ABSTRACT
Macroautophagy/autophagy is a catabolic process that is essential for cellular homeostasis. Studies on autophagic degradation of cytoplasmic components have generated interest in nuclear autophagy. Although its mechanisms and roles have remained elusive, tremendous progress has been made toward understanding nuclear autophagy. Nuclear autophagy is evolutionarily conserved in eukaryotes that may target various nuclear components through a series of processes, including nuclear sensing, nuclear export, autophagic substrate encapsulation and autophagic degradation in the cytoplasm. However, the molecular processes and regulatory mechanisms involved in nuclear autophagy remain largely unknown. Numerous studies have highlighted the importance of nuclear autophagy in physiological and pathological processes such as cancer. This review focuses on current advances in nuclear autophagy and provides a summary of its research history and landmark discoveries to offer new perspectives.
Introduction
Autophagy is a catabolic process that is essential to maintaining cellular homeostasis in various eukaryotes, from yeasts to mammals. Autophagy plays important roles in differentiation, development, immunity and life span. Dysfunctional autophagic regulation has been associated with various human diseases, including cancer, neurodegenerative diseases, metabolic disorders and microbial infections.Citation1 Thus, understanding autophagic mechanisms has important physiological and therapeutic implications.
Autophagy is an intracellular degradation process that is utilized to recycle intracellular components by lysosomal/vacuolar hydrolysis when cells encounter stress, such as during nutrient starvation.Citation2 Autophagosomal formation initiates from a cup-shaped double-membrane structure, known as the phagophore. The phagophore gradually elongates and expands by recruiting proteins, such as the lipidated LC3B protein of the ATG family. This is followed by phagophore closure and the formation of a double-membrane autophagosome, which collects cytoplasmic components destined for degradation. The autophagosome undergoes progressive maturation by fusing with lysosomes to form autolysosomes. The captured macromolecular contents are ultimately degraded by lysosomal/vacuolar proteases within the acidic environment of the autolysosome/vacuole. The degraded monomeric molecules (e.g., amino acids) are exported to the cytosol for reuse by the cell. Autophagy's recycling role can “kill 2 birds with one stone”Citation3 by eliminating harmful or excessive cellular components while providing new materials for cellular and tissue remodeling. Thus, autophagy is a mechanism for cellular macromolecular recycling, architectural remodeling, growth regulation and self-protection against viral and bacterial infections and is a process that supports cell survival and cell death.
Since the discovery of autophagy in the 1950s, most studies in this field have focused on degradation of the cytoplasmic components. Autophagy typically involves the nonselective sequestration of cytoplasm or the targeted removal of undesirable cytoplasmic components. Selective autophagy is generally categorized as follows depending on the cargo: mitophagy (mitochondria),Citation4 pexophagy (peroxisomes),Citation5 reticulophagy (endoplasmic reticulum),Citation6 ribophagy (ribosomes),Citation7 lipophagy (lipid droplets),Citation8 aggrephagy (protein aggregates)Citation9 and xenophagy (invasive microbes).Citation10 However, the degradation of nuclear materials has been largely ignored. As an essential part of the cell, the nucleus encounters various physiological and pathological types of stress, such as in genotoxicity and oncogenesis. Selective nuclear autophagy (nucleophagy) has recently been observed in eukaryotic cells, and the discovery of nuclear autophagy has extended our understanding of the complex biological processes that occur within and between cells. This review focuses on current advances in nuclear autophagy and summarizes its research history and landmark discoveries to provide new perspectives for nuclear autophagy.
Discovery of nuclear autophagy in yeast
The cell nucleus is the central organelle of the eukaryotic cell. The primary roles of the nucleus are to maintain genomic integrity from generation to generation and to spatially and temporally control gene expression. To maintain nuclear homeostasis, cells require a strategy for eliminating undesirable nuclear components in response to stress. Autophagy is the ideal removal process because it can either selectively or nonselectively degrade nuclear components in bulk. Nuclear autophagy was initially documented in yeast in 2003.Citation11 Nucleus-vacuole (NV) junctions were observed to promote the sequestration of small pieces of the nucleus into vacuolar membrane invaginations for subsequent degradation, a process initially termed piecemeal microautophagy of the nucleus (PMN). Carbon and nitrogen starvation induces PMN through the target of rapamycin (TOR) kinase pathway. Core macroautophagy factors, such as Atg3, Atg5, Atg8, Atg12 and Atg16, are necessary autophagic components for effective PMN.Citation12 Furthermore, electrochemical potential and lipid-modifying enzymes are needed to form PMN vesicles.Citation13 In fact, similar to the NV in yeast, association between the lysosome and dissolution of the nuclear envelope, or germinal vesicle breakdown is observed during maturation of the mammalian oocyte.Citation14 However, any connection of the latter to nuclear autophagy remains unclear. In addition, late nucleophagy (LN), a process unlike PMN, has been proposed to occur in yeast. LN can be detected after prolonged periods of nitrogen starvation (20–24 h) using a dual-labeling approach involving an outer nuclear membrane reporter (Nvj1-EYFP) and a nucleoplasm reporter (NAB35-DsRed.T3).Citation15 LN can also occur in nvj1- and vac8-deficient cells, and it utilizes some of the core macroautophagic machinery. PMN requires the formation of nuclear-vacuole junction structures whereas LN does not, indicating a major mechanistic difference between PMN and LN. The molecular pathways that underlie PMN and LN remain to be explored.
Recently, 2 novel proteins, Atg39 and Atg40, have recently been identified in yeast.Citation16,17 Mochida and colleagues provide evidence for selective autophagy of the nucleus by showing that Atg39 localization to the perinuclear endoplasmic reticulum (ER)/nuclear envelope induces the autophagic sequestration of the nuclear envelope and some intranuclear components in response to nitrogen deprivation. Additionally, Atg39-regulated nuclear autophagy may participate in nuclear membrane reconstruction during mitosis or in nuclear membrane repair following damage.Citation18 These observations indicate that autophagy targets nuclear components to the vacuole for degradation. Autophagic recycling of these components maintains energy homeostasis and provides intranuclear quality control in response to stress (i.e., starvation and injury).
