ABSTRACT
Prion protein modulates many cellular functions including the secretion of trophic factors by astrocytes. Some of these factors are found in exosomes, which are formed within multivesicular bodies (MVBs) and secreted into the extracellular space to modulate cell-cell communication. The mechanisms underlying exosome biogenesis were not completely deciphered. Here, we demonstrate that primary cultures of astrocytes and fibroblasts from prnp-null mice secreted lower levels of exosomes than wild-type cells. Furthermore, prnp-null astrocytes exhibited reduced MVB formation and increased autophagosome formation. The reconstitution of PRNP expression at the cell membrane restored exosome secretion in PRNP-deficient astrocytes, whereas macroautophagy/autophagy inhibition via BECN1 depletion reestablished exosome release in these cells. Moreover, the PRNP octapeptide repeat domain was necessary to promote exosome secretion and to impair the formation of the CAV1-dependent ATG12–ATG5 cytoplasmic complex that drives autophagosome formation. Accordingly, higher levels of CAV1 were found in lipid raft domains instead of in the cytoplasm in prnp-null cells. Collectively, these findings demonstrate that PRNP supports CAV1-suppressed autophagy to protect MVBs from sequestration into phagophores, thus facilitating exosome secretion.
Introduction
PRNP (prion protein) is a glycoprotein found attached to the cell membrane through a glycosylphosphatidylinositol anchor.Citation1 Major conformational changes that convert PRNP to its abnormal form (PrPSc) are associated with the generation of prions, the infectious agents that cause transmissible spongiform encephalopathies.Citation2 The mechanisms associated with PrPSc in neurodegeneration underlying transmissible spongiform encephalopathies and the physiological functions of PRNP in the nervous system have been largely explored.Citation3,4 PRNP cellular functions are attributed to its interaction with a set of protein complexes such as LAMA1/laminin-1-GRM/metabotropic glutamate receptors and STIP1 (stress-induced phosphoprotein 1)-CHRNA7/α7 nicotinic acetylcholine receptors.Citation5-7 Some PRNP interactions take place in lipid raft microdomains such as those involving NCAM (neural cell adhesion molecule) and CAV1/caveolin-1, which couple PRNP to the tyrosine kinase FYN (Fyn proto-oncogene).Citation8,9 These complexes modulate neuroprotection against cellular and systemic insults,Citation10-12 participate in cell signaling cascades,Citation13 promote neuritogenesis,Citation14 regulate neuronal plasticity, excitability, memory formation, and consolidation,Citation15 and mediate synaptic vesicle transmission.Citation16
PRNP is abundantly expressed in neuronal cells, but has also been observed in non-neuronal cells.Citation17,18 In astrocytes, PRNP modulates survival and differentiation via the PRKA/protein kinase A and MAPK1/ERK2-MAPK3/ERK1 pathways.Citation19 Some studies have addressed the role of PRNP in neuron-astrocyte communication. For instance, glutamate uptake by astrocytes is dependent on PRNP expression, and may influence neuronal survival.Citation20 In conditioned media from astrocytes, STIP1 is found within MVB-derived extracellular vesicles (EVs) containing classical exosome markers.Citation21 STIP1-containing exosomes are responsible for PRNP-dependent neuronal survival. Accordingly, glioblastomas, which are tumors derived from astrocyte precursors, also secrete STIP1 whose interaction with PRNP at the cell surface modulates the in vitro and in vivo proliferation of these tumor cells.Citation22 Remarkably, conditioned media from prnp-null astrocytes promote a reduced neuronal survival compared to media obtained from wild-type astrocytes.Citation19 Nonetheless, the role of PRNP in the secretion of trophic components, particularly those sorted to exosomes, is still unknown.
EV secretion from cells represents a remarkable system for biomolecule transfer that serves as a general mode of cell-cell communication. EVs are classified into 3 main classes based on their biogenesis: exosomes, microvesicles, and apoptotic bodies.Citation23 In contrast to microvesicles, which are generated by budding from the plasma membrane, exosomes are derived from the endolysosomal pathway.Citation24 The biogenesis of exosomes begins by invagination and budding of the limiting membrane of late endosomes, generating vesicle-laden endosomes, or multivesicular bodies (MVBs).Citation25 The proteins of the ESCRT/Endosomal Sorting Complex Required for Transport are the best-described components contributing to MVB and intralumenal vesicle (ILV) biogenesis. The first complex, ESCRT-0, binds to the cargo on endosomes and with the help of the ESCRTs-I, -II, and –III, promotes cargo accumulation on the endosomal membrane. At the end of sorting, an AAA-type ATPase, VPS4A (vacuolar protein sorting 4 homolog A), disrupts the ESCRT complexes, and the cargo-containing membrane invaginates into the maturing endosome.Citation26,27
Different subpopulations of MVBs coexist simultaneously in cells. MVB-derived ILVs can be secreted as exosomes by MVB fusion to the plasma membrane or can serve as transporters for the delivery of proteins to the lysosome for degradation.Citation24,28 A specific degradation of damaged proteins and organelles occurs in autophagic-lysosomal compartments in a catabolic process called autophagy. This process is controlled by specialized set of gene products that promote the genesis of autophagosomes, a well-characterized structure of the autophagy pathway, substrate recruitment, lysosome–autophagosome fusion, and final degradation of autolysosome contents.Citation29 BECN1/Beclin 1 and ULK1 (unc-51 like kinase 1)-ULK2, are key autophagy-promoting factors that are activated by a complex phosphorylation cascade downstream of AMP-activated protein kinase (AMPK) and MTOR (mechanistic target of rapamycin [serine/threonine kinase]).Citation30 Remarkably, autophagosomes can fuse with endocytic structures such as MVBs.Citation31
prnp-null cells display increased autophagy,Citation32 and several lines of evidence indicate a close relationship between the autophagy pathway and exosome biogenesis and secretion.Citation33,34 Together with the fact that conditioned media from prnp-null astrocytes was only capable of weak neuroprotection,Citation35 these observations guided us to explore the role of PRNP in exosome secretion. Herein, we used primary cultures from astrocytes and fibroblasts as well as plasma from wild-type, prnp-null and PRNP-overexpressing mice to evaluate exosome secretion. PRNP reconstitution in prnp-null cells or PRNP knockdown in wild-type cells was also conducted to address our hypothesis. The analysis of endocytic pathways and specific PRNP partners made it possible to decipher the mechanism associated with the participation of PRNP in the regulation of exosome secretion.
Results
PRNP expression levels directly correlate with exosome secretion
To evaluate if the expression status of PRNP affects exosome secretion, we analyzed conditioned media (CM) samples from primary cultures of astrocytes () and fibroblasts () collected from wild-type, prnp-null, and PRNP-overexpressing mice. We previously demonstrated that EVs isolated from astrocyte CM typically express exosomal markers and exhibit classical exosome morphology and size as evidenced by electron microscopy and nanoparticle tracking analysis (NTA).Citation21 Additionally, our previous data showed that a dominant-negative variant of VPS4A, an AAA-ATPase essential for MVB biogenesis, reduces the release of EVs into astrocyte CM by 85%, indicating that a large fraction of these EVs are composed of exosomes.Citation21 Our present results pointed out that CM from prnp-null astrocytes derived from ZrchI (prnp0/0) or Npu (prnp−/−) mice contained reduced levels of exosomes compared to the respective controls (Prnp+/+, Prnpwt/wt)(). Conversely, exosome levels in the CM of astrocytes isolated from mice overexpressing PRNP (TG20) were considerably higher than those of wild-type CM ().
Figure 1. Exosome secretion correlates with PRNP expression. (A) Nanoparticle tracking analysis (NTA) measurements of exosome concentration in the CM of astrocytes isolated from Zrchl mice (Prnp+/+, prnp0/0, TG20) and Npu mice (Prnpwt/wt, prnp−/−). (B) NTA measurements of exosome concentration in the CM of primary MEFs isolated from Prnp+/+, prnp0/0, and TG20 mice. Western blot analysis of exosomes (Exo) for FLOT1 and TSG101 in astrocytes (C) and MEFs (D). Cell lysates (CL) were used as positive control. (E) Transmission electron microscopy images of exosomes derived from astrocytes and MEFs. Cup-shaped structures, with 150 nm average size were identified as being exosomes. (F) NTA measurements of exosome concentration in purified plasma from Prnp+/+, prnp0/0, and TG20 mice. Data shown represent the mean ± SD of at least 3 independent experiments. One-way ANOVA and Tukey's post hoc test were used to assess statistical significance. *P < 0 .05, **P < 0 .01, ***P < 0 .001.
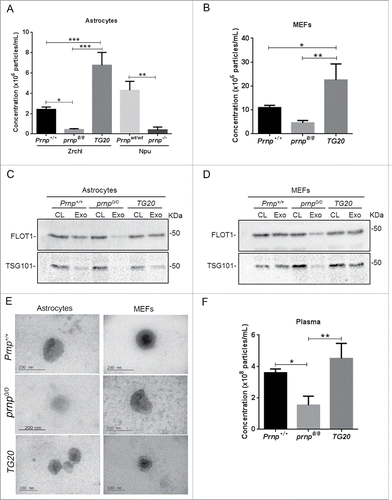
Exosomes in the CM of mouse embryonic fibroblasts (MEFs) isolated from Prnp+/+, prnp0/0, and TG20 mice were also analyzed. In agreement with the results obtained in astrocytes, prnp0/0 MEFs secreted fewer exosomes, whereas TG20 cells secreted greater levels of exosomes compared to Prnp+/+ MEFs (). Remarkably, MEFs secreted about 4 times more exosomes than astrocytes.
