ABSTRACT
Autophagy is important for degradation and recycling of intracellular components. In a diversity of genera and species, orthologs and paralogs of the yeast Atg4 and Atg8 proteins are crucial in the biogenesis of double-membrane autophagosomes that carry the cellular cargoes to vacuoles and lysosomes. Although many plant genome sequences are available, the ATG4 and ATG8 sequence analysis is limited to some model plants. We identified 28 ATG4 and 116 ATG8 genes from the available 18 different plant genome sequences. Gene structures and protein domain sequences of ATG4 and ATG8 are conserved in plant lineages. Phylogenetic analyses classified ATG8s into 3 subgroups suggesting divergence from the common ancestor. The ATG8 expansion in plants might be attributed to whole genome duplication, segmental and dispersed duplication, and purifying selection. Our results revealed that the yeast Atg4 processes Arabidopsis ATG8 but not human LC3A (HsLC3A). In contrast, HsATG4B can process yeast and plant ATG8s in vitro but yeast and plant ATG4s cannot process HsLC3A. Interestingly, in Nicotiana benthamiana plants the yeast Atg8 is processed compared to HsLC3A. However, HsLC3A is processed when coexpressed with HsATG4B in plants. Molecular modeling indicates that lack of processing of HsLC3A by plant and yeast ATG4 is not due to lack of interaction with HsLC3A. Our in-depth analyses of ATG4 and ATG8 in the plant lineage combined with results of cross-kingdom ATG8 processing by ATG4 further support the evolutionarily conserved maturation of ATG8. Broad ATG8 processing by HsATG4B and lack of processing of HsLC3A by yeast and plant ATG4s suggest that the cross-kingdom ATG8 processing is determined by ATG8 sequence rather than ATG4.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a major process for degradation and recycling of intracellular materials from yeast to human. Autophagy plays fundamental roles in growth and differentiation, development, and response to various stresses.Citation1-6 Recent studies have revealed that autophagy is also involved in nitrogen remobilization, abiotic stress responses, starch degradation during the night-time, and innate immunity including programmed cell death (PCD).Citation3,7,8 In the autophagy pathway, a double-membrane vesicle called autophagosome encloses intracellular cargoes to deliver them into the vacuole or lysosome.Citation9,10 More than 30 ATG genes are directly involved in autophagy in yeastCitation11 and most of these genes are conserved in other organisms including plants. Among ATG proteins, a ubiquitin-like Atg8 in yeast and its orthologs and paralogs in other species have an important role in the early stage of the autophagic pathway.Citation12 Cleavage of members of the ATG8 family by the ATG4 cysteine protease results in the exposure of the conserved glycine (Gly) residue at the C terminus of ATG8. This processed ATG8 conjugates to the phagophore (the autophagosome precursor) membrane through lipidation with phosphatidylethanolamine (PE) and takes part in autophagosome formation. In Arabidopsis thaliana, 2 conjugation systems involved in autophagy biogenesis are also highly conserved.Citation4 Ubiquitin-like ATG12 is transferred to a target ATG5 that is mediated by a chain reaction through E1-like ATG7 activity and E2-like ATG10 conjugation reaction.Citation10 The ATG5 complex bound with ATG12 further interacts with ATG16L1. The assembled ATG5 complex appears to play a role as an E3-like enzyme for the lipidation of ATG8-family members, such as the LC3 mammalian orthologs of yeast Atg8.Citation13 The conserved Gly of the processed ATG8 is implicated in adduct formation with PE after the other ubiquitin-like conjugation reaction by ATG7, E2-like ATG3, and ATG12–ATG5-ATG16L1.Citation9,10,13
While yeast has single Atg4 and Atg8 proteins, genes in higher eukaryotes such as animals and plants contain large families of ATG4 and ATG8, suggesting their diverse functions.Citation3,14-20 In humans, 4 ATG4s (HsATG4s) and 7 ATG8s (HsATG8s) have been identified based on sequence similarity to the yeast ATG4 (ScATG4) and ATG8 (ScATG8), respectively.Citation15,21 Rescue of autophagy-defective mutant strains of Saccharomyces cerevisiae such as atg4/aut2Citation21 by HsATG4 suggests that ATG4-mediated processing of ATG8 is evolutionarily conserved. HsATG8s are classified into 2 subfamilies; GABARAP (GABA type A receptor-associated protein), along with the GABARAPL1 and GABARAPL2 isoforms, and 4 MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3) isoforms (LC3A/B/B2/C).Citation15 Previously, it has been suggested that ATG8 isoforms might have similar functions in autophagosome biogenesis.Citation22,23 However, recent evidence indicates a specific function for ATG8 isoforms.Citation19,24 Human GABARAP sequence is more similar to GABARAPL2 than LC3. Overexpression of LC3 fails to rescue the autophagy defect in GABARAP or GABARAPL2 knockdown cells, and vice versa. LC3 overexpression in GABARAP knockdown cells causes accumulation of large autophagosomes, and overexpression of GABARAPL2 in LC3 knockdown cells decreases the size of autophagosomes.Citation25 Although human ATG8 orthologs are involved in delivery of phospholipids to the autophagy membrane, LC3 isoforms cannot complement the mutant phenotype of GABARAP or GABARAPL2.Citation18,25 Based on these results, it has been proposed that HsLC3s function in PAS elongation during early autophagosome biogenesis in mammalian cells and GABARAP proteins play a role in maturation of late autophagosomes.Citation25 Recently, Caenorhabditis elegans ATG8 orthologs LGG-1 and LGG-2 have been shown to have differential and nonredundant functions in autophagy.Citation19,26,27 Crystallography studies of LGG-1 and LGG-2 indicate that they have distinct substrate binding pockets.Citation19
In A. thaliana, 2 ATG4s (AtATG4) and 9 ATG8s (AtATG8) have been identified.Citation28,29 AtATG8s are classified into 3 groups and they show different spatiotemporal expression patterns in response to a variety of conditions.Citation16 Recently, we have shown specificity and redundancy for AtATG4-mediated processing of different AtATG8 isoforms.Citation30,31 However, little is known about ATG genes or autophagy mechanism in other plant species. Although a few ATG8 orthologs have been used as markers in some plants,Citation32-37 there is no systematic analysis of ATG4 and ATG8 in agronomically important crop plants.
Here, we report genome-wide analyses of genes encoding reported and potential ATG4 and ATG8 proteins from the currently available 18 plant genomes including green algae.Citation38 Phylogenetic relationships and evolutionary features of ATG4s and ATG8s indicate the conserved mechanism of autophagosome biogenesis in the plant kingdom. To determine the cross-kingdom ATG4 processing properties, we designed synthetic substrates containing AtATG8a,Citation30 tomato ATG8 (SlATG8), ScAtg8, and HsLC3A and investigated the processing efficiency by ATG4s from different species in vitro and in vivo. We found broad catalytic activity of HsATG4B for ATG8 orthologs compared to yeast and plant ATG4s. Structure-based modeling results suggest a major conformational change between HsLC3A and AtATG8a at specific regions of these 2 proteins. These findings suggest that the kinetic activity of ATG4s is determined by ATG8 sequence rather than ATG4.
Results
Identification of ATG4s and ATG8s in the plant kingdom
We used various publicly available plant genome sequences from single cell Chlamydomonas reinhardtii (green algae), Physcomitrella patens (moss; bryophyte), Selaginella moellendorffii (spike moss; lycophyte), and 15 different flowering plants to identify plant orthologs of ATG4 and ATG8 (Table S1). We performed BLASTP searches using the amino acid sequence of AtATG4s and AtATG8s and Hidden Markov Model (HMM) searches with HMM profiles of ATG4 (PF03416) and ATG8 (PF02991). BLASTP and HMM search results were combined (see Materials and methods for detail). From our analyses, we removed genes that encode proteins with an incomplete domain or the predicted protein that lacks functionally important residue such as the Gly at the C terminus of ATG8s required for PE-conjugation. Based on these analyses, a total of 28 ATG4s and 116 ATG8s were identified from 18 plant genomes ( and Table S2).
Table 1. The number of ATG4 and ATG8 orthologs in 18 plant genomes.
The numbers of ATG4s in most plants varied from one to 2 except in Brassica rapa that contains 3 ATG4s (). The amino acid sequence similarity of plant ATG4s with the yeast Atg4 is low (27% to 32% identity), however, the core amino acids required for the cysteine protease function such as the catalytic cysteine (Cys) residue in the active site is conserved (Fig. S1). In the case of ATG8, unicellular green algae C. reinhardtii contains a single copy of ATG8 like yeast while the other plants including mosses have 3 to 12 ATG8s, indicating gene duplication events have been occurred during the speciation and evolution of plants (). However, there is no correlation between the number of ATG8s and evolution of plant lineages.