Nuclear autophagy is evolutionarily conserved in eukaryotes
Nuclear autophagy occurs in yeast and in numerous eukaryotes, including unicellular organisms (i.e., Tetrahymena thermophila) and mammals. Increasing evidence supports the notion that nuclear autophagy is evolutionarily conserved in eukaryotes.
Tetrahymena thermophila contains 2 functionally distinct nuclei—the polyploidy somatic macronucleus and the diploid germline micronucleus. During sexual reproduction, Tetrahymena undergoes a unique process of programmed nuclear death (PND) to degrade its parental macronucleus. Several studies indicate that PND is associated with autophagy. Alterations to the composition and distribution of the lipids and proteins within the nuclear membrane during PND may be responsible for the membrane's interaction with the lysosome.Citation19 Additionally, 2 ATG8 genes involved in PND have been identified.Citation20 Both ATG8 proteins are primarily targeted to the nuclear periphery of the degrading nucleus, and ATG8 knockout studies show a marked delay in nuclear degradation.Citation20 Further dissection of the PND-associated molecular pathways and the PND connection to autophagy will provide insights into nuclear autophagy in Tetrahymena.
Nuclear autophagy also occurs in numerous fungal species. Magnaporthe grisea causes rice blast, and appressorial morphogenesis in M. grisea involves nuclear degeneration. One study has shown that autophagic cell death of the fungal spore is responsible for its infectivity.Citation21 A genome-wide analysis of autophagy indicates that there are 23 ATG genes in Magnaporthe oryzae and 38 in Fusarium oxysporum that mediate autophagic initiation, nucleation, phagophore, expansion and recycling.Citation22,23 Deletion mutants of the F. oxysporum FoATG8 gene show that autophagy regulates the number of nuclei per hyphal compartment during vegetative growth.Citation23 In addition, autophagy-dependent degradation of the entire nucleus has been observed with EGFP-labeled HTB/histone H2B in the basal cells of Aspergillus oryzae.Citation24 These data provide new ideas for the development of antifungal technologies through manipulating nuclear autophagy.
Nuclear autophagy has been observed in the intestinal cells of aging C. elegans, implying that nuclear autophagy is associated with the pathophysiology of aging.Citation25 The autophagosomal nature of the 4-layered membrane structure has been observed in murine macrophages during HSV-1 infection.Citation26 Further immunoelectron microscopy for LC3B, an autophagosome marker, confirms that the autophagosome originates from the nuclear envelope.Citation26,27 Autophagosomal/autolysosomal features have been observed in the perinuclear vacuolar structures in laminopathies that are caused by LMN/lamin and EMD/emerin mutations.Citation28 Based on these observations, the term nucleophagy is proposed to describe nuclear degradation-associated autophagic degradation.Citation28 Recent studies have highlighted the importance of nuclear autophagy in physiological and pathological processes such as cancer.Citation29-34 Nevertheless, the underlying molecular mechanisms remain to be explored.
Types of nuclear autophagy
How autophagic substrates in the nucleus are sequestered and transferred into the cytoplasm for autophagic degradation remains unknown. Although numerous forms of nuclear autophagy have been observed, current data indicate that nuclear autophagy can be mainly divided into 4 primary types: micronucleophagy, macronucleophagy, gigantic nuclear macroautophagy and unconventional nucleophagy.
Piecemeal microautophagy of the nucleus in yeast is a typical form of micronucleophagy ().Citation12 Components of the autophagic machinery, such as Atg11, Atg15, Atg17 and Atg18, are necessary for effective PMN. PMN undergoes a series of morphologically distinct changes. The interaction between Nvj1 in the outer nuclear membrane and Vac8 in the vacuolar membrane produces contact nucleus-vacuole (NV) junctions, which cause small pieces of the nucleus to be sequestered into vacuolar membrane invaginations for subsequent degradation. Vac8 interacts with Nvj1, which then recruits at least 2 proteins, Tsc13 and Osh1, to the NV junctions to form the autophagosomal membrane.Citation35-38 It is worth investigating whether PMN occurs in other eukaryotic organisms.
Figure 1. Schematic models for nuclear autophagy. (A) In yeast, there are 2 types of nuclear autophagy. i) PMN undergoes a series of morphologically distinct changes, such as nucleus-vacuole contact, nuclear bulges into invaginations of the vacuole, and isolation of PMN vesicles. Vac8 interacts with Nvj1, which then recruits at least 2 additional proteins, Tsc13 and Osh1, to the NV junctions. Figure adapted from Citation12. Nuclear components are finally degraded in the vacuolar lumen. ii) Atg39-mediated selective macroautophagy degrades the partial nucleus in yeast.Citation16,17 Atg39 binds to Atg8 on forming autophagosomal membranes. Partial nuclear materials are encapsulated within an autophagosome. (B) In mammals, there are also 2 types of nuclear autophagy. i) Nuclear materials to be degraded will be encapsulated through a process similar to exocytosis. The encapsulated nuclear materials will go through the process of autophagic degradation. LMNB1 can interact with LC3B, which probably induces the nuclear autophagosome formation.Citation30 ii) Micronuclear autophagy. Genotoxicity can result in the generation of some micronuclei in affected cells, which contain damaged chromosome fragments. Micronuclei can be sequestered and degraded by autophagy.Citation40 γH2AFX is a sensitive molecular marker of DNA damage and repair. (C) Tetrahymena undergoes a unique process of programmed nuclear death for degradation of its parental macronucleus through nuclear autophagy.Citation19 Alteration of lipid and protein composition and distribution in the nuclear membrane in PND is probably responsible for its interaction with the lysosome. The digestive vesicles fuse with the nuclear membrane and release their contents into the nucleus for final degradation. (D) Autophagy-mediated degradation of the entire nucleus in basal cells in fungi.Citation24 The entire nucleus is taken up into vacuoles through a ring-like autophagosomal precursor to encircle apparently the entire nucleus under starvation conditions. AoAtg8 probably participates in the nucleus-vacuole formation.