In order to confirm exosome isolation, we analyzed in our samples the presence of well-established exosome markers. FLOT1 (flotilin 1) and TSG101 (tumor susceptibility gene 101), proteins which are secreted associated with exosomes in several cell typesCitation25 were detected by western blot in exosomes derived from astrocytes () and fibroblasts (). However, as expected prnp0/0 astrocytes and fibroblasts showed lower levels of these exosomal markers due to a reduced number of exosomes in the conditioned media from these cells.
Furthermore, we also analyzed the morphology of isolated exosomes from Prnp+/+, prnp0/0 and TG20 astrocytes and MEFs by electron microscopy. The phase-contrast electron micrographs of the exosomes revealed rounded and double-membraned structures with a size of approximately 100–150 nm ().
Exosomes can also be detected in bodily fluids. Therefore, we measured circulating exosomes in plasma isolated from the mice described above. prnp0/0 animals show a significant decrease in the levels of circulating exosomes, whereas greater levels of exosomes were detected in the plasma of TG20 mice (). Taken together, our data indicate a correlation between PRNP expression and levels of secreted exosomes.
The NTA software identified and measured particles of the expected exosomal size, averaging 150 nm, and there were no significant differences in exosome size distribution between groups (Fig. S1).
To confirm that exosome secretion is associated with PRNP levels, we knocked down PRNP expression in Prnp+/+ astrocytes using small interfering RNA (siRNA) () or restored PRNP expression in prnp0/0 astrocytes (). The levels of exosomes released by PRNP-depleted cells were markedly reduced compared to control cells (), whereas PRNP re-expression almost completely rescued the secretion of exosomes to Prnp+/+ levels (). The expression of a PRNP mutant that is correctly inserted into the plasma membrane, but is unable to be endocytosed (*N-PrP-3F4),Citation36 also restored exosome secretion in prnp0/0 astrocytes (). Conversely, a PRNP mutant lacking the amino-terminal leader peptide resulting in its intracellular retention (EGFP-PRNP[Δ1–22]) failed to restore exosome secretion in prnp0/0 astrocytes (). Collectively, these results demonstrate that PRNP is required for regulating exosome secretion, and must be localized to the plasma membrane to properly perform this function.
Figure 2. Modulation of PRNP expression affects exosomal release. (A) Western blot analysis of PRNP protein expression in cell extracts isolated from Prnp+/+ astrocytes transfected with scrambled (Scr) or PRNP-specific siRNAs. TUB1A1 was used as a loading control. (B) Histogram shows densitometry analysis of PRNP expression relative to TUB1A1 expression. (C) NTA measurements of exosome concentration in the CM of Prnp+/+ astrocytes transfected with scrambled (Scr) or Prnp-specific siRNAs. (D) Western blot analysis of PRNP protein expression in cell extracts isolated from Prnp+/+ and prnp0/0 astrocytes transfected with full-length and truncation mutants of PRNP. pCDNA3 (empty vector control), PRNP-3F4 (full-length PRNP), *N-PRNP-3F4 (membrane localized, but internalization impaired), pEGFP-C1 (empty vector control), EGFP-PRNP(Δ1–22) (lacks membrane localization signal).TUB1A1 was used as a loading control. (E) NTA of exosome concentration in the CM of Prnp+/+ and prnp0/0 astrocytes transfected with full-length and truncation mutants of PRNP (described in [C]). Data shown represent the mean ± SD of at least 3 independent experiments. One-way ANOVA and Tukey's post hoc test were used to assess statistical significance. *P < 0.01, **P < 0 .001, ***P < 0 .0001.
![Figure 2. Modulation of PRNP expression affects exosomal release. (A) Western blot analysis of PRNP protein expression in cell extracts isolated from Prnp+/+ astrocytes transfected with scrambled (Scr) or PRNP-specific siRNAs. TUB1A1 was used as a loading control. (B) Histogram shows densitometry analysis of PRNP expression relative to TUB1A1 expression. (C) NTA measurements of exosome concentration in the CM of Prnp+/+ astrocytes transfected with scrambled (Scr) or Prnp-specific siRNAs. (D) Western blot analysis of PRNP protein expression in cell extracts isolated from Prnp+/+ and prnp0/0 astrocytes transfected with full-length and truncation mutants of PRNP. pCDNA3 (empty vector control), PRNP-3F4 (full-length PRNP), *N-PRNP-3F4 (membrane localized, but internalization impaired), pEGFP-C1 (empty vector control), EGFP-PRNP(Δ1–22) (lacks membrane localization signal).TUB1A1 was used as a loading control. (E) NTA of exosome concentration in the CM of Prnp+/+ and prnp0/0 astrocytes transfected with full-length and truncation mutants of PRNP (described in [C]). Data shown represent the mean ± SD of at least 3 independent experiments. One-way ANOVA and Tukey's post hoc test were used to assess statistical significance. *P < 0.01, **P < 0 .001, ***P < 0 .0001.](/cms/asset/33f78b9f-0381-4105-ba02-f3467035dad5/kaup_a_1226735_f0002_b.gif)
PRNP-deficient astrocytes lack MVB-containing ILVs, but accumulate larger lysosomes
In the endocytic pathway, as early endosomes transition into late endosomes, exosomes form by inward budding of the late endosome/MVB.Citation37,38 Therefore, we examined if PRNP levels affect the distribution of different endocytic compartments. We assessed the expression of RAB5, an early endosomal marker, RAB7, a late endosomal marker and RAB11A, a recycling endosome marker in astrocytes isolated from Prnp+/+ and prnp0/0 mice by immunofluorescence. RAB5 staining in early endosomes appeared punctate and diffuse throughout the cytoplasm (Fig. S2A), and RAB7 staining in late endosomes was more concentrated around the nucleus (Fig. S2B), whereas RAB11A appeared in a punctate pattern in the cell periphery (Fig. S2C). There was no alteration in the distribution of these endosomal compartments between Prnp+/+ and prnp0/0 astrocytes.
We further analyzed the distribution of the late endosomal phospholipid lysobisphosphatidic acid (LBPA), which is incorporated into the ILVs of MVBs.Citation39 Confocal imaging revealed that Prnp+/+ astrocytes exhibited LBPA staining in a punctate pattern, especially in perinuclear areas, suggesting that LBPA is abundant in ILVs and therefore labels mature MVBs in these cells. Conversely, poor staining was observed in prnp0/0 cells (). Transmission electron microscopy confirmed the presence of multivesicular structures with the characteristic MVB morphology in Prnp+/+ cells (). However, MVBs were not found in prnp0/0 cells.
Figure 3. PRNP regulates ILV abundance and lysosomal morphology. Immunofluorescence analysis of ILV marker lysobisphosphatidic acid (LBPA) within MVBs (A), lysosomal/late endosome marker LAMP1 (B) and lysosomal marker LAMP2 (C) of representative Prnp+/+ and prnp0/0 astrocytes, with nuclear staining (DRAQ5, blue). Histograms (right side) represent quantifications of fluorescent signal (mean ± SD). Scale bars: 30 μm. Electron microscopy analysis of ultrathin sections of representative Prnp+/+ (D) and prnp0/0 (E) cells. Red boxes in the left correspond to magnified right images. MVB containing exosomes inside are evident in Prnp+/+ cells. Phagophore (red arrowhead), double-membrane autophagosomes (yellow arrowhead) and late-autophagic compartments (green arrowhead) are evident in prnp0/0 cells. Scale bars: 10 μm left images, 1 μm (D) and 2 μm (E) right images. Data shown are from at least 3 independent experiments. Fluorescence levels for each marker were quantified using a single in-focus plane. A.U., arbitrary units. A Student t test was used to assess statistical significance. *P < 0 .001.
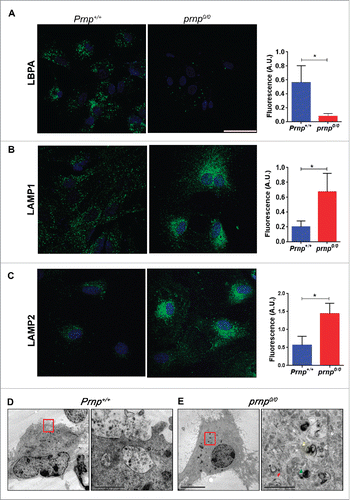
The fusion of the MVB with the plasma membrane allows the release of exosomes into the extracellular space, but alternatively, MVBs may fuse with the lysosome to facilitate degradation of their content.Citation40 Staining of Prnp+/+ cells with antibodies against LAMP1 (lysosomal-associated membrane protein 1), a lysosomal/late endosome marker and LAMP2, a specific lysosomal marker, showed that lysosomes were widely dispersed throughout the cell. Conversely, lysosomes from prnp0/0 cells were larger and aggregated in the perinuclear region. Additionally, there was a large increase in the amount of LAMP1 and LAMP2 in prnp0/0 cells (). These results suggest that PRNP decreases the late endosomal trafficking of ILV-containing MVBs toward lysosomes. It has been demonstrated that under conditions of serum deprivation, autophagy is activated in prnp0/0 hippocampal neurons compared to Prnp+/+ neurons.Citation41 Furthermore, lysosome activity during the course of autophagy requires the fusion between autophagosomes and lysosomes.Citation42 Transmission electron microscopy confirmed that a large number of autophagic structures (phagophores, autophagosomes, and late-autophagic compartments) were present in prnp0/0 astrocytes compared to Prnp+/+ cells ().