The amino acid sequences of ATG8s between species are more conserved than those of ATG4s (Fig. S1 and Fig. S2). Amino acid sequence identity of plant ATG8s with yeast Atg8 is between 47% and 77%. Some ATG8s have 100% amino acid sequence identity between species such as in Solanaceae (Sl02g080590.2.1 with StPGSC0003DMP400028821 and Sl07g064680 with StPGSC0003DMP400025414, StPGSC0003DMP400038670, and Ca07g18990). Previously, identical ATG8 sequences in maize genome have been reported.Citation32 We found more examples of identical ATG8 sequences in potato (StPGSC0003DMP400025414 with StPGSC0003DMP400038670) and soybean (Gm09g00630 with Gm15g11510). These results indicate that ATG4 and ATG8 genes are conserved among plant lineages.
Gene structures and locations of plant ATG4s and ATG8s
The coding regions of ATG4s and ATG8s were compared among 18 plant genomes. The numbers of exons of ATG4s were generally conserved among the plant kingdom even in lower plants such as green algae (Table S2). In addition, the length of each exon was also conserved except for green algae. Among them, an ortholog of ATG4 in B. rapa has 3 exons while most other orthologs have 8 exons (Fig. S3). This could be the result of duplication during the emergence of this species because B. rapa has one more ortholog of ATG4 compared to its closest relatives A. thaliana and A. lyrata ().
Gene structures of ATG8 are also extremely similar among different plant species. Most members of the ATG8 gene family consist of 5 exons except few genes having alternative spliced transcripts. However, all ATG8 genes in P. patens have 4 exons (Table S2). Further analysis revealed that second exon of PpATG8s is split in most ATG8s in other plants (Fig. S4). However, genes lacking introns such as human MAP1LC3B2/LC3B2 were not found in plant ATG8s. We also found an ATG8 ortholog in B. rapa (Br018503) that has additional domain at the C terminus (putative ATP-synthase). We explored the physical location of ATG4s and ATG8s in the genome. While most orthologs in other plants are dispersed on their genome, ATG8s in P. patens showed tandem and proximal duplication (Fig. S5).
Phylogenetic relationships and sequence analysesof plant ATG4s and ATG8s
To understand the evolutionary relationships of ATG4s and ATG8s in the plant kingdom, phylogenetic trees were constructed using ATG4 and ATG8 domain regions of 18 plants, human, and yeast sequences ( and ). The alignments were performed using nucleotide sequences, and the nucleotide alignments based on the alignments with deduced amino acids were also considered for the phylogenetic construction (Fig. S1 and S2). Results suggest that ATG4 appears to be diverged according to the lineage (). They were branched according to the speciation with high bootstrap support. These data suggest that most divergence of ATG4 occurred during their speciation and paralogs by duplication have occurred in some plants. ATG4 orthologs were split among Brassicaceae plants suggesting that the divergence of members might have occurred before the divergence of Brassicaceae. In addition, we identified a putative novel motif in plant ATG4s compared to ScAtg4 and HsATG4 ( and Fig. S1). This novel motif contains 13 conserved amino acids among plant ATG4s () but not in yeast and human ATG4s (Fig. S1), suggesting that this novel motif may have specific roles in plant autophagy.
Figure 1. Analysis of ATG4s from different plant species. (A) A phylogenetic tree was constructed using nucleotide sequences and amino acid alignments of ATG4 domains. The maximum likelihood model was used and ATG4s of human and yeast were used as outgroup. Bootstrap analysis was performed with 1000 replicates to support each branch. Same colors mean related species. Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Pt, Populus trichocarpa; Me, Manihot esculenta; Vv, Vitis vinifera; Mt, Medicago truncatula; Gm, Glycine max; Nb, Nicotiana benthamiana; Ca, Capsicum annum; Sl, Solanum lycopersicum; At, Arabidopsis thaliana; Br, Brassica rapa; Os, Oryza sativa; Bd, Brachypodium distachyon; Zm, Zea mays; Sm, Seleginella moellendorffii; Pp, Physcomitrella patens; Cr, Chlamydomonas reinhardtii. (B) Plant-specific motif in ATG4. Amino acid sequences of plant ATG4s were aligned and plant-specific motif logo was created using the alignment. The height of symbols indicates the sequence conservation and residue prevalence of multiple alignment positions.
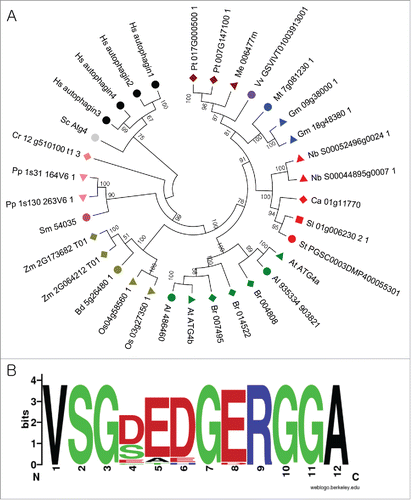
Figure 2. Analysis of ATG8s from different plant species. (A) A phylogenetic tree was constructed using nucleotide sequences and amino acid alignments of ATG8 domains. Maximum likelihood model was used and bootstrap analysis was performed with 1000 replicates to support each branch. Same colors mean related species and the 3 ATG8 subgroups (a to d, e to g, and h and i) were marked as arcs. For different species abbreviations, see legend. (B) Domain sequences in each subgroup of ATG8. Amino acid sequences of plant ATG8s of each subgroup were aligned and the domain sequences were extracted according to PFAM database (PF02991). Single insertion sequence was removed and the motif logo was created using the alignment. The height of symbols indicates the sequence conservation and residue prevalence of multiple alignment positions.
Divergence pattern of ATG8s is distinct from that of ATG4s. The ATG8 genes seem to have a common ancestor (green algae) and have diverged into 3 groups during the speciation of plants (). The phylogenetic relationship was consistent with the evolutionary history among 3 groups. HsATG8s were branched out as an outgroup. Most ATG8s in ancestral land plants such as P. patens and S. moellendorffii formed a monophyletic group, but we did not include them in the subgroups because of relatively low bootstrap support. However, their domain sequences are more similar to that of the ATG8 a-d and e-g group than of the h-i group, indicating distinct evolution of genes in the h-i group in vascular plants. Interestingly, while monocot plants have lost the e-g group ATG8s ( and ), Capsicum annuum and Zea mays have lost the h-i group. In Fabaceae, M. truncatula does not contain the a-d group of ATG8s while G. max has 6 ATG8s in the group. Together, these results suggest that ATG4 and ATG8 genes might have been not only duplicated but also lost during evolution, resulting in the extinction and expansion of some subgroups in their specific lineages.
We compared domain sequences of each ATG8 group and found that each group has some distinct sequence features (). The genes in the a-d group contain the most conserved domain. Sequences between the a-d and the e-g group are similar than between those of the h-i group. The sequences in the h-i group are less conserved, suggesting their rapid evolution processes. Some positions have been only conserved in each group of which positions are less conserved among the groups. For example, position 53 and 54 in the a-d and the e-g group are conserved as basic residues (arginine and lysine) while the basic amino acids are often changed to a polar amino acid such as serine in the h-i group (). It appears that the change in the amino acid sequences among different ATG8s might cause functional differences between the groups. In addition, all ATG8s in the h-i group from Brassicaceae have the exposed Gly like AtATG8h and AtATG8i. However, some h-i group ATG8s in other plant species have additional residues after the Gly (Fig. S6).
Duplication of ATG4s and ATG8s in the kingdom Plantae
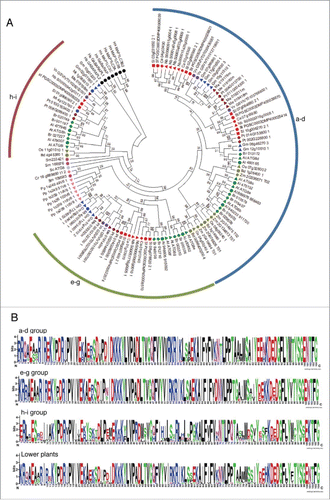
Gene duplication is thought to be one of the most important driving forces for the evolution of the species. All angiosperms have undergone ancient whole genome duplications (WGDs), including multiple WGDs in many angiosperms.Citation39,40 There are different modes of duplication such as tandem, WGD/segmental, and rearrangements by transposon.Citation41,42 We analyzed the mode of duplication for the ATG4 and ATG8 families in 18 plant genomes using MCScanX.Citation43 Each gene was assigned into 5 categories: singleton, dispersed, proximal, tandem, and WGD/segmental duplication (). As a result, most genes were duplicated and retained from WGD/segmental or dispersed duplication. However, some ATG8s in P. patens showed tandem or proximal duplication as discussed above (Fig. S5). In addition, all ATG4s and ATG8s in soybean were duplicated by WGD/segmental duplication suggesting recent lineage-specific WGD. In the case of Brassicaceae, B. rapa has experienced genome triplication since the divergence occurred from the common ancestor Arabidopsis.Citation44 This may explain why B. rapa has more ATG4s and ATG8s than Arabidopsis. On the other hand, genes of ATG4s and ATG8s by segmental duplication were not found in N. benthamiana, M. truncatula, and Brachypodium distachyon. Although we should consider the possibility of incomplete genome assembly of these species, they showed species-specific duplication patterns.