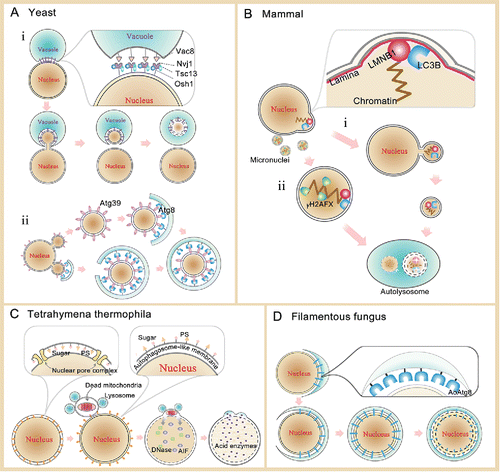
Macronucleophagy is a major and common type of nuclear autophagy (i.e., nucleophagy) that is primarily observed in mammalian cells. A generation model for the nuclear autophagosome is proposed here (). The nuclear materials destined for degradation are encapsulated by the membrane structure. The nuclear autophagosomal generation process may be similar to the exocytosis process that utilizes the nuclear membrane. Alternatively, a sequestering membrane for encapsulating the nuclear materials may form near the nuclear membrane. Because the active form of the LC3B autophagy protein, LC3B-II, is present in the nucleus/nuclear membrane, LC3B-II participates in the formation of the nuclear autophagosome. Macronucleophagy has also been detected in yeast (). For example, Atg39-mediated selective macroautophagy can degrade the partial nucleus in yeast.Citation16,17 Atg39 binds to Atg8 on the developing autophagosomal membranes, allowing partial nuclear materials to be encapsulated into the autophagosome.
Hagen and colleagues have recently presented a mechanism for vesicular formation at the inner nuclear membrane through docking, budding and nuclear egress complex formation in herpesvirus-infected cells.Citation39 This model may apply to nuclear autophagosome formation because it provides a mechanistic basis for the nucleo-cytoplasmic transport of large cargoes. Nevertheless, future investigations into these models and further dissections of the molecular processes involved in nuclear autophagosome formation are needed. LC3B antibody-based immunoelectron microscopy would be an appropriate approach to test these models in various cell types.
Micronuclear autophagy is another type of nuclear macroautophagy in mammalian cells (). Genotoxicity often promotes the generation of extranuclear bodies, known as micronuclei, in affected cells. These micronuclei contain damaged chromosomal fragments that are enriched with molecular indicators of DNA damage and repair, such as γH2AFX. Micronuclei can fuse with the lysosome to be degraded by autophagy.Citation40 Both the envelope and chromatin are autophagic substances. Micronuclear autophagy is probably an important mechanism in the maintenance of genomic stability.
Gigantic nuclear macroautophagy is a special type of nuclear autophagy in Tetrahymena thermophila (). During sexual reproduction, Tetrahymena undergoes a unique process of programmed nuclear death for degradation of its parental macronucleus through nuclear autophagy.Citation19 The overall composition and distribution of the lipids and proteins (e.g., sugars and phosphatidylserine) in the nuclear membrane of a cell undergoing PND is different from that of a cell in the absence of PND. This PND-associated molecular alteration facilitates the nuclear membrane interaction with the lysosome. The digestive vesicles subsequently fuse with the nuclear envelope to release their contents (i.e., acidic enzymes) into the nucleus. In this way, the parental macronucleus undergoes macroautophagy-mediated degradation. The autophagic degradation of the entire nucleus has also been detected in fungal basal cells.Citation24 The entire nucleus is taken up into a vacuole through a ring-like autophagosomal precursor to encirclement (). It is likely that Aspergillus oryzae (Ao)Atg8 participates in autophagosomal formation. Moreover, AoAtg15 is necessary for the degradation of autophagic substances. Fungi use extracellular nutrients to support growth, making the recycling of basal cell components by autophagic degradation important when environmental nutrients are scarce. Autophagy of the entire nucleus has not been observed in more complex eukaryotes, such as mammals, suggesting that this autophagic process may be limited to lower eukaryotes, such as fungi and Tetrahymena.
Other types of unconventional nucleophagy are certain to exist. For example, when cells undergo a prolonged period of starvation, the autophagic process and its machinery would likely be different from both macronucleophagy and micronucleophagy. The LN observed in yeast may be an unconventional nucleophagic process because LN occurs in the absence of Nvj1 and Vac8 and does not require Vps30/Atg6 or Atg11. However, functional Atg8 and Atg10 proteins are essential for efficient LN.Citation15 In addition to pursuing the multiple nuclear autophagy types, questions pertaining to the molecularly distinct processes and mechanisms of the various types of nuclear autophagy remain to be addressed.
Roles of nuclear membrane in nuclear autophagy
What components are eliminated from the nucleus by autophagy? Several studies show that chromatin and the nuclear membrane are nuclear autophagic targets. Nuclear components protrude from the nucleus during nuclear autophagy, and the micronuclei or tiny nuclei subsequently form in the cytoplasm, adjacent to the nuclear membrane, for degradation. These micronuclei can be detected easily using DNA and/or histone markers. To detect their autophagic degradation, the autophagy machinery (i.e., the LC3B-II protein) should appear in the micronuclei, which then gradually mature to become autophagosomes (possibly by fusing with de novo autophagosomes). These nuclear component-containing autophagosomes ultimately fuse with the lysosome to form the autolysosome. Increasing evidence shows that autophagy targets nuclei in mammalian cells.Citation29,33,40-45 Nuclear autophagy is induced by mutations in genes that encode nuclear membrane proteins, such as LMNs/lamins and EMD/emerin,Citation28 indicating that the nuclear membrane plays a key role in nuclear autophagy.
