrinogen decarboxylase), cyp27a7 (cytochrome P450, family 27, subfamily A, polypeptide 7), and gbgt1l4 (globoside α-1,3-N-acetylgalactosaminyltransferase 1, like 4) ( and Table S3), involved in thermogenic, energy metabolism, heme synthesis, oxidoreductase activity and immune response, are all altered, implicating metabolic roles of Nr1d1 in zebrafish.Citation20,56,65-67 Further, as mitochondrial activities are intimately related with autophagy activities,Citation54 low mitochondrial activities and contents as revealed by JC-1 staining, and downregulation of ppargc1a expression in nr1d1 mutant fish (), suggesting that Nr1d1 is essential for maintaining zebrafish autophagy rhythms and altered autophagy activities in nr1d1 mutant in turn results in off-balance of the cellular mitochondrial energy.Citation22 In addition, gria3a (glutamate receptor, ionotropic, AMPA 3a), slc5a1 (solute carrier family 5 (sodium/glucose cotransporter), member 1), fn1b (fibronectin 1b) and wdr3 (WD repeat domain 3), involved in signal transduction or cellular functions, also are disrupted in nr1d1 mutant zebrafish ( and Table S3).Citation68-70 Even though the exact mechanisms underlying how nr1d1 regulates these genes need detailed investigation in the future, our transcriptome analysis reveals that in addition to circadian and autophagy functions, zebrafish Nr1d1 contributes extensively to numerous life processes, setting the stage for investigation of its novel functions in the future.
In summary, our study provides, for the first time, clear evidence of how the circadian clock regulates autophagy rhythms, i.e., both E-box-mediated and RORE-mediated transcriptional regulatory mechanisms underlie circadian regulation of autophagy rhythms. Expression of autophagy genes and proteins and the number of mature autolysosomes are all upregulated without rhythmicity in per1b and TALEN-generated nr1d1 mutant fish, suggesting Per1b and Nr1d1 are critical for maintaining autophagy rhythms. Disruption of expression of core circadian clock genes and circadian behaviors in nr1d1 mutant indicates that Nr1d1 is also essential for the zebrafish circadian clock. Fasting results in altered expression of both circadian clock genes and autophagy genes in zebrafish adult peripheral organs. Further, transcriptome analysis implicates extensive roles of Nr1d1 in numerous life processes. Because altered autophagy changes normal triglyceride hydrolysis in lipid droplets and causes lipid metabolism disorders,Citation1 our study implicates that dysregulated autophagy derived from circadian misalignment or mutations of circadian clock genes such as nr1d1 likely contributes to common metabolic diseases such as obesity.
Materials and methods
Animals
Wild-type AB strain zebrafish (Danio rerio), nr1d1, per1b mutant zebrafish were maintained on a 14 h/10 h light/dark cycle at the Soochow University Zebrafish Facility according to standard protocols. Embryos were collected from group crosses of 2 males and 2 females. All animal protocols were approved by the Soochow University Animal Ethics and Use Committee.
TALEN construction, mRNA synthesis and microinjection
The right and left arms of TALEN targeting nr1d1 were designed using a web-tool ‘TALEN-NT’,Citation40 and constructed using the ‘unit assembly’ method with Sharkey-AS and Sharkey-R forms of FokI cleavage domains as described previously.Citation41 Capped mRNAs of the TALEN expression vectors were in vitro transcribed with Sp6 mMACHINE Kit (Thermo Fisher, AM1340), and microinjected in pair into one-cell zebrafish embryos at a concentration of 250 pg.