PRNP levels correlated with autophagy and exosome secretion
To further explore the role of PRNP in autophagy and its correlation with exosome secretion, we monitored autophagosome biogenesis under different cellular conditions in cells expressing different levels of PRNP and compared it with the levels of exosome secretion. Autophagosome biogenesis was evaluated by fluorescence microscopy () of MAP1LC3B (microtubule-associated protein 1 light chain 3), a protein involved in phagophore formation.Citation43 During autophagy, the cytosolic form of MAP1LC3B (MAP1LC3B-I) is conjugated to phosphatidylethanolamine to generate MAP1LC3B-II, which is then recruited to phagophore membranes.Citation44 The expression construct encoding EGFP-MAP1LC3B was transfected into cells to visualize MAP1LC3B without any additional labeling. The number of EGFP-MAP1LC3B-II puncta was quantified in Prnp+/+, prnp0/0 and TG20 astrocytes maintained in 10% fetal bovine serum (FBS), under serum-free conditions (starved), or after treatment with 100 nM rapamycin. Cellular extracts from cells treated with the same conditions were used to evaluate MAP1LC3-II expression by western blot ().
Figure 4. PRNP expression controls autophagy and exosome secretion. (A) Fluorescence microscopy analysis of EGFP-MAP1LC3B puncta (green) with nuclear staining (DRAQ5, blue) in representative Prnp+/+, prnp0/0 and TG20 cells transfected with a EGFP-MAP1LC3B expression vector and cultured under serum-replete (10% FBS), serum-free, or rapamycin-treated (100 nM) conditions. Scale bar: 10 μm. (B) Histogram represents the average number of EGFP-MAP1LC3B puncta/cell ± SD from 3 independent experiments. At least 100 cells were analyzed per condition. (C) Western blot analysis of MAP1LC3-II (14 kDa) expression in cell extracts isolated from Prnp+/+, Prnp0/0 and TG20 astrocytes grown in 10% FBS, serum-free (0% FBS), or rapamycin-treated (100 nM) conditions for 24 h. TUB1A1 was used as a loading control. (D) Histogram shows densitometry analysis of MAP1LC3-II expression relative to TUB1A1 expression. (E) NTA quantification of exosome concentration in the CM of Prnp+/+ and prnp0/0 astrocytes cultured in 10% FBS, serum-free or rapamycin-treated (100 nM) conditions. Data represent the mean ± SD from at least 3 independent experiments. Two-way ANOVA and Tukey's post hoc test were used to assess statistical significance. *P < 0 .05, **P < 0 .01, *** P < 0 .001.
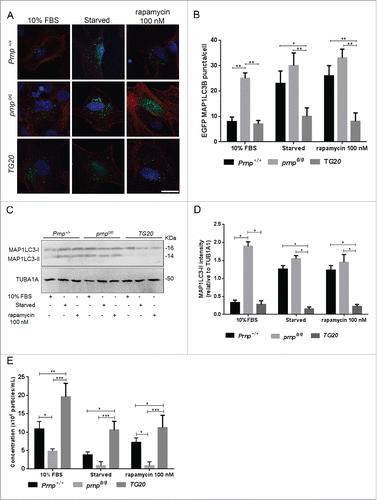
In normal culture conditions (10% FBS), prnp0/0 astrocytes showed higher levels of EGFP-MAP1LC3B puncta and MAP1LC3-II levels, whereas lower levels of these markers were observed in TG20 cells when compared to wild-type astrocytes (). Under serum-starved and rapamycin-treated conditions, Prnp+/+ astrocytes exhibited a large increase in EGFP-MAP1LC3B puncta () as well as higher expression of MAP1LC3-II (), which is indicative of autophagy induction. The number of EGFP- MAP1LC3B puncta () and MAP1LC3-II levels () did not change in prnp0/0 or TG20 cells after starvation and rapamycin treatments. In prnp0/0 cells autophagy levels were already high even in unstimulated conditions, suggesting that these cells already achieved a plateau in autophagy induction. Conversely, TG20 cells were completely resistant to stress-induced autophagy. These results highlight the fundamental role of PRNP in the induction of autophagy.
Remarkably, autophagy levels () in cells under normal growth or under stress conditions were always inversely correlated with exosome secretion (), confirming the existence of a PRNP-dependent autophagy-exosome axis. These results indicate that PRNP levels are inversely correlated with autophagosome formation and autophagy.
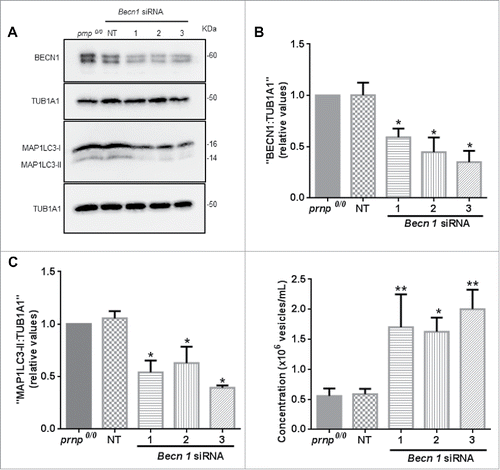
To confirm these observations, we depleted BECN1 in prnp0/0 cells to decrease the rate of autophagy. BECN1 participates in the regulation of autophagy and is required with its partner PIK3C3/VPS34 for the initiation of autophagosome formation.Citation45 We confirmed the knockdown of BECN1 in prnp0/0 astrocytes transfected with 3 different Becn1-specific siRNAs (). In parallel, we assessed the expression of MAP1LC3B-II, which correlates with the number of autophagosomes, in BECN1-depleted cells by western blot. MAP1LC3-II levels were decreased in BECN1-depleted astrocytes compared to nontransfected or control siRNA-transfected prnp0/0 cells (), confirming that reduced BECN1 expression impairs autophagy. Remarkably, BECN1-depleted cells exhibited increased levels of exosomes secreted into the CM compared to control prnp-null cells (). Taken together, these results indicate that PRNP is a negative modulator of autophagy, and that autophagy induction in prnp-null astrocytes disrupts exosome secretion.
Figure 5. PRNP is a negative regulator of autophagy. (A) Western blot analysis of BECN1 (60 kDa) and MAP1LC3-II (14 kDa) protein expression in cell extracts isolated from prnp0/0 cells transfected with Becn1-specific siRNA. NT, nontarget. TUB1A1 was used as a loading control. (B) Histogram shows densitometry analysis of BECN1 expression relative to TUB1A1//alpha;-tubulin expression. (C) Histogram shows densitometry analysis of MAP1LC3-II expression relative to TUB1A1 expression. (D) NTA of exosome concentration in the CM of Prnp0/0 astrocytes transfected with Becn1-specific siRNAs. Data information: (B-D) Data shown represent the mean ± SD of at least 3 independent experiments. One-way ANOVA and Tukey's post hoc test were used to assess statistical significance. *P < 0 .05, **P < 0 .01 when compared to prnp0/0 cells.
The octapeptide repeat domain of PRNP is required for regulating autophagy induction and exosome secretion
Recently, it was reported that the octapeptide repeat domain of PRNP may play a pivotal role in controlling autophagy in prnp-null neuronal cells.Citation41 To test if this region is also important for exosome secretion in astrocytes, prnp0/0 cells were transfected with constructs expressing GFP fused to full-length PRNP (PRNP-GFP), truncated PRNP mutants lacking the octapeptide repeat domain (GFP-PRNP[Δ51–90] and GFP-PRNP[Δ32–135]), and a truncated PRNP mutant lacking the hydrophobic domain (GFP-PRNP[Δ105–128]). The expression of each GFP-PRNP construct was confirmed by western blot (). Cells expressing full-length PRNP or PRNP(Δ105–128) showed a reduction in autophagy as measured by MAP1LC3A/B-II levels () and by the formation of the ATG12 (autophagy-related 12)–ATG5 complex (), which is involved in expansion of the phagophore, and is negatively regulated by CAV1.Citation46 Remarkably, restoration of full-length PRNP or PRNP lacking the hydrophobic domain rescued the ability of prnp0/0 cells to secrete exosomes (). Conversely, autophagy inhibition was not observed in cells expressing PRNP(Δ51–90) or PRNP(Δ32–135) (), and these mutants failed to rescue exosome secretion (). These results suggest that the octapeptide repeat domain of PRNP is important for attenuating autophagic flux and facilitating exosome secretion.
Figure 6. The octapeptide repeat domain of PRNP is necessary for regulating autophagy and exosome secretion. (A) Western blot analysis of PRNP, ATG12–ATG5, and MAP1LC3-II (14 kD) protein levels in cell extracts isolated from prnp0/0 cells transfected with full-length or truncation mutants of PRNP. pEGFP-C1 (empty vector control), PRNP(Δ51–90) and PRNP(Δ32–135) (lack the octapeptide repeat domain), PRNP(Δ105–128) (lacks hydrophobic domain). TUB1A1 was used as a loading control. (B) Histogram shows densitometry analysis of MAP1LC3-II expression relative to TUB1A1/α-tubulin expression. (C) Histogram shows densitometry analysis of ATG12–ATG5 expression relative to TUB1A1 expression. (D) NTA of exosome concentration in the CM of prnp0/0 cells transfected with full-length or truncation mutants of PRNP (described in [A]). Data information: (B-D) Data shown represent the mean ± SD from at least 3 independent experiments. One-way ANOVA and Tukey's post hoc test were used to assess statistical significance. *P < 0 .05, **P < 0 .01, ***P < 0 .001, #P < 0 .0001.