Evidence of the autophagic targeting of micronuclear membranes is based on their pre-autophagosomal nature. Nuclear membrane proteins, chromatin and the LC3B protein have been detected in mammalian cell micronuclei under various stress conditions, such as oncogene-induced senescence and starvation.Citation26,29,41 An accumulation of LMNA/lamin A and LMNB1/lamin B1 has also been observed in corresponding regions of nuclear degradation in differentiating keratinocytes.Citation45 A recent study shows that autophagy degrades nuclear LMNB1 to drive cellular senescence in response to oncogenic stress, which is mediated by the LC3B interaction with LMNB1.Citation30 Nuclear membrane degradation may drive senescence and suppress tumorigenesis. Thus, nuclear autophagy acts as a guarding mechanism that protects cells from tumorigenesis. Multiple studies show a close association between the nuclear and sequestering membrane.Citation26,27,32,46,47 Together, these data indicate that the nuclear membrane may contribute to the autophagosomal membrane in addition to being an autophagic target. Future studies will characterize the molecular features of the nuclear autophagosomal membrane and the underlying molecular mechanisms that drive autophagic targeting of the nuclear membrane.
Regulation of nuclear autophagy
Although the molecular mechanisms of nuclear autophagic regulation remain largely unknown, important steps have been taken toward highlighting the interface between the nucleus and the autophagic machinery. First, transcription regulation plays an important role in autophagy processes. Many transcription factors regulate spatiotemporal expressions of autophagy-related genes. Of particular importance is autophagy regulation of development. For example, MYBL2 regulates the spatiotemporal expression of VDAC2, an autophagy suppressor, in the developing ovary.Citation48 Nevertheless, transcription regulation of genes related to nuclear autophagy remains to be characterized. Second, LC3B/Atg8 is a key regulator of autophagic substrate delivery and membrane trafficking. Thus, identifying LC3B interacting partners will provide insight into the regulation of nuclear autophagy.
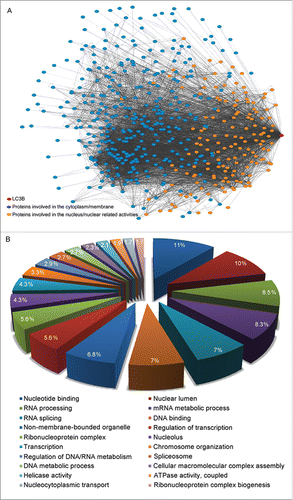
Behrends and colleagues have discovered numerous autophagy-associated interacting proteins in human 293T cells and provided an autophagy interaction network for basal autophagic conditionsCitation49 that serves as a comprehensive resource for the analysis of autophagic regulation. According to interaction data from Behrends et al. and biological general repository for interaction data sets (BioGRID; thebiogrid.org), there are approximately 500 candidate interacting proteins for LC3B. Interestingly, one-third of the interacting proteins are associated with nuclear components and pathways, adding a level of complexity to the mechanisms of nuclear autophagic regulation (). A gene ontology-based analysis of the LC3B-interacting proteins indicates that nuclear autophagy is involved in several pathways, including nucleotide binding, nuclear lumen activities, RNA processing, the mRNA metabolic process and RNA splicing (). These analyses suggest that nuclear autophagy plays an important role in the post-transcriptional RNA modification process in addition to maintaining nuclear DNA homeostasis.
Figure 2. Network map of LC3B and its interacting-proteins with GO categories. (A) There are approximately 500 candidate interacting proteins for LC3B. One-third of the interacting proteins are associated with nuclear components and pathways. The interaction data are from Behrends and colleagues,Citation49 and BioGRID (thebiogrid.org). The program of Cytoscape (version 3.2.1)Citation70 is used to visualize and analyze the interaction network. Gene relationships are obtained from BioGrid (version 3.2)Citation71 and input into Cytoscape with undirected connection. Finally, the network is displayed using force-directed layout algorithms. Red dot, LC3B; orange dots, proteins involved in the nucleus and nuclear related activities; blue dots, other interacting proteins. (B) GO categories of LC3B interacting-proteins involved in the nucleus and nuclear related activities. The subsets of gene ontology terms for LC3B and interacting-proteins are analyzed according to the DAVID program.Citation72 Top 20 categories with interacting-protein numbers are listed.
LC3B localizes to both the cytoplasm and the nucleus.Citation29,50 Moreover, nuclear- and nuclear membrane-associated LC3B proteins seem to be primarily in the LC3B-II form. Additionally, the nucleocytoplasmic transport and activation of LC3B is dependent on its starvation-associated acetylation/deacetylation activities.Citation50 These results indicate that nuclear autophagy is a complicated process that involves a broad biological context within the nucleus. Future studies will focus on nuclear autophagic regulation with a special emphasis on RNA modification and regulation. Further studies should also focus on how LC3B-II participates in nuclear autophagy. The underlying mechanisms of nuclear autophagy will be revealed by dissecting the various processes involved in the nuclear activities associated with LC3B-II and by categorizing the nuclear components (including the large molecules) involved in nuclear autophagy.
In addition to LC3B and its partners, a group of nuclear autophagy-associated genes have been identified in fungi, yeast, tetrahymena and mammals (). Although nuclear autophagic regulation remains poorly understood, there are different subgroups of nuclear autophagy-associated genes and various types of nuclear autophagy, indicating the existence of distinct species- and type-specific mechanisms that underlie nuclear autophagy. Nevertheless, specific regulatory similarities do exist. First, LC3B is a key player in nuclear autophagy, having been detected in vacuoles associated with multiple nuclear autophagy types and shown to target the nuclear membrane. Second, some of the core autophagic machinery may be shared in nuclear autophagy, including proteins involved in forming the phagophore, such as ATG3, ATG5, BECN1, ATG7, ATG10, ATG12 and ATG16L1. Although they have not been identified in every species, these proteins comprise key ubiquitin-like conjugation systems that are involved in macroautophagy.
