ABSTRACT
Autophagy is a fast-moving field with an enormous impact on human health and disease. Understanding the complexity of the mechanism and regulation of this process often benefits from the use of simple experimental models such as the social amoeba Dictyostelium discoideum. Since the publication of the first review describing the potential of D. discoideum in autophagy, significant advances have been made that demonstrate both the experimental advantages and interest in using this model. Since our previous review, research in D. discoideum has shed light on the mechanisms that regulate autophagosome formation and contributed significantly to the study of autophagy-related pathologies. Here, we review these advances, as well as the current techniques to monitor autophagy in D. discoideum. The comprehensive bioinformatics search of autophagic proteins that was a substantial part of the previous review has not been revisited here except for those aspects that challenged previous predictions such as the composition of the Atg1 complex. In recent years our understanding of, and ability to investigate, autophagy in D. discoideum has evolved significantly and will surely enable and accelerate future research using this model.
Introducing D. discoideum, a simple model for autophagy with striking similarities to animal cells
Dictyostelium discoideum is a protist that belongs to the Amoebozoa, a sister group of animals and fungi. They are known as social amoebae since the motile cells are able to aggregate and form a simple multicellular organism. Under starvation, groups of approximately 100,000 cells form a mound that goes through different stages of development to give rise to a fruiting body, in which vacuolated dead cells form a stalk that supports the spores (). Vegetative D. discoideum cells are more similar to animal cells than fungi or plant cells in many respects. For example, they lack rigid cell walls that might restrict movement, allowing the cells to perform typical animal-like processes such as phagocytosis, macropinocytosis, pseudopod-based cell motility and chemotaxis. The parallels with animal cells extends to the evolutionary conservation of many genes that have been lost during fungal evolution.Citation1
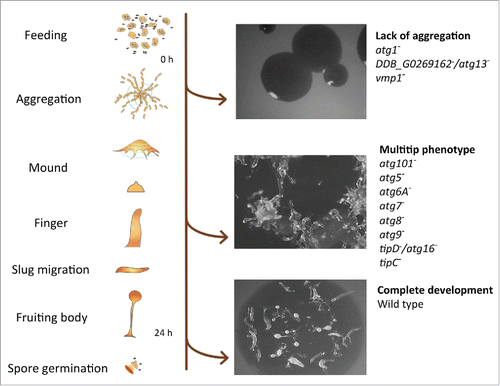
Figure 1. D. discoideum's life cycle and representative phenotypes associated with the lack of autophagy. Mutant strains have been described for the genes depicted at the right side (see details in the main text). The pictures correspond from top to bottom to the strains DDB_G0269162−/atg13−, tipC− and wild-type AX4.
The similarity to animal cells makes D. discoideum a suitable model to address the study of genes or processes relevant to human disease. These include the study of pathogen infections,Citation2 cell motility-related pathologies,Citation3 mitochondrial diseases,Citation4 the study of the mechanism of action of certain drugs,Citation5 cancer,Citation6 neurodegenerative disordersCitation7 and lysosomal-related disordersCitation8 among others. D. discoideum also provides useful ways to study the mechanisms and the physiological roles of autophagy.Citation9 As in in other organisms, autophagy is required for D. discoideum to survive starvation, for the turnover of proteins, to remove protein aggregates and is also fundamental during infection by intracellular pathogens. The first description of autophagic vacuoles in D. discoideum (defined at that time simply as “bodies containing partially degraded cytoplasmic material”) comes from transmission electron microscopy (TEM) studies of germinating spores in 1969,Citation10 shortly after Christian de Duve coined the term autophagy.Citation11 We have come a long way from these initial morphological observations and now begin to understand the significance and the mechanisms of this crucial cellular process in the context of a whole organism.
During starvation, autophagy is responsible for the liberation of nutrients to maintain viability. Therefore, defects in autophagy drastically reduce the ability of cells to survive prolonged periods of nutrient deprivation. Typically, wild-type D. discoideum cells survive for more than 10 d under amino acid starvation, whereas autophagy-deficient mutants start losing viability after just 24 h.Citation12,13 During starvation, autophagy is also responsible for a reduction in both cell volume and total protein.Citation12,13 The D. discoideum developmental program takes place in the absence of nutrients; therefore, the intracellular degradation and recycling of the cell´s own material by autophagy becomes essential as a source of energy and simple metabolites required for the biosynthetic pathways taking place during development. Thus, it is not surprising that autophagic dysfunction leads to abnormal development, which facilitates the identification of autophagy mutants in the laboratory. The severity of the phenotype ranges from complete lack of aggregation to the formation of multi-tipped mounds, which are unable to form normal stalks and viable spores ().Citation13 It is unclear how much of the defects in development are simply the result of imbalanced nutrient homeostasis or whether more specific signaling events are involved. However, the signaling peptide SDF-2 (spore differentiation factor 2) is essential for spore formation, and its precursor protein, AcbA, is secreted by an unconventional mechanism that requires autophagy.Citation14 Moreover, terminal stalk differentiation also seems to require autophagy for the formation of the vacuoles that accompany stalk cell differentiation.Citation15 Nonrecyling roles for autophagy are therefore important for the terminal differentiation of both spores and stalk cells.
Three forms of autophagy have been described that differ in the mechanism by which the cargo is delivered to the lysosome. Chaperone-mediated autophagy,Citation16 microautophagy,Citation17 and macroautophagy. Macroautophagy (denoted here as autophagy for simplicity) is the best known, the most conserved autophagic pathway and the only one described in D. discoideum so far. Although autophagy was initially thought to simply be a mechanism to recycle nutrients by nonselective self-degradation (so called bulk autophagy), the enormous importance of selective autophagy is now becoming clear. The selective process is devoted to the specific degradation of abnormal protein aggregates (aggrephagy), organelles (mitophagy, pexophagy, ribophagy, reticulophagy, nucleophagy) or pathogens (xenophagy). Impaired selective autophagy is therefore thought to underlie the etiology of numerous diseases.Citation18
The hallmark of autophagy is the formation of intracellular double-membrane vesicles (autophagosomes), which eventually fuse with lysosomes, leading to degradation of the cargo and the inner membrane. The basic molecular machinery that controls autophagy was initially discovered in Saccharomyces cerevisiae, in which the proteins involved were named Atg, for autophagy-related. These proteins form different complexes that are required for the induction, elongation and completion of the autophagic process. This has been extensively reviewed in the past few years.Citation19-21 Most of these proteins and complexes can be recognized in D. discoideum by sequence homology as summarized in . A more detailed description of the putative D. discoideum Atg protein homologs was included in a previous review.Citation9 The generation and analysis of a number of mutants from each complex have subsequently confirmed the functional conservation of the autophagic machinery in D. discoideum (the available mutants and their phenotypes are depicted in ). Lack of aggregation is the phenotype observed for atg1−,Citation22 DDB_G0269162−/atg13−,Citation23 and vmp1−.Citation24 Formation of multiple tips in mounds is the phenotype observed in atg101−,Citation23 atg5−,Citation13 atg6A−,Citation22 atg7−,Citation13 atg8−,Citation22 atg9−,Citation25 tipD−/atg16−Citation26-28 and tipC−.Citation27 Importantly, additional non-Atg proteins have been found to be involved in Dictyostelium autophagy such as the ER transmembrane protein Vmp1 (vacuole membrane protein 1), which demonstrates the usefulness of Dictyostelium as an alternative simple genetic system to study autophagy.
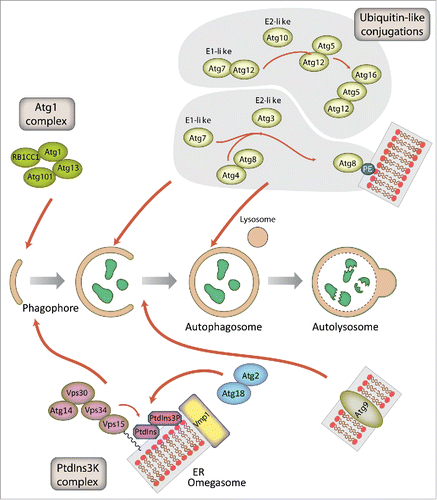
Figure 2. Schematic representation of autophagosome formation and the proteins involved in D. discoideum. The autophagic proteins were identified by sequence homology with those described in yeast and mammalian cells. The predicted complexes have been organized using the information available from the yeast S. cerevisiae, mammalian cells and D. discoideum. The inductive stage depends on the serine/threonine Atg1 kinase complex and the class III PtdIns3K complex. The latter complex generates the signaling lipid PtdIns3P, which is essential for the recruitment of Atg18 and Atg2. The elongation of the phagophore membrane requires 2 ubiquitin-like (Ubl) conjugation systems (upper right). In the first conjugation reaction Atg12 is covalently bound to Atg5. Atg12–Atg5 interacts with Atg16 and localizes to the phagophore membrane, from which it regulates the second conjugation reaction that attaches the protein Atg8 (known as LC3 in mammals) to phosphatidylethanolamine (PE) of the expanding phagophore membrane. Atg8 initially localizes to both sides of the phagophore and remains inside the completed autophagosome until fusion with the lysosome occurs; Atg8 on the cytosolic side of the autophagosome is cleaved from PE by Atg4 and recycled, whereas Atg8–PE of the lumenal leaflet of the bilayer is degraded. Atg9-containing vesicles supply membrane for phagophore expansion. Recent advances have clarified the composition of the Atg1 complex and the role of Vmp1 as a key protein in the origin of the autophagosomes from the ER.
Methods and tools to study autophagy in D. discoideum
Over recent years the number of tools available to study autophagy in D. discoideum has expanded greatly. TEM remains the gold standard for identifying the double-membrane autophagic structures, but it is difficult and time-consuming to obtain quantitative data. In particular, as each D. discoideum cell only contains approximately 5 autophagosomes under conditions of amino acid starvation,Citation29 very few are present in each of the thin sections required for TEM.Citation13 However, with recent advances in 3D electron microscopy it should now be possible to reconstruct complete amoeba, although quantification of autophagosomes has yet to be demonstrated. In contrast, light microscopy has been extensively used, allowing rapid quantification of large numbers of cells, and dynamic information to be captured.
It is often experimentally desirable to induce autophagy in a defined manner. Typically in mammalian cell cultures autophagy is induced using either starvation e.g. Hank's balanced salt solution or drug treatment, e.g., rapamycin, which inhibits MTORC1 (mechanistic target of rapamycin [serine/threonine kinase] complex 1) function. In D. discoideum, complete starvation can be induced using non-nutrient buffers but this initiates development, complicating interpretation. To avoid this, it is preferable to use a defined medium such as SIH medium lacking arginine and lysine (available from ForMedium for use in SILAC). Within 5 min this gives a comparable induction of autophagy to complete starvation, without cells entering development.Citation29 Although D. discoideum has a functional rapamycin-sensitive TORC1 complex,Citation30 short-term rapamycin treatment is unable to activate autophagy.
Several markers have been used to study autophagy in D. discoideum. In mammalian systems the most commonly used markers are MAP1LC3/LC3 (microtubule associated protein 1 light chain 3) family proteins, the ubiquitin-like (Ubl) orthologs of yeast Atg8 that associate with autophagic entities from initiation until recycling.Citation31 D. discoideum possess 2 Atg8 paralogs (Atg8a and b), which are both good markers of autophagosomes although they play partially nonredundant roles (discussed below).Citation32 Another useful marker is Atg18, the ortholog of mammalian WIPI2. Atg18 localizes to omegasome-anchored phagophores (the precursors to autophagosomes) at initiation events but, in contrast to Atg8, dissociates immediately after autophagosome formation is completed.Citation29,33,34 Atg18 is therefore specific for the formation of new autophagosomes.