![Figure 6. The octapeptide repeat domain of PRNP is necessary for regulating autophagy and exosome secretion. (A) Western blot analysis of PRNP, ATG12–ATG5, and MAP1LC3-II (14 kD) protein levels in cell extracts isolated from prnp0/0 cells transfected with full-length or truncation mutants of PRNP. pEGFP-C1 (empty vector control), PRNP(Δ51–90) and PRNP(Δ32–135) (lack the octapeptide repeat domain), PRNP(Δ105–128) (lacks hydrophobic domain). TUB1A1 was used as a loading control. (B) Histogram shows densitometry analysis of MAP1LC3-II expression relative to TUB1A1/α-tubulin expression. (C) Histogram shows densitometry analysis of ATG12–ATG5 expression relative to TUB1A1 expression. (D) NTA of exosome concentration in the CM of prnp0/0 cells transfected with full-length or truncation mutants of PRNP (described in [A]). Data information: (B-D) Data shown represent the mean ± SD from at least 3 independent experiments. One-way ANOVA and Tukey's post hoc test were used to assess statistical significance. *P < 0 .05, **P < 0 .01, ***P < 0 .001, #P < 0 .0001.](/cms/asset/15391c2d-a107-4f5d-a9ef-aa5540a97a8e/kaup_a_1226735_f0006_b.gif)
CAV1 arrest in lipid raft domains induces autophagy in the absence of PRNP
The PRNP octapeptide repeat domain is critical for some important biological functions, such as Cu2+ bindingCitation47 and PRNP internalization.Citation48 It has been reported that the interaction between PRNP and CAV1 at the plasma membrane occurs through this region.Citation9,49 Interestingly, CAV1 impairs the interaction between ATG5 and ATG12 in the cytoplasm, thereby suppressing autophagy.Citation46 We thus investigated if CAV1 is affected by PRNP expression.
CAV1 protein levels were similar in astrocytes derived from Prnp+/+ or prnp0/0 mice (). Lipid rafts are often enriched in CAV1, which is thought to play roles in cholesterol movement and the scaffolding of signaling molecules.Citation50 Despite being an integral membrane protein a fraction of CAV1 is present in cells as a soluble cytoplasmic protein or associated with other organelles.Citation51 Thus, depending on the cell location, CAV1 has distinct functions. Therefore, we characterized the cellular distribution of CAV1 by isolating lipid rafts via discontinuous sucrose density gradient ultracentrifugation. In Prnp+/+ cells, CAV1 was located in lipid raft domains (fractions 4–5) identified by the presence of FLOT1. CAV1 was also detected in high-density nonlipid raft fractions (fractions 7–12) corresponding to the cytoplasm, suggesting that CAV1 is shuttled back and forth between the cell surface and cytoplasm (). Conversely, CAV1 was enriched in lipid raft domains (fractions 3–5), but was absent in high-density nonlipid raft fractions in prnp0/0 cells (). Quantification of CAV1 levels in lipid raft domains revealed significantly higher levels in prnp0/0 cells than in Prnp+/+ cells (). This result suggests that in the absence of PRNP, CAV1 may be arrested in lipid rafts of the plasma membrane instead of internalized. This would directly affect the inhibitory role of cytoplasmic CAV1 on ATG12–ATG5 engagement and autophagy stimulation, as observed in .
Figure 7. CAV1 is arrested in lipid rafts in the absence of PRNP. (A) Western blot analysis of CAV1 protein expression in cell extracts isolated from Prnp+/+ and prnp0/0 astrocytes. TUB1A1 was used as a loading control. Histogram (right side) shows densitometry analysis of CAV1 expression relative to TUB1A1 expression. (B) Western blot analysis of CAV1, FLOT1, and PRNP protein levels in lipid rafts isolated by sucrose density fractionation from Prnp+/+ and prnp0/0 astrocytes. An equal volume of each fraction was analyzed. Histogram (below) shows densitometry analysis of lipid raft CAV1 expression relative to total CAV1 expression. (C) Fluorescence microscopy analysis of CTxB (green) in representative Prnp+/+ and prnp0/0 astrocytes incubated with Alexa Fluor 488-labeled CTxB for the indicated time points. Nuclear staining (DRAQ5, red). Scale bar: 10 μm. (D) Quantification of the average CTxB fluorescent signal in C. A.U., arbitrary units. Data information: (A,B,D) Data shown represent the mean ± SD from at least 3 independent experiments. A Student t test was used to assess statistical significance.*P < 0 .05, **P < 0 .001.
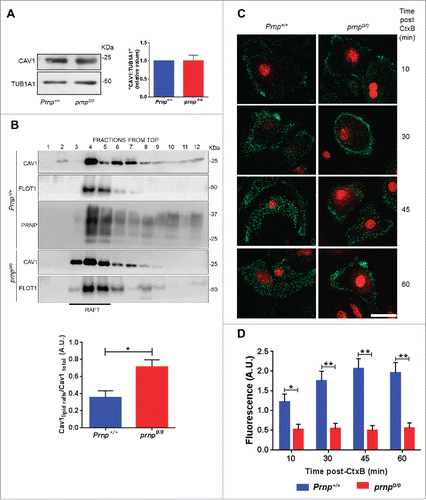
To confirm impaired CAV1 internalization in prnp0/0 cells, we examined the uptake of cholera enterotoxin B subunit (CTxB), which uses caveolae membrane invaginations to become internalized.Citation50 Labeling of cells with Alexa Fluor 488-conjugated CTxB revealed it to be bound to the plasma membrane and internalized as punctate endosomal structures in Prnp+/+ cells (). However, CTxB remained closer to the cell surface in prnp0/0 cells (). Quantification of the fluorescent signal confirmed that prnp0/0 cells internalized less CTxB than Prnp+/+ cells (). Taken together, these results suggest that PRNP regulates the internalization and localization of CAV1 in lipid raft domains, which ultimately affects autophagy activation.
Discussion
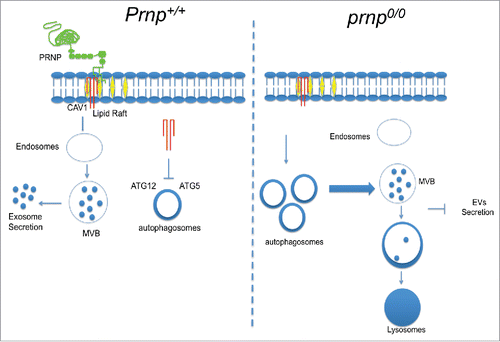
In this study, we demonstrated that PRNP is a positive regulator of exosome secretion. PRNP appears to control the distribution of CAV1 between lipid raft domains on the cell membrane and the cytoplasm where CAV1 can function to impair the ATG12–ATG5 complex and thus to inhibit autophagy progression. Under these conditions, MVBs form and exosomes are secreted. In the absence of PRNP at the membrane, CAV1 internalization is inhibited, ATG12–ATG5 complexes are formed, and autophagy is stimulated. Autophagosomes fuse to MVBs and both are delivered to lysosomes, thus sequestering the cellular source of exosomes and abolishing their secretion into the extracellular milieu ().
Figure 8. Model of PRNP function in exosome secretion. (Left panel) Membrane-bound PRNP (green) organizes CAV1 (red) into lipid rafts (yellow) and allows its internalization and subsequent inhibition of cytoplasmic ATG12–ATG5 engagement. Under these conditions, autophagy progression is impaired, and MVBs fuse with the cell membrane to release exosomes (EVs) into the extracellular milieu (right panel). In the absence of PRNP, CAV1 internalization is prevented, allowing ATG12–ATG5 complex formation and autophagy induction. Autophagosomes fuse with MVBs and are delivered to lysosomes for degradation, leading to a drastic reduction in exosome secretion (left panel).
We investigated the underlying mechanisms by which PRNP regulates exosome secretion. We first found that this process requires the presence of PRNP at the plasma membrane, as a PRNP mutant lacking the membrane localization signal was incapable of restoring exosome secretion in prnp-null cells. Conversely, the internalization of PRNP into intracellular compartments was unnecessary for this function, thus indicating that exosome secretion regulation primarily occurs at the cell surface. In fact, the role of membrane proteins in exosome secretion has been recently explored. The transmembrane heparansulphate proteoglycans of the syndecan family are connected by the cytoplasmic soluble protein SDCBP/syntenin (syndecan binding protein) to PDCD6IP/ALIX (programmed cell death 6 interacting protein), an auxiliary component of the ESCRT machinery that supports endosomal membrane budding.Citation52
Our initial observations suggested that in the absence of PRNP, the endocytic compartments of astrocytes were irregularly distributed, as seen by the reduction in ILVs and MVBs, the enhancement of lysosomal morphology, and the presence of autophagosomes. These data comply with the findings of other groups demonstrating that autophagy is upregulated in PRNP-deficient cells.Citation53,54 Moreover, our findings are consistent with a connection between the endocytic pathway, autophagy, and exosome biogenesis and secretion.Citation33,34 It is well known that autophagosome maturation requires ESCRT function, and that autophagosomes are able to fuse with endocytic vesicles (as MVBs) or lysosomes, which contain the hydrolytic enzymes required to degrade autophagosomal content.Citation31,55-57 Additionally, diverse conditions that stimulate autophagy, such as serum starvation, rapamycin treatment, or MAP1LC3-II overexpression, inhibit exosome release.Citation33 Our experiments demonstrating that exosome secretion in prnp-null cells was rescued by either PRNP re-expression or autophagy inhibition via BECN1 knockdown highlight, for the first time, that PRNP regulates exosome secretion by modulating autophagy induction.