Table 1. Nucleophagy-related genes identified in diverse species
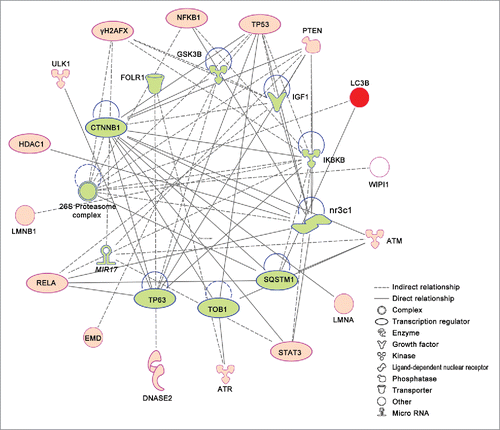
To investigate regulatory relationships among autophagy proteins, we used causal network analysis and software Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com). Upstream regulators can be connected to autophagy proteins based on a set of direct or indirect interaction relationships. Our IPA-based evaluation of mammalian nuclear autophagic pathways shows that most nuclear autophagic proteins can be organized into a single network (). For example, CTNNB1 regulates nuclear autophagic proteins ATM, LMNA, PTEN, STAT3 and TP53, and MIR17 may regulate H2AFX, PTEN, STAT3 and ATM during nuclear autophagy. This network plays potential roles in tissue morphology, DNA replication, recombination and repair, and gene expression, in addition to cell death. Further dissections of these pathways will uncover the regulatory mechanisms of nuclear autophagy.
Figure 3. Ingenuity pathway analysis (IPA) networks of nuclear autophagy proteins in mammals. Nuclear autophagy proteins are represented on the outer circle in red. Predicted regulators are represented on the inner circle filled in green. Different shape denotes the nature of the molecules. A logic network that involves more than one link is determined using causal network analysis. The network is implemented and available within the datasets of software Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com).
How does the nucleus sense autophagic signaling?
The mechanism that promotes nuclear sensing and autophagic initiation is unknown. After experiencing cellular stress (i.e., oncogenic stress or pathological induction), cells respond by repairing DNA damage that would otherwise trigger cell death. Nucleic sensors recognize self DNA and RNA for autophagic degradation. The ATM and ATR pathways are well-known sensors of double-stranded breaks.Citation56 A connection among the ATM/ATR pathway, DNA repair and autophagy has been established.Citation55,57 ATM phosphorylates PTEN and regulates its translocation into the nucleus. PTEN induces autophagy together with the JUN-SESN2-AMPK pathway in response to DNA damage.Citation57 There are also numerous stress-responsive transcription factors that regulate autophagy, including TP53, STAT3 and NFKB.Citation58 Multiple TP53 target genes are responsible for regulating autophagy including MTOR. STAT3 regulates transcription of several autophagy genes and most of them inhibit autophagy. NFKB and autophagy are intricately interconnected via a complex network of transcription-dependent signals.Citation58 In addition, nucleases, such as DNASE2/DNASE2A, can clear cells of excess nucleic acids through TMEM173/STING-dependent DNA-sensing, and prevent cells from accumulating self DNA via autophagic degradation.Citation42 These data show that the nucleus senses and triggers autophagy through a DNA-binding process. However, crosstalk between autophagy and RNA processes has remained unexplored. In addition to answering how the nucleus senses and initiates autophagy, future studies will need to address the contribution of RNA to nuclear autophagy.
Diseases relevant to nuclear autophagy
Autophagic impairment is implicated in multiple clinical ailments, including neurodegenerative diseases,Citation59 cancersCitation60 and aging.Citation61 Nevertheless, a few of the diseases have been reported to be involved in nuclear autophagy (). In cancers, the interaction between 2 key proteins, LC3B and LMNB1, mediates nuclear lamina degradation in response to tumorigenic stress.Citation30 Thus nuclear autophagy may protect cells from tumorigenesis. In fact, integrity of the nuclear membrane is closely connected to aging. LMNA and EMD mutations cause laminopathies and emerinopathies respectively, and are associated with nuclear autophagy.Citation28 EMD deficiency causes cardiac dysfunctions with nuclear-associated vacuoles and a structural fragility of myonuclei.Citation62 Thus, these genes are associated with relevant diseases involving muscle dysfunction, aging and cancer, potentially through nuclear autophagy. One report shows that nuclear autophagy is constitutively active in the epidermal granular layer, and a lack of nucleophagy in psoriatic skin disease is associated with parakeratosis.Citation45 To efficiently discover more nuclear autophagic genes and identify their functions and associated pathways, it will be necessary to develop high-throughput screening strategies, one of which would involve CRISPR-Cas9-mediated gene editing. Future studies will be necessary to develop novel therapeutics to modulate nuclear autophagy in human diseases. For example, small regulatory molecules, such as MIR17, may hold clinical promise for relevant diseases. In addition, nuclear autophagy probably plays a role in senescence. Intestinal nuclei loss was observed in old worms, and long-lived DAF-2 worms have been previously shown to be associated with autophagy,Citation25,63 indicating a potential role of DAF-2 in nuclear autophagy.
Table 2. Diseases associated with nuclear autophagy
Future perspectives
To date, most autophagy studies have focused on the degradation of cytoplasmic components, but recent progress has been made toward understanding nuclear autophagy. Nuclear autophagy is a mechanism by which cells can maintain cellular homeostasis and ensure nuclear integrity, stability, and correctness of gene expression. Thus, proper nuclear autophagy is beneficial to cellular health in response to various types of stress. Additionally, autophagy induces cellular senescence in response to oncogenic stress by degrading the nuclear lamina. Autophagy can also promote degradation of the entire nucleus to ultimately kill cells, and autophagic degradation is a mechanism used by fungi to recycle basal cell components. However, the existence of nuclear autophagy pathways that trigger an excessive nuclear degradation remain to be identified.