Mutagenesis frequencies and identification of nr1d1 mutants
The microinjected embryos were maintained in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) at 28.5°C. The survival rate of injected embryos was recorded at 1 d post fertilization (Table S2). Genomic DNAs were extracted from approximately 10 embryos at 2 dpf. A 426-bp DNA fragment containing the nr1d1 target fragment was amplified by PCR (Primers used are listed in Table S1), and digested using BslI (NcoI). Estimated mutagenesis frequencies were calculated by software ImageJ as described previously.Citation71 The uncleaved bands were cloned into pMD 19T vector (Takara, D102C) for sequencing analysis. To identify the germline-transmitted mutations, the injected founder embryos were raised to adulthood and then outcrossed with wild types. Genomic DNAs were extracted from 8 F1 embryos each cross, and analyzed via PCR amplification and sequencing as described above. Siblings of the F1 embryos that carry heritable mutations were raised to adulthood and individual F1 fish was reidentified via PCR amplification and sequencing with fin-clipped DNAs. Two nr1d1-null mutation lines were established.
Behavioral analysis for zebrafish
Locomotor activity analysis was performed as described previously with modifications.Citation72 After the fourth d postfertilization, zebrafish larvae were placed into each well of 96-well plate singly. Locomotor activities of larvae were monitored and recorded for 4 consecutive d (14 h light and 10 h dark for LD or 24 h dark for DD) using an automate video-tracking system (Noldus, DanioVision, Wageningen, Netherlands). Locomotor activities were recorded from d 5 to d 8 postfertilization as the total distance moved by one larva during 10-min time duration. Data are presented as a moving average distance for each group (N = 48). The period length of each larval locomotor trace was retrieved. Statistical analysis of period length differences between the treatment groups was performed with one-way analysis of variance (ANOVA) followed by the Dunnett post-hoc test comparing each sample group with the control group.
RNA extraction and quantitative real-time PCR
Total RNAs were extracted from approximately 50 larvae of homozygous nr1d1 mutants or wild types at 4-h intervals from 72 to 96 h postfertilization (hpf) under LD or 96 to 120 hpf under DD, and adult organs (brain, liver, heart, eyes and testis, at least 5 fish for each sample) under the LD using TriZol (Invitrogen, 15596018). Quantitative real-time PCR (Thermo Fisher, StepOnePlus, 437660, Waltham, USA) was performed in ABI thermal profiles with the SYBR green detection system (Takara, RR420A) and a thermal profile of 40 cycles of 95°C, 10 s, 60°C, 30 s. Data represent mean ± s .d. of the 3 independent experiments. All primers used were listed in Table S1.
DNA site-directed mutagenesis and vector constructs
Mutagenized DNA vectors were constructed by PCR-based site-directed mutagenesis. PCR was performed with KOD plus DNA polymerase (TOYOBO, KOD-201). A DpnI (NEB, R0176) restriction enzyme-treated PCR product was transformed into E. coli. Positive clones were randomly selected and verified by sequencing. A 2,000-bp fragment of the zebrafish ulk1a promoter region containing 2 RORE motifs and one Rev-DR2 motif was isolated and cloned into the luciferase reporter containing pGL 4.17 vector (Promega, 9PIE672), cloned into pcDNA 3.1 vector (Invitrogen, V790-20) or the pcDNA 3.1-FLAG vector (Addgene, 20011; deposited by Stephen Smale). All DNA constructs were confirmed by DNA sequencing. The primers used are listed in Table S1.
Protein extraction and western blotting
Total proteins of zebrafish embryos were extracted with the tissue lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% NP-40, Thermo Fisher, 28324) and protein inhibitors (Roche, 04693116001) and then separated in 12% sodium dodecyl sulfate-PAGE gel. Western blotting was performed following the CST (Cell Signaling Technology) western blotting instruction. Protein bands were then transferred to a nitrocellulose member and incubated with desired antibodies as follows: Anti-mouse LC3B antibody (Cell Signaling Technology, 4108), SQSTM1/p62 (Cell Signaling Technology, 5114), anti-zebrafish Tuba1 antibody (Cell Signaling Technology, 2148) at a dilution of 1:1, 000 and then blocking with secondary antibodies at a dilution of 1:3, 000. Chemiluminescence Detection Kit (Biological Industries, 20-500-120) visualized the membranes. Tuba1 was used as a loading control. Data represent mean ± s .d. of the 3 independent studies.