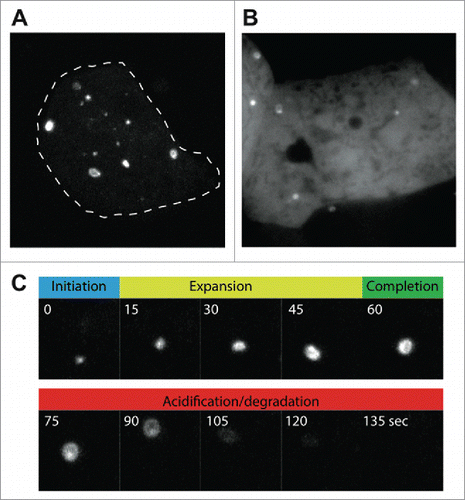
Immunolabeling of endogenous autophagy proteins allows a variety of markers to be observed, and several good antibodies against D. discoideum Atg8 have been generated.Citation32,35 While these antibodies are useful for many experiments, fixed samples lack dynamic information. It is therefore often useful to perform live-cell imaging and track markers in real-time to dissect the different phases of autophagosome formation. GFP-Atg8 fusions permit visualization from initiation through expansion, closure and fusion with the lysosome, whereupon the GFP fluorescence becomes quenched. Movement in the Z-axis, however, makes following individual autophagosomes over time challenging. To reduce this problem, sheets of agarose can be placed upon the cells to compress them, reducing cell height and improving image quality. This approach allows individual autophagosomes to be followed from an initial punctum, to a cup and then a complete circular autophagosome ( and Movie S1). The timing of each phase can be quantified from such movies, as well as the time it takes for the GFP fluorescence to be quenched, giving an indirect combined measure of acidification and proteolysis. This is a powerful method to directly quantify autophagosome dynamics, but does not represent basal autophagy levels, as the compression itself potently induces autophagosome initiation.Citation29
Figure 3. Live-cell imaging of autophagosome formation in D. discoideum, using (A) GFP-Atg8a, and (B) GFP-Atg18. Images were obtained under compression to improve clarity, and to allow individual autophagosomes to be followed over time. (C) Shows the different stages of autophagosome formation from the cell in (A) marked by GFP-Atg8a. The autophagosome shown in (C) has been marked by an arrow in Movie S1.
An additional way to differentiate autolysosomes (the product of autophagosomes fusing with lysosomes) is by fusing Atg8 to both GFP and RFP in tandem.Citation36 As RFP is less sensitive to quenching by low pH, only the red signal can be observed in mammalian autolysosomes, whereas phagophores and autophagosomes emit both green and red fluorescence. Unfortunately, D. discoideum lysosomes have a much lower pH than their mammalian counterparts (3.5 compared to 4.5-5.0), which is sufficient to quench both GFP and RFP. Treatment with lysosomotropic agents such as ammonium chloride (NH4Cl), however, can be used to partially elevate lysosomal pH, and reveal autolysosomes, with the caveat that this treatment itself will induce autophagy and lead to osmotically distended compartments with dilute content.Citation37,38
Autophagic flux refers to the net amount of material that is captured and degraded by this pathway over time. This is an important but complex and dynamic measurement and is not a simple reflection of the number of autophagosomes. Currently, the best method to measure flux is based on observing the autophagic cleavage of GFP from its fusion with cytosolic proteins.Citation38 Although GFP fluorescence is rapidly quenched within acidic lysosomes, the GFP protein itself is relatively resistant to lysosomal proteolysis. When GFP is fused to cytosolic proteins, captured and delivered to lysosomes by autophagy, GFP fragments accumulate while the rest of the fusion protein is degraded.Citation39 GFP cleavage fragments can therefore be quantified by western blot analysis of whole cell lysates probed with anti-GFP antibodies. As D. discoideum are professional phagocytes, they possess an extremely efficient proteolytic machinery and free GFP is barely detectable due to rapid degradation. Therefore NH4Cl treatment is again required to inhibit proteolysis sufficiently for cleaved GFP fragments to accumulate.Citation37,38 Band intensities for GFP-marker and free-GFP can then be quantified to give a measure of autophagic flux. It is again important to note that the concentrations of NH4Cl required are very high (0.2-0.3 M), and likely to induce autophagy themselves. Nonetheless, this system provides a good measure of maximum autophagic capacity, and is useful to demonstrate gross effects of mutations on the pathway.Citation37,38
The array of techniques available, coupled with orthologs of most mammalian autophagy genes, makes D. discoideum a robust model for the study of autophagy. While a system for studying autophagic flux in noninduced conditions is yet to be established, significant progress has been made and D. discoideum provides many useful means in which to study this pathway.
Phagophore nucleation: The Atg1 complex in D. discoideum is closely related to themammalian ULK1 complex
The Atg1/ULK1 protein kinase complex works at the initiation step in both selective and nonselective autophagy.Citation40 In mammals, ULK1 phosphorylates BECN1 to activate the PIK3C3/VPS34 lipid kinase,Citation41 and phosphorylation of the transmembrane protein Atg9 by its budding yeast homolog Atg1 plays a key role in the recruitment of Atg18 and Atg8 to the phagophore assembly site (PAS).Citation42 Initial studies of nonselective autophagy in yeast identified the different components of the Atg1 complex: the Atg1 protein kinase itself, the regulatory subunit Atg13 and the Atg17-Atg31-Atg29 scaffolding subcomplex.Citation43 Selective autophagy processes, such as the cytoplasm-to-vacuole targeting (Cvt) pathway, involve a different scaffold protein, Atg11 (). Although not absolutely required in nonselective autophagy, Atg11 interacts with Atg29Citation44 and, thus, it is still unclear whether Atg11 and the Atg17-Atg31-Atg29 subcomplex form mutually exclusive complexes with Atg1. Dephosphorylation of the TOR kinase substrate Atg13 upon nutrient starvation promotes Atg1 complex assembly in nonselective autophagy.Citation45 In contrast, under nutrient-rich conditions, selective autophagy receptor-bound targets activate Atg1 via the scaffold protein Atg11.Citation46 Autophagy receptors have been identified in mitophagy (Atg32), pexophagy (Atg36), nucleophagy (Atg39), reticulophagy (Atg40) and the Cvt pathway (Atg19 and Atg34).Citation47
Figure 4. The Atg1/ULK1 complex in yeast, mammalian cells and D. discoideum. Proteins are colored according to their conservation, and protein size is proportional to the molecular weight. The inhibition of the Atg1/ULK1 kinase activity by TORC1, and its activation through binding of the autophagy receptors to the Atg11 subunit, are indicated.
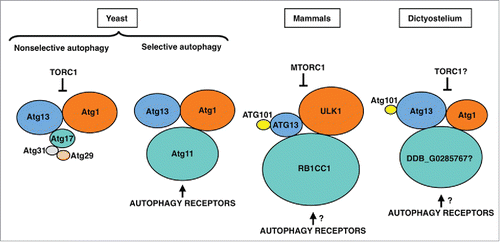
The ULK1 protein complex in mammals differs from the budding yeast Atg1 complex in several aspects. This complex consists of the ULK1 protein kinase and 3 proteins, ATG13, ATG101 and the scaffold protein RB1CC1/FIP200 (). ATG101, which is absent in budding yeast, binds to and stabilizes Atg13.Citation48,49 The crystal structure of the Atg13-Atg101 complex revealed that the HORMA domains present in these 2 proteins heterodimerize.Citation50,51 In budding yeast, Atg13 contains a 3-strand β-sheet insertion in the HORMA domain that stabilizes this protein in the absence of ATG101. The scaffold protein RB1CC1 is the functional counterpart of both Atg17 and Atg11 in yeast, and results from phylogenetic analysis suggest that Atg17 arose via gene duplication of Atg11 and loss of the C-terminal region.Citation52 This gene duplication event and functional specialization appears to be essentially restricted to fungi. In mammals, the HTT (huntingtin) protein also shares structural similarity to yeast Atg11 and, like RB1CC1, functions as a scaffold protein in the ULK1 complex.Citation53
The Atg1 complex in D. discoideum is evolutionarily closer to the mammalian ULK1 complex than its budding yeast counterpart. In addition to the Atg1 protein kinase,Citation54 Atg13 and Atg101 homologs have been identifiedCitation23 and interaction studies have shown that Atg13, like its mammalian homolog, binds to both Atg1 and Atg101 (). The likely RB1CC1/Atg11 homolog in D. discoideum is DDB_G0285767.Citation52 A putative Atg17 homolog has also been reported but functional studies argue against a role in autophagy,Citation23 thus supporting the idea that Atg17 proteins are restricted to fungi. Similarities between mammals and D. discoideum are not limited to the core components of the complex, as shown by the conservation of the mammalian selective autophagy receptor and Atg8-binding protein Sqstm1/p62 in D. discoideum.Citation23,55 Interestingly, D. discoideum Atg1 kinase also interacts with the pentose phosphate pathway (PPP) enzyme transketolase (Tkt).Citation23 Although no direct phosphorylation has been reported, the activity of TKT is altered in strains lacking or overexpressing Atg1, suggesting a possible crosstalk between autophagy and the PPP. Interestingly, a recent report shows that another component of the PPP, the ribose-5-phosphate isomerase (RpiA), regulate autophagy in HeLa cells.Citation56
The origin of the autophagosomal membrane, function of Vmp1 and regulation of PtdIns3P
The origin and elongation of the phagophore membrane are still the subject of intense research. In mammalian cells the ER, establishing close contacts with other organelles such as mitochondria and lipid droplets, forms a specialized cradle-like structure known as the omegasome.Citation57 This structure is enriched in the signaling lipid PtdIns3P, generated by the class III phosphatidylinositol 3-kinase (PtdIns3K) PIK3C3/VPS34. It is thought that the phagophore expands by fusing with vesicles that originate from different cellular compartments. Some of these vesicles contain the transmembrane protein ATG9 and emanate from the Golgi and recycling endosomes,Citation58,59 whereas others can originate from the ER-Golgi intermediate compartment (ERGIC).Citation60-62
D. discoideum autophagosomes form simultaneously in different locations of the ERCitation29 and are thus, in this respect, more similar to those in mammalian cells than yeast, in which autophagosomes originate from the PAS, a single spot near the vacuole. It has been proposed that S. cerevisiae has become highly specialized during evolution. As a result, some processes such as autophagy have diverged from a more universally conserved mechanism, which has remained unaltered in other organisms such as D. discoideum.Citation63 A valuable example of these differences is the presence in D. discoideum of the conserved protein Vmp1 that is required for correct formation of the omegasome, but absent in S. cerevisiae and other fungi.
The phenotype of Vmp1-deficient cells differs from that of cells lacking classical Atg proteins because it affects a wide array of processes apparently not connected to autophagy as described in D. discoideum and Chlamydomonas reinhardtii.Citation24,64 Autophagic flux is blocked in Vmp1-deficient D. discoideum but PtdIns3P production and subsequent recruitment of the autophagy machinery to the ER still occurs. Indeed, PtdIns3P is abnormally high in the mutants and correlates with the accumulation of enlarged omegasomes and LC3-containing structures in D. discoideum,Citation65 C. elegansCitation66 and mammalian cells.Citation67 Taken together, these studies suggest that Vmp1 might be required for the correct structure of the omegasome and/or the capacity of the phagophore to elongate and become a functional autophagosome. Since Vmp1 is an ER-resident protein it is tempting to speculate that Vmp1 generates an ER microenvironment that orchestrates the autophagic machinery to allow the correct expansion of the phagophore rather than the recruitment of Atg proteins ().
Figure 5. Vmp1 is essential for correct PtdIns3P signaling and omegasome formation in D. discoideum and mammalian cells. Vmp1 accumulates in subdomains of the ER where the autophagic machinery is recruited. PtdIns3P is formed at these domains to regulate omegasome formation and phagophore elongation. In the absence of Vmp1, PtdIns3P is aberrantly generated leading to persistent recruitment of autophagy proteins and unproductive autophagosome formation.