Future studies will address autophagic molecular targets in the nucleus and their molecular processes. How autophagic substrates in the nucleus are sequestered and transferred into the cytoplasm for autophagic degradation remains to be studied. Interestingly, RNA autophagy will be a promising direction in this field. The molecular and regulatory mechanisms of nuclear autophagy are poorly understood. Future studies that address the physiological and pathological implications of nuclear autophagy will help elucidate the pathogenesis of multiple human diseases and will promote the development of effective therapeutic strategies that involve manipulating nuclear autophagy.
Abbreviations
| AIF | = | apoptosis inducing factor, mitochondria associated |
| AoAtg | = | Aspergillus oryzae Atg |
| ATG | = | autophagy associated |
| ATM | = | ATM serine/threonine kinase |
| ATR | = | ATR serine/threonine kinase |
| BioGRID | = | biological general repository for interaction datasets |
| Cas9 | = | CRISPR associated protein 9 |
| CRISPR | = | clustered regularly interspaced short palindromic repeats |
| CTNNB1 | = | catenin β 1 |
| DAVID | = | the database for annotation, visualization and integrated discovery |
| DNASE2 | = | deoxyribonuclease II, lysosomal |
| EGFP | = | enhanced green fluorescent protein |
| EMD | = | emerin |
| ER | = | endoplasmic reticulum |
| EYFP | = | enhanced yellow fluorescent protein |
| FoAtg | = | Fusarium oxysporum Atg |
| FOLR1 | = | folate receptor 1 |
| γH2AFX | = | H2A histone family member X phosphorylated on serine 139 |
| GO | = | gene ontology |
| GSK3B | = | glycogen synthase kinase 3 β |
| HDACs | = | histone deacetylase |
| HSV-1 | = | herpes simplex virus 1 |
| Hsv2 | = | homolog with Svp1 |
| IGF1 | = | insulin like growth factor 1 |
| IKBKB | = | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase β |
| IPA | = | ingenuity pathway analysis |
| LC3B | = | microtubule associated protein 1 light chain 3 β |
| LMNA | = | lamin A/C |
| LMNB1 | = | lamin B1 |
| LN | = | late nucleophagy |
| MIR17 | = | microRNA 17 |
| MoAtg | = | Magnaporthe oryzae Atg |
| NFKB | = | nuclear factor kappa B |
| NR3C1 | = | nuclear receptor subfamily 3 group C member 1 |
| NV | = | nucleus-vacuole |
| PMN | = | piecemeal microautophagy of the nucleus |
| PND | = | programmed nuclear death |
| PTEN | = | phosphatase and tensin homolog |
| RPS19 | = | ribosomal protein S19 |
| SQSTM1 | = | sequestosome 1 |
| STAT3 | = | signal transducer and activator of transcription 3 |
| TOB1 | = | transducer of ERBB2, 1 |
| TP53 | = | tumor protein p53 |
| TP63 | = | tumor protein p63 |
| ULK1 | = | unc-51 like autophagy activating kinase 1 |
| WIPI1 | = | WD repeat domain, phosphoinositide interacting 1. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Natural Science Foundation of China, National Key Technologies R&D Program, Hubei Science & Tech Project and the Chinese 111 Project Grant B06018.
References
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814-22; PMID:20811353; http://dx.doi.org/10.1038/ncb0910-814
- Stromhaug PE, Klionsky DJ. Approaching the molecular mechanism of autophagy. Traffic 2001; 2:524-31; PMID:11489210; http://dx.doi.org/10.1034/j.1600-0854.2001.20802.x
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728-41; PMID:22078875; http://dx.doi.org/10.1016/j.cell.2011.10.026
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011; 12:9-14; PMID:21179058; http://dx.doi.org/10.1038/nrm3028
- Scott SV, Klionsky DJ. Delivery of proteins and organelles to the vacuole from the cytoplasm. Curr Opin Cell Biol 1998; 10:523-9; PMID:9719874; http://dx.doi.org/10.1016/S0955-0674(98)80068-9
- Cebollero E, Reggiori F, Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol 2012; 2012:182834; PMID:22481944; http://dx.doi.org/10.1155/2012/182834
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 2008; 10:602-10; PMID:18391941; http://dx.doi.org/10.1038/ncb1723
- Weidberg H, Shvets E, Elazar Z. Lipophagy: selective catabolism designed for lipids. Dev Cell 2009; 16:628-30; PMID:19460339; http://dx.doi.org/10.1016/j.devcel.2009.05.001
- Klionsky DJ, Cuervo AM, Dunn WA, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy 2007; 3:413-6; PMID:17568180; http://dx.doi.org/10.4161/auto.4377
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005; 120:159-62; PMID:15680321
- Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell 2003; 14:129-41; PMID:12529432; http://dx.doi.org/10.1091/mbc.E02-08-0483
- Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen EL, Millen J, Goldfarb DS, Thumm M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell 2008; 19:4492-505; PMID:18701704; http://dx.doi.org/10.1091/mbc.E08-04-0363
- Dawaliby R, Mayer A. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions. Mol Biol Cell 2010; 21:4173-83; PMID:20943953; http://dx.doi.org/10.1091/mbc.E09-09-0782
- Ezzell RM, Szego CM. Luteinizing hormone-accelerated redistribution of lysosome-like organelles preceding dissolution of the nuclear envelope in rat oocytes maturing in vitro. J Cell Biol 1979; 82:264-77; PMID:573271; http://dx.doi.org/10.1083/jcb.82.1.264