Transmission electron microscopy (TEM)
Zebrafish liver samples were fixed in 2.5 % glutaraldehyde buffer and embedded for 24 h, and their TEM images were acquired with a transmission electron microscope (Hitachi, H-600, Tokyo, Japan) at the Soochow University Electron Microscopy Core Facility. Autophagosome and autolysosome numbers were counted manually, and the cytoplasmic areas were quantified with software ImageJ.
JC-1 staining
JC-1 mitochondrial membrane potential dye (MesGen Biotechnology, MG9621) was dissolved in DMSO to the concentration of 200 μM. Zebrafish larvae at 56 hpf were incubated with 10 μM of JC-1 dye at 37°C for 30 min. Three independent treatments were conducted. The JC-1 dye-stained larvae were photographed with a microscope (Olympus confocal microscope system, FV1200, Tokyo, Japan).
Cell transfection and luciferase reporter assay
A 2,129-bp zebrafish cebpb promoter fragment or a 1,999-bp zebrafish ulk1a promoter fragment was PCR amplified and cloned into pGL4.17 vector (Promega, 9PIE672), respectively. Human embryonic kindey (HEK) 293T cells were cultured in DMEM containing 10% serum and penicillin-streptomycin in a 24-well plate. Transfection was done with Lipofectamine 2000 (Invitrogen, 11668027) according the manufacturer's instructions. Luciferase reporter assays were performed with a Dual-Report assay system (Promega, E1980). 100 ng each of cebpb-luc, ulk1a-luc, clocka/clock1a, arntl1b/bmal1b and cry1ab were used; for the dosage-dependent experiment, 100, 200 and 300 ng of nr1d1 or roraa, were used. The total DNA amounts were equalized by pcDNA 3.1 vector. Three independent experiments were conducted for each assay.
Chromatin immunoprecipation (ChIP) assays
ChIP assays were performed with Chromatin Immuno-precipitation Assay Kit (Millipore, 17-295). For the Arntl1b ChIP assay, the larvae were collected at 120 hpf and 132 hpf; for the FLAG-Nr1d1 ChIP assay, the microinjected larvae were collected at 72 hpf and 84 hpf. The larval samples were cross-linked in 2% formaldehyde for 10 min, and then fractionated with protein A Agarose (EMDmillipore, 16-157). Chromatin lysates were immunoprecipitated with Arntl1b antibodies (Abgent, customized, peptide PSSQLTQSPESDR) or negative control mouse IgG (Invitrogen, 10400C) in the presence of protein A Agarose-salmon sperm DNA (50% Slurry, EMDmillipore, 16-157).Citation30 Then DNA was purified using a purification kit (OMEGA, D2500) and subsequently analyzed by qRT-PCR using primers flanking the predicted binding sites in the promoter regions. Primers used for the ChIP assay are listed in Table S1.
Pharmacological treatment
GSK4112 (Sigma, G0673) were dissolved in DMSO at concentrations of 100 μM or 50 μM, respectively. Zebrafish larvae at 72 hpf and 84 hpf were incubated in 20 μM of GSK4112 for 24 h at 28.5°C, respectively. Three independent treatments for each drug were performed.
Restricting feeding experiment
Four-mo-old wild-type and nr1d1 mutant zebrafish were starved for 2 consecutive wk for the restricting feeding experiment as previously described.Citation73 The brains, the livers, the hearts, the eyes, and the intestines of at least 5 males each from the nr1d1 mutant, fasted nr1d1 mutant, fasted wild-type and wild-type fish were then dissected out at 12 time points with 4-h interval for 2 consecutive days for RNA extraction and qRT-PCR analysis. At least 3 independent experiments were done for each time point under examination.