Figure 3. Duplication types of ATG4s and ATG8s in plants. Duplication events of ATG4 and ATG8 families in 18 plant genomes with phylogenetic relationship. The MCScanX algorithm was used to analyze modes of duplication. The 5 different modes of duplication events were observed in ATG8 families while tandem and proximal duplication have not been occurred in ATG4 families. The different duplication modes are color-coded and the box plot indicates ATG4 or ATG8 in each species.
To investigate the evolution of ATG4 and ATG8 by duplication, Ka (nonsynonymous substitutions per site), Ks (synonymous substitutions per site), and Ka/Ks values were estimated using the duplicated pairs of genes in each species (Table S3 and S4). As mentioned above, we could not find duplicated pairs in some genome data because of incomplete assembly. All Ka/Ks values in the duplicated gene pairs of ATG4 and ATG8 were less than 1 (Table S3 and S4), indicating that purifying selection might play a role in the evolution of ATG4s and ATG8s. Together, ATG4 and ATG8 genes in plants have been duplicated and retained well by purifying selection but they have lineage-specific duplication patterns.
Duplication and loss of ATG8 orthologs have also been occurred in metazoan during evolution. Orthologs of human LC3, GABARAP, and GABARAPL2 have been identified in earlier animal linages such as cnidarians and sponges. However, in honeybee, Apis mellifera, the GABARAPL2 ortholog is not present and fruit fly, Drosophila melanogaster, lacks GABARAPL2 and LC3 orthologs.Citation15 Therefore, expansion and extinction of ATG8 orthologs may be common phenomena in all kingdoms during evolution.
In order to assess the expression patterns of duplicated plant paralogs, we analyzed the publicly available gene expression data of ATG4s and ATG8s from various organs in some plants (Fig. S7). Expression of both ATG4s and ATG8s was detected in various organs, indicating that they might be constitutively expressed. In addition, we found several pairs of paralogs with similar expression patterns in the tissues. This indicates that functional redundancy exists among these paralogs. However, there was no correlation between duplicated pairs and expression patterns suggesting that the duplicated genes could achieve independent expression patterns.
Endogenous yeast Atg4 can process plant ATG4 but not human LC3A
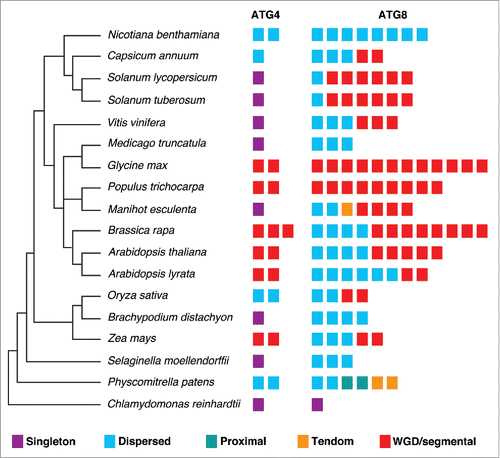
Although our phylogenetic analyses of ATG4s and ATG8s showed HsATG4s and HsATG8s were diverged in an early evolution period and classified to the outgroup, it has been shown that HsATG4s could rescue the phenotypes of the yeast atg4/aut2 mutant.Citation21 This suggests that ATG4 protease-mediated processing of ATG8s may be conserved in the course of evolution. Therefore, we investigated processing of plant AtATG8a and human LC3A (HsLC3A) using yeast-based cleavable transcription activator and reporter approach.Citation45 In this approach, the cytoplasmic death domain (DD) of the FAS/CD95 type I transmembrane receptor is replaced with a chimeric transcription activator (TA) comprised of the LexA DNA-binding domain and B42 activation domain (FAS-DDΔ-TA) (). We inserted ATG8 or LC3A between FAS-DDΔ and TA as a translational fusion. If ATG8 or LC3A is cleaved at the Gly residue, then chimeric TA will enter the nucleus and activate expression of a reporter gene such as LacZ (). To determine if yeast Atg4 could process plant AtATG8a and human HsLC3A, we expressed FAS-DDΔ-AtATG8a-TA or FAS-DDΔ-HsLC3A-TA in the EGY48 yeast strain. Interestingly, the endogenous yeast Atg4 could process AtATG8a but not HsLC3A (, top panel). In addition, we tested if an AtATG4 proteolytic-resistant mutant of AtATG8a in which the conserved Gly residue in the C terminus is changed to alanine (G117A) could be processed by endogenous yeast Atg4 by expressing FAS-DDΔ-AtATG8aG117A-TA in the EGY48 yeast strain. The AtATG8aG117A mutant was not processed by endogenous yeast Atg4 (, top panel). Furthermore, AtATG8a was not processed in an Scatg4-deleted yeast strain (EGY48 Scatg4Δ) (, bottom panel). To further confirm these results, we transformed AtATG4a, AtATG4b, and ScATG4 individually into the Scatg4Δ yeast strain expressing the FAS-DDΔ-AtATG8a-TA construct. Both AtATG4s and ScAtg4 were able to process wild type AtATG8a but not the mutant AtATG8aG117A (). Together, these results indicate that yeast Atg4 could process plant AtATG8a but not human HsLC3A.
Figure 4. Yeast Atg4 can process plant AtATG8a but not human HsLC3A. (A) Schematics of yeast-based cleavable transcription activator and reporter approach.Citation45 Chimeric transcription activator (TA) comprised of LexA DNA binding domain (LexA DNA-BD) and B42 activation domain (B42-AD) was fused to the C terminus of death domain deleted FAS type I transmembrane receptor (FAS-DDΔ). ATG8 or LC3A is inserted as a translational fusion between FAS-DDΔ and TA. ATG4-mediated processing of ATG8 or LC3A at the Gly residue (not shown) will lead to translocation of TA into the nucleus to induce LacZ expression. (B) FAS-DDΔ-AtATG8a-TA, FAS-DDΔ-AtATG8aG117A-TA, and FAS-DDΔ-HsLC3A-TA plasmids were transformed into EGY48 and Scatg4-deleted EGY48 (EGY48 Scatg4Δ) yeast strains. The selected yeast cells were spotted onto media plates with X-gal substrate. Blue color indicates that endogenous yeast Atg4 can process plant AtATG8a (top panel, column 2). White colony indicates that the endogenous yeast Atg4 cannot process mutant AtATG4aG117A and human HsLC3A (top panel). The deletion of ScATG4 from EGY48 results in lack of processing of AtATG8a (bottom panel, column 2). (C) EGY48 Scatg4Δ yeast cells lacking ScATG4 were transformed with plasmid encoding FAS-DDΔ-AtATG8a-TA and AtATG4a or AtATG4b and FAS-DDΔ-AtATG8a-TA with ScATG4. The transformed cells were grown on media plates with X-gal substrate. Both yeast and plant ATG4s could process plant AtATG8a (middle column). The empty vector and the mutant AtATG8aG117A were used as controls (first and last columns).
Human ATG4 can process yeast and plant ATG8 in vitro
To determine if plant ATG4 could process ScAtg8 and HsLC3A, we performed our recently described synthetic substrate-based in vitro cleavage assay that enabled examination of ATG4-mediated processing of ATG8.Citation30,31 In this ATG8 synthetic substrate, Citrine fluorescent protein (C) and modified Renilla luciferase SuperhRLUC (ShR) were fused to the N and C termini of ATG8, respectively (C-ATG8-ShR). We expressed and purified recombinant ATG4s and synthetic ATG8 substrates from Escherichia coli and used for in vitro cleavage assay (see Materials and methods for detail).Citation30 ATG4s efficiently processed corresponding species ATG8 compared to the MBP-alone control (). Yeast and plant ATG4s were able to process both yeast and plant ATG8 synthetic substrates but they were not able to process HsLC3A ( to 5C). Interestingly, HsATG4B could process yeast and plant ATG8s, and human LC3A synthetic substrates (). These results indicate that HsATG4B has broad ATG8 processing activity compared to yeast and plant ATG4s.