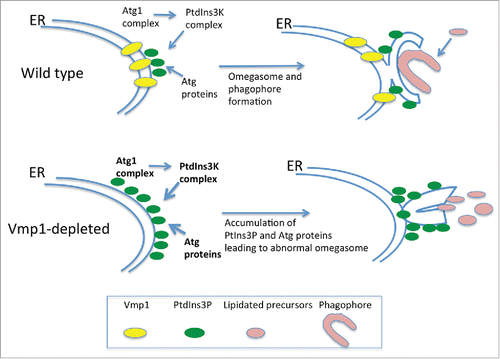
Another interesting phenotype of the Vmp1 mutant is the defect in macropinocytosis, a mechanism of nutrient uptake that allows D. discoideum to grow in liquid media. The abnormal accumulation of PtdIns3P in the mutant impairs macropinocytosis indirectly and prevents growth in liquid media. This conclusion is based on experiments that preclude the accumulation of PtdIns3P. Atg1 is upstream of the PtdIns3K and therefore a double mutant in Vmp1 and Atg1 lacks this abnormal signaling.Citation65 Remarkably, macropinocytosis and cell growth in liquid media is recovered in the double mutant, suggesting that additional nonautophagy-related defects may occur as a result of an abnormal regulation of PtdIns3P signaling at the omegasome.Citation65 It will be interesting to determine if such a phenomenon also occurs in mammalian cells.
The 2 ubiquitin-like (Ubl) conjugation systems are highly conserved in D. discoideum
The proteins involved in the 2 Ubl conjugation systems are highly conserved from yeast to mammalian cells and very likely function in an analogous manner in all eukaryotes including D. discoideum.Citation9 However, while yeast harbors only one isoform for each of the 8 different proteins of the 2 Ubl conjugation systems there are 2 paralogs for ATG16 (ATG16L1, L2), 4 for ATG4 (ATG4A, B, C, D) and 7 for ATG8/LC3 in mammals.Citation9,68,69 In particular, the large number of ATG8/LC3 paralogs in humans and mice complicates functional studies in these organisms. These proteins are grouped into the MAP1LC3 (microtubule associated protein 1 light chain 3) subfamily with 4 members (LC3A, B, B2 and C), and the GABARAP (GABA type A receptor-associated protein) subfamily with 3 members (GABARAP, GABARAPL1 and GABARAPL2/GATE16) protein.Citation68 The GABARAPL2 protein is approximately equally distant from the LC3 and the GABARAP subfamilies () and the precise functions of the individual proteins are largely unresolved.Citation70 In contrast, fungi possess only a single ATG8 gene, while plants, insects, nematodes and amoebozoa, like D. discoideum, usually have 2 genes for Atg8.
Figure 6. Evolutionary relationship of Atg8/LC3 family members. Phylogenetic analysis of Atg8/LC3 family proteins from animals (red), amoebozoa (orange), fungi (blue) and plants (green). A CLUSTALX alignment was used to create a phylogenetic tree with the TreeView program. The scale bar indicates amino acid substitutions per site. Bootstrap values are provided at the node of each branch. GenBank, SwissProt or dictyBase (http://dictybase.org/) accession numbers are provided on the right of the tree. Hs, Homo sapiens; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Nc, Neurospora crassa; Sc, Saccharomyces cerevisiae; Ac, Acanthamoeba castellanii; Dd, D. discoideum. LC3, microtubule-associated protein 1 light chain 3; GABARAP, GABA type A receptor-associated protein; GABARAPL2/GATE16, GABA type A receptor associated protein like 2; lgg: LC3, GABARAP and GATE-16 family.
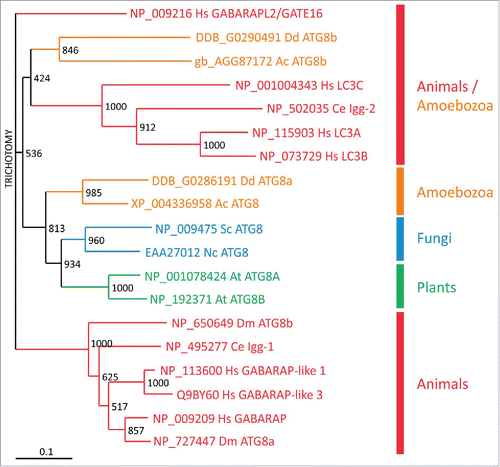
Phylogenetic analysis showed that D. discoideum Atg8a and Acanthamoeba castellanii Atg8 are situated in the same evolutionary branch as the orthologs from fungi and plants. In contrast, the 2 Atg8b proteins from D. discoideum and A. castellanii appear evolutionarily more closely related to the LC3 subfamily from animals (). Live cell imaging of D. discoideum cells expressing RFP-Atg8a and GFP-Atg8b showed that Atg8b associates with autophagosomes before ATG8a.Citation32 In mammals, LC3 subfamily members are involved in the elongation of the phagophore, whereas the GABARAP subfamily seem to be essential for a later stage in maturation.Citation71,72 Thus, Atg8b is likely the functional ortholog of the mammalian LC3 subfamily, which is also supported by the grouping in the evolutionary tree (), and Atg8a the ortholog of the GABARAP subfamily.Citation32 It appears therefore, that duplication of Atg8 early in eukaryotic evolution allowed specialization of the paralogs during autophagosome maturation. Expansion of the respective subfamilies in mammals likely led to the acquisition of further specialized functions in more complex animals.Citation70 Similarly, the Atg4 paralogs and the Atg16 paralog in the 2 Ubl conjugation systems may enable the fine-tuning of the activation of the different Atg8/LC3 family members.
Atg9 studies in D. discoideum reveal new functions
The multimembrane spanning Atg9 protein is the only known integral membrane protein of the core autophagic machinery and thought to be involved in the delivery of membrane lipids to the growing autophagosome.Citation73 Atg9 orthologs exist in all eukaryotic species so far examined and the protein is essential for autophagy. For example, in Drosophila melanogaster, Atg9 depletion reduces both the number and size of autophagosomes, and blocks the fusion of autophagosomes with lysosomes.Citation74 Modulation of Atg9 levels in yeast strains directly correlates with the frequency of autophagosome formation and autophagic activity.Citation75
The PAS in yeast originates from Atg9-positive clusters, called Atg9 peripheral sites.Citation76 The origin of these clusters, however, has remained poorly understood. They localize in proximity to the mitochondrial membraneCitation77 and may represent vesicles derived from mitochondrial-ER contact sites. Alternatively, based on partial colocalization between Atg9 and the late Golgi protein marker Sec7, the Atg9 peripheral sites could emerge as a new organelle through the delivery of newly synthesized Atg9 to these sites via the secretory pathway.Citation76 Upon delivery of the lipids, Atg9 is no longer required and may be retrieved to the peripheral sites by an as yet unknown mechanism.
In mammals, there are 2 ATG9 paralogs, namely ATG9A/ATG9L1 and ATG9B/ATG9L2. ATG9A (ATG9 hereafter) is ubiquitously expressed in human adult tissues, whereas ATG9B is only expressed in placenta and pituitary gland.Citation78 Mammalian ATG9 is localized at the ER, the trans-Golgi network and early, late and recycling endosomes.Citation79,80 Recently, it was shown that in D. melanogaster and in human cells the Atg1/ULK1 protein kinase phosphorylates a MYLK/myosin light chain kinase, which in turn activates MYL/myosin II regulatory light chain. This is followed by myosin II activation, which apparently drives transport of ATG9-containing membranes to the phagophore.Citation81 D. discoideum Atg9 does not colocalize with mitochondria, the ER or lysosomes; however, there is a partial colocalization with the Golgi apparatus and many Atg9-GFP-containing vesicles localize along microtubules and accumulate around the microtubule organizing center.Citation25 Disruption of Atg9 results in a pleiotropic phenotypes with severe impairments in development, slug migration, vegetative growth, phagocytosis, clearance of Legionella pneumophila and proteasomal activity.Citation25 A similar pleiotropic phenotype is observed in tipD−/atg16− cells.Citation26
Unexpectedly, some of the defects of the atg9 knockout mutant can be partially or fully restored by expression of wild-type or point-mutated CdcD/p97, an evolutionarily highly conserved member of the AAA-ATPase family.Citation82 VCP/p97, the mammalian homologue, is a key player in ER-associated protein degradation, in the ubiquitin-proteasome system for protein degradation system, in aggresome formation and also in autophagosome maturation.Citation83-85 Heterozygous mutations in the human VCP/p97 gene cause autosomal-dominant inclusion body myopathy with early onset Paget disease of bone and frontotemporal dementia and further neurological disorders;Citation86,87 however, the molecular basis of the pathogenesis remains unknown. VCP is essential for autophagosome maturation in mouse embryonic fibroblasts and, moreover, disease-causing VCP point mutations impair autophagy.Citation88
Interestingly, atg9−, tipD−/atg16− and atg9− tipD−/atg16− double mutant D. discoideum cells have a severe and counterintuitive defect in proteasomal activity. Since the amounts of proteasomal subunits are not altered in knockouts of atg9 or tipD/atg16, this result suggests that intact autophagy is required for optimal proteasomal activity.Citation26,82 Furthermore, this phenotype is fully or partially suppressed by overexpression of wild-type or point-mutated cdcD/p97 in atg9− cells. This result provides evidence that in D. discoideum CdcD/p97 and Atg9 are functionally directly or indirectly linked and supports the existence of a delicate balance between the 2 major protein degradation pathways, proteasomal degradation and autophagy. Disruption of this balance causes severe impairment of essential cellular functions in lower eukaryotes such as D. discoideum but ultimately may cause late-onset complex diseases in humans. How exactly the ubiquitin-proteasome system and autophagy are interconnected needs to be unraveled in the future.