Deep sequencing-based transcriptome analysis of nr1d1 mutant fish
1. RNA quantification and qualification. Total RNAs from 2 stages, 122 hpf (CT2) and 134 hpf (CT14) were used. RNAs were examined on 1% agarose gels, purity and concentrations was checked with a Nanodrop 2000 spectrophotometer (Thermo Fisher, Waltham, USA). 2. Library preparation for sequencing. A total amount of 3 μg RNA per sample was used for the RNA sample preparations. Sequencing libraries were generated using NEBNext→Ultra™ RNA Library Prep Kit (NEB, USA) following the manufacturer's instructions. 3. Clustering and sequencing. Clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit (Illumia, PE-401-3001) according to the manufacturer's instructions. After clustering, the library preparations were sequenced on an Illumina Hiseq 2000 platform. 4. Quality control. We used Perl scripts for removing the adapter for clean reads, calculated the Q20, Q30, GC-content, duplication data and then generated the Raw reads. All the following analyses were based on clean data with high quality. 5. Transcriptome assembly. Transcriptome assembly was performed according to the protocol described previously.Citation74 6. Gene functional annotation. For the Gene Ontology (GO) enrichment analysis, the differentially expressed genes (DEGs) was implemented by the GOseq R packages based Wallonia noncentral hyper-geometric distribution;Citation75 For the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, we used KOBAS software to test the statistical enrichment of DEGs in KEGG pathways to predict and classify functions of the assembled sequences (Mao et al, 2005).Citation76 7. Quantification of gene expression levels. Gene expression levels were estimated by RSEM for each RNA sample.Citation77
Heatmaps
For the behavior assays, heatmaps were constructed by an automated video-tracking system software EthoVision XT10 (Noldus, DanioVision, Wageningen, Netherlands). For the ADJ.P values and correlation efficiencies in different organs of wild-type-fed, wild-type fasted, nr1d1-fed and nr1d1 fasted, heatmaps were made with custom scripts and implemented in R.
Statistical analysis
Values are means ± s.d. of the indicated number of measurements. Statistical significance was determined using the 2-tailed unpaired Student t test or one-way ANOVA or 2-way ANOVA with a significance of P ≤ 0 .05. In particular, multivariate experiments (e.g., time points and mutants) were analyzed using 2-way ANOVA with the Tukey post hoc test. A repeated-measure adjustment was employed when appropriate (e.g., daily onset profiles). An F test was used to assess the differences in variability to circadian-mutant shifts or time points between genotypes. For the JTK-CYCLE analysis, each parameter was recorded and analyzed as previously described by R2. All statistical analyses were performed using SPSS 16.0 software.
Abbreviations
| atg4a | = | autophagy-related 4 homolog A |
| atp6v1d | = | ATPase H+ transporting V1 subunit D |
| becn1/beclin1 | = | myosin-like BCL2 interacting protein |
| bnip3 | = | BCL2/adenovirus E1B interacting protein 3 |
| arntl/bmal | = | brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like |
| CCGs | = | circadian controlled genes |
| clock | = | clock circadian regulator |
| ChIP | = | chromatin immunoprecipitation |
| cry | = | cryptochrome circadian clock |
| DD | = | dark/dark |
| DEGs | = | differentially expressed genes |
| gabarapa | = | GABA(A) receptor-associated protein a |
| gsk4112 | = | agonist of nr1d1 |
| period | = | period circadian clock |
| LD | = | light/dark |
| Map1lc3b | = | microtubule-associated protein 1 light chain 3 β |
| RT-PCR | = | reverse transcriptase-polymerase chain reaction |
| ppargc1a/pgc1a | = | peroxisome proliferator-activated receptor gamma, coactivator 1 α |
| Sqstm1/p62 | = | sequestosome 1 |
| TALEN | = | transcription activator-like effector nucleases |
| TEM | = | transmission electron microscopy |
| Ulk1 | = | unc-51 like autophagy activating kinase 1 |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1183843_Supplemental_Material.zip
Download Zip (7.4 MB)Acknowledgments
We wish to thank Bo Zhang from Peking University for providing TALEN Unit Assembly vectors, Weijun Pan from Shanghai Institutes for Biological Sciences, the Chinese Academy of Sciences for providing the mCherry-map1lc3b plasmid, Yilin Yan from the University of Oregon and Yingbin Zhong of our lab for discussion and critical reading of the manuscript, and Zhaomin Zhong for maintaining fish lines and assistance in behavioral assays.