Figure 5. Human ATG4B can cleave cross-kingdom ATG8s in vitro. Purified recombinant maltose binding protein (MBP) fused to different ATG4s (MBP-ATG4) and MBP-alone were incubated with various C-ATG8- or LC3A-ShR synthetic substrates as described in the Materials and Methods section. The reaction mixtures were separated on SDS-PAGE gel and blots were probed with anti-ShR. Arrows and arrowheads indicate full-length synthetic substrate (C-ATG8-ShR) and cleaved byproduct ShR, respectively. The yeast Atg4 (A), Arabidopsis ATG4a (B), and tomato ATG4 (C) could process both yeast and plant ATG8s but not human HsLC3A. HsATG4B could process all species ATG8 synthetic substrates (D). The tomato ATG4 mutant (SlATG4178-188Δ) in which the plant specific domain is deleted decreased the efficiency of processing of plant ATG8s and completely inhibited processing of yeast Atg8 (E). Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; At, Arabidopsis thaliana; Sl, Solanum lycopersicum.
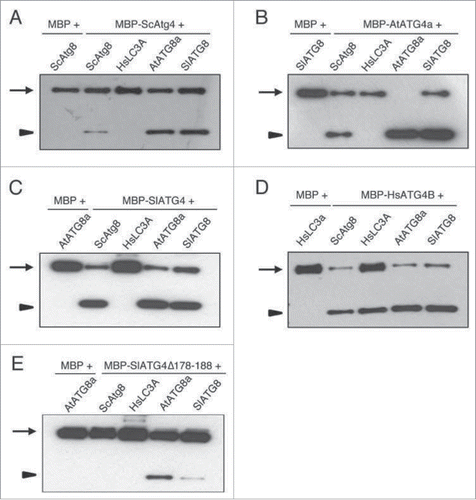
Since we found plant specific motif in ATG4 homologs (), we hypothesized that this motif could influence either plant ATG4 kinetic activity or ATG8 substrate recognition. To test this, we generated a tomato ATG4 mutant in which amino acids 178 to 188 were deleted (SlATG4178-188Δ). The processing of plant ATG8s is significantly reduced with SlATG4178–188Δ compared to the wild-type SlATG4 (comparing with 5C). Interestingly, processing of yeast ScAtg8 is completely abolished with the SlATG4178-188Δ mutant (). These results indicate that the plant specific motif is required for full activity of tomato ATG4 and broad substrate processing for ScAtg8.
Yeast and plant ATG4s bind HsLC3A but cannot process it
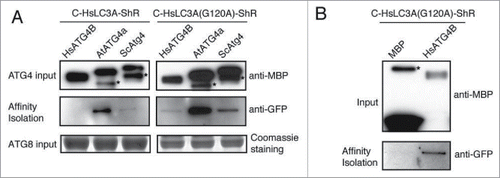
Lack of processing of HsLC3A by yeast and plant ATG4 ( to 5C) could be due to lack of binding to the LC3A substrate. To test this, we performed in vitro affinity isolation with MBP-tagged yeast, Arabidopsis, and human ATG4 and the C-HsLC3A-ShR substrate. We detected the HsLC3A affinity isolation product with yeast and plant ATG4s (; left panels) suggesting that the lack of processing of HsLC3A by yeast and plant ATG4s is not due to lack of interaction. However, we failed to detect the affinity isolate of HsLC3A with HsATG4B (, left panels). A simple explanation could be that the efficient cleavage of HsLC3A may destabilize the interaction between HsATG4B and HsLC3A. To test this possibility, the noncleavable mutant HsLC3AG120A was used for affinity isolation. The HsLC3AG120A was detected in the affinity isolation sample with HsATG4B and also with yeast and plant ATG4s (; right panels) but not with the MBP-alone control (). Together, these results indicate that failure to process HsLC3A by yeast and plant ATG4s is not due to lack of interaction but may be due to the structural differences between HsLC3A and yeast and plant ATG8s.
Figure 6. HsLC3A directly binds to yeast, Arabidopsis, and human ATG4s. (A) C-HsLC3A-ShR and catalytic resistant C-HsLC3AG120A-ShR were used for in vitro affinity isolation with various ATG4 homologs. The yeast and Arabidopsis ATG4s affinity isolated C-HsLC3A-ShR but HsATG4B failed to affinity isolate HsLC3A due to kinetic activity of HsATG4B (left, middle panel). All tested ATG4 homologs affinity isolated the catalytic resistant C-HsLC3AG120A-ShR (right, middle panel). Anti-MBP and Coomassie staining were used for inputs of ATG4 and the ATG8 substrates, respectively. Asterisks indicate degraded ATG4s. (B) C-HsLC3AG120A-ShR is not affinity isolated by the MBP-alone control (bottom panel). Same amount of C-HsLC3AG120A-ShR was used for affinity isolation as shown in (A). Anti-MBP was used for inputs. Asterisks indicate nonspecific binding products with amylose resin.
Since plant ATG4 and ATG8 structures are not available, we performed homology modeling based on the known structures of HsATG4B (PDB, 2zzp) and HsLC3A (PDB, 2zzp).Citation46 Although we generated a reliable homology model of Arabidopsis AtATG8a (Fig. S8A), the homology model of AtATG4a contained an uncertain region that includes a plant-specific motif ( and S8B). However, the overall fold of the AtATG4a active site is well aligned with the template structure of HsATG4B (Fig. S8C). The AtATG4a putative catalytic triad sequence (C170/D264/H268) is well aligned with the corresponding HsATG4B catalytic triad sequence (C74/D278/H280) (Fig. S8C)Citation47 suggesting that the processing mechanism of ATG4 homologs might be similar.
To examine the binding interfaces between HsATG4B and AtATG8a, the homology model of AtATG8a was overlaid with the HsLC3 fold in the cocrystal structure of HsATG4B and HsLC3 (PDB, 2zzp)Citation46 (Fig. S8D). Many of the key residues that interact across the interface between HsATG4B and HsLC3 are conserved in the plant AtATG8a. For example, a basic residue, Arg66 in the AtATG8a exists at the corresponding Arg68 in HsLC3 that forms a salt bridge with Asp171 in HsATG4B and Phe78 in AtATG8a has the same position to Phe80 of HsLC3 that makes a hydrophobic interaction with HsATG4B (Fig. S8E). These computational analyses support our in vitro results that HsATG4B can directly bind AtATG8a.
Examination of the AtATG8a homology model and the HsATG8 structure indicated a loop (PDB, 2zzp; residues 83 to 94) that is predicted to have significant conformational change (almost 4Å shift in corresponding residues) between the HsLC3 structure and the AtATG8a model. We have high confidence in the AtATG8a model at this region due to several available ATG8 crystal structures (Fig. S8A). The difference in the loop regions is most likely stems from the successive Proline residues at 86 and 87 positions in the AtATG8a sequence (Fig. S2 and S8F). Interestingly, the HsLC3 ortholog HsGABARAP has a similar sequence at this loop region, with yeast and plant ATG8s. Furthermore, the loop structure of HsGABARAP (PDB, 1gnu) is much closer to yeast and plant ATG8 than HsLC3 (Fig. S8F). These analyses suggest that human ATG8 homologs have 2 distinct sequences at the loop structure (Fig. S8F). However, yeast and plant ATG8 homologs have a similar loop structure. The difference in the loop structure could be one possible reason for lack of cleavage of HsLC3 by yeast and plant ATG4s.
Endogenous plant ATG4 can process yeast ATG8 efficiently but not human LC3A
We further investigated if plant ATG4 could process yeast and human ATG8s using Agrobacterium-mediated expression in N. benthamiana plants. The yeast Atg8 synthetic substrate expressed in N. benthamiana under the control of the 35S promoter was efficiently processed under normal conditions and the cleavage product was detectable in the presence of concanamycin A (Conc A) that inhibits degradation of autophagic bodies in the vacuole (, lane 1). In contrast, the cleavage byproduct of HsLC3A was under the detection limit (, lane 2). These results suggest that the endogenous N. benthamiana ATG4s could efficiently process ScAtg8 but not HsLC3A under normal conditions. To test if expression of HsATG4B can process HsLC3A inside plant cells, we coexpressed HsLC3A and HsATG4B in N. benthamiana leaves. We observed efficient cleavage of HsLC3A when HsATG4B was coexpressed (, lane 3). Consistent with these results, Citrine-HsLC3A colocalized with HsATG4B compared to the Citrine control (Fig. S9). These results indicate that endogenous N. benthamiana ATG4s cannot process HsLC3A efficiently to the level detectable under normal conditions.