Autophagy and disease: New leads from the social amoeba in human genetic disorders
Autophagic dysfunction may lead to defects in protein and organelle homeostasis associated with numerous human diseases.Citation89 A very relevant example is the connection of autophagy with major neurodegenerative diseases (including Alzheimer, Parkinson and Huntington diseases), often associated with the accumulation of abnormal protein aggregates or nonfunctional organelles. Autophagy dysfunction in D. discoideum often leads to the accumulation of large ubiquitinated protein aggregates.Citation55 Ubiquitin is a small regulatory protein that can be covalently linked to proteins to target them for proteasomal or autophagic degradation. These protein aggregates can be observed by both immunofluorescence microscopy using anti-ubiquitin antibodies and by western blot. As these aggregates are insoluble in 1% Triton X-100, they can rapidly be separated from whole cell lysates by centrifugation, separated by SDS-PAGE and detected using commercially available anti-ubiquitin antibodies. Ubiquitinated proteins are dramatically increased in the Triton-insoluble fraction of autophagy-deficient mutants.Citation55 The size and number of these aggregates correlates well with the severity of the phenotypes of the different autophagy mutants.Citation55 Interestingly, the protein aggregates formed in the Vmp1 mutant also accumulate the receptor protein Sqstm1,Citation55 which is involved in the clearance of several ubiquitinated cargos in mammalian cells and has been implicated in aggregation disorders and infectious diseases.Citation90
Packing proteins into aggregates, if properly degraded by autophagy, could function as a regulated cellular mechanism to handle abnormal proteins that otherwise would be toxic. However, several studies in D. discoideum suggest that the protein aggregation phenotype might be deleterious for cell function. Overexpression of the actin-binding protein VasP fused to an endosomal targeting signal leads to the formation of huge actin aggregates reminiscent of Hirano bodies,Citation91 structures that are often present in neurodegenerative diseases. These actin aggregates sequester a number of actin binding and endosomal proteins promoting their disappearance from their normal location in the cytoplasmCitation91 leading to cytoskeletal defects. These findings open the possibility that protein sequestration might also contribute to neuronal malfunction in these pathologies. Hirano body-like aggregates can also be induced in D. discoideum by the overexpression of a truncated form of a 34-kDa actin-binding protein.Citation92 Another report showed that both autophagy and the proteasome pathway contribute to the degradation of Hirano bodies in D. discoideum.Citation91 The autophagosome marker protein GFP-Atg8 colocalizes with Hirano bodies in wild-type D. discoideum cells, but not in cells deficient in the autophagic proteins Atg5 or Atg1.Citation93 Interestingly, Hirano bodies can be released by exocytosis following autophagy, which could explain the presence of these aggregates in both intracellular and extracellular spaces in the brain.Citation93
Another example using D. discoideum to study autophagy-related pathologies was the discovery of a possible connection of the rare disease Chorea-acanthocytosis (ChAc) with autophagy. ChAc is a neurodegenerative disease associated with abnormally shaped erythrocytes known as acanthocytes, leading to disability and premature death. ChAc is caused by loss-of-function mutations in VPS13A, most of which lead to a decrease or absence of the VPS13A protein (also known as Chorein).Citation94,95 The molecular function of this protein is not known and the literature provides fragmented and nonconclusive results. Proposed functions for VPS13A include membrane trafficking, membrane morphogenesis, phagocytosis and actin cytoskeleton regulation.Citation96-99 There are 3 additional VPS13 proteins in humans (VPS13B,C and D)Citation100 and mutations in 2 of them, VPS13B and C, lead to Cohen syndrome and Parkinson disease, respectively.Citation101,102 D. discoideum has 6 VPS13-related proteins, which are more similar to human VPS13A and C than to VPS13B or D. One of these proteins was initially named TipC since the phenotype of an insertional mutant was the formation of multiple tips in large mounds, a typical phenotype of autophagy mutants as pointed out above.Citation28 Several years later this mutant was revisited and the presence of a strong autophagy blockade was confirmed.Citation27 Subsequently, additional studies of the autophagic pathway in VPS13A-depleted human HeLa cells confirmed that autophagy could play a role in the etiology of this devastating disease.Citation27
Defects in either the retromer or WASH (WAS protein family homolog) complexes also lead to endosomal trafficking and autophagy defects associated with human diseases. The WASH complex regulates the formation of actin filaments on endosomes, driving multiple endocytic protein sorting and recycling events,Citation103 and mutations in the gene encoding KIAA0196/strumpellin, a component of the WASH complex, cause a form of autosomal dominant hereditary spastic paraplegia.Citation104 Furthermore, an aspartate to asparagine substitution (D620N) in VPS35, a component of the retromer sorting complex that recruits WASH to endosomes, results in autosomal dominant familial Parkinson disease.Citation105 D. discoideum have all components of the WASH complex, and mutants in WASH also have a defect in autophagy. However, while defects either in the retromer or WASH lead to an early inhibition of autophagosome formation due to defective trafficking of ATG9-containing vesicles and deficient BECN1 activation in mammals,Citation105,106 D. discoideum WASH null cells do not have defects in the induction of autophagosome formation.Citation12 The observed blockade in autophagy is due to impaired recycling of vacuolar-type H+-translocating ATPase (V-ATPase) subunits and lysosomal hydrolases that leads to a late defect in lysosome recycling that blocks degradation of the cargo.Citation12
Mitochondrial diseases are complex degenerative disorders caused by mutations in mitochondrial proteins encoded in the nuclear or mitochondrial genome. D. discoideum has been used to study the cytopathology involved in mitochondrial disease, specially those involving alterations in pathways that affect AMP-activated protein kinase (AMPK), the kinase required to sense cellular energy levels and regulate autophagy through MTOR, ULK1 and BECN1.Citation4,107,108 Unexpected connections with autophagy are observed in D. discoideum cells deficient in components of the NADH:ubiquinone oxidoreductase or complex I, the first complex of the respiratory chain. Mutants affecting complex I assembly factors Ndufaf5 and MidA/Ndufaf7 have a strong activation of bulk autophagy,Citation109,110 but not mitophagy, which, in the case of MidA could be mediated by a chronic activation of AMPK. Given the importance of autophagy in cellular homeostasis, this connection might have relevance in human pathology and subsequently a similar phenomenon has been observed in skin fibroblast cultures derived from patients with mitochondrial disease.Citation111,112
Autophagy and bacterial infectious diseases in D. discoideum
Autophagy also represents an efficient mechanism for eukaryotic cells to capture and kill invading pathogens. The killing of intracellular pathogens by autophagy, termed “xenophagy,” is therefore emerging as a major regulator of both cytosolic and vacuolar pathogens. Many bacterial pathogens are able to manipulate, either by inhibition or induction, the host xenophagic response to take advantage of autophagy for their own benefits.Citation113 Since D. discoideum lives in the soil and feeds on bacteria and yeasts, it likely developed mechanisms to discriminate between food and pathogenic organisms early during evolution. Due to its easy experimental manipulation, D. discoideum has become a useful model to study host-pathogen interactions and xenophagyCitation9 and in the past 5 y has been used to study interactions of the host autophagic machinery with pathogenic microbes such as Salmonella enterica serovar Typhimurium (S. enterica), L. pneumophila, Francisella noatunensis, Staphylococcus aureus and Mycobacterium marinum.Citation26,35,114-118 Furthermore, D. discoideum has also been used to identify and dissect the mechanisms of action of drugs with potential utility in medical therapy against pathogens.Citation119-122
M. marinum infects mainly fish and frogs, but it can also produce skin lesions in humans. Due to safety reasons and to the faster growth rate than its close relative M. tuberculosis,Citation123 M. marinum represents an easier tool to research mycobacterial pathogenesis. In mammalian cells and zebrafish, M. marinum induces an autophagic response that includes upregulation of some autophagy-related genes and LC3 recruitment to the mycobacteria-containing vacuole (MCV).Citation124-128 This recognition by the autophagy machinery requires the activity of the mycobacterial pore-forming toxin ESAT-6, and engagement of the host autophagy-related proteins SQSTM1, MYD88 (myeloid differentiation primary response 88), DRAM1 (DNA damage regulated autophagy modulator protein 1) and TMEM173/STING (transmembrane protein 173).Citation124,125,129,130 ESAT-6 is secreted by the type VII secretion system ESX-1, and induces permeabilization of the MCV membrane, exposing M. marinum to the cytosol.Citation131 As a consequence, the ESX-1-dependent membrane damage allows the host autophagy machinery to recognize bacterial products including DNA, and to induce an autophagic response dependent on the cytosolic DNA-sensing TMEM173/STING pathway.Citation125,129 However, apart from its indirect role in MCV permeabilization, ESX-1 seems to be dispensable for ubiquitination of M. marinum in macrophages.Citation130
It has been recently shown that, in D. discoideum, the autophagy machinery is not only recruited to cytosolic M. marinum, but it also targets the bacteria escaping out of hosts cell by the nonlytic mechanism called “ejection.”Citation35,132 During this process, ubiquitin, Atg8, Sqstm1 and Atg18 localize at the distal pole of the ejecting bacteria, with a proposed function in sealing the plasma membrane wound.Citation35 This recruitment of autophagy to the ejecting bacteria seems to be independent on ESX-1, as well as on other M. marinum virulence factors. However, host Atg1, Atg5, Atg6A and Atg7 are all required for Atg8 association with the ejecting bacterium suggesting that this process may be entirely driven by the host. Furthermore, even though Atg1 is not necessary for ejectosome formation, it is essential for the tight membrane sealing during ejection, avoiding cytosolic leakage and, thereby, death upon bacterial egress.Citation35
Interestingly, disruption of Sqstm1, encoding the only selective autophagy receptor protein described so far in D. discoideum, seems to have no effect on Atg8 recruitment to M. marinum. Gerstenmaier and collaborators thus suggest that other receptor proteins must exist in this amoeba.Citation35 Ubiquitin-binding CUE-domain containing proteins have been recently described to act as autophagy recepptors for protein aggregates in yeast and humans. Consistent with the evolutionary conservation of these proteins, 2 D. discoideum CUE domain-containing proteins, CnrD and DDB_G0274365/CueA also localize to MCVs (Drs. J. King and T. Soldati, unpublished results). In addition to the domain for ubiquitin binding, these proteins present LIR motifs in their sequences (), making them ideal selective autophagy receptor candidates.
Figure 7. Xenophagy in D. discoideum. (A) The autophagosomal marker Atg8 is recruited to the compartment harboring both S. aureus and S. enterica, which might be a response to the putative membrane damage produced by these bacteria. As a consequence, the bacteria are engulfed in phagophores and killed in autolysosomes, where the lumenal leaflet Atg8 is digested. After uptake by D. discoideum, F. noatunensis escapes phagosomal maturation and resides in a compartment lacking the V-ATPase. The majority of F. noatunensis replicates in the cytosol, while a small proportion of bacteria succumb to autophagy; M. marinum also prevents phagosomal acidification by the V-ATPase via WASH-induced actin polymerization. After proliferation inside the MCV, M. marinum is released to the cytosol, where it recruits the autophagy machinery, but does not seem to be fully engulfed in autophagosomes (unpublished observations). Finally, M. marinum is ejected through the D. discoideum plasma membrane in an autophagy-dependent manner; Atg9 controls L. pneumophila infection, whereas the autophagy activator kinase AMPK enhances its proliferation. Therefore, the function of autophagy during L. pneumophila in D. discoideum remains unclear. (B) Domain and motif organization of the already characterized D. discoideum autophagy receptor (DdAR) Sqstm1 and the 2 additional putative adaptors CnrD and DDB_G0274365/CueA. PB1, Phox and Bem1 domain; ZZ, zinc finger, ZZ-type; N_BRCA1, Next to BRCA1, central domain; UBA, ubiquitin-associated domain; CUE, ubiquitin system component Cue; U/C, UBA and CUE domains. Protein domains and LIR motifs were predicted with the web prediciton resources InterPro (http://www.ebi.ac.uk/interpro) and iLIR, respectively.
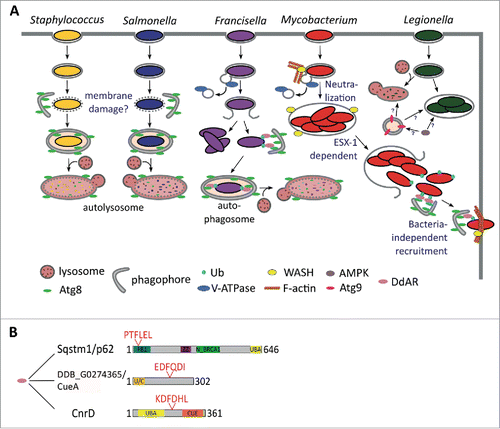
The WASH complex is also necessary for M. marinum ejection and is possibly involved in D. discoideum xenophagy.Citation118 During infection, WASH-induced actin polymerization prevents MCV acidification by promoting recycling of the V-ATPase, rendering the compartment more permissive to infection.Citation118 This correlates with previous results in mammalian cells, in which M. marinum modifies the composition of its MCV by depleting the V-ATPase and the lysosomal protease CTSD (cathepsin D), as also observed during infection of D. discoideum.Citation124,133
D. discoideum has also been recently established as a model host for another fish pathogen, F. noatunensis.Citation116 This Gram-negative bacterium exploits autophagy in human and murine macrophages.Citation134,135 In D. discoideum, the mRNA levels of atg8 and sqstm1 slightly increase during F. noatunensis infection, and Atg8, Sqstm1 and ubiquitin are recruited to the compartment containing this bacterium, suggesting an induction of autophagy. Furthermore, autophagy controls intracellular proliferation of F. noatunensis, since deletion of atg1 in D. discoideum increases the bacterial burden after 24 h of infection.Citation116
In contrast to growth of F. noatunensis, L. pneumophila shows similar proliferation kinetics when infecting wild-type and atg1−, atg5−, atg6A−, atg7− and atg8− D. discoideum cells. Likewise, this intracellular pathogen of amoebae and macrophages, rarely recruits Atg8, suggesting that xenophagy is not involved in amoebal immunity against L. pneumophila.Citation136 Whether L. pneumophila actively escapes autophagic capture in D. discoideum by uncoupling Atg8 from the phagophore membrane, via action of the RavZ toxin, as it does in human cells,Citation137 remains to be studied. Interestingly, Atg9 might be involved in controlling L. pneumophila, because its expression rises after 24 h of infection, and bacterial clearance decreases in atg9− cells. Uptake of this bacterium is also decreased in atg9−, tipD−/atg16− and atg9− tipD−/atg16− double-mutant D. discoideum cells.Citation25,26 However, the mechanism is unclear, because, contrary to atg9, the expression of TipD/Atg16 and Atg8a is downregulated after infection with L. pneumophila.Citation25 Interestingly, the intracellular proliferation of L. pneumophila in D. discoideum increases in cells overexpressing the catalytic subunit of AMPK. Since AMPK regulates ATG9 localization via the activation of ULK1 in mammalian cells,Citation138 the different L. pneumophila proliferation rates observed during infection of atg1−, atg9− and AMPK-overexpressing cells may indicate autophagy-independent functions for Atg9 and TipD/Atg16 as suggested by Xiong and collaborators.Citation26
Finally, it is controversial whether S. enterica, one of the Gram-negative bacteria more commonly used in laboratories to study the host innate immune responses at the molecular level,Citation139 is pathogenic or not for D. discoideum. S. enterica survives and proliferates inside very diverse human cell types,Citation140 but both its survival and death inside amoebae have been reported.Citation114,141,142 Sillo and collaborators proposed that the discrepancies observed might be due to the use of the antibiotic gentamicin, normally employed in S. enterica infection protocols for mammalian cells. Gentamicin is likely to be taken up in large quantities by macropinocytosis during D. discoideum infection, thus affecting intracellular bacteria.Citation114 Regardless, early during infection of D. discoideum, Atg8 is recruited to the compartment harboring S. enterica,Citation142 and concomitant cleavage of GFP-Atg8 suggests an enhanced autophagic response.Citation115 Together with the fact that atg1−, atg6A− and atg7− cells are more permissive for S. enterica growth,Citation142 this observation implies a rapid autophagy-dependent killing of these bacteria by D. discoideum cells.