Funding
This work was supported by the grants from National Basic Research Program of China (973 Program) (2012CB947600), the National Natural Science Foundation of China (NSFC) (31030062), National High Technology Research and Development Program of China (863 Program)(2011AA100402-2), the Jiangsu Distinguished Professorship Program (SR13400111), and the Natural Science Foundation of Jiangsu Province (BK2012052), the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions (YX13400214), the High-Level Innovative Team of Jiangsu Province, and the “333” project of Jiangsu Province (BRA2015328).
References
- Walther TC, Farese RV, Jr. Lipid droplets and cellular lipid metabolism. Ann Rev Biochem 2012; 81:687-714; PMID:22524315; http://dx.doi.org/10.1146/annurev-biochem-061009-102430
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 2010; 25:372-80; PMID:20876817; http://dx.doi.org/10.1177/0748730410379711
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature 2002; 417:329-35; PMID:12015613; http://dx.doi.org/10.1038/417329a
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature 2002; 418:534-9; PMID:12152080; http://dx.doi.org/10.1038/nature00906
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genetics 2006; 15 Spec No 2:R271-7; http://dx.doi.org/10.1093/hmg/ddl207
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012; 485:123-7; PMID:22460952; http://dx.doi.org/10.1038/nature11048
- Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR, et al. Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc Natl Acad Sci U S A 2008; 105:14946-51; PMID:18815372; http://dx.doi.org/10.1073/pnas.0802636105
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/10.1038/nature06639
- Rabinowitz JD, White E. Autophagy and metabolism. Science 2010; 330:1344-8; PMID:21127245; http://dx.doi.org/10.1126/science.1193497
- Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy 2013; 9:124-37; PMID:23295650; http://dx.doi.org/10.4161/auto.23323
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009; 284:12297-305; PMID:19258318; http://dx.doi.org/10.1074/jbc.M900573200
- Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Seminars Cell Dev Biol 2010; 21:683-90; http://dx.doi.org/10.1016/j.semcdb.2010.03.002
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672-6; PMID:10604474; http://dx.doi.org/10.1038/45257
- Pfeifer U, Scheller H. A morphometric study of cellular autophagy including diurnal variations in kidney tubules of normal rats. J Cell Biol 1975; 64:608-21; PMID:1171105; http://dx.doi.org/10.1083/jcb.64.3.608
- Reme CE, Sulser M. Diurnal variation of autophagy in rod visual cells in the rat. Albrecht Von Graefes Archiv Klin Exp Ophthalmol 1977; 203:261-70; PMID:303474; http://dx.doi.org/10.1007/BF00409832
- Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J 2011; 30:4642-51; PMID:21897364; http://dx.doi.org/10.1038/emboj.2011.322
- Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Baugé E, Havinga R, Bloks VW, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterol 2008; 135:689-98; http://dx.doi.org/10.1053/j.gastro.2008.05.035
- Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, Gervois P, Soncin F, Mandrup S, Fruchart JC, Fruchart-Najib J, et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem 2003; 278:37672-80; PMID:12821652; http://dx.doi.org/10.1074/jbc.M304664200
- Fontaine C, Rigamonti E, Pourcet B, Duez H, Duhem C, Fruchart JC, Chinetti-Gbaguidi G, Staels B. The nuclear receptor Rev-erbalpha is a liver X receptor (LXR) target gene driving a negative feedback loop on select LXR-induced pathways in human macrophages. Mol Endocrinol 2008; 22:1797-811; PMID:18511497; http://dx.doi.org/10.1210/me.2007-0439
