ABSTRACT
Autophagy helps to maintain cellular homeostasis by removing misfolded proteins and damaged organelles, and generally acts as a cytoprotective mechanism for neuronal survival. Here we showed that mice deficient in the Vici syndrome gene Epg5, which is required for autophagosome maturation, show accumulation of ubiquitin-positive inclusions and SQSTM1 aggregates in various retinal cell types. In epg5−/− retinas, photoreceptor function is greatly impaired, and degenerative features including progressively reduced numbers of photoreceptor cells and increased numbers of apoptotic cells in the outer nuclear layer are observed, while the morphology of other parts of the retina is not severely affected. Downstream targets of the unfolded protein response (UPR), including the death inducer DDIT3/CHOP, and also levels of cleaved CASP3 (caspase 3), are elevated in epg5−/− retinas. Thus, apoptotic photoreceptor cell death in epg5−/− retinas may result from the elevated UPR. Our results reveal that Epg5-deficient mice recapitulate key characteristics of retinitis pigmentosa and thus may provide a valuable model for investigating the molecular mechanism of photoreceptor degeneration.
Introduction
Autophagy is a lysosome-mediated degradation process, involving the engulfment of cytosolic contents in a double-membrane autophagosome, maturation of the autophagosome by fusion with endocytic vesicles, and eventual fusion of the autophagosome with lysosomes for degradation.Citation1-3 Constitutive basal autophagy acts as a quality control system by removing misfolded or aggregation-prone proteins and damaged organelles to maintain cellular homeostasis.Citation4,5 Mice with neural-specific deficiency in genes essential for autophagosome formation, including Atg5, Atg7 and Ei24, exhibit accumulation of ubiquitin-positive inclusions and SQSTM1 aggregates in neurons and also massive neuronal death.Citation6-8 However, these knockout mice fail to exhibit the selective vulnerability of a certain population of neurons which is associated with neurodegenerative diseases.
The mammalian retina is composed of the outer retina, which includes the photoreceptors and the adjacent retinal pigment epithelium (RPE), and the inner retina, which includes different neuronal types and glial cells. The rod and cone photoreceptors are responsible for night and daylight vision, respectively. Retinitis pigmentosa (RP) is a collection of retinal diseases characterized by loss of function and progressive degeneration of photoreceptors, leading to visual loss and eventually to blindness.Citation9,10 Photoreceptor loss in RP may result from apoptotic cell death induced by disruption of protein homeostasis.Citation10,11 For example, the majority of autosomal dominant RP (adRP)-linked rhodopsin mutations, such as Pro23His (P23H) and Thr17Met (T17M), cause misfolding and aggregation of rhodopsin, which activates the unfolded protein response (UPR) to promote apoptotic cell death.Citation10,12,13
Autophagy has diverse functions in different types of retinal cell. It is involved in maintenance of normal homeostasis in the RPE.Citation14,15 Inactivation of the essential autophagy gene Rb1cc1/Fip200 causes age-dependent degeneration of the RPE and a secondary loss of photoreceptors.Citation14 A subset of autophagy proteins, including ATG5 and BECN1/Beclin 1, act in LC3-associated phagocytosis in the RPE to digest photoreceptor outer segments, which further contributes to the retinoid supply for the photoreceptors.Citation16 Autophagy has a cytoprotective role in retinal ganglion cells (RGCs) after optic nerve axotomy,Citation17,18 while autophagy has a detrimental effect on crush-induced acute axonal degeneration of the optic nerve.Citation19 The role of autophagy in maintaining photoreceptors remains largely unknown. Mice with neuron-specific Atg5-deficiency exhibit apoptotic cell death in the photoreceptor layer.Citation20 Deleting Atg5 specifically in rods (Atg5ΔRod) leads to progressive degeneration of rod photoreceptors.Citation21 The phototransduction protein transducin-α accumulates in Atg5ΔRod retinas.Citation21 However, depletion of Atg7 in rods causes no abnormality in retinal architecture, but leads to increased susceptibility to light-induced retinal degeneration.Citation22 Atg5 and Atg7 also act in autophagy-independent processes, which may contribute to their function in maintaining photoreceptor homeostasis. In a mouse RP model caused by a missense mutation in the Pde6b gene, lysosomal function is impaired and photoreceptor cell death is exacerbated by autophagy stimulation and ameliorated by autophagy downregulation.Citation23
The Vici syndrome gene Epg5 is required for maturation of autophagosomes into degradative autolysosomes.Citation24,25 Although autophagy activity is systemically impaired in Epg5-deficient mice and SQSTM1 aggregates accumulate in many regions of the brain and spinal cord, these animals exhibit selective damage of cortical layer 5 pyramidal neurons and spinal cord motor neurons.Citation25 Here we demonstrated that epg5 knockout mice also exhibit visual loss and display progressive photoreceptor degeneration, recapitulating key features of RP.