Autophagy has also been implicated during infection of lipopolysaccharide-treated D. discoideum cells with S. aureus.Citation115 The fate of this Gram-positive bacterium inside amoebae is affected by autophagy, since its uptake is reduced in atg9− cells.Citation25,115 Treatment with lipopolysaccharide, which also induces bacteria clearance in an Atg1- and Atg9-dependent manner, further increases the localization of Atg8 to the vacuoles containing S. aureus.Citation115 Because S. aureus localizes to LC3-labeled compartments and subverts the autophagy pathway in mammalian cells,Citation143-145 manipulating autophagy with protease inhibitors or compounds blocking autolysosome formation is needed to confirm the induction or blockade of autophagy by these bacteria.Citation146
In conclusion, different bacterial species differentially subvert autophagy in D. discoideum, as occurs in mammalian cells. The D. discoideum autophagy machinery is recruited to S. aureus, S. enterica, F. noatunensis and M. marinum at different stages during infection (). This recruitment is normally induced by the damage or rupture of the bacteria-containing compartments and, as a consequence, the bacteria are killed in autolysosomes. However, both F. noatunensis and M. marinum are able to prevent phagosomal acidification by the V-ATPase, therefore, bypassing xenophagy.
Autophagy and cell death pathways in D. discoideum
In addition to caspase-mediated apoptosis in animal cells, an increasing number of nonapoptotic cell death types are being reported within and outside the animal kingdom. In these multiple different cell death types there might be common, conserved cell death core elements that can be analyzed through studying cell death in alternative model organismsCitation147 such D. discoideum.
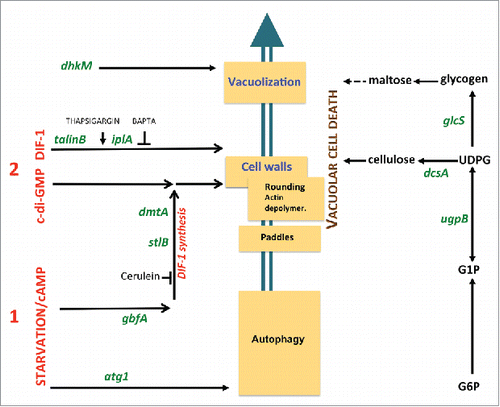
The D. discoideum fruiting body includes a stalk made of vacuolized, cellulose-walled dead cells.Citation148 This developmental cell death can be mimicked in vitro in monolayer conditionsCitation149-151 that include a stereotyped sequence of eventsCitation9,150 such as vacuolization and cellulose encasing (). The induction of this process requires 2 exogenous signals. The first signal, starvation and cAMP, sensitizes the cells. Only cells which have received this first signal can be induced to die by a second signal, classically the polyketide DIF-1.Citation152 In starved cells subjected to DIF-1, autophagy determines the type of cell death. In the presence of autophagy, DIF-1 induces a normal vacuolar cell death. However, in the absence of autophagy, DIF-1 induces necrotic cell death.Citation153,154 This necrotic death includes mitochondrial uncouplingCitation155-157 and depends not only on the absence of autophagy upon starvation, but also on given targets of the DIF-1 molecule, distinct from those that induce vacuolar cell death.Citation158 These results are in line with the known mitochondrial uncoupling effects of DIF-1 treatmentCitation159 and with the recently demonstrated multiplicity of DIF-1-induced effects.Citation160 Importantly, although both pathways result in programmed death, while vacuolar cell death is part of normal development, necrotic cell death is incompatible with it.Citation153
Figure 8. Genes and pathways governing D. discoideum vacuolar cell death. The cascade of subcellular cell death events is schematically depicted (middle). These include the formation of “paddle” cells, following by their rounding, biogenesis of a cellulose shell and extensive vacuolization. Random insertional mutagenesis and targeted mutagenesis identified a number of genes, here shown in green letters, encoding molecules required for this cell death. Details are given in the main text. These genes in turn helped define pathways to cell death, such as a polysaccharide pathway (right), a first signal pathway induced by starvation and cAMP (lower left) and second signal pathways (upper left). The latter include a DIF-1-induced autonomous pathway and a c-di-GMP pathway requiring endogenously synthesized or exogenous DIF-1. Ca2+-related drugs such as thapsigargin and BAPTA affect DIF-1 signaling leading to cell death.
Whether autophagy is also involved at the execution stage of vacuolar cell death has not been unambiguously proven. However, a large amount of debris is observed in the death-accompanying vacuoles indicating active degradation. Autophagy has also been proposed to have a structural role in terminal stalk differentiation.Citation15 Stalk-cell vacuoles seem to originate from acidic vesicles and autophagosomes, which fuse to form autolysosomes. Their repeated fusion expands the vacuole, accompanied by cellulose wall formation enabling the stalk cells to make the stalk rigid and hold the spores aloft.Citation15
DIF-1-induced vacuolar cell death in monolayers has been studied by random insertional mutagenesis. This led to the identification of a number of molecules required for this pathway, including the ITPR/IP3R homolog Ca2+ flux-controlling IplA,Citation161 the UDP-glucose pyrophosphorylase UgpB, the glycogen synthase GlcS,Citation162 the histidine kinase DhkM,Citation163 the transcription factor GbfA and the integrin-cytoskeleton intermediate TalB/talinB.Citation157 These and other studies have contributed to defining molecular pathways required for DIF-1-induced vacuolar cell death ().
An important recent discovery was the demonstration that not only DIF-1, but also the cyclic dinucleotide c-di-GMP are able to induce D. discoideum cell death.Citation164,165 c-di-GMP is a universal bacterial second messengerCitation166 and can trigger innate immunity in bacterially-infected animal cells.Citation167 Unexpectedly, experiments using both inducers show marked synergy,Citation168 although c-di-GMP alone is insufficient to induce cell death in D. discoideum cell monolayers, requiring either DIF-1 or a DIF-1-related polyketide. The required DIF-1 can be endogenous, as shown by the inability of c-di-GMP to induce cell death when DIF-1 synthesis was blocked pharmacologically (by cerulenin), and by its rescue upon complementation by exogenous DIF-1.Citation168 This was confirmed genetically using mutants in the polyketide synthase stlB and the methylase dmtA genes that are required for DIF-1 biosynthesis.Citation169,170 The corresponding stlB− or dmtA− mutant cells cannot be induced to die by exogenous c-di-GMP, and this can be complemented by exogenous DIF-1.Citation168 Thus, c-di-GMP can only trigger cell death in the presence of DIF1 or DIF-1-related polyketides, providing an additional level of control to this, and perhaps other c-di-GMP-dependent pathways.
How c-di-GMP synergizes with endogenous DIF-1, to drive cell death is not clear. However, at least part of the c-di-GMP pathway seems to be distinct from the autonomous DIF-1 pathway, since mutants of the latter, talB− and iplA−, do not impair the former.Citation168 Among several cytosolic molecules able to sense c-di-GMP in animal cells, DDX41Citation171 is especially well conserved and has a clear D. discoideum ortholog. Attempts to inactivate D. discoideum DDX41 by targeted mutagenesis have not led to alterations of cell death so far (Y. Song, unpublished) although several other c-di-GMP binding proteins have been reported.Citation172 Studying not only DIF-1- but also c-di-GMP-induced cell death in D. discoideum may provide important detail on the molecular mechanisms, including the role of autophagy, as well as suggestions as to how c-di-GMP acts in animal cells.
Concluding remarks
The strong conservation of the autophagic pathway between D. discoideum and mammalian cells justifies the use of this social amoeba as a model system for the study of the molecular mechanisms of autophagy and autophagy-related diseases. For basic studies D. discoideum can beautifully complement the enormous wealth of information obtained in S. cerevisiae, in particular those genes not conserved in yeast. In recent years, this potential has also become a reality in a wide range of biomedical conditions as exemplified in the use of D. discoideum as a surrogate host for studying the role of autophagy in infectious diseases.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1226737_Supplemental_Material.docx
Download MS Word (11.6 KB)1226737_Supplemental_Movie.mov
Download QuickTime Video (1.1 MB)Funding
This work was supported by grant BFU2015-64440-P (MINECO/FEDER) to R.E. and O.V. from the Spanish Ministerio de Ciencia e Innovación. LE acknowledges support of this work by the German Research Foundation (Deutsche Forschungsgemeinschaft: CRC670, TP01) and by Köln Fortune.
References
- Muñoz-Braceras S, Mesquita A, Escalante R. Dictyostelium discoideum as a model in biomedical research. In: Romeralo M, Baldauf S, Escalante R, eds. Dictyostelids Evolution, Genomics and Cell Biology. Berlin: Springer, 2013:1-34.