Autophagy can be regulated by many different mechanisms,Citation58 but the identification of the link between autophagy induction and PRNP is essential for understanding how PRNP facilitates exosome secretion. Our group proposed that PRNP acts to organize signaling molecules into lipid rafts due to its ability to interact with and modulate the activity of several membrane proteins.Citation59 CAV1 is one well-described PRNP ligand that interacts with the octapeptide repeat domain to activate signal transduction events downstream of the kinases FYN and LYN (Lyn proto-oncogene Src family tyrosine kinase).Citation49,60 Therefore, our findings that CAV1 is localized primarily within lipid rafts instead of the cytoplasm in prnp-null cells are in accordance with these previous studies. Remarkably, CAV1 has been described as an important negative regulator of autophagosome formation,Citation61-62 given its ability to impair ATG12–ATG5 engagement.Citation46 The introduction of full-length PRNP, but not the deletion mutants lacking the octapeptide repeat domain, in prnp-null astrocytes prevented cytoplasmic CAV1 function. Consequently, ATG12–ATG5 complex formation was impaired leading to diminished autophagy and increased exosome release. Finally, it is important to note that PRNP is internalized by caveolae.Citation63 However, our results suggest that PRNP internalization is not necessary for controlling exosome secretion.
Primary astrocytes actively participate in neural development and communicate extensively with neurons.Citation64 Exosomes secreted by glial cells not only contribute to the development of the central nervous system (CNS), but also promote the repair of neuronal injuries and regeneration of peripheral nerves.Citation65,66 Glial cells may release SYN1 (synapsin I) into exosomes in response to stressful conditions, such as the oxidative stressor ischemia, to modulate neuronal outgrowth and neuron-glia interactions.Citation67 In the peripheral nervous system, Schwann cells release exosomes that are specifically internalized by axons, resulting in increased axonal regeneration after sciatic nerve injury.Citation68 Additionally, exosomes derived from multipotent mesenchymal stromal cells (MSCs) improve the functional recovery of neurons after stroke in a rat model.Citation69
Notably, our present findings demonstrate that astrocytes and fibroblasts derived from mice overexpressing PRNP exhibited higher levels of secreted exosomes, indicating a general role for PRNP in regulating exosome secretion independent of cell type. Furthermore, plasma from these mice contained altered levels of exosomes, implying that exosome secretion in other cell types is also affected by PRNP expression.
Over the past few years, exosomes have emerged as important mediators of intercellular communication that facilitate the transmission of physiological signals between cells to regulate a diverse range of biological processes. Exosomes are also prominent mediators of neurodegenerative diseases. These vesicles can carry aggregated proteins or prions to recipient neurons to induce toxic effects. PrPSc is actively released into the extracellular environment associated with exosomes, and promotes the distribution of prions throughout the organism to cause prion diseases.Citation70 Additionally, autophagy can play a protective role in the clearance of pathological PrPSc accumulated within neurons. Remarkably, defective autophagy may contribute to the formation of spongiform changes in prion disease.Citation32 Interestingly, TG20 mice succumb to terminal disease more rapidly than wild-type mice,Citation71 which is consistent with the lower autophagy activity observed in cells from these animals. Moreover, decreased autophagy in TG20 cells may contribute to the regulation of infection and spreading of PrPSc due to their improved secretion of exosomes.
In conclusion, our findings show for the first time that PRNP modulates the endocytic pathways that control exosome secretion. Given the pivotal role of exosomes in physiological and pathological conditions, targeting the mechanisms by which exosomes are secreted offers important therapeutic possibilities. Furthermore, the unpredicted function of PRNP in facilitating exosome secretion in addition to its convenient localization at the cell membrane highlight its potential promise as an amenable candidate for the development of therapeutic strategies seeking to target exosome delivery.
Materials and methods
Reagents
A polyclonal antibody against recombinant mouse PRNP was produced in prnp-null (prnp0/0) miceCitation72 and was used at a dilution of 1:500. Primary antibodies against anti-LBPA (6C4, 1:250; Echelon Biosciences Inc., Z-SLBPA), anti-TUB1A1 (1:10,000; Sigma-Aldrich,T-9026), anti-RAB5 (1:100; Abcam, ab18211), RAB7 (1:100; Abcam, ab50533), Anti-RAB11A antibody (1:100; Abcam, ab65200) anti-LAMP1 (1:250; Abcam, ab25245), anti- LC3A/B (D3U4C) (1:1,000; Cell Signaling Technology, 12741), anti-BECN1 (1:1,000; Cell Signaling Technology, 3738), anti-ATG12 (D88H11, 1:1,000; Cell Signaling Technology, 4180), anti-LAMP2 (ABL-93, 1:50; Abcam, ab25339), anti-TSG101 (4A10, 1:500, Abcam, ab83) and anti-FLOT1 (1:1000; Merck-Millipore, Ab9292) were used in western blot and immunofluorescence experiments. Secondary antibodies included: goat anti-mouse Alexa Fluor 488 (1:1000; ThermoFisher Scientific, A-11001), goat anti-rabbit Alexa Fluor 488 (1:1000; ThermoFisher Scientific, A-11034) and goat anti-rat Alexa Fluor 488 (1:1000; ThermoFisher Scientific, A-11006). Expression vectors encoding GFP-PRNP, GFP-PRNP(Δ1–22), GFP-PRNP(Δ32–135), GFP-PRNP(Δ51–90), and GFP-PRNP(Δ105–128) were constructed and expressed as previously described.Citation73,74 Plasmids for PRNP-3F4 and *N-PRNP-3FCitation36 were kindly provided by Dr. R. Morris (King's College, London, England).
Mice
Two independent lines of prnp-null mice were used for astrocyte isolation, designated in this study as prnp0/0 (ZrchI)Citation75 and prnp−/− (Npu).Citation76 ZrchI prnp0/0 mice were provided by Dr. C. Weissmann (The Scripps Research Institute, Jupiter, FL, USA). Control mice (Prnp+/+) were generated by crossing F1 siblings from matings between 129/Sv and C57BL/6J mice. Npu prnp−/− mice were provided by Drs. B. Chesebro and R. Race (Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, Hamilton, MT, USA), and are descendants from the Npu line backcrossed to C57BL/10 mice over at least 20 generations. Prnpwt/wt mice were used as the respective control. PRNP-overexpressing mice (TG20) originally describedCitation77 were provided by Dr. C. Weissmann (The Scripps Research Institute, Jupiter, FL, USA). The Principles of Laboratory Animal Care (National Institutes of Health publication number 85–23, 1996) was strictly followed in all experiments, and all protocols were approved by the A.C. Camargo Cancer Center Animal Care and Use Committee (project number 058/13).
Primary cortical astrocyte culture
Astrocyte primary cultures were prepared from the cerebral hemispheres of embryonic day (E) 17 wild-type, prnp-null, and TG20 mice. Briefly, single-cell suspensions were obtained by dissociating cerebral hemispheres in Dulbecco's modified Eagle's medium (DMEM-low glucose) (ThermoFisher Scientific, 31600034) supplemented with 40 mg garamycin (Nova Farma, 42859) and 3 mM sodium bicarbonate (Sigma Aldrich, S5761). Cells were seeded on pre-coated poly-L-lysine plates and grown in DMEM containing 10% FBS (ThermoFisher Scientific, 12657). The medium was changed every 2 d. For transfection experiments, confluent astrocytes were incubated with 10 µg of Lipofectamine 2000 (ThermoFisher Scientific, 11668019) and 5 µg of plasmid DNA in OPTI-MEM (ThermoFisher Scientific, 51985034) for 4 h, and afterwards, the medium was changed to DMEM containing 10% FBS.
Primary MEF culture
MEFs were prepared from E14 embryos. The head and organs were dissected, and fetal tissue samples were rinsed in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4), minced, trypsinized for 10 min at 37°C, and subsequently dissociated in DMEM. Cells were seeded on pre-coated poly-L-lysine plates and grown in DMEM containing 10% FBS. The medium was changed every 2 d.
Preparation of conditioned medium (CM) and exosome isolation
CM was obtained as previously described.Citation35 Confluent astrocytes or fibroblasts grown in 100-mm culture dishes were washed 3 times with PBS and incubated with serum-free medium for 48 h. CM was collected on ice and pre-cleared from cell debris by sequential centrifugation (1,500 xg for 10 min, 4,500 xg for 10 min, and 10,000 xg for 30 min). Exosomes were obtained by ultracentrifugation at 100,000 xg for 16 h using a SW 40 Ti rotor (Beckman-Coulter, 331302). The pelleted fraction was resuspended in PBS and centrifuged again at 100,000 xg for 16 h.
Exosome isolation from mouse plasma
Mice were bled by cardiac puncture, and blood samples were centrifuged twice at 1,500 xg for 15 min at room temperature to obtain platelet-free plasma. The plasma was further centrifuged at 10,000 x g for 30 min at 4°C to remove cell debris, followed by ultracentrifugation at 100,000 xg for 2 h. The pellet was resuspended in PBS and ultracentrifuged again at 100,000 xg for 2 h.