Figure 7. Nicotiana benthamiana plant ATG4 efficiently process yeast Atg8 but weakly process human LC3A. (A) C-ScAtg8-ShR (lane 1), C-HsLC3A-ShR (lane 2), and C-HsLC3A-ShR with HsATG4B (lane 3) were expressed in N. benthamiana plant leaves and cleavage byproduct (ShR) was detected with anti-ShR antibodies (top panel). The cleaved ShR byproduct was observed in C-ScAtg8-ShR-expressing tissue (lane 1) but not in C-HsLC3A-ShR (lane 2). Coexpression of C-HsLC3A-ShR with HsATG4B resulted in accumulation of the cleaved ShR byproduct (lane 3). Arrow and arrowhead indicate full-length ATG8 sensors and cleaved byproducts, respectively. Anti-MYC was used to detect the HsATG4B input (middle panel, lane 3). Anti-PEPC was used for input loading control (bottom panel). (B to D) C-ScAtg8-ShR sensor was processed by endogenous N. benthamiana ATG4s and the mature form of C-ScAtg8 was incorporated into autophagic bodies in the vacuole (B). Coexpression of C-HsLC3A-ShR with HsATG4B showed enhanced accumulation of autophagic bodies in the vacuole (D) compared to the expression of C-HsLC3A-ShR alone (C) in N. benthamiana leaves. Scale bar: 20 µm. (E) Quantification of autophagic bodies observed in (B to D). One-way ANOVA test indicates a statistically different number of autophagic bodies accumulated in the vacuole of cells expressing ScATG8 alone and HsLC3A with HsATG4B compared to HsLC3A alone. Lowercase letters indicate statistical differences (P < 0 .0001). (F) C-ScAtg8-ShR (lane 1), C-HsLC3A-ShR (lane 2), C-HsLC3A-ShR with HsATG4B (lane 3), and Citrine alone with HsATG4B (lane 4) were expressed in N. benthamiana plant leaves. After dark-induced autophagy, the isolated proteins were separated on SDS-PAGE and probed with anti-ShR antibodies (top panel). C-HsLC3A-ShR synthetic substrate (top panel, lane 2) could not be processed by endogenous N. benthamiana ATG4s compared to ScAtg8 (top panel, lane 1) in the dark-induced autophagy condition. The HsLC3A was efficiently processed when coexpressed with HsATG4B (top panel, lane 3). Anti-GFP antibody was used to detect free Citrine fluorescent protein and mature processed C-ATG8A or -LC3A (second panel). Anti-MYC was used to detect the HsATG4B input (third panel). Anti-PEPC was used for input loading control (bottom panel). Arrows and arrowheads indicate full-length synthetic sensors and cleaved byproducts, respectively. Asterisk represents free Citrine.
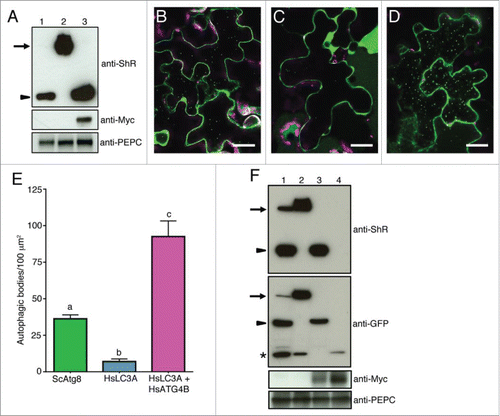
To determine if the ATG8 processing is altered under autophagy-induced condition, we tested the ATG8 cleavage under dark-induced autophagy. For this, detached leaves or entire plants were incubated in the dark for 12 h before imaging or harvesting tissue for immunoblot analyses. The yeast synthetic substrate C-ScAtg8-ShR in the presence of ConcA was processed by N. benthamiana endogenous ATG4s and accumulated as puncta representing autophagic bodies in the vacuole (). In contrast, we observed very few autophagic bodies in the vacuole of cells expressing human synthetic substrate C-HsLC3A-ShR (). However, when C-HsLC3A-ShR was coexpressed with HsATG4B in N. benthamiana plants, large number of autophagic bodies accumulated in the vacuole (). Therefore, even under autophagy-induced conditions, the endogenous N. benthamiana ATG4 could not efficiently process HsLC3A.
Although we failed to detect the processed mature form of HsLC3A in our immunoblot analyses (, lane 2 and , top panel, lane 2), few autophagic puncta were visible in the vacuole of HsLC3A-expressing cells (). It is possible that plant ATG4 may cleave HsLC3A weakly and the processed mature HsLC3A is under the detection level of our immunoblot assay. In GFP-AtATG8A transgenic plants, free GFP has been shown to accumulate under nutrient starvation conditions.Citation48 It has been hypothesized that this free GFP is a stable breakdown product of GFP-AtATG8A in the vacuole.Citation48 Therefore, we probed the blot with an anti-GFP antibody. Free Citrine was observed in protein isolated from both C-ScAtgG8-ShR and C-HsLC3A-ShR-expressing cells (, middle panel; lanes 1 and 2). However, we failed to detect the processed mature form of HsLC3A compared to ScAtg8 (; middle panel; compare lanes 1 and 2). These results suggest that a low level of HsLC3A could be processed in plant cells and targeted to the vacuole ( and S9) but the mature form of HsLC3A is under the detection level in the immunoblot assay. Consistent with these observations, coexpression of HsATG4B in N. benthamiana efficiently processed C-HsLC3A-ShR synthetic substrate () and the mature HsLC3A form is readily detectable (). Together these results indicate that endogenous N. benthamiana plant ATG4 efficiently processes ScAtg8 but weakly processes HsLC3A whose active catalysis requires HsATG4 in a heterologous expression system.
Discussion
The ubiquitin-like ATG8 plays an important role in autophagosome initiation and biogenesis.Citation10,49 Furthermore, ATG8 serves as a docking site for receptor proteins and also plays a key role in autophagic cargo recruitment into autophagosomes.Citation10,50 Emerging evidence also suggests that ATG8 plays an important role in selective autophagy.Citation50-52 The ATG4-mediated processing of ATG8 followed by lipidation is an important step in autophagosome initiation and maturation. ATG4 also plays a role in the recycling of ATG8 once the autophagosome fuses with the vacuole or lysosome. Currently, analyses of ATG4 and ATG8 genes are limited to some model plant species. Here, we implemented genome-wide analyses of ATG4 and ATG8 from 18 published and sequenced plant genomes including agriculturally important crop plants such as tomato, potato, rice, maize, cassava, and other plants. Our results identified a total of 28 ATG4s and 116 ATG8s from the available plant genome sequences providing most updated analyses of ATG4 and ATG8 genes from different plant species. We found an additional ATG8 in soybean genome based on BLAST and HMM search compared to 11 GmATG8s that were reported previously.Citation37 In contrast, we have removed one ATG8 ortholog from 5 previously reported ATG8s from the rice genome.Citation32
Our analyses indicate that at least one ATG4 and ATG8 gene is present in all analyzed plant genomes including lower plant lineages such as S. moellendorffii, P. patens, and C. reinhardtii suggesting that the autophagic pathway is conserved among all plant lineages. However, the number of gene family members differs in various plants. Gene expansion of ATG8 has occurred in ancestral land plant lineage. These varying numbers of ATG4 and ATG8 genes within flowering plants might be attributed to ancient polyploidy events and additional recent lineage-specific WGDs (discussed below). Paralogs by duplication suggests both subfunctionalization and neofunctionalization. Consistent with this, the Arabidopsis ATG4A and ATG4B showed different affinity to ATG8s.Citation30 In addition, Arabidopsis ATG8 gene family members display subtle differences in their expression patterns indicating tissue-specific roles.Citation16 The gene structures of ATG4 and ATG8 are retained well in plant genomes, suggesting that they are one of the most conserved proteins in eukaryotes.Citation53 The amino acid sequence of different ATG8s is also highly conserved indicating that they may play important roles in autophagy process across species. In addition, as mentioned above, we found identical genes within and between species representing newly duplicated paralogs and orthologs, respectively.