- Mijaljica D, Prescott M, Devenish RJ. A late form of nucleophagy in Saccharomyces cerevisiae. PLoS One 2012; 7:e40013; PMID:22768199; http://dx.doi.org/10.1371/journal.pone.0040013
- Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015; 522:359-62; PMID:26040717; http://dx.doi.org/10.1038/nature14506
- Nakatogawa H, Mochida K. Reticulophagy and nucleophagy: New findings and unsolved issues. Autophagy 2015; 11:2377-8; PMID:26566146; http://dx.doi.org/10.1080/15548627.2015.1106665
- Rubinsztein DC. Cell biology: Receptors for selective recycling. Nature 2015; 522:291-2; PMID:26040721; http://dx.doi.org/10.1038/nature14532
- Akematsu T, Pearlman RE, Endoh H. Gigantic macroautophagy in programmed nuclear death of Tetrahymena thermophila. Autophagy 2010; 6:901-11; PMID:20798592; http://dx.doi.org/10.4161/auto.6.7.13287
- Liu ML, Yao MC. Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila. Eukaryot Cell 2012; 11:494-506; PMID:22366125; http://dx.doi.org/10.1128/EC.05296-11
- Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006; 312:580-3; PMID:16645096; http://dx.doi.org/10.1126/science.1124550
- Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci U S A 2009; 106:15967-72; PMID:19717456; http://dx.doi.org/10.1073/pnas.0901477106
- Corral-Ramos C, Roca MG, Di Pietro A, Roncero MI, Ruiz-Roldan C. Autophagy contributes to regulation of nuclear dynamics during vegetative growth and hyphal fusion in Fusarium oxysporum. Autophagy 2015; 11:131-44; PMID:25560310; http://dx.doi.org/10.4161/15548627.2014.994413
- Shoji JY, Kikuma T, Arioka M, Kitamoto K. Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PLoS One 2010; 5:e15650; PMID:21187926
- McGee MD, Weber D, Day N, Vitelli C, Crippen D, Herndon LA, Hall DH, Melov S. Loss of intestinal nuclei and intestinal integrity in aging C. elegans. Aging Cell 2011; 10:699-710; PMID:21501374; http://dx.doi.org/10.1111/j.1474-9726.2011.00713.x
- English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 2009; 10:480-7; PMID:19305394; http://dx.doi.org/10.1038/ni.1720
- English L, Chemali M, Desjardins M. Nuclear membrane-derived autophagy, a novel process that participates in the presentation of endogenous viral antigens during HSV-1 infection. Autophagy 2009; 5:1026-9; PMID:19556870; http://dx.doi.org/10.4161/auto.5.7.9163
- Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, Nishino I. Autophagic degradation of nuclear components in mammalian cells. Autophagy 2009; 5:795-804; PMID:19550147; http://dx.doi.org/10.4161/auto.8901
- Chen K, Huang C, Yuan J, Cheng H, Zhou R. Long-term artificial selection reveals a role of TCTP in autophagy in mammalian cells. Mol Biol Evol 2014; 31:2194-211; PMID:24890374; http://dx.doi.org/10.1093/molbev/msu181
- Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al. Autophagy mediates degradation of nuclear lamina. Nature 2015; 527:105-9; PMID:26524528; http://dx.doi.org/10.1038/nature15548
- Strzyz P. Autophagy: Nuclear autophagy in tumour suppression. Nat Rev Mol Cell Biol 2015; 16:700-1; PMID:26530389; http://dx.doi.org/10.1038/nrm4091
- Lenain C, Gusyatiner O, Douma S, van den Broek B, Peeper DS. Autophagy-mediated degradation of nuclear envelope proteins during oncogene-induced senescence. Carcinogenesis 2015; 36:1263-74; PMID:26354777; http://dx.doi.org/10.1093/carcin/bgv124
- Sica V, Galluzzi L, Bravo-San Pedro JM, Izzo V, Maiuri MC, Kroemer G. Organelle-specific initiation of autophagy. Mol Cell 2015; 59:522-39; PMID:26295960; http://dx.doi.org/10.1016/j.molcel.2015.07.021
- Mijaljica D, Devenish RJ. Nucleophagy at a glance. J Cell Sci 2013; 126:4325-30; PMID:24013549; http://dx.doi.org/10.1242/jcs.133090
- Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell 2000; 11:2445-57; PMID:10888680; http://dx.doi.org/10.1091/mbc.11.7.2445
- Kvam E, Goldfarb DS. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy 2007; 3:85-92; PMID:17204844; http://dx.doi.org/10.4161/auto.3586
- Kvam E, Goldfarb DS. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J Cell Sci 2004; 117:4959-68; PMID:15367582; http://dx.doi.org/10.1242/jcs.01372
- Kvam E, Gable K, Dunn TM, Goldfarb DS. Targeting of Tsc13p to nucleus-vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol Biol Cell 2005; 16:3987-98; PMID:15958487; http://dx.doi.org/10.1091/mbc.E05-04-0290
- Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, Bauerlein FJ, et al. Structural basis of vesicle formation at the inner nuclear membrane. Cell 2015; 163:1692-701; PMID:26687357; http://dx.doi.org/10.1016/j.cell.2015.11.029
- Rello-Varona S, Lissa D, Shen S, Niso-Santano M, Senovilla L, Marino G, Vitale I, Jemaa M, Harper F, Pierron G, et al. Autophagic removal of micronuclei. Cell Cycle 2012; 11:170-6; PMID:22185757; http://dx.doi.org/10.4161/cc.11.1.18564
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. Lysosome-mediated processing of chromatin in senescence. J Cell Biol 2013; 202:129-43; PMID:23816621; http://dx.doi.org/10.1083/jcb.201212110
- Lan YY, Londono D, Bouley R, Rooney MS, Hacohen N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep 2014; 9:180-92; PMID:25284779; http://dx.doi.org/10.1016/j.celrep.2014.08.074
- Erenpreisa J, Huna A, Salmina K, Jackson TR, Cragg MS. Macroautophagy-aided elimination of chromatin: sorting of waste, sorting of fate? Autophagy 2012; 8:1877-81; PMID:22935563; http://dx.doi.org/10.4161/auto.21610