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Sci 2007; 318:1786-9; http://dx.doi.org/10.1126/science.1150179
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Structural Mol Biol 2007; 14:1207-13; http://dx.doi.org/10.1038/nsmb1344
- Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MK, Paquet C, Delhaye S, Shin Y, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med 2013; 19:1039-46; PMID:23852339; http://dx.doi.org/10.1038/nm.3213
- He C, Klionsky DJ. Analyzing autophagy in zebrafish. Autophagy 2010; 6:642-4; PMID:20495344; http://dx.doi.org/10.4161/auto.6.5.12092
- He C, Bartholomew CR, Zhou W, Klionsky DJ. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy 2009; 5:520-6; PMID:19221467; http://dx.doi.org/10.4161/auto.5.4.7768
- Appelbaum L, Wang GX, Maro GS, Mori R, Tovin A, Marin W, Yokogawa T, Kawakami K, Smith SJ, Gothilf Y, et al. Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc Natl Acad Sci U S A 2009; 106:21942-7; PMID:19966231; http://dx.doi.org/10.1073/pnas.906637106
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 2011; 29:699-700; PMID:21822242; http://dx.doi.org/10.1038/nbt.1939
- Wang D, Jao LE, Zheng N, Dolan K, Ivey J, Zonies S, Wu X, Wu K, Yang H, Meng Q, et al. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc Natl Acad Sci U S A 2007; 104:12428-33; PMID:17640903; http://dx.doi.org/10.1073/pnas.0705502104
- Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E. Efficient target-selected mutagenesis in zebrafish. Genome Res 2003; 13:2700-7; PMID:14613981; http://dx.doi.org/10.1101/gr.1725103
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31:227-9; PMID:23360964; http://dx.doi.org/10.1038/nbt.2501
- Huang J, Zhong Z, Wang M, Chen X, Tan Y, Zhang S, He W, He X, Huang G, Lu H, et al. Circadian modulation of dopamine levels and dopaminergic neuron development contributes to attention deficiency and hyperactive behavior. J Neurosci 2015; 35:2572-87; PMID:25673850; http://dx.doi.org/10.1523/JNEUROSCI.2551-14.2015
- Tan Y, DeBruyne J, Cahill GM, Wells DE. Identification of a mutation in the Clock1 gene affecting zebrafish circadian rhythms. J Neurogenetics 2008; 22:149-66; http://dx.doi.org/10.1080/01677060802049738
- Wang M, Zhong Z, Zhong Y, Zhang W, Wang H. The Zebrafish period2 protein positively regulates the circadian clock through mediation of retinoic acid receptor (RAR)-related orphan receptor α (Roralpha). J Biol Chem 2015; 290:4367-82; PMID:25544291; http://dx.doi.org/10.1074/jbc.M114.605022
- Kyostila K, Syrja P, Jagannathan V, Chandrasekar G, Jokinen TS, Seppala EH, Becker D, Drögemüller M, Dietschi E, Drögemüller C, et al. A missense change in the ATG4D gene links aberrant autophagy to a neurodegenerative vacuolar storage disease. PLoS Genetics 2015; 11:e1005169; PMID:25875846; http://dx.doi.org/10.1371/journal.pgen.1005169
- Eskelinen EL. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy 2008; 4:257-60; PMID:17986849; http://dx.doi.org/; http://dx.doi.org/10.4161/auto.5179
- Delaunay F, Thisse C, Marchand O, Laudet V, Thisse B. An inherited functional circadian clock in zebrafish embryos. Science 2000; 289:297-300; PMID:10894777; http://dx.doi.org/10.1126/science.289.5477.297
- Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab 2010; 11:47-57; PMID:20074527; http://dx.doi.org/10.1016/j.cmet.2009.11.002
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Ann Rev Genetics 2009; 43:67-93; http://dx.doi.org/10.1146/annurev-genet-102808-114910
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009; 137:1001-4; PMID:19524504; http://dx.doi.org/10.1016/j.cell.2009.05.023