Results
epg5−/− mice show progressive photoreceptor degeneration
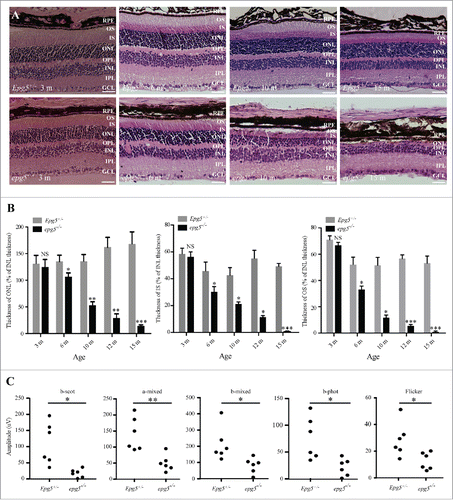
To investigate retinal development in epg5 knockout (KO) mice, we performed histological analysis of retinal sections from epg5−/− mice and age-matched littermate controls by hematoxylin and eosin (H&E) staining. A ∼25% reduction in the thickness of the outer nuclear layer (ONL) and a ∼30% reduction in the thickness of the inner segment (IS) and outer segment (OS) were detected in epg5−/− mice at the age of 6 mo (). The ONL consists of photoreceptor nuclei, and the IS and OS contain the inner segments and outer segments of photoreceptors, respectively. At 10 mo, the thickness of the ONL and IS decreased to ∼50% of that observed in heterozygote mice, and the thickness of the OS was reduced by ∼75% compared with control mice. The OS and IS were barely detected at 15 mo in KO mice. The number of RPE cells was not obviously reduced in 10-mo-old epg5−/− mice compared with controls (Fig. S1A). These results show that epg5 KO mice exhibit progressive photoreceptor loss.
Figure 1. Retinal degeneration in epg5 knockout mice. (A) Histological analysis of retinas from epg5−/− and Epg5+/− mice at the age of 3, 6, 10 and 15 mo. All sections were stained with hematoxylin-eosin and photographed from the first dorsal location (∼0.18 mm from the optic nerve head) at the same magnification (200 ×). Scale bars: 50 µm. RPE, retinal pigment epithelium; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; IS, inner segments; OS, outer segments. As a result of embedding, the RPE layer occasionally separates from the photoreceptor segments. (B) Temporal changes in retinal degeneration, presented as thickness of ONL, IS and OS normalized to INL thickness to correct for sectioning artifacts, in epg5−/− and Epg5+/− mice at the age of 3, 6, 10, 12 and 15 mo. Progressive decrease in the thickness of ONL, IS and OS is observed in epg5−/− retinas from 6 mo onwards. At 15 mo, the IS and OS are barely detected in epg5−/− mice. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical comparisons were performed with 2-tailed, unpaired Student t tests. (C) The amplitudes of scotopic (dark-adapted) b-wave, mixed a-wave and mixed b-wave, and photopic (light-adapted) b-wave and flicker responses from epg5−/− and Epg5+/− mice at 6 mo (n = 6). *P < 0.05, **P < 0.01. Data were compared with 2-tailed, unpaired Student t tests.