Phylogenetic analyses presented here provide insights into the evolution of ATG4s and ATG8s. In the case of ATG8, most of them were divided into 3 subgroups according to A. thaliana ATG8s (a-d, e-g, and h-i). Our results suggest that the expansion of ATG8 genes occurred in the common ancestor of angiosperms. Although these groups underwent further expansion in plant lineages, the e-g group was lost in monocot plants. In addition, we also found that pepper and maize lost the h-i group while Medicago lost the a-d group of ATG8 subfamily members. We found in silico evidence for the ATG8 h-i group in pepper, but this gene was ruled out based on the pepper annotation pipeline.Citation54
Duplication events are one of the most important mechanisms for the evolution of life. There are different types of duplications, and these duplications might explain the copy number variation of ATG4 and ATG8 in plants.Citation42 Our result suggests that segmental and dispersed duplications were primary influence in the ATG4 and ATG8 families' expansion in higher plants. For example, the recent lineage-specific WGD events led to the specific expansion observed in B. rapa and soybean. In addition, genome rearrangement, gene loss, and transposon might affect dispersed duplication. On the other hand, tandem duplication was observed in P. patens, which has been rarely found in higher plants. These mechanisms of duplication may lead to genetic and functional redundancies. In fact, some duplicated pairs of ATG8s showed similar expression patterns in their organs. However, other pairs showed different expression patterns indicating changed expression context by other factors affecting their functions. Together, ATG4 and ATG8 genes might be retained or lost in a biased manner caused by their mode of duplication.
Sequence and phylogenetic analyses revealed that sequences of ATG8s in yeast, Arabidopsis, and tomato are similar while a divergent sequence of HsLC3A placed HsLC3A into the outgroup in the phylogenetic tree. It appears that the distinct sequence feature of HsLC3 causes a structural difference in a local region despite conservation of overall protein folding of ATG8 orthologs (Fig. S8F). This could be one of the reasons that HsLC3A can bind to yeast and plant ATG4s but not processed (). According to the cocrystal structure of HsLC3 and a catalytic mutant of HsATG4B in which the catalytic Cys74 is changed to Ser, the Arg68 residue in HsLC3 form a salt bridge with HsATG4B.Citation46 In addition, Phe80 and Leu82 residues of HsLC3 form a hydrophobic interaction with HsATG4B. In our computational analyses, the Arg68 and Phe80 residues of HsLC3A are conserved in AtATG8a as the residues of Arg66 and Phe78. However, the Leu82 residue of HsLC3A is changed to Phe80 in AtATG8a. This substitution may not affect the hydrophobic contact between HsLC3 and AtATG4a. Therefore, the binding interface appears to be similar between HsATG4B and AtATG4a, as well as with HsLC3A. Consistent with the computational analyses, our affinity isolation results showed that AtATG4a directly binds to HsLC3A. The lack of processing could be due to a distinct loop structure that is present in our model between ATG8 homologs and HsLC3 (Fig. S8F). Interestingly, this loop structure of HsLC3 is quite different compared to HsGABARAP that is more similar to yeast and plant ATG8s (Fig. S8F). It is possible that the different loop structures of ATG8 orthologs may result in repositioning of the conserved Gly in the active site of the cysteine proteases and hence yeast and plant ATG4s cannot cleave HsLC3A but they can bind HsLC3A. The broad catalytic activity of HsATG4B could be that it has been evolved to recognize and process 2 types of human ATG8 orthologs such as the LC3 family as well as GABARAP, GABARAPL1 and GABARAPL2. Therefore, ATG4-mediated processing may be determined by ATG8 protein sequence rather than by ATG4s. In addition, cleavage compatibility of ScAtg8 by plant ATG4s may result from high sequence and structural similarity as well as evolutionary closeness to plant ATG8s ().
In contrast, evolutionally distinct features of HsLC3A appear to affect a kinetic rate by yeast and plant ATG4s, resulting in loss and/or inefficient cleavage activity by plant ATG4s and yeast Atg4 (). The observed lack of processing of HsLC3A by endogenous yeast and plant ATG4s is not due to the LC3A synthetic sensor structure because the sensor gets cleaved efficiently upon coexpression with HsATG4B in plant cells. Nonetheless, heterologous expression of HsLC3A in plant cells showed weak formation of autophagic bodies in the vacuole even though we failed to detect the mature processed form of HsLC3A in immunoblot assay ().
In conclusion, our analyses indicate conservation of ATG4s and ATG8s from 18 different plant genomes but expansion of the numbers of ATG4 and ATG8 genes from single cell green algae to crop plants such as rice, maize, Nicotiana, tomato, soybean, and others. Interestingly, yeast and plant ATG4 protease activity is highly conserved and can process ATG8s from either species but not from human. Furthermore, the human ATG4B has a broader substrate kinetic property that can process both yeast and plant ATG8s. The results described here should provide a basis for understanding the unique and redundant functions of ATG4 and ATG8 orthologs in model and crop plants.
Materials and methods
Identification of ATG4 and ATG8 in plant genomes
Sequences of ATG4 and ATG8 in yeast, human, and A. thaliana were obtained from NCBI. Amino acids and nucleotide sequences of annotated gene model within 18 plant genomes were retrieved from Phytozome (www.phytozome.net) portal that contains genome sequences of various plant species, SGN (solgenomics.net) portal contains genome sequences of various Solanaceae sp, and pepper genome database (peppergenome.snu.ac.kr) (Table S1). To identify homologs of ATG4 and ATG8 in the plant genomes, both BLAST and Hidden Markov model (HMM) searches were performed. Possible homolog candidates were searched using BLASTP with the amino acid sequences of A. thaliana ATG4s and ATG8s (with e-value cutoff 1.0e−04). The matched proteins were used for HMM search using Pfam domain (PF03416 for ATG4 and PF02991 for ATG8). Genes having incomplete domain were removed. In case of alternative splicing, representative gene was selected. For the analysis of exon-intron structure and location, general feature format (gff) files of 18 plant genomes were downloaded from Phytozome or from each genome sequencing consortium. To depict the genomic structures of genes, Exon-Intron Graphic Maker was used (http://wormweb.org/exonintron).
Phylogenetic analysis and data visualization
ATG4 and ATG8 domain regions were parsed and used for alignment. Multiple alignments of amino acid sequences were performed by clustalW2 with default options, and the resulting amino acid sequence alignments were used to guide the alignments of the nucleotide coding sequences using translatorX.Citation55 Then, the alignments were corrected manually and used for phylogenetic analyses. Phylogenetic trees of ATG4s and ATG8s were constructed using maximum likelihood method using MEGA 6.0.Citation56 The K2+G+I model was used to establish the best tree and a total of 1000 rapid bootstrap replicates were performed. To visualize the conserved sequences, multiple alignments of amino acid sequences were implemented and gaps were removed. The alignments were used to create sequence logos using WEBLOGO.Citation57
Duplication analysis of ATG4s and ATG8s
Duplicated pairs of genes were predicted and Ka and Ks value was calculated using McScanX.Citation43 For this analysis, all-by-all BLASTP hits, gff information and cds sequences from each species were used. To compare the expression patterns of duplicated pairs, absolute expression values of ATG4s and ATG8s were obtained from The Bio-Analytic Resource for Plant Biology (http://bar.utoronto.ca/welcome.htm) and they were shown as heatmap using R package.
Monitoring ATG4 activity in living yeast cells
Open reading frames (ORFs) of AtATG8a, AtATG8aG117A, and HsLC3A were inserted into the FAS-DDΔ-TA plasmid.Citation45 The plasmids were introduced into yeast strains EGY48 or EGY48 Scatg4−/−. For the introduction of AtATG4a, AtATG4b, and ScATG4 into the EGY48 Scatg4Δ yeast strain, the ATG4s were cloned into JG4-5 or pGADT7 vector backbone (Takara Bio Inc., 630442) in which the nuclear localization sequence and activation domain were removed. The transformed yeast cells were grown on the media plates with X-gal (Takara Bio Inc., 9031; Madison, USA) to detect expression of the LacZ reporter gene.