- Lattante S, de Calbiac H, Le Ber I, Brice A, Ciura S, Kabashi E. Sqstm1 knock-down causes a locomotor phenotype ameliorated by rapamycin in a zebrafish model of ALS/FTLD. Hum Mol Genetics 2015; 24:1682-90; http://dx.doi.org/10.1093/hmg/ddu580
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 2012; 40:W117-22; PMID:22693217; http://dx.doi.org/10.1093/nar/gks608
- Huang P, Xiao A, Tong X, Zu Y, Wang Z, Zhang B. TALEN construction via “Unit Assembly” method and targeted genome modifications in zebrafish. Methods 2014; 69:67-75; PMID:24556555; http://dx.doi.org/10.1016/j.ymeth.2014.02.010
- del Pozo A, Sanchez-Ferez JA, Sanchez-Vazquez FJ. Circadian rhythms of self-feeding and locomotor activity in zebrafish (Danio Rerio). Chronobiol Int 2011; 28:39-47; PMID:21182403; http://dx.doi.org/10.3109/07420528.2010.530728
- Chung S, Lee EJ, Yun S, Choe HK, Park SB, Son HJ, Kim KS, Dluzen DE, Lee I, Hwang O, et al. Impact of circadian nuclear receptor REV-ERBalpha on midbrain dopamine production and mood regulation. Cell 2014; 157:858-68; PMID:24813609; http://dx.doi.org/10.1016/j.cell.2014.03.039
- Sasaki T, Lian S, Qi J, Bayliss PE, Carr CE, Johnson JL, Guha S, Kobler P, Catz SD, Gill M, et al. Aberrant autolysosomal regulation is linked to the induction of embryonic senescence: differential roles of Beclin 1 and p53 in vertebrate Spns1 deficiency. PLoS Genetics 2014; 10:e1004409; PMID:24967584; http://dx.doi.org/10.1371/journal.pgen.1004409
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Topics Microbiol Immunol 2009; 335:1-32.
- Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem 2012; 287:39107-14; PMID:22992773; http://dx.doi.org/10.1074/jbc.M112.412569
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 1997; 91:1043-53; PMID:9428526; http://dx.doi.org/10.1016/S0092-8674(00)80494-8
- Dekens MP, Whitmore D. Autonomous onset of the circadian clock in the zebrafish embryo. EMBO J 2008; 27:2757-65; PMID:18800057; http://dx.doi.org/10.1038/emboj.2008.183
- Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Ann Rev Physiol 2010; 72:579-603; http://dx.doi.org/10.1146/annurev-physiol-073109-130051
- Kakizawa T, Nishio S, Triqueneaux G, Bertrand S, Rambaud J, Laudet V. Two differentially active alternative promoters control the expression of the zebrafish orphan nuclear receptor gene Rev-erbalpha. J Mol Endocrinol 2007; 38:555-68; PMID:17496157; http://dx.doi.org/10.1677/JME-06-0063
- Grant D, Yin L, Collins JL, Parks DJ, Orband-Miller LA, Wisely GB, Joshi S, Lazar MA, Willson TM, Zuercher WJ. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbalpha. ACS Chem Biol 2010; 5:925-32; PMID:20677822; http://dx.doi.org/10.1021/cb100141y
- Amaral IP, Johnston IA. Circadian expression of clock and putative clock-controlled genes in skeletal muscle of the zebrafish. Am J Physiol Regulatory, Integrative Comparative Physiol 2012; 302:R193-206; http://dx.doi.org/10.1152/ajpregu.00367.2011
- Sanchez JA, Sanchez-Vazquez FJ. Feeding entrainment of daily rhythms of locomotor activity and clock gene expression in zebrafish brain. Chronobiol Int 2009; 26:1120-35; PMID:19731109; http://dx.doi.org/10.3109/07420520903232092
- Mai S, Muster B, Bereiter-Hahn J, Jendrach M. Autophagy proteins LC3B, ATG5 and ATG12 participate in quality control after mitochondrial damage and influence lifespan. Autophagy 2012; 8:47-62; PMID:22170153; http://dx.doi.org/10.4161/auto.8.1.18174
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002; 110:251-60; PMID:12150932; http://dx.doi.org/10.1016/S0092-8674(02)00825-5
- Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, Bugge A, Hou C, Ferrara C, Seale P, et al. The nuclear receptor Rev-erbalpha controls circadian thermogenic plasticity. Nature 2013; 503:410-3; PMID:24162845; http://dx.doi.org/10.1038/nature12642
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science 2015; 348(6242):1488-92
- Sakaki K, Yoshina S, Shen X, Han J, DeSantis MR, Xiong M, Mitani S, Kaufman RJ. RNA surveillance is required for endoplasmic reticulum homeostasis. Proc Natl Acad Sci U S A 2012; 109:8079-84; PMID:22562797; http://dx.doi.org/10.1073/pnas.1110589109
- Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci U S A 2006; 103:17696-700; PMID:17062752; http://dx.doi.org/10.1073/pnas.0607993103
- Guo C, Tang TS, Friedberg EC. SnapShot: nucleotide excision repair. Cell 2010; 140:754-4 e1; PMID:20211143; http://dx.doi.org/10.1016/j.cell.2010.02.033
- Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, Imanishi Y, Palczewski K, von Lintig J. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab 2008; 7:258-68; PMID:18316031; http://dx.doi.org/10.1016/j.cmet.2008.01.009
- Ahn SG, Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev 2003; 17:516-28; PMID:12600944; http://dx.doi.org/10.1101/gad.1044503
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 2007; 5:e34; PMID:17298173; http://dx.doi.org/10.1371/journal.pbio.0050034
- Li W, Giles C, Li S. Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res 2014; 42:7069-83; PMID:24813444; http://dx.doi.org/10.1093/nar/gku333
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 2013; 498:511-5; PMID:23728303; http://dx.doi.org/10.1038/nature12209
- Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA 2012; 109:582-7; PMID:22184247; http://dx.doi.org/10.1073/pnas.1106750109
- Lee MS. Role of islet β cell autophagy in the pathogenesis of diabetes. Trends Endocrinol Metabol 2014; 25:620-7; http://dx.doi.org/10.1016/j.tem.2014.08.005
- Wu Y, Arai AC, Rumbaugh G, Srivastava AK, Turner G, Hayashi T, Suzuki E, Jiang Y, Zhang L, Rodriguez J, et al. Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc Natl Acad Sci USA 2007; 104:18163-8; PMID:17989220; http://dx.doi.org/10.1073/pnas.0708699104
- Hoppmann V, Wu JJ, Soviknes AM, Helvik JV, Becker TS. Expression of the eight AMPA receptor subunit genes in the developing central nervous system and sensory organs of zebrafish. Dev Dynamics 2008; 237:788-99; http://dx.doi.org/10.1002/dvdy.21447
- Cheng P, Andersen P, Hassel D, Kaynak BL, Limphong P, Juergensen L, Kwon C, Srivastava D. Fibronectin mediates mesendodermal cell fate decisions. Development 2013; 140:2587-96; PMID:23715551 http://dx.doi.org/10.1242/dev.089052
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 2012; 30:460-5; PMID:22484455; http://dx.doi.org/10.1038/nbt.2170
- Smadja Storz S, Tovin A, Mracek P, Alon S, Foulkes NS, Gothilf Y. Casein kinase 1delta activity: a key element in the zebrafish circadian timing system. Plos One 2013; 8:e54189; PMID:23349822; http://dx.doi.org/10.1371/journal.pone.0054189
- Craig PM, Moon TW. Fasted zebrafish mimic genetic and physiological responses in mammals: a model for obesity and diabetes?. Zebrafish 2011; 8:109-17; PMID:21854210; http://dx.doi.org/10.1089/zeb.2011.0702
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 2011; 29:644-52; PMID:21572440; http://dx.doi.org/10.1038/nbt.1883
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010; 11:R14; PMID:20132535; http://dx.doi.org/10.1186/gb-2010-11-2-r14
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res 2008; 36:D480-4; PMID:18077471; http://dx.doi.org/10.1093/nar/gkm882
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011; 12:323; PMID:21816040; http://dx.doi.org/10.1186/1471-2105-12-323