Exosome quantification and analysis
Pellets of exosomes isolated from CM or plasma were resuspended in 1 ml of PBS. The number of particles and particle size were measured using a nanoparticle tracking analysis device NanoSight LM10 coupled to a CCD camera and a laser emitting a 60-mW beam at 405 nm (Malvern, United Kingdom). Video acquisitions were performed in 5 records of 60 s using the following parameters: shutter = 604, gain = 100, and threshold = 10. At least 1,000 particles were tracked in each sample.
siRNA transfection
To silence PRNP, wild-type astrocytes were transfected with target-specific siRNAs and a scrambled siRNA as a control. The target sequence for mouse Prnp was: 5′GGUUUUUGGUUUGCUGGGCTT3′. The scrambled control siRNA sequence was: 5′UGUUGUUGGCGUUUGUGGCTT3′. To silence BECN1 3 different siRNAs targeting mouse Becn1 mRNA and nontarget siRNA were used (Life Technologies, 4390771). Astrocytes were seeded on 100-mm plates and were 70–90% confluent at the time of transfection. 200 pmol of RNA duplexes were transfected using Lipofectamine 2000 reagent according to the manufacturer's instructions. The efficacy of siRNA-mediated knockdown was confirmed by isolating total cell lysates from siRNA-transfected or nontransfected cells 24 h after transfection and performing SDS-PAGE and western blot analysis with the specific antibodies listed under Reagents.
Transmission electron microscopy
Cells
Astrocytes in culture were fixed in 4% paraformaldehyde (PFA; Polysciences Inc., 18814):2% glutaraldehyde (Sigma-Aldrich, 340855) in 0.1 M phosphate buffer, pH 7.4. After fixation, the samples were rinsed several times with PBS followed by post-fixation in 1% osmium tetroxide (EMS, 19150) in phosphate buffer for 1 h. The samples were rinsed with PBS for 15 min and dehydrated through a series of graded ethanol washes ranging from 70% to 100%. The samples were then immersed in an ethanol:epon mixture (1:1) and polymerized in pure epon (Polysciences, Inc. 02334-500) at 60°C for 48 h. Ultrathin sections were stained with uranyl acetate and lead citrate and imaged on a JEOL 1200 EX II transmission electron microscope (JEOL, USA).
Exosomes
Isolated exosomes were deposited onto formvar-carbon-coated electron microscopy grids (EMS, FCF200H-Cu), fixed with a mixture of 2% paraformaldehyde and 0.125% glutaraldehyde for 20 min. A 100-µL drop of PBS was placed on a sheet of parafilm and grids were transferred with the sample membrane side facing down using clean forceps for 2 min. The grids were kept wet on the side of the membrane during all steps, but dry on the opposite side. The grids were transferred to a 50-µL drop of 1% glutaraldehyde for 5 min before transferring to a 100-µL drop of distilled water for 2 min. This was repeated 7 times for a total of 8 water washes. To contrast the samples, grids were transferred to a 50-µL drop of uranyl-oxalate solution (4% uranyl acetate [EMS, 22400-4]; 0.15 M oxalic acid [Sigma-Aldrich, 75688] pH 7), for 5 min before transferring to a 50-µL drop of methyl-cellulose-UA, (a mixture of 4% uranyl acetate and 2% methyl cellulose [Sigma-Aldrich, M6385] in a ratio of 100 µL:900 µL), for 10 min, placing the grids on a glass dish covered with parafilm on ice. The grids were removed with stainless steel loops and excess fluid blotted gently on Whatman no.1 filter paper. Grids were left to dry and stored in appropriate grid storage boxes, then observed under a JEOL 1200 EX II transmission electron microscope.
Immunofluorescence and confocal microscopy
Cells were cultured on glass coverslips, fixed in 4% PFA for 20 min at room temperature, washed in PBS, blocked in a 5% bovine serum albumin (Sigma-Aldrich, A4503) solution in PBS, permeabilized in 0.5% Triton X-100 (Sigma-Aldrich, T8787), incubated with the primary antibodies listed under Reagents (diluted in PBS containing 0.5% bovine serum albumin), and then stained with the appropriate secondary antibodies, DRAQ5 (1:500; ThermoFisher Scientific, 62252), and rhodamine-phalloidin (1:50; ThermoFisher Scientific, R415). Coverslips were mounted onto glass slides using FluorSave reagent (Calbiochem, 345789). All fluorescent images were acquired on a Leica TCS SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany). To quantify fluorescence levels, a single in-focus plane was acquired. An outline was drawn around each cell and circularity, area, mean fluorescence, and several adjacent background readings were measured using ImageJ (v1.48, NIH). The total corrected cellular fluorescence (TCCF) was calculated as the integrated density (area of selected cell × mean fluorescence of background readings).Citation78
Autophagy induction
Cells were washed 3 times with pre-warmed PBS and then incubated in serum-free DMEM at 37°C for 24 h in the presence or absence of 100 nM rapamycin (Sigma-Aldrich, R0395). The medium was collected and exosomes were isolated as described above. Cell extracts were used for western blot analyses.
Quantification of GFP-MAP1LC3B puncta formation
Cells were cultured on glass coverslips and tranfected with EGFP-LC3 plasmid (AddGene, 11546, deposited by Karla Kirkegaard). Transfected cells were fixed in 4% PFA for 20 min at room temperature and then rinsed with PBS. The nuclei were stained with DRAQ5. Slides were mounted with FluorSave mounting medium and examined by fluorescence microscopy. To quantify autophagy activation, at least 150 EGFP-LC3-expressing cells were analyzed and the number of puncta per cell, based on EGFP expression, was determined by using the ‘analyze particles' function in ImageJ.
Lipid raft isolation
Primary astrocytes were lysed for 10 min in cold TNE buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA) containing 0.5% Triton X-100, and Protease Inhibitor Cocktail (Roche, 11836153001). Extracts were centrifuged at 1,300 xg for 5 min. One ml of the cleared supernatant fraction was mixed with 85% sucrose (Merck Millipore, 107651) in TNE buffer and layered on the bottom of a Polyallomer 12-ml centrifuge tube (Beckman Coulter, 331372). The lysate was overlaid with 4 ml of 35% sucrose in TNE buffer, and finally with 4 ml of 5% sucrose in TNE buffer. The samples were centrifuged in a SW41 Ti rotor at 260,110 xg for 18 h at 4°C. At the end of the run, 1-ml fractions were collected from the top of the gradient and proteins were precipitated with 15% trichloroacetic acid (Sigma-Aldrich, T6399). The samples were then subjected to 10% SDS PAGE and western blot analysis.
CTxB uptake assay
Lipid raft labeling was performed using the Vybrant Alexa Fluor 488 Lipid Raft labeling kit according to the manufacturer's protocol (Thermo Fisher Scientific, V34403). Briefly, live cells were washed with ice cold PBS, labeled with Alexa Fluor 488-conjugated CTx-B, and crosslinked with anti-CTx-B antibody in serum-free media for 15 min at 4°C. Cells were shifted to 37°C for 10, 30, 45, and 60 min, washed with PBS twice, fixed in 4% PFA, stained with DRAQ5 to mark nuclei, and mounted onto glass slides before imaging on the Leica SP5 confocal system. For quantitative analysis, the fluorescence of internalized CTx-B was measured using ImageJ software (n = 100 cells).
Statistical analysis
All experiments were repeated 3 to 5 times. Statistical analyses were performed using GraphPad Prism 5 (La Jolla, CA). Experimental groups were compared using one-way or 2-way ANOVA followed by Tukey's post hoc test or the Student t test.
Abbreviations
| ATG5 | = | autophagy-related 5 |
| ATG7 | = | autophagy- related 7 |
| BECN1 | = | Beclin 1, autophagy related |
| CAV1 | = | caveolin 1, caveolae protein |
| CM | = | conditioned medium |
| CTxB | = | cholera enterotoxin B subunit |
| DMEM | = | Dulbecco's modified Eagle's medium |
| EGFP | = | enhanced green fluorescent protein |
| ESCRT | = | endosomal sorting complex required for transport |
| EV | = | extracellular vesicles |
| FBS | = | fetal bovine serum |
| FLOT1 | = | flotilin 1 |
| GFP | = | green fluorescent protein |
| ILV | = | intraluminal vesicle |
| LAMP1 | = | lysosomal-associated membrane protein 1 |
| LAMP2 | = | lysosomal-associated membrane protein 2 |
| LBPA | = | lysobisphosphatidic acid |
| MAP1LC3B | = | microtubule-associated protein 1 light chain 3 β |
| MEF | = | mouse embryonic fibroblast |
| MVB | = | multivesicular body |
| NTA | = | nanoparticle tracking analysis |
| PBS | = | phosphate-buffered saline |
| PFA | = | paraformaldehyde |
| PRKA/PKA | = | protein kinase, AMP-activated |
| PRNP | = | prion protein |
| PrPSc | = | pathological scrapie isoform |
| RAB5 | = | RAB5, member RAS oncogene family |
| RAB7 | = | RAB7, member RAS oncogene family |
| RAB11A | = | RAB11A, member RAS oncogene family |
| siRNA | = | small interfering RNA |
| TSG101 | = | tumor susceptibility gene 101 |
| TUBA1A | = | tubulin, α 1A |
| VPS4 | = | vacuolar protein sorting 4 |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary files
Download Zip (10.8 MB)Acknowledgments
We are very grateful to Dr. Luis Lamberti da Silva for reagents and scientific discussion, Dr. Renato Arruda Mortara for LAMP2 antibody and Dr. Richard Morris for PRNP plasmids.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 09/14027-2) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 467566/2014-3). Fellowships from FAPESP to MS (2010/19200-1) B.R.R (2012/19019-0), are gratefully acknowledged.