Plasmid construction and purification of ATG4s and ATG8 synthetic substrates
All plasmid construction was performed as previously described.Citation30 In brief, ORFs of ScATG4 (NM_001183061.1), HsATG4B (XM_005574890.1), AtATG4a (AT2G44140), and SlATG4 (Solyc01g006230) were amplified and cloned into pMal-C2 expression vector (NEW ENGLAND BIOLABS, current version of this vector is pMal-c5X, N8108), resulting in maltose binding protein (MBP)-ATG4. For synthetic substrates, ScATG8 (NM_001178318.1), HsLC3A (XM_005568770.1), AtATG8a (AT4G21980), and SlATG8a (Solyc07g064680) ORFs were amplified and cloned into BamHI (NEW ENGLAND BIOLABS, R0136S) and SalI (NEW ENGLAND BIOLABS, R0138S)-restricted pET28-Citrine-ShR plasmid.Citation30 Inserts in all plasmids were confirmed by sequencing. Primer sequences used for cloning are available upon request. Plasmids of (HIS)6-C-ATG8-ShR synthetic substrates and MBP-ATG4 were transformed into E. coli strain BL21 (DE3). Purification of all recombinant proteins, western blot analyses, and in vitro cleavage assay were performed as described.Citation30 To generate plant expression plasmids, inserts were amplified from bacterial expression vector and cloned into pYL400 containing 2x35S promoter and a NOS terminator. For transient expression in N. benthamiana, Agrobacterium strain GV2260 harboring the plant expression constructs were used as described previously.Citation58
In vitro affinity isolation assay
One μg of purified MBP-tagged ATG4s was precleared with 25 μL of amylose resin (NEW ENGLAND BIOLABS, E8021L) in 1 mL of affinity isolation buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.5% Triton X-100 [SIGMA-ALDRICH, X-100-100ML], 35 μM β-mercaptoethanol (SIGMA-ALDRICH, M6250-100ML) for 1 h. For affinity isolation, 1 μg of C-HsLC3A-ShR or C-HsLC3AG120A-ShR was added and incubated for 1 h at room temperature with gentle shaking. Fresh 25 μL of amylose resin was incubated for collection of the affinity isolation products for 1 h. Collected amylose resin was washed with the affinity isolation buffer for 3 times and the affinity isolation products were analyzed by immunoblotting with anti-MBP (SIGMA-ALDRICH, SAB4200082; St Louis, USA) or anti-GFP antibody (Santa Cruz Biotechnology, sc-5384).
Structure-based model generation
A 3D structural model of AtATG8a was generated using the RosettaCM protocol.Citation59 Templates were identified using the HMMER3 online server,Citation60 and the 10 best matches based on E-value were used as templates for molecular modeling (PDB: 2kwc, 2li5, 3vh3, 3rui, 3vxw, 2kq7, 1eo6, 4co7, 1gnu, 2r2q ). Alignments were generated using Promals3D,Citation61fragments sets were generatedCitation62 and 3D evolutionary constraints were generated using the templates.Citation63 RosettaCM was used to generate 1000 decoys using the threaded templates as input. The lowest energy model was selected to represent the AtATG8a structure. A 3D structural model of AtATG4a was generated by following similar approach as for the AtATG8a. Several different templates were found using HMMER3; however, previous workCitation46 suggests that the Cys74Ser catalytic mutant structure of HsATG4B (PDB 2zzp) is the most reliable model of the active site. Therefore, we used this template for generating a model. The lack of local sequence overlap in the template resulted in a number of loop regions failing to structurally converge during sampling. The AtATG4 homology model did converge at the active site region.
Colocalization and quantification of autophagosomes
Agrobacterium with corresponding ATG4 or ATG8 constructs were infiltrated onto 3- to 4-wk old N. benthamiana plant leaves and 2 d post-infiltration leaf pieces were used to obtain autophagosome images under confocal microscope as described.Citation30 To stabilize autophagic bodies in the vacuole, 1 µM concanamycin A (Santa Cruz Biotechnology, sc-202111A) was treated for 12 h before imaging. For colocalization, AtATG4a-tagCFP or HsATG4A-tagCFP were coinfiltrated with C-HsLC3A-ShR, or Citrine alone into N. benthamiana. Confocal images were obtained using Zeiss LSM710 confocal microscope equipped with a LDC-apochromat 40X/1.1W Korr M27 (N.A. 1. ) water immersion objective. Images were captured using 5% of 405 nm Diode laser excitation and 460 to 510 nm spectral detection for tagCFP (Evrogen, FP111) and 5% of 514 nm helium laser excitation and 519 to 568 nm spectral detection for Citrine. For quantification of the autophagosomes, images were captured only for the Citrine fluorescence. Autophagic bodies in the vacuole of cells were counted by cell counter plugin in ImageJ (National Institutes of Health, Bethesda, MD USA), followed by graphical visualization. Total area was over 3 mm2 from at least 9 independent images. Prism 6 (Graphpad Software Inc., La Jolla, CA USA) was used for statistical analyses.
Statistical analysis
Statistical analysis was performed to compare accumulation of autophagic bodies in using one-way ANOVA (GraphPad PRISM 6.0 software). All values were represented as average ± SEM. The observed P-value less than 0.05 were considered statistically significant.
Major reagents and antibodies used
Vectors: pGADT7 (Takara Bio USA, Inc., 630442), pMal-C2 (New England Biolabs, current version of this vector, pMal-c5X, N8108), and tagCFP (EVROGEN, FP111). Restriction enzymes: BamHI (New England Biolabs, R0136S) and SalI (New England Biolabs, R0138S). Chemicals and reagents: X-gal (Takara Bio USA, Inc., 9031), amylose resin (New England Biolabs, E8021L), Tris-HCl (Sigma-Aldrich, T5941-500G), NaCl (Sigma-Aldrich, S3014-1KG), Triton X-100 (Sigma-Aldrich, X-100- 100ML, St. Louis, MO USA), β-mercaptoethanol (SIGMA-ALDRICH, M6250-100ML), and Concanamycin A (Santa Cruz Biotechnology, sc-202111A). Antibodies: anti-MBP (SIGMA-ALDRICH, SAB4200082), anti-GFP antibody (Santa Cruz Biotechnology, sc-5384), anti-RLUC antibody (EMD Millipore). Softwares: ImageJ (National Institutes of Health, Bethesda, USA) and Prism 6 (Graphpad software Inc., La Jolla, USA).
Abbreviations
| ATG | = | autophagy |
| GABARAPL2/GATE-16 | = | GABA type A receptor associated protein like 2 |
| GABARAP | = | GABA type A receptor-associated protein |
| HMM | = | Hidden Markov model |
| MAP1LC3/LC3 | = | microtubule associated protein 1 light chain 3 |
| PCD | = | programmed cell death |
| PE | = | phosphatidylethanolamine |
| ShR | = | SuperhRLUC. |
| WGDs | = | whole genome duplications |
Disclosure of potential conflicts of interest
No potential conflicts of interest.
KAUP_A_1217373_supplementarymaterials.zip
Download Zip (17.9 MB)Acknowledgments
We thank Drs. Hideki Hayashi and John Reed for FAS-DDΔ-TA vector.
Funding
National Science Foundation Molecular and Cellular Biosciences-1355459 and 1549580 funds (to SPD-K) and National Research Foundation of the Korea Ministry of Education, Science and Technology, project number NRF-2015R1A2A1A01002327 (to ES and DC) supported this work.