- Steinert M. Pathogen-host interactions in Dictyostelium, Legionella, Mycobacterium and other pathogens. Semin Cell Dev Biol 2011; 22:70-6; PMID:21109012; http://dx.doi.org/10.1016/j.semcdb.2010.11.003
- Carnell MJ, Insall RH. Actin on disease - Studying the pathobiology of cell motility using Dictyostelium discoideum. Semin Cell Dev Biol 2010; 22:82-8; PMID:21145982; http://dx.doi.org/10.1016/j.semcdb.2010.12.003
- Francione LM, Annesley SJ, Carilla-Latorre S, Escalante R, Fisher PR. The Dictyostelium model for mitochondrial disease. Semin Cell Dev Biol 2011; 22:120-30; PMID:21129494; http://dx.doi.org/10.1016/j.semcdb.2010.11.004
- Ludtmann MH, Boeckeler K, Williams RS. Molecular pharmacology in a simple model system: implicating MAP kinase and phosphoinositide signalling in bipolar disorder. Semin Cell Dev Biol 2011; 22:105-13; PMID:21093602; http://dx.doi.org/10.1016/j.semcdb.2010.11.002
- Alexander S, Alexander H. Lead genetic studies in Dictyostelium discoideum and translational studies in human cells demonstrate that sphingolipids are key regulators of sensitivity to cisplatin and other anticancer drugs. Semin Cell Dev Biol 2011; 22:97-104; PMID:20951822; http://dx.doi.org/10.1016/j.semcdb.2010.10.005
- Myre MA. Clues to gamma-secretase, huntingtin and Hirano body normal function using the model organism Dictyostelium discoideum. J Biomed Sci 2012; 19:41; PMID:22489754; http://dx.doi.org/10.1186/1423-0127-19-41
- Maniak M. Dictyostelium as a model for human lysosomal and trafficking diseases. Semin Cell Dev Biol 2011; 22:114-9; PMID:21056680; http://dx.doi.org/10.1016/j.semcdb.2010.11.001
- Calvo-Garrido J, Carilla-Latorre S, Kubohara Y, Santos-Rodrigo N, Mesquita A, Soldati T, Golstein P, Escalante R. Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy 2010; 6:686-701; PMID:20603609; http://dx.doi.org/10.4161/auto.6.6.12513
- Cotter DA, Miura-Santo LY, Hohl HR. Ultrastructural changes during germination of Dictyostelium discoideum spores. J Bacteriol 1969; 100:1020-6; PMID:5391047
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol 1966; 28:435-92; PMID:5322983; http://dx.doi.org/10.1146/annurev.ph.28.030166.002251
- King JS, Gueho A, Hagedorn M, Gopaldass N, Leuba F, Soldati T, Insall RH. WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol Biol Cell 2013; 24:2714-26; PMID:23885127; http://dx.doi.org/10.1091/mbc.E13-02-0092
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem 2003; 278:17636-45; PMID:12626495; http://dx.doi.org/10.1074/jbc.M212467200
- Cabral M, Anjard C, Malhotra V, Loomis WF, Kuspa A. Unconventional secretion of AcbA in Dictyostelium discoideum through a vesicular intermediate. Eukaryot Cell 2010; 9:1009-17; PMID:20472692; http://dx.doi.org/10.1128/ec.00337-09
- Uchikawa T, Yamamoto A, Inouye K. Origin and function of the stalk-cell vacuole in Dictyostelium. Dev Biol 2011; 352:48-57; PMID:21256841; http://dx.doi.org/10.1016/j.ydbio.2011.01.014
- Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res 2014; 24:92-104; PMID:24281265; http://dx.doi.org/10.1038/cr.2013.153
- Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci 2012; 69:1125-36; PMID:22080117; http://dx.doi.org/10.1007/s00018-011-0865-5
- Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol 2016; 482:1714-24; http://dx.doi.org/10.1016/j.jmb.2016.02.004
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/10.1038/cr.2013.168
- Parzych KR, Klionsky DJ. An Overview of autophagy: Morphology, mechanism, and regulation. Antioxid Redox Signal 2014; 20:460-73; PMID:23725295; http://dx.doi.org/10.1089/ars.2013.5371
- Ktistakis NT, Tooze SA. Digesting the expanding mechanisms of autophagy. Trends Cell Biol 2016; Apr. 2:S0962-8924(16)00045-3
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J Biol Chem 2004; 279:15621-9; PMID:14736886; http://dx.doi.org/10.1074/jbc.M311139200
- Mesquita A, Tabara LC, Martinez-Costa O, Santos-Rodrigo N, Vincent O, Escalante R. Dissecting the function of Atg1 complex in Dictyostelium autophagy reveals a connection with the pentose phosphate pathway enzyme transketolase. Open Bio 2015; 5:pii: 150088.
- Calvo-Garrido J, Carilla-Latorre S, Lazaro-Dieguez F, Egea G, Escalante R. Vacuole membrane protein 1 is an endoplasmic reticulum protein required for organelle biogenesis, protein secretion, and development. Mol Biol Cell 2008; 19:3442-53; PMID:18550798; http://dx.doi.org/10.1091/mbc.E08-01-0075
- Tung SM, Unal C, Ley A, Pena C, Tunggal B, Noegel AA, Krut O, Steinert M, Eichinger L. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell Microbiol 2010; 12:765-80; PMID:20070309; http://dx.doi.org/10.1111/j.1462-5822.2010.01432.x
- Xiong Q, Unal C, Matthias J, Steinert M, Eichinger L. The phenotypes of ATG9, ATG16 and ATG9/16 knock-out mutants imply autophagy-dependent and -independent functions. Open Bio 2015; 5:150008; http://dx.doi.org/10.1098/rsob.150008
- Munoz-Braceras S, Calvo R, Escalante R. TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells. Autophagy 2015; 11:918-27; PMID:25996471; http://dx.doi.org/10.1080/15548627.2015.1034413
- Stege JT, Laub MT, Loomis WF. tip genes act in parallel pathways of early Dictyostelium development. Dev Genet 1999; 25:64-77; PMID:10402673; http://dx.doi.org/10.1002/(SICI)1520-6408(1999)25:1%3c64::AID-DVG7%3e3.0.CO;2-1
- King JS, Veltman DM, Insall RH. The induction of autophagy by mechanical stress. Autophagy 2011; 7:1490-9; PMID:22024750; http://dx.doi.org/10.4161/auto.7.12.17924
- Rosel D, Khurana T, Majithia A, Huang X, Bhandari R, Kimmel AR. TOR complex 2 (TORC2) in Dictyostelium suppresses phagocytic nutrient capture independently of TORC1-mediated nutrient sensing. J Cell Sci 2012; 125:37-48; PMID:22266904; http://dx.doi.org/10.1242/jcs.077040
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; PMID:11060023; http://dx.doi.org/10.1093/emboj/19.21.5720
- Matthias J, Messling S, Eichinger L. The two Dictyostelium autophagy eight proteins, ATG8a and ATG8b, associate with the autophagosome in succession. Eur J Cell Biol 2016; 95:15-25; PMID:26697781; http://dx.doi.org/10.1016/j.ejcb.2015.10.007
- Krick R, Henke S, Tolstrup J, Thumm M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy 2008; 4:896-910; PMID:18769150; http://dx.doi.org/10.4161/auto.6801
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010; 6:506-22; PMID:20505359; http://dx.doi.org/10.4161/auto.6.4.11863
- Gerstenmaier L, Pilla R, Herrmann L, Herrmann H, Prado M, Villafano GJ, Kolonko M, Reimer R, Soldati T, King JS, et al. The autophagic machinery ensures nonlytic transmission of mycobacteria. Proc Natl Acad Sci U S A 2015; 112:E687-92; PMID:25646440; http://dx.doi.org/10.1073/pnas.1423318112
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3:452-60; PMID:17534139; http://dx.doi.org/10.4161/auto.4451
- Mesquita A, Calvo-Garrido J, Carilla-Latorre S, Escalante R. Monitoring autophagy in Dictyostelium. Methods Mol Biol 2013; 983:461-70; PMID:23494324; http://dx.doi.org/10.1007/978-1-62703-302-2_26
- Calvo-Garrido J, Carilla-Latorre S, Mesquita A, Escalante R. A proteolytic cleavage assay to monitor autophagy in Dictyostelium discoideum. Autophagy 2011; 7:1063-8; PMID:21876387; http://dx.doi.org/10.4161/auto.7.9.16629
- Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy 2010; 6:794-7; PMID:20523132; http://dx.doi.org/10.4161/auto.6.6.12348
- Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Curr Opin Cell Biol 2016; 39:61-8; PMID:26921696; http://dx.doi.org/10.1016/j.ceb.2016.02.010
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15(7):741-50; PMID:23685627
- Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell 2014; 53:471-83; PMID:24440502; http://dx.doi.org/10.1016/j.molcel.2013.12.011
- Noda NN, Fujioka Y. Atg1 family kinases in autophagy initiation. Cell Mol Life Sci 2015; 72(16):3083-96; PMID:25948417
- Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, Yip CK, Klionsky DJ. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A 2013; 110:E2875-84; PMID:23858448; http://dx.doi.org/10.1073/pnas.1300064110
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 2010; 30:1049-58; PMID:19995911; http://dx.doi.org/10.1128/mcb.01344-09
- Kamber RA, Shoemaker CJ, Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol Cell 2015; 59:372-81; PMID:26166702; http://dx.doi.org/10.1016/j.molcel.2015.06.009
- Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol 2016; 26:6-16; PMID:26437584; http://dx.doi.org/10.1016/j.tcb.2015.08.010
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009; 5:973-9; PMID:19597335; http://dx.doi.org/10.4161/auto.5.7.9296
- Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009; 5:649-62; PMID:19287211; http://dx.doi.org/10.4161/auto.5.5.8249
- Suzuki H, Kaizuka T, Mizushima N, Noda NN. Structure of the Atg101-Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat Struct Mol Biol 2015; 22:572-80; PMID:26030876; http://dx.doi.org/10.1038/nsmb.3036
- Qi S, Kim do J, Stjepanovic G, Hurley JH. Structure of the human Atg13-Atg101 HORMA heterodimer: an interaction hub within the ULK1 complex. Structure 2015; 23:1848-57; PMID:26299944; http://dx.doi.org/10.1016/j.str.2015.07.011
- Li F, Chung T, Vierstra RD. Autophagy-related 11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 2014; 26:788-807; PMID:24563201; http://dx.doi.org/10.1105/tpc.113.120014
- Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol 2015; 17:262-75; PMID:25686248; http://dx.doi.org/10.1038/ncb3101
- Tekinay T, Wu MY, Otto GP, Anderson OR, Kessin RH. Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot Cell 2006; 5:1797-806; PMID:17031001; http://dx.doi.org/10.1128/ec.00342-05
- Calvo-Garrido J, Escalante R. Autophagy dysfunction and ubiquitin-positive protein aggregates in Dictyostelium cells lacking Vmp1. Autophagy 2010; 6:100-9; PMID:20009561; http://dx.doi.org/10.4161/auto.6.1.10697
- Heintze J, Costa JR, Weber M, Ketteler R. Ribose 5-phosphate isomerase inhibits LC3 processing and basal autophagy. Cell Signal 2016; 28:1380-8; PMID:27328773; http://dx.doi.org/10.1016/j.cellsig.2016.06.015
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685-701; PMID:18725538; http://dx.doi.org/10.1083/jcb.200803137
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013; 154:1285-99; PMID:24034251; http://dx.doi.org/10.1016/j.cell.2013.08.044
- He S, Ni D, Ma B, Lee JH, Zhang T, Ghozalli I, Pirooz SD, Zhao Z, Bharatham N, Li B, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol 2013; 15:1206-19; PMID:24056303; http://dx.doi.org/10.1038/ncb2848
- Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife 2014; 3:e04135; PMID:25432021; http://dx.doi.org/10.7554/eLife.04135
- Carlsson SR, Simonsen A. Membrane dynamics in autophagosome biogenesis. J Cell Sci 2015; 128:193-205; PMID:25568151; http://dx.doi.org/10.1242/jcs.141036
- Lemus L, Ribas JL, Sikorska N, Goder V. An ER-localized SNARE protein is exported in specific COPII vesicles for autophagosome biogenesis. Cell Rep 2016; 14:1710-22; PMID:26876173; http://dx.doi.org/10.1016/j.celrep.2016.01.047
- King JS. Autophagy across the eukaryotes: Is S. cerevisiae the odd one out? Autophagy 2012; 8:1159-62; PMID:22722653; http://dx.doi.org/10.4161/auto.20527