- Kung HJ, Changou CA, Li CF, Ann DK. Chromatophagy: autophagy goes nuclear and captures broken chromatin during arginine-starvation. Autophagy 2015; 11:419-21; PMID:25650867; http://dx.doi.org/10.1080/15548627.2015.1009789
- Akinduro O, Sully K, Patel A, Robinson DJ, Chikh A, McPhail G, Braun KM, Philpott MP, Harwood CA, Byrne C, et al. Constitutive autophagy and nucleophagy during epidermal differentiation. J Invest Dermatol 2016; 136:1460-70; PMID:27021405; http://dx.doi.org/10.1016/j.jid.2016.03.016
- Deroyer C, Renert AF, Merville MP, Fillet M. New role for EMD (emerin), a key inner nuclear membrane protein, as an enhancer of autophagosome formation in the C16-ceramide autophagy pathway. Autophagy 2014; 10:1229-40; PMID:24819607; http://dx.doi.org/10.4161/auto.28777
- Mijaljica D, Prescott M, Devenish RJ. The intricacy of nuclear membrane dynamics during nucleophagy. Nucleus 2010; 1:213-23; PMID:21327066; http://dx.doi.org/10.4161/nucl.11738
- Yuan J, Zhang Y, Sheng Y, Fu X, Cheng H, Zhou R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy 2015; 11:1081-98; PMID:26060891; http://dx.doi.org/10.1080/15548627.2015.1040970
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/10.1038/nature09204
- Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell 2015; 57:456-66; PMID:25601754; http://dx.doi.org/10.1016/j.molcel.2014.12.013
- He M, Kershaw MJ, Soanes DM, Xia Y, Talbot NJ. Infection-associated nuclear degeneration in the rice blast fungus Magnaporthe oryzae requires non-selective macro-autophagy. PLoS One 2012; 7:e33270; PMID:22448240; http://dx.doi.org/10.1371/journal.pone.0033270
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol 2010; 190:965-73; PMID:20855502; http://dx.doi.org/10.1083/jcb.201002075
- Krick R, Henke S, Tolstrup J, Thumm M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy 2008; 4:896-910; PMID:18769150; http://dx.doi.org/10.4161/auto.6801
- Leger-Silvestre I, Caffrey JM, Dawaliby R, Alvarez-Arias DA, Gas N, Bertolone SJ, Gleizes PE, Ellis SR. Specific role for yeast homologs of the diamond blackfan anemia-associated rps19 protein in ribosome synthesis. J Biol Chem 2005; 280:38177-85; PMID:16159874; http://dx.doi.org/10.1074/jbc.M506916200
- Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 2011; 471:74-9; PMID:21368826; http://dx.doi.org/10.1038/nature09803
- Awasthi P, Foiani M, Kumar A. ATM and ATR signaling at a glance. J Cell Sci 2015; 128:4255-62; PMID:26567218; http://dx.doi.org/10.1242/jcs.169730
- Chen JH, Zhang P, Chen WD, Li DD, Wu XQ, Deng R, Jiao L, Li X, Ji J, Feng GK, et al. ATM-mediated PTEN phosphorylation promotes PTEN nuclear translocation and autophagy in response to DNA-damaging agents in cancer cells. Autophagy 2015; 11:239-52; PMID:25701194; http://dx.doi.org/10.1080/15548627.2015.1009767
- Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol 2013; 23:310-22; PMID:23726895; http://dx.doi.org/10.1016/j.semcancer.2013.05.008
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med 2013; 19:983-97; PMID:23921753; http://dx.doi.org/10.1038/nm.3232
- Hoare M, Young AR, Narita M. Autophagy in cancer: having your cake and eating it. Semin Cancer Biol 2011; 21:397-404; PMID:21945348
- Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, et al. Autophagy maintains stemness by preventing senescence. Nature 2016; 529:37-42; PMID:26738589; http://dx.doi.org/10.1038/nature16187
- Ozawa R, Hayashi YK, Ogawa M, Kurokawa R, Matsumoto H, Noguchi S, Nonaka I, Nishino I. Emerin-lacking mice show minimal motor and cardiac dysfunctions with nuclear-associated vacuoles. Am J Pathol 2006; 168:907-17; PMID:16507906; http://dx.doi.org/10.2353/ajpath.2006.050564
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003; 301:1387-91; PMID:12958363; http://dx.doi.org/10.1126/science.1087782
- Dou Z, Ivanov A, Adams PD, Berger SL. Mammalian autophagy degrades nuclear constituents in response to tumorigenic stress. Autophagy 2016; 12:1416-7; PMID:26654219
- Finnsson J, Sundblom J, Dahl N, Melberg A, Raininko R. LMNB1-related autosomal-dominant leukodystrophy: Clinical and radiological course. Ann Neurol 2015; 78:412-25; PMID:26053668; http://dx.doi.org/10.1002/ana.24452
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 2013; 27:1787-99; PMID:23934658; http://dx.doi.org/10.1101/gad.223834.113
- Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev 2011; 25:2579-93; PMID:22155925; http://dx.doi.org/10.1101/gad.179515.111
- Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci U S A 2000; 97:5214-9; PMID:10779560; http://dx.doi.org/10.1073/pnas.090525897
- Luzhna L, Kathiria P, Kovalchuk O. Micronuclei in genotoxicity assessment: from genetics to epigenetics and beyond. Front Genet 2013; 4:131; PMID:23874352; http://dx.doi.org/10.3389/fgene.2013.00131
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2007; 2:2366-82; PMID:17947979; http://dx.doi.org/10.1038/nprot.2007.324
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006; 34:D535-D9; PMID:16381927; http://dx.doi.org/10.1093/nar/gkj109
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/10.1038/nprot.2008.211