References
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nature Cell Biol 2007; 9:1102-9; PMID:17909521; http://dx.doi.org/10.1038/ncb1007-1102
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280-93; PMID:20965422; http://dx.doi.org/10.1016/j.molcel.2010.09.023
- Hayward AP, Dinesh-Kumar SP. What can plant autophagy do for an innate immune response? Annu Rev Phytopathol 2011; 49:557-76; PMID:21370973; http://dx.doi.org/10.1146/annurev-phyto-072910-095333
- Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 2012; 17:526-37; PMID:22694835; http://dx.doi.org/10.1016/j.tplants.2012.05.006
- Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 2014; 157:65-75; PMID:24679527; http://dx.doi.org/10.1016/j.cell.2014.02.049
- Yang X, Bassham DC. New Insight into the Mechanism and Function of Autophagy in Plant Cells. Int Rev Cell Mol Biol 2015; 320:1-40; PMID:26614870; http://dx.doi.org/10.1016/bs.ircmb.2015.07.005
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C. Autophagy, plant senescence, and nutrient recycling. J Exp Bot 2014; 65:3799-811; PMID:24687977; http://dx.doi.org/10.1093/jxb/eru039
- Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, et al. Autophagy contributes to leaf starch degradation. Plant Cell 2013; 25:1383-99; PMID:23564204; http://dx.doi.org/10.1105/tpc.112.108993
- Feng YC, He D, Yao ZY, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/10.1038/cr.2013.168
- Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nature Struct Mol Biol 2014; 21:336-45; PMID:24699082; http://dx.doi.org/10.1038/nsmb.2787
- Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 2013; 194:341-61; PMID:23733851; http://dx.doi.org/10.1534/genetics.112.149013
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007; 130:165-78; PMID:17632063; http://dx.doi.org/10.1016/j.cell.2007.05.021
- Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nature Struct Mol Biol 2013; 20:59-66; PMID:23202584; http://dx.doi.org/10.1038/nsmb.2431
- Ketelaar T, Voss C, Dimmock SA, Thumm M, Hussey PJ. Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett 2004; 567:302-6; PMID:15178341; http://dx.doi.org/10.1016/j.febslet.2004.04.088
- Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 2011; 12:226; PMID:21867568; http://dx.doi.org/10.1186/gb-2011-12-7-226
- Slavikova S, Shy G, Yao Y, Glozman R, Levanony H, Pietrokovski S, Elazar Z, Galili G. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot 2005; 56:2839-49; PMID:16157655; http://dx.doi.org/10.1093/jxb/eri276
- Slobodkin MR, Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem 2013; 55:51-64; PMID:24070471; http://dx.doi.org/10.1042/bse0550051
- Weidberg H, Shpilka T, Shvets E, Elazar Z. Mammalian Atg8s: one is simply not enough. Autophagy 2010; 6:808-9; PMID:20581472; http://dx.doi.org/10.4161/auto.6.6.12579
- Wu F, Watanabe Y, Guo XY, Qi X, Wang P, Zhao HY, Wang Z, Fujioka Y, Zhang H, Ren JQ, et al. Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy. Mol Cell 2015; 60:914-29; PMID:26687600; http://dx.doi.org/10.1016/j.molcel.2015.11.019
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004; 16:2967-83; PMID:15494556; http://dx.doi.org/10.1105/tpc.104.025395
- Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem 2003; 278:3671-8; PMID:12446702; http://dx.doi.org/10.1074/jbc.M208247200
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 2008; 15:344-57; PMID:18804433; http://dx.doi.org/10.1016/j.devcel.2008.08.012
- Cann GM, Guignabert C, Ying L, Deshpande N, Bekker JM, Wang L, Zhou B, Rabinovitch M. Developmental expression of LC3alpha and β: absence of fibronectin or autophagy phenotype in LC3beta knockout mice. Dev Dyn 2008; 237:187-95; PMID:18069693; http://dx.doi.org/10.1002/dvdy.21392
- Michaeli S, Honig A, Levanony H, Peled-Zehavi H, Galili G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 2014; 26:4084-101; PMID:25281689; http://dx.doi.org/10.1105/tpc.114.129999
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 2010; 29:1792-802; PMID:20418806; http://dx.doi.org/10.1038/emboj.2010.74
- Manil-Segalen M, Lefebvre C, Jenzer C, Trichet M, Boulogne C, Satiat-Jeunemaitre B, Legouis R. The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev Cell 2014; 28:43-55; PMID:24374177; http://dx.doi.org/10.1016/j.devcel.2013.11.022
- Lin L, Yang PG, Huang XX, Zhang H, Lu Q, Zhang H. The scaffold protein EPG-7 links cargo receptor complexes with the autophagic assembly machinery. J Cell Biol 2013; 201:113-29; PMID:23530068; http://dx.doi.org/10.1083/jcb.201209098
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 2002; 277:33105-14; PMID:12070171; http://dx.doi.org/10.1074/jbc.M204630200
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 2002; 129:1181-93; PMID:12114572; http://dx.doi.org/10.1104/pp.011024
- Woo J, Park E, Dinesh-Kumar SP. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci USA 2014; 111:863-8; PMID:24379391; http://dx.doi.org/10.1073/pnas.1318207111
- Park E, Woo J, Dinesh-Kumar SP. Arabidopsis ATG4 cysteine proteases specificity toward ATG8 substrates. Autophagy 2014; 10:926-7; PMID:24658121; http://dx.doi.org/10.4161/auto.28280
- Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 2009; 149:220-34; PMID:18790996; http://dx.doi.org/10.1104/pp.108.126714
- Kuzuoglu-Ozturk D, Cebeci Yalcinkaya O, Akpinar BA, Mitou G, Korkmaz G, Gozuacik D, Budak H. Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta 2012; 236:1081-92; PMID:22569921; http://dx.doi.org/10.1007/s00425-012-1657-3
- Pei D, Zhang W, Sun H, Wei X, Yue J, Wang H. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep 2014; 33:1697-710; PMID:24996626; http://dx.doi.org/10.1007/s00299-014-1648-x
- Perez-Perez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 2010; 152:1874-88; PMID:20107021; http://dx.doi.org/10.1104/pp.109.152520
- Shibuya K, Niki T, Ichimura K. Pollination induces autophagy in petunia petals via ethylene. J Exp Bot 2013; 64:1111-20; PMID:23349142; http://dx.doi.org/10.1093/jxb/ers395
- Xia T, Xiao D, Liu D, Chai W, Gong Q, Wang NN. Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS One 2012; 7:e37217; PMID:22629371; http://dx.doi.org/10.1371/journal.pone.0037217
- Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nuc Acids Res 2012; 40:D1178-D86; PMID:22110026; http://dx.doi.org/10.1093/nar/gkr944
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003; 422:433-8; PMID:12660784; http://dx.doi.org/10.1038/nature01521
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science 2008; 320:486-8; PMID:18436778; http://dx.doi.org/10.1126/science.1153917
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 2009; 60:433-53; PMID:19575588; http://dx.doi.org/10.1146/annurev.arplant.043008.092122
- Wang Y, Wang X, Paterson AH. Genome and gene duplications and gene expression divergence: a view from plants. Ann N Y Acad Sci 2012; 1256:1-14; PMID:22257007; http://dx.doi.org/10.1111/j.1749-6632.2011.06384.x
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nuc Acids Res 2012; 40:e49; PMID:22217600; http://dx.doi.org/10.1093/nar/gkr1293
- Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res 2005; 15:516-25; PMID:15781573; http://dx.doi.org/10.1101/gr.3531105
- Hayashi H, Cuddy M, Shu VCW, Yip KW, Madiraju C, Diaz P, Matsuyama T, Kaibara M, Taniyama K, Vasile S, et al. Versatile assays for high throughput screening for activators or inhibitors of intracellular proteases and their cellular regulators. PLoS One 2009; 4: PMID:19876397; http://dx.doi.org/10.1371/journal.pone.0007655
- Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 2009; 28:1341-50; PMID:19322194; http://dx.doi.org/10.1038/emboj.2009.80
- Kumanomidou T, Mizushima T, Komatsu M, Suzuki A, Tanida I, Sou YS, Ueno T, Kominami E, Tanaka K, Yamane T. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J Mol Biol 2006; 355:612-8; PMID:16325851; http://dx.doi.org/10.1016/j.jmb.2005.11.018
- Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 2010; 62:483-93; PMID:20136727; http://dx.doi.org/10.1111/j.1365-313X.2010.04166.x
- Jin M, Klionsky DJ. Regulation of autophagy: modulation of the size and number of autophagosomes. FEBS Lett 2014; 588:2457-63; PMID:24928445; http://dx.doi.org/10.1016/j.febslet.2014.06.015
- Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 2014; 53:167-78; PMID:24462201; http://dx.doi.org/10.1016/j.molcel.2013.12.014
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell 2009; 34:259-69; PMID:19450525; http://dx.doi.org/10.1016/j.molcel.2009.04.026
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol Cell 2015; 58:1053-66; PMID:26004230; http://dx.doi.org/10.1016/j.molcel.2015.04.023
- Shemi A, Ben-Dor S, Vardi A. Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy 2015; 11:701-15; PMID:25915714; http://dx.doi.org/10.1080/15548627.2015.1034407
- Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, Seo E, Choi J, Cheong K, Kim KT, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nature Genet 2014; 46:270-8; PMID:24441736; http://dx.doi.org/10.1038/ng.2877
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nuc Acids Res 2010; 38:W7-13; PMID:20435676; http://dx.doi.org/10.1093/nar/gkq291
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/10.1093/molbev/mst197
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res 2004; 14:1188-90; PMID:15173120; http://dx.doi.org/10.1101/gr.849004
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 2008; 132:449-62; PMID:18267075; http://dx.doi.org/10.1016/j.cell.2007.12.031
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. High-resolution comparative modeling with RosettaCM. Structure 2013; 21:1735-42; PMID:24035711; http://dx.doi.org/10.1016/j.str.2013.08.005
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nuc Acids Res 2011; 39:W29-37; PMID:21593126; http://dx.doi.org/10.1093/nar/gkr367
- Pei J, Grishin NV. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol Biol 2014; 1079:263-71; PMID:24170408; http://dx.doi.org/10.1007/978-1-62703-646-7_17
- Gront D, Kulp DW, Vernon RM, Strauss CE, Baker D. Generalized fragment picking in Rosetta: design, protocols and applications. PLoS One 2011; 6:e23294; PMID:21887241; http://dx.doi.org/10.1371/journal.pone.0023294
- Thompson J, Baker D. Incorporation of evolutionary information into Rosetta comparative modeling. Proteins 2011; 79:2380-8; PMID:21638331; http://dx.doi.org/10.1002/prot.23046