- Tenenboim H, Smirnova J, Willmitzer L, Steup M, Brotman Y. VMP1-deficient Chlamydomonas exhibits severely aberrant cell morphology and disrupted cytokinesis. BMC Plant Biol 2014; 14:121; PMID:24885763; http://dx.doi.org/10.1186/1471-2229-14-121
- Calvo-Garrido J, King JS, Munoz-Braceras S, Escalante R. Vmp1 regulates PtdIns3P signaling during autophagosome formation in Dictyostelium discoideum. Traffic 2014; 15:1235-46; PMID:25131297; http://dx.doi.org/10.1111/tra.12210
- Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 2010; 141:1042-55; PMID:20550938; http://dx.doi.org/10.1016/j.cell.2010.04.034
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010; 6:764-76; PMID:20639694; http://dx.doi.org/10.4161/auto.6.6.12709
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; PMID:21801009; http://dx.doi.org/10.1146/annurev-cellbio-092910-154005
- Zhang L, Li J, Ouyang L, Liu B, Cheng Y. Unraveling the roles of Atg4 proteases from autophagy modulation to targeted cancer therapy. Cancer Lett 2016; 373:19-26; PMID:26805760; http://dx.doi.org/10.1016/j.canlet.2016.01.022
- Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 2011; 12:226; PMID:21867568; http://dx.doi.org/10.1186/gb-2011-12-7-226
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011; 80:125-56; PMID:21548784; http://dx.doi.org/10.1146/annurev-biochem-052709-094552
- Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell 2011; 20:444-54; PMID:21497758; http://dx.doi.org/10.1016/j.devcel.2011.02.006
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102-9; PMID:17909521; http://dx.doi.org/10.1038/ncb1007-1102
- Bader CA, Shandala T, Ng YS, Johnson IR, Brooks DA. Atg9 is required for intraluminal vesicles in amphisomes and autolysosomes. Bio Open 2015; 4:1345-55; http://dx.doi.org/10.1242/bio.013979
- Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol 2014; 24:1314-22; PMID:24881874; http://dx.doi.org/10.1016/j.cub.2014.04.048
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 2010; 190:1005-22; PMID:20855505; http://dx.doi.org/10.1083/jcb.200912089
- Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 2005; 1:101-9; PMID:16874040; http://dx.doi.org/10.4161/auto.1.2.1840
- Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 2005; 280:18283-90; PMID:15755735; http://dx.doi.org/10.1074/jbc.M413957200
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006; 119:3888-900; PMID:16940348; http://dx.doi.org/10.1242/jcs.03172
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. ATG16L1 meets ATG9 in recycling endosomes: additional roles for the plasma membrane and endocytosis in autophagosome biogenesis. Autophagy 2014; 10:182-4; PMID:24257061; http://dx.doi.org/10.4161/auto.27174
- Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J 2011; 30:636-51; PMID:21169990; http://dx.doi.org/10.1038/emboj.2010.338
- Arhzaouy K, Strucksberg KH, Tung SM, Tangavelou K, Stumpf M, Faix J, Schroder R, Clemen CS, Eichinger L. Heteromeric p97/p97(R155C) complexes induce dominant negative changes in wild-type and autophagy 9-deficient Dictyostelium strains. PLoS ONE 2012; 7:e46879; PMID:23056506; http://dx.doi.org/10.1371/journal.pone.0046879
- Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol 2001; 3:740-4; PMID:11483959; http://dx.doi.org/10.1038/35087056
- Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A 2005; 102:14296-301; PMID:16186509; http://dx.doi.org/10.1073/pnas.0505014102
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 2004; 429:841-7; PMID:15215856; http://dx.doi.org/10.1038/nature02656
- Kazamel M, Sorenson EJ, McEvoy KM, Jones LK, Jr., Leep-Hunderfund AN, Mauermann ML, Milone M. Clinical spectrum of valosin containing protein (VCP)-opathy. Muscle Nerve 2016; 54:94-9; PMID:26574898; http://dx.doi.org/10.1002/mus.24980
- Kimonis VE, Mehta SG, Fulchiero EC, Thomasova D, Pasquali M, Boycott K, Neilan EG, Kartashov A, Forman MS, Tucker S, et al. Clinical studies in familial VCP myopathy associated with Paget disease of bone and frontotemporal dementia. Am J Med Genet A 2008; 146A:745-57; PMID:18260132; http://dx.doi.org/10.1002/ajmg.a.31862
- Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 2010; 6:217-27; PMID:20104022; http://dx.doi.org/10.4161/auto.6.2.11014
- Schneider JL, Cuervo AM. Autophagy and human disease: emerging themes. Curr Opin Genet Dev 2014; 26C:16-23; http://dx.doi.org/10.1016/j.gde.2014.04.003
- Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J 2015; 282:4672-8; PMID:26432171; http://dx.doi.org/10.1111/febs.13540
- Schmauch C, Claussner S, Zoltzer H, Maniak M. Targeting the actin-binding protein VASP to late endosomes induces the formation of giant actin aggregates. Eur J Cell Biol 2009; 88:385-96; PMID:19324455; http://dx.doi.org/10.1016/j.ejcb.2009.02.185
- Maselli AG, Davis R, Furukawa R, Fechheimer M. Formation of Hirano bodies in Dictyostelium and mammalian cells induced by expression of a modified form of an actin-crosslinking protein. J Cell Sci 2002; 115:1939-49; PMID:11956325
- Kim DH, Davis RC, Furukawa R, Fechheimer M. Autophagy contributes to degradation of Hirano bodies. Autophagy 2009; 5:44-51; PMID:18989098; http://dx.doi.org/10.4161/auto.5.1.7228
- Rampoldi L, Dobson-Stone C, Rubio JP, Danek A, Chalmers RM, Wood NW, Verellen C, Ferrer X, Malandrini A, Fabrizi GM, et al. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat Genet 2001; 28:119-20; PMID:11381253; http://dx.doi.org/10.1038/88821
- Ueno S, Maruki Y, Nakamura M, Tomemori Y, Kamae K, Tanabe H, Yamashita Y, Matsuda S, Kaneko S, Sano A. The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat Genet 2001; 28:121-2; PMID:11381254; http://dx.doi.org/10.1038/88825
- Foller M, Hermann A, Gu S, Alesutan I, Qadri SM, Borst O, Schmidt EM, Schiele F, vom Hagen JM, Saft C, et al. Chorein-sensitive polymerization of cortical actin and suicidal cell death in chorea-acanthocytosis. FASEB J 2012; 26:1526-34; PMID:22227296; http://dx.doi.org/10.1096/fj.11-198317
- Alesutan I, Seifert J, Pakladok T, Rheinlaender J, Lebedeva A, Towhid ST, Stournaras C, Voelkl J, Schaffer TE, Lang F. Chorein sensitivity of actin polymerization, cell shape and mechanical stiffness of vascular endothelial cells. Cell Physiol Biochem 2013; 32:728-42; PMID:24080826; http://dx.doi.org/10.1159/000354475
- Park JS, Neiman AM. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J Cell Sci 2012; 125:3004-11; PMID:22442115; http://dx.doi.org/10.1242/jcs.105114
- Samaranayake HS, Cowan AE, Klobutcher LA. Vacuolar protein sorting protein 13A, TtVPS13A, localizes to the tetrahymena thermophila phagosome membrane and is required for efficient phagocytosis. Eukaryot Cell 2011; 10:1207-18; PMID:21764909; http://dx.doi.org/10.1128/ec.05089-11
- Velayos-Baeza A, Vettori A, Copley RR, Dobson-Stone C, Monaco AP. Analysis of the human VPS13 gene family. Genomics 2004; 84:536-49; PMID:15498460; http://dx.doi.org/10.1016/j.ygeno.2004.04.012
- Balikova I, Lehesjoki AE, de Ravel TJ, Thienpont B, Chandler KE, Clayton-Smith J, Traskelin AL, Fryns JP, Vermeesch JR. Deletions in the VPS13B (COH1) gene as a cause of Cohen syndrome. Hum Mutat 2009; 30:E845-54; PMID:19533689; http://dx.doi.org/10.1002/humu.21065
- Wang L, Cheng L, Li NN, Yu WJ, Sun XY, Peng R. Association of four new candidate genetic variants with Parkinson's disease in a Han Chinese population. Am J Med Genet B 2016; 171:342-7; http://dx.doi.org/10.1002/ajmg.b.32410
- Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton 2010; 67:193-206; PMID:20175130
- Freeman C, Seaman MN, Reid E. The hereditary spastic paraplegia protein strumpellin: characterisation in neurons and of the effect of disease mutations on WASH complex assembly and function. Biochim Biophys Acta 2013; 1832:160-73; PMID:23085491; http://dx.doi.org/10.1016/j.bbadis.2012.10.011
- Zavodszky E, Seaman MN, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC. Mutation in VPS35 associated with Parkinson's disease impairs WASH complex association and inhibits autophagy. Nat Commun 2014; 5:3828; PMID:24819384; http://dx.doi.org/10.1038/ncomms4828
- Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J 2013; 32:2685-96; PMID:23974797; http://dx.doi.org/10.1038/emboj.2013.189
- Sarkar S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans 2013; 41:1103-30; PMID:24059496; http://dx.doi.org/10.1042/BST20130134
- Francione L, Smith PK, Accari SL, Taylor PE, Bokko PB, Bozzaro S, Beech PL, Fisher PR. Legionella pneumophila multiplication is enhanced by chronic AMPK signalling in mitochondrially diseased Dictyostelium cells. Dis Model Mech 2009; 2:479-89; PMID:19638422; http://dx.doi.org/10.1242/dmm.003319
- Carilla-Latorre S, Gallardo ME, Annesley SJ, Calvo-Garrido J, Grana O, Accari SL, Smith PK, Valencia A, Garesse R, Fisher PR, et al. MidA is a putative methyltransferase that is required for mitochondrial complex I function. J Cell Sci 2010; 123:1674-83; PMID:20406883; http://dx.doi.org/10.1242/jcs.066076
- Carilla-Latorre S, Annesley SJ, Munoz-Braceras S, Fisher PR, Escalante R. Ndufaf5 deficiency in the Dictyostelium model: new roles in autophagy and development. Mol Biol Cell 2013; 24:1519-28; PMID:23536703; http://dx.doi.org/10.1091/mbc.E12-11-0796
- Peng M, Ostrovsky J, Kwon YJ, Polyak E, Licata J, Tsukikawa M, Marty E, Thomas J, Felix CA, Xiao R, et al. Inhibiting cytosolic translation and autophagy improves health in mitochondrial disease. Hum Mol Genet 2015; 24:4829-47; PMID:26041819; http://dx.doi.org/10.1093/hmg/ddv207
- Moran M, Delmiro A, Blazquez A, Ugalde C, Arenas J, Martin MA. Bulk autophagy, but not mitophagy, is increased in cellular model of mitochondrial disease. Biochim Biophys Acta 2014; 1842:1059-70; PMID:24704045; http://dx.doi.org/10.1016/j.bbadis.2014.03.013
- Escoll P, Rolando M, Buchrieser C. Modulation of host autophagy during bacterial infection: Sabotaging host munitions for pathogen nutrition. Frontiers Immunol 2016; 7:81; http://dx.doi.org/10.3389/fimmu.2016.00081
- Sillo A, Matthias J, Konertz R, Bozzaro S, Eichinger L. Salmonella typhimurium is pathogenic for Dictyostelium cells and subverts the starvation response. Cell Microbiol 2011; 13:1793-811; PMID:21824247; http://dx.doi.org/10.1111/j.1462-5822.2011.01662.x
- Pflaum K, Gerdes K, Yovo K, Callahan J, Snyder ML. Lipopolysaccharide induction of autophagy is associated with enhanced bactericidal activity in Dictyostelium discoideum. Biochem Biophys Res Commun 2012; 422:417-22; PMID:22575510; http://dx.doi.org/10.1016/j.bbrc.2012.05.006
- Lampe EO, Brenz Y, Herrmann L, Repnik U, Griffiths G, Zingmark C, Sjostedt A, Winther-Larsen HC, Hagedorn M. Dissection of Francisella - host cell interactions in Dictyostelium discoideum. Appl Environ Microbiol 2015; 82:1586-98; PMID:26712555; http://dx.doi.org/10.1128/aem.02950-15
- Barisch C, Paschke P, Hagedorn M, Maniak M, Soldati T. Lipid droplet dynamics at early stages of Mycobacterium marinum infection in Dictyostelium. Cell Microbiol 2015; 17:1332-49; PMID:25772333; http://dx.doi.org/10.1111/cmi.12437
- Kolonko M, Geffken AC, Blumer T, Hagens K, Schaible UE, Hagedorn M. WASH-driven actin polymerization is required for efficient mycobacterial phagosome maturation arrest. Cell Microbiol 2014; 16:232-46; PMID:24119059; http://dx.doi.org/10.1111/cmi.12217