References
- Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 1987; 51:229-40; PMID:2444340; http://dx.doi.org/10.1016/0092-8674(87)90150-4
- Prusiner SB, Groth DF, Bolton DC, Kent SB, Hood LE. Purification and structural studies of a major scrapie prion protein. Cell 1984; 38:127-34; PMID:6432339; http://dx.doi.org/10.1016/0092-8674(84)90533-6
- Prusiner SB. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet [Internet] 2013; 47:601-23. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4010318&tool=pmcentrez&rendertype=abstract; PMID:24274755; http://dx.doi.org/10.1146/annurev-genet-110711-155524
- Martins VR, Beraldo FH, Hajj GN, Lopes MH, Lee KS, Prado MA, Linden R. Prion protein: Orchestrating neurotrophic activities. Curr Issues Mol Biol 2010; 12:63-86; PMID:19767651
- Beraldo FH, Arantes CP, Santos TG, Machado CF, Roffe M, Hajj GN, Lee KS, Magalhães AC, Caetano FA, Mancini GL, et al. Metabotropic glutamate receptors transduce signals for neurite outgrowth after binding of the prion protein to laminin γ1 chain. FASEB J 2011; 25:265-79; PMID:20876210; http://dx.doi.org/10.1096/fj.10-161653
- Um J, Kaufman A, Kostylev M, Heiss J, Stagi M, Takahashi H, Kerrisk M, Vortmeyer A, Wisniewski T, Koleske A, et al. Metabotropic Glutamate Receptor 5 Is a Coreceptor for Alzheimer Aβ Oligomer Bound to Cellular Prion Protein. Neuron [Internet] 2013; 79:887-902; PMID:24012003; http://dx.doi.org/10.1016/j.neuron.2013.06.036
- Beraldo FH, Arantes CP, Santos TG, Queiroz NGT, Young K, Rylett RJ, Markus RP, Prado MA, Martins VR. Role of ??7 nicotinic acetylcholine receptor in calcium signaling induced by prion protein interaction with stress-inducible protein. J Biol Chem 2010; 285:36542-50; PMID:20837487; http://dx.doi.org/10.1074/jbc.M110.157263
- Santuccione A, Sytnyk V, Leshchyns'ka I, Schachner M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J Cell Biol 2005; 169:341-54; PMID:15851519; http://dx.doi.org/10.1083/jcb.200409127
- Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, Launay JM, Kellermann O. Signal transduction through prion protein. Science 2000; 289:1925-8; PMID:10988071; http://dx.doi.org/10.1126/science.289.5486.1925
- McLennan NF, Brennan PM, McNeill A, Davies I, Fotheringham A, Rennison KA, Ritchie D, Brannan F, Head MW, Ironside JW, et al. Prion protein accumulation and neuroprotection in hypoxic brain damage. Am J Pathol 2004; 165:227-35; PMID:15215178; http://dx.doi.org/10.1016/S0002-9440(10)63291-9
- Lopes MH, Hajj GNM, Muras AG, Mancini GL, Castro RMPS, Ribeiro KCB, Brentani RR, Linden R, Martins VR. Interaction of cellular prion and stress-inducible protein 1 promotes neuritogenesis and neuroprotection by distinct signaling pathways. J Neurosci 2005; 25:11330-9; PMID:16339028; http://dx.doi.org/10.1523/JNEUROSCI.2313-05.2005
- Hajj GNM, Lopes MH, Mercadante AF, Veiga SS, da Silveira RB, Santos TG, Ribeiro KCB, Juliano MA, Jacchieri SG, Zanata SM, et al. Cellular prion protein interaction with vitronectin supports axonal growth and is compensated by integrins. J Cell Sci 2007; 120:1915-26; PMID:17504807; http://dx.doi.org/10.1242/jcs.03459
- Roffé M, Beraldo FH, Bester R, Nunziante M, Bach C, Mancini G, Gilch S, Vorberg I, Castilho BA, Martins VR, et al. Prion protein interaction with stress-inducible protein 1 enhances neuronal protein synthesis via mTOR. Proc Natl Acad Sci U S A 2010; 107:13147-52; http://dx.doi.org/10.1073/pnas.1000784107
- Graner E, Mercadante AF, Zanata SM, Forlenza OV, Cabral ALB, Veiga SS, Juliano MA, Roesler R, Walz R, Minetti A, et al. Cellular prion protein binds laminin and mediates neuritogenesis. Mol Brain Res 2000; 76:85-92; PMID:10719218; http://dx.doi.org/10.1016/S0169-328X(99)00334-4
- Coitinho AS, Lopes MH, Hajj GNM, Rossato JI, Freitas AR, Castro CC, Cammarota M, Brentani RR, Izquierdo I, Martins VR. Short-term memory formation and long-term memory consolidation are enhanced by cellular prion association to stress-inducible protein 1. Neurobiol Dis 2007; 26:282-90; PMID:17329112; http://dx.doi.org/10.1016/j.nbd.2007.01.005
- Robinson SW, Nugent ML, Dinsdale D, Steinert JR. Prion protein facilitates synaptic vesicle release by enhancing release probability. Hum Mol Genet 2014; 23:4581-96; PMID:24722203; http://dx.doi.org/10.1093/hmg/ddu171
- Manson J, West JD, Thomson V, McBride P, Kaufman MH, Hope J. The prion protein gene: a role in mouse embryogenesis? Development 1992; 115:117-22; PMID:1353438
- Hajj GNM, Santos TG, Cook ZSP, Martins VR. Developmental expression of prion protein and its ligands stress-inducible protein 1 and vitronectin. J Comp Neurol 2009; 517:371-84; PMID:19760599; http://dx.doi.org/10.1002/cne.22157
- Arantes C, Nomizo R, Lopes MH, Hajj GNM, Lima FRS, Martins VR. Prion Protein and Its Ligand Stress Inducible Protein 1 Regulate Astrocyte Development. 2009; 1449:1439-49
- Brown DR, Mohn CM. Astrocytic glutamate uptake and Prion Protein Expression. Glia 1999; 25:282-92
- Hajj GNM, Arantes CP, Dias MVS, Roffé M, Costa-Silva B, Lopes MH, Porto-Carreiro I, Rabachini T, Lima FR, Beraldo FH, et al. The unconventional secretion of stress-inducible protein 1 by a heterogeneous population of extracellular vesicles. Cell Mol Life Sci 2013; 70:3211-27; PMID:23543276; http://dx.doi.org/10.1007/s00018-013-1328-y
- Lopes MH, Santos TG, Rodrigues BR, Cunha IW, Wasilewska-sampaio AP. Disruption of prion protein - HOP engagement impairs glioblastoma growth and cognitive decline and improves overall survival. Oncogene 2015; 34(25):3305-14. PMID: 25151961; http://dx.doi.org/10.1038/ onc.2014.261
- EL Andaloussi S, Mäger I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov [Internet] 2013; 12:347-57; PMID:23584393; http://dx.doi.org/10.1038/nrd3978
- Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol [Internet] 2014; 30:255-89. Available from: http://www.annualreviews.org/doi/abs/10.1146/annurev-cellbio-101512-122326; PMID:25288114; http://dx.doi.org/10.1146/annurev-cellbio-101512-122326
- Février B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol 2004; 16:415-21; http://dx.doi.org/10.1016/j.ceb.2004.06.003
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature [Internet] 2010; 464:864-9. Available from: http://dx.doi.org/10.1038/nature08849
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol 2010; 11:556-66; PMID:20588296; http://dx.doi.org/10.1038/nrm2937
- Bobrie A, Colombo M, Raposo G, Théry C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 2011; 12:1659-68; PMID:21645191; http://dx.doi.org/10.1111/j.1600-0854.2011.01225.x
- Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol [Internet] 2013; 25:455-60; http://dx.doi.org/10.1016/j.ceb.2013.03.004
- Russell RC, Tian Y, Yuan H, Park HW, Chang Y, Kim H, Neufeld TP, Dillin A, Guan K. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. NIH Public Access 2014; 15
- Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ 2009; 16:70-8; PMID:19008921; http://dx.doi.org/10.1038/cdd.2008.168
- Yao H, Zhao D, Khan SH, Yang L. Role of autophagy in prion protein-induced neurodegenerative diseases molecular mechanism of autophagy and its regulation autophagy. Acta Biochim Biophys Sin 2013; 45:494-502; PMID:23459558; http://dx.doi.org/10.1093/abbs/gmt022
- Fader CM, Sánchez D, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic [Internet] 2008 [cited 2014 Sep 19]; 9:230-50. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17999726
- Baixauli F, López-otín C, Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol 2014; 5:1-6
- Lima FRS, Arantes CP, Muras AG, Nomizo R, Brentanià RR, Martins VR. Cellular prion protein expression in astrocytes modulates neuronal survival and differentiation. J Neurochem 2007; 103:2164-76
- Sunyach C, Jen A, Deng J, Fitzgerald KT, Frobert Y, Grassi J, Mccaffrey MW, Morris R. The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J 2003; 22:3591-601
- Candelario KM, Steindler DA. The role of extracellular vesicles in the progression of neurodegenerative disease and cancer. Trends Mol Med [Internet] 2014; 20:368-74; PMID:24835084; http://dx.doi.org/10.1016/j.molmed.2014.04.003
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol 2004; 5:317-23; PMID:15071556; http://dx.doi.org/10.1038/nrm1360
- Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 1998; 392:193-7; PMID:9515966
- Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol [Internet] 2007; 8:622-32. Available from: http://www.nature.com/doifinder/10.1038/nrm2217; PMID:17637737; http://dx.doi.org/10.1038/nrm2217
- Oh JM, Shin HY, Park SJ, Kim BH, Choi JK, Choi EK, Carp RI, Kim YS. The involvement of cellular prion protein in the autophagy pathway in neuronal cells. Mol Cell Neurosci 2008; 39:238-47; PMID:18674620; http://dx.doi.org/10.1016/j.mcn.2008.07.003
- Zhou J, Tan S, Nicolas V, Bauvy C, Yang N, Zhang J, Xue Y, Codogno P, Shen H. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Nat Publ Gr [Internet] 2013; 23:508-23. Available from: http://dx.doi.org/10.1038/cr.2013.11
- Kimura S, Fujita N, Noda T, Yoshimori T. Chapter 1 monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol [Internet] 2009; 452:1-12 Available from: http://dx.doi.org/10.1016/S0076-6879(08)03201-1
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/10.4161/auto.19496
- Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al. XEGFR-mediated beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell [Internet] 2013; 154:1269-84. Available from: http://dx.doi.org/10.1016/j.cell.2013.08.015
- Chen ZH, Cao JF, Zhou JS, Liu H, Che LQ, Mizumura K, Li W, Choi AMK, Shen HH. Interaction of caveolin-1 with ATG12-ATG5 system suppresses autophagy in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol [Internet] 2014; 306:L1016-25. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24727585; PMID:24727585; http://dx.doi.org/10.1152/ajplung.00268.2013
- Younan ND, Klewpatinond M, Davies P, Ruban AV, Brown DR, Viles JH. Copper ( II ) -induced secondary structure changes and reduced folding stability of the prion protein. J Mol Biol [Internet] 2011; 410:369-82; PMID:21619885; http://dx.doi.org/10.1016/j.jmb.2011.05.013
- Perera WS, Hooper NM. Ablation of the metal ion-induced endocytosis of the prion protein by disease-associated mutation of the octarepeat region. Curr Biol 2001; 11:519-23
- Shi Q, Wang YJS, Chen C, Sun H. PrP octarepeats region determined the interaction with caveolin-1 and phosphorylation of caveolin-1 and Fyn. Med Microbiol Immunol 2013; 1:215-27
- Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol 2007; 8:185-94; PMID:17318224; http://dx.doi.org/10.1038/nrm2122
- Quest AFG, Leyton L, Párraga M. Caveolins, caveolae, and lipid rafts in cellular transport, signaling, and disease. Biochem Cell Biol 2004; 82:129-44; PMID:15052333; http://dx.doi.org/10.1139/o03-071
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat Cell Biol [Internet] 2012; 14:677-85; http://dx.doi.org/10.1038/ncb2502
- Oh JM, Choi EK, Carp RI, Kim YS. Oxidative stress impairs autophagic flux in prion protein-deficient hippocampal cells. Autophagy 2012; 8:1448-61; PMID:22889724; http://dx.doi.org/10.4161/auto.21164
- Barbieri G, Palumbo S, Gabrusiewicz K, Azzalin A, Marchesi N, Spedito A, Biggiogera M, Sbalchiero E, Mazzini G, Miracco C, et al. Silencing of cellular prion protein (PrP c) expression by DNA-antisense oligonucleotides induces autophagy-dependent cell death in glioma cells. Autophagy 2011; 7:840-53; PMID:21478678; http://dx.doi.org/10.4161/auto.7.8.15615
- Rusten TE, Stenmark H. How do ESCRT proteins control autophagy? J Cell Sci 2009; 122:2179-83; PMID:19535733; http://dx.doi.org/10.1242/jcs.050021
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts E a, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol [Internet] 2011; 13:453-60; http://dx.doi.org/10.1038/ncb2204
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol [Internet] 2010; 12:823-30; http://dx.doi.org/10.1038/ncb0910-823
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol [Internet] 2010; 221:3-12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20225336; PMID:20225336; http://dx.doi.org/10.1002/path.2697
- Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR. Physiology of the prion protein. Physiol Rev 2008; 88:673-728; PMID:18391177; http://dx.doi.org/10.1152/physrev.00007.2007
- Toni M, Spisni E, Griffoni C, Santi S, Riccio M, Lenaz P, Tomasi V. Cellular prion protein and caveolin-1 interaction in a neuronal cell line precedes Fyn/Erk 1/2 signal transduction. J Biomed Biotechnol 2006; 2006:1-13; http://dx.doi.org/10.1155/JBB/2006/69469
- Le Lay S, Briand N, Blouin CM, Chateau D, Prado C, Lasnier F, Le Liepvre X, Hajduch E, Dugail I. The lipoatrophic caveolin-1 deficient mouse model reveals autophagy in mature adipocytes. Autophagy 2010; 6:754-63; PMID:20574167; http://dx.doi.org/10.4161/auto.6.6.12574
- Shiroto T, Romero N, Sugiyama T, Sartoretto JL, Kalwa H, Yan Z, Shimokawa H, Michel T. Caveolin-1 is a critical determinant of autophagy, metabolic switching, and oxidative stress in vascular endothelium. PLoS One 2014; 9:e87871
- Peters PJ, Mironov A, Peretz D, Van Donselaar E, Leclerc E, Erpel S, DeArmond SJ, Burton DR, Williamson RA, Vey M, et al. Trafficking of prion proteins through a caveolae-mediated endosomal pathway. J Cell Biol 2003; 162:703-17; PMID:12925711; http://dx.doi.org/10.1083/jcb.200304140
- Allen NJ, Barres BA. Neuroscience: Glia - more than just brain glue. Nature 2009; 457:675-7; PMID:19194443; http://dx.doi.org/10.1038/457675a
- Sharma P, Schiapparelli L, Cline HT. Exosomes function in cell-cell communication during brain circuit development. Curr Opin Neurobiol [Internet] 2013; 23:997-1004. Available from: http://www.sciencedirect.com/science/article/pii/S0959438813001530
- Kingham P, Ching R. The role of exosomes in peripheral nerve regeneration. Neural Regen Res [Internet] 2015; 10:743. Available from: http://www.nrronline.org/text.asp?2015/10/5/743/156968; PMID:26109947; http://dx.doi.org/10.4103/1673-5374.156968
- Wang S, Cesca F, Loers G, Schweizer M, Buck F, Benfenati F, Schachner M, Kleene R. Synapsin I is an oligomannose-carrying glycoprotein, acts as an oligomannose-binding lectin, and promotes neurite outgrowth and neuronal survival when released via glia-derived exosomes. J Neurosci [Internet] 2011 [cited 2014 Sep 29]; 31:7275-90. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21593312; PMID:21593312; http://dx.doi.org/10.1523/JNEUROSCI.6476-10.2011
- Lopez-Verrilli MA, Picou F, Court FA. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia 2013; 61:1795-806; PMID:24038411; http://dx.doi.org/10.1002/glia.22558
- Xin H, Li Y, Cui Y, Yang JJ, Zhang ZG, Chopp M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J Cereb Blood Flow Metab [Internet] 2013; 33:1711-5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3824189&tool=pmcentrez&rendertype=abstract; PMID:23963371; http://dx.doi.org/10.1038/jcbfm.2013.152
- Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A [Internet] 2004; 101:9683-8. Available from: http://www.pnas.org/content/101/26/9683.full; PMID:15210972; http://dx.doi.org/10.1073/pnas.0308413101
- Sandberg MK, Al-Doujaily H, Sharps B, Clarke a R, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature [Internet] 2011; 470:540-2. Available from: http://dx.doi.org/10.1038/nature09768; PMID:21350487
- Chiarini LB, Freitas ARO, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO J 2002; 21:3317-26; PMID:12093733; http://dx.doi.org/10.1093/emboj/cdf324
- Lee KS, Magalhães AC, Zanata SM, Brentani RR, Martins VR, Prado MA. Internalization of mammalian fluorescent cellular prion protein and N-terminal deletion mutants in living cells. J Neurochem 2001; 79:79-87; PMID:11595760; http://dx.doi.org/10.1046/j.1471-4159.2001.00529.x
- Zanata SM, Lopes MH, Mercadante AF, Hajj GNM, Chiarini LB, Nomizo R, Freitas ARO, Cabral ALB, Lee KS, Juliano MA, et al. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J 2002; 21:3307-16; PMID:12093732; http://dx.doi.org/10.1093/emboj/cdf325
- Büeler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 1992; 356:577-82; PMID:1373228; http://dx.doi.org/10.1038/356577a0
- Manson JC, Clarke AR, Hooper ML, Aitchison L, McConnell I, Hope J. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol Neurobiol 1994; 8:121-7; PMID:7999308; http://dx.doi.org/10.1007/BF02780662
- Fischer M, Rülicke T, Raeber a, Sailer a, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J 1996; 15:1255-64; PMID:8635458
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 2014; 13:1400-12; PMID:24626186; http://dx.doi.org/10.4161/cc.28401