- Schiebler M, Brown K, Hegyi K, Newton SM, Renna M, Hepburn L, Klapholz C, Coulter S, Obregon-Henao A, Henao Tamayo M, et al. Functional drug screening reveals anticonvulsants as enhancers of mTOR-independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO Mol Med 2014; 7:127-39; PMID:25535254; http://dx.doi.org/10.15252/emmm.201404137
- Jaiswal P, Soldati T, Thewes S, Baskar R. Regulation of aggregate size and pattern by adenosine and caffeine in cellular slime molds. BMC Dev Biol 2012; 12:5; PMID:22269093; http://dx.doi.org/10.1186/1471-213X-12-5
- Kicka S, Trofimov V, Harrison C, Ouertatani-Sakouhi H, McKinney J, Scapozza L, Hilbi H, Cosson P, Soldati T. Establishment and validation of whole-cell based fluorescence assays to identify anti-mycobacterial compounds using the Acanthamoeba castellanii-Mycobacterium marinum host-pathogen system. PLoS ONE 2014; 9:e87834; PMID:24498207; http://dx.doi.org/10.1371/journal.pone.0087834
- Swer PB, Lohia R, Saran S. Analysis of rapamycin induced autophagy in Dictyostelium discoideum. Indian J Exp Biol 2014; 52:295-304; PMID:24772931
- Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol 2008; 10:1027-39; PMID:18298637; http://dx.doi.org/10.1111/j.1462-5822.2008.01133.x
- Lerena MC, Colombo MI. Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system. Cell Microbiol 2011; 13:814-35; PMID:21447143; http://dx.doi.org/10.1111/j.1462-5822.2011.01581.x
- van der Vaart M, Korbee CJ, Lamers GE, Tengeler AC, Hosseini R, Haks MC, Ottenhoff TH, Spaink HP, Meijer AH. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLP-MYD88 to authophagic defense. Cell Host Microbe 2014; 15:753-67; PMID:24922577; http://dx.doi.org/10.1016/j.chom.2014.05.005
- Sato E, Imafuku S, Ishii K, Itoh R, Chou B, Soejima T, Nakayama J, Hiromatsu K. Vitamin D-dependent cathelicidin inhibits Mycobacterium marinum infection in human monocytic cells. Journal Dermatol Sci 2013; 70:166-72; http://dx.doi.org/10.1016/j.jdermsci.2013.01.011
- Hosseini R, Lamers GE, Hodzic Z, Meijer AH, Schaaf MJ, Spaink HP. Correlative light and electron microscopy imaging of autophagy in a zebrafish infection model. Autophagy 2014; 10:1844-57; PMID:25126731; http://dx.doi.org/10.4161/auto.29992
- Mohanty S, Jagannathan L, Ganguli G, Padhi A, Roy D, Alaridah N, Saha P, Nongthomba U, Godaly G, Gopal RK, et al. A mycobacterial phosphoribosyltransferase promotes bacillary survival by inhibiting oxidative stress and autophagy pathways in macrophages and zebrafish. J Biol Chem 2015; 290:13321-43; PMID:25825498; http://dx.doi.org/10.1074/jbc.M114.598482
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012; 150:803-15; PMID:22901810; http://dx.doi.org/10.1016/j.cell.2012.06.040
- Collins CA, De Maziere A, van Dijk S, Carlsson F, Klumperman J, Brown EJ. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog 2009; 5:e1000430; PMID:19436699; http://dx.doi.org/10.1371/journal.ppat.1000430
- Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, McDonald KL, Szyk A, LaRonde-LeBlanc N, Gao LY. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun 2008; 76:5478-87; PMID:18852239; http://dx.doi.org/10.1128/iai.00614-08
- Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 2009; 323:1729-33; PMID:19325115; http://dx.doi.org/10.1126/science.1169381
- Hagedorn M, Soldati T. Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell Microbiol 2007; 9:2984; http://dx.doi.org/10.1111/j.1462-5822.2007.01064.x
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A 2006; 103:14578-83; PMID:16983090; http://dx.doi.org/10.1073/pnas.0601838103
- Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T. Francisella tularensis Harvests Nutrients Derived via ATG5-Independent Autophagy to Support Intracellular Growth. PLoS Pathog 2013; 9:e1003562; PMID:23966861; http://dx.doi.org/10.1371/journal.ppat.1003562
- Otto GP, Wu MY, Clarke M, Lu H, Anderson OR, Hilbi H, Shuman HA, Kessin RH. Macroautophagy is dispensable for intracellular replication of Legionella pneumophila in Dictyostelium discoideum. Mol Microbiol 2004; 51:63-72; PMID:14651611; http://dx.doi.org/10.1046/j.1365-2958.2003.03826.x
- Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 2012; 338:1072-6; PMID:23112293; http://dx.doi.org/10.1126/science.1227026
- Mack HI, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy 2012; 8:1197-214; PMID:22932492; http://dx.doi.org/10.4161/auto.20586
- LaRock DL, Chaudhary A, Miller SI. Salmonellae interactions with host processes. Nat Rev Microbiol 2015; 13:191-205; PMID:25749450; http://dx.doi.org/10.1038/nrmicro3420
- Garai P, Gnanadhas DP, Chakravortty D. Salmonella enterica serovars Typhimurium and Typhi as model organisms: revealing paradigm of host-pathogen interactions. Virulence 2012; 3:377-88; PMID:22722237; http://dx.doi.org/10.4161/viru.21087
- Skriwan C, Fajardo M, Hagele S, Horn M, Wagner M, Michel R, Krohne G, Schleicher M, Hacker J, Steinert M. Various bacterial pathogens and symbionts infect the amoeba Dictyostelium discoideum. Int J Med Microbiol 2002; 291:615-24; PMID:12008915; http://dx.doi.org/10.1078/1438-4221-00177
- Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A 2009; 106:14564-9; PMID:19667176; http://dx.doi.org/10.1073/pnas.0813319106
- Mestre MB, Fader CM, Sola C, Colombo MI. Alpha-hemolysin is required for the activation of the autophagic pathway in Staphylococcus aureus-infected cells. Autophagy 2010; 6:110-25; PMID:20110774; http://dx.doi.org/10.4161/auto.6.1.10698
- Schnaith A, Kashkar H, Leggio SA, Addicks K, Kronke M, Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem 2007; 282:2695-706; PMID:17135247; http://dx.doi.org/10.1074/jbc.M609784200
- O'Keeffe KM, Wilk MM, Leech JM, Murphy AG, Laabei M, Monk IR, Massey RC, Lindsay JA, Foster TJ, Geoghegan JA, et al. Manipulation of Autophagy in Phagocytes Facilitates Staphylococcus aureus Bloodstream Infection. Infect Immun 2015; 83:3445-57; PMID:26099586; http://dx.doi.org/10.1128/iai.00358-15
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016; 12:1-222; PMID:26799652; http://dx.doi.org/10.1080/15548627.2015.1100356
- Golstein P, Aubry L, Levraud JP. Cell-death alternative model organisms: why and which? Nature Rev Mol Cell Biol 2003; 4:798-807; http://dx.doi.org/10.1038/nrm1224
- Whittingham WF, Raper KB. Non-viability of stalk cells in Dictyostelium. Proc Natl Acad Sci USA 1960; 46:642-9; PMID:16590653; http://dx.doi.org/10.1073/pnas.46.5.642
- Kay RR. Cell differentiation in monolayers and the investigation of slime mold morphogens. Methods Cell Biol 1987; 28:433-48; PMID:3600415; http://dx.doi.org/10.1016/S0091-679X(08)61661-1
- Cornillon S, Foa C, Davoust J, Buonavista N, Gross JD, Golstein P. Programmed cell death in Dictyostelium. J Cell Sci 1994; 107:2691-704; PMID:7876338
- Giusti C, Tresse E, Luciani MF, Golstein P. Autophagic cell death: analysis in Dictyostelium. Biochim Biophys Acta 2009; 1793:1422-31; PMID:19133302; http://dx.doi.org/10.1016/j.bbamcr.2008.12.005
- Morris HR, Taylor GW, Masento MS, Jermyn KA, Kay RR. Chemical structure of the morphogen differentiation inducing factor from Dictyostelium discoideum. Nature 1987; 328:811-4; PMID:3627228; http://dx.doi.org/10.1038/328811a0
- Luciani MF, Giusti C, Harms B, Oshima Y, Kikuchi H, Kubohara Y, Golstein P. Atg1 allows second-signaled autophagic cell death in Dictyostelium. Autophagy 2011; 7:501-8; PMID:21301205; http://dx.doi.org/10.4161/auto.7.5.14957
- Kosta A, Roisin-Bouffay C, Luciani MF, Otto GP, Kessin RH, Golstein P. Autophagy gene disruption reveals a non-vacuolar cell death pathway in Dictyostelium. J Biol Chem 2004; 279:48404-9; PMID:15358773; http://dx.doi.org/10.1074/jbc.M408924200
- Kosta A, Laporte C, Lam D, Tresse E, Luciani MF, Golstein P. How to assess and study cell death in Dictyostelium discoideum. Methods Mol Biol 2006; 346:535-50; PMID:16957313
- Laporte C, Kosta A, Klein G, Aubry L, Lam D, Tresse E, Luciani MF, Golstein P. A necrotic cell death model in a protist. Cell Death Differ 2007; 14:266-74; PMID:16810325; http://dx.doi.org/10.1038/sj.cdd.4401994
- Giusti C, Luciani MF, Klein G, Aubry L, Tresse E, Kosta A, Golstein P. Necrotic cell death: From reversible mitochondrial uncoupling to irreversible lysosomal permeabilization. Exp Cell Res 2009; 315:26-38; PMID:18951891; http://dx.doi.org/10.1016/j.yexcr.2008.09.028
- Luciani MF, Kubohara Y, Kikuchi H, Oshima Y, Golstein P. Autophagic or necrotic cell death triggered by distinct motifs of the differentiation factor DIF-1. Cell Death Differ 2009; 16:564-70; PMID:19079140; http://dx.doi.org/10.1038/cdd.2008.177
- Shaulsky G, Loomis WF. Mitochondrial DNA replication but no nuclear DNA replication during development of Dictyostelium. Proc Natl Acad Sci USA 1995; 92:5660-3; PMID:7777565; http://dx.doi.org/10.1073/pnas.92.12.5660
- Sugden C, Urbaniak MD, Araki T, Williams JG. The Dictyostelium prestalk inducer differentiation-inducing factor-1 (DIF-1) triggers unexpectedly complex global phosphorylation changes. Mol Biol Cell 2015; 26:805-20; PMID:25518940; http://dx.doi.org/10.1091/mbc.E14-08-1319
- Lam D, Golstein P. A specific pathway inducing autophagic cell death is marked by an IP3R mutation. Autophagy 2008; 4:349-50; PMID:18196962; http://dx.doi.org/10.4161/auto.5521
- Tresse E, Kosta A, Giusti C, Luciani MF, Golstein P. A UDP-glucose derivative is required for vacuolar autophagic cell death. Autophagy 2008; 4:680-91; PMID:18424909; http://dx.doi.org/10.4161/auto.6084
- Giusti C, Luciani MF, Ravens S, Gillet A, Golstein P. Autophagic cell death in Dictyostelium requires the receptor histidine kinase DhkM. Mol Biol Cell 2010; 21:1825-35; PMID:20375146; http://dx.doi.org/10.1091/mbc.E09-11-0976
- Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature 2012; 488:680-3; PMID:22864416; http://dx.doi.org/10.1038/nature11313
- Chen ZH, Schaap P. Secreted cyclic-di-GMP induces stalk cell differentiation in the eukaryote Dictyostelium discoideum. J Bacteriol 2016; 198:27-31; PMID:26013485; http://dx.doi.org/10.1128/jb.00321-15
- Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 2013; 77:1-52; PMID:23471616; http://dx.doi.org/10.1128/mmbr.00043-12
- Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell 2013; 154:962-70; PMID:23993090; http://dx.doi.org/10.1016/j.cell.2013.08.014
- Song Y, Luciani MF, Giusti C, Golstein P. c-di-GMP induction of Dictyostelium cell death requires the polyketide DIF-1. Mol Biol Cell 2015; 26:651-8; PMID:25518941; http://dx.doi.org/10.1091/mbc.E14-08-1337
- Thompson CRL, Kay RR. The role of DIF-1 signaling in Dictyostelium development. Mol Cell 2000; 6:1509-14; PMID:11163223; http://dx.doi.org/10.1016/S1097-2765(00)00147-7
- Saito T, Kato A, Kay RR. DIF-1 induces the basal disc of the Dictyostelium fruiting body. Dev Biol 2008; 317:444-53; PMID:18402932; http://dx.doi.org/10.1016/j.ydbio.2008.02.036
- Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol 2012; 13:1155-61; PMID:23142775; http://dx.doi.org/10.1038/ni.2460
- Chou SH, Galperin MY. Diversity of c-di-GMP-binding proteins and mechanisms. J Bacteriol 2015; 198:32-46; http://dx.doi.org/10.1128/jb.00333-15