epg5−/− mice display impaired retinal function
We next examined the visual function in epg5−/− mice. We performed full-field electroretinogram (ERG) recordings to measure retinal function in epg5−/− mice at the age of 6 mo, when the photoreceptor degeneration becomes evident by histological analysis. The b-wave amplitudes of scotopic ERGs measure the rod response. The dark-adapted mixed a-wave and mixed b-wave amplitudes measure combined rod and cone function, while the photopic b-wave and the photopic flicker at 30 Hz performed against a light background measure cone responses only. The amplitude of the ERG response was lower in epg5−/− mice than Epg5+/− mice in the scotopic b-wave test and the mixed a-wave and mixed b-wave tests, revealing impairments in rod function and in mixed rod and cone function, respectively (). The epg5−/− mice also showed a reduced response in the photopic tests (photopic b-wave and 30 Hz flicker), indicating that cone function is impaired (). Therefore, epg5 KO mice show severely impaired retinal function.
The retinas of epg5−/− mice exhibit defective autophagic flux
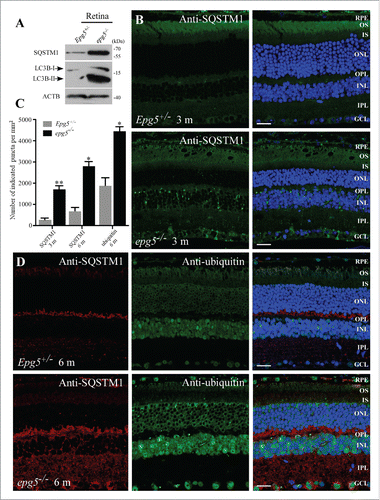
We examined the autophagic flux in epg5−/− retinas by determining the level and distribution of SQSTM1 and ubiquitin, 2 well-characterized autophagy substrates that are elevated after autophagy inhibition.Citation26 Compared to control littermates, extracts of epg5−/− retinas from 6-mo-old animals showed dramatic accumulation of SQSTM1 (). At 3 mo, no cytoplasmic SQSTM1 aggregates were present in control retinas, while epg5−/− retinas had numerous SQSTM1 aggregates in GCL, INL and a few in the ONL (), even though histological analysis showed no evident retinal change (). At the age of 6 mo, many SQSTM1 aggregates and ubiquitin-positive inclusions accumulated in GCL, IPL, INL, OPL, ONL and RPE in epg5 KO mice (). Depletion of Epg5 causes accumulation of nondegradative autophagic vacuoles in worm and mammalian cells and thus an increase in levels of LC3-II, a lipidated form of LC3 associated with autophagic structures.Citation24,25 Levels of LC3-II were also dramatically increased in epg5−/− retinas (). SQSTM1 aggregates became more abundant with age and were detected in all layers of the epg5−/− retina by 15 mo compared with age-matched Epg5+/− controls (). These results indicate that autophagy flux is impaired throughout the epg5−/− retina.
Figure 2. Accumulation of LC3-II, SQSTM1 aggregates and ubiquitin-positive inclusions in the retinas of epg5 knockout mice. (A) Immunoblot of LC3 and SQSTM1 in retinal extracts from epg5−/− and Epg5+/− mice aged 6 mo. (B) SQSTM1 aggregates dramatically accumulate in the GCL, OPL and INL of 3-mo-old epg5−/− mice, but not littermate controls. (C) Quantification of the number of cytoplasmic SQSTM1 and ubiquitin-positive aggregates in the retinas of epg5−/− and Epg5+/− mice aged 3 and 6 mo. Means ± SEM of 4 mice are shown. *P < 0.05, **P < 0.01. Statistical comparisons were performed with 2-tailed, unpaired Student t tests. (D) SQSTM1 aggregates and ubiquitin-positive inclusions dramatically accumulate and are colocalized in Epg5+/− retinas aged 6 mo. Scale bars in ((B)and D): 20 µm.
Photoreceptors in epg5−/−retinas undergo apoptotic cell death
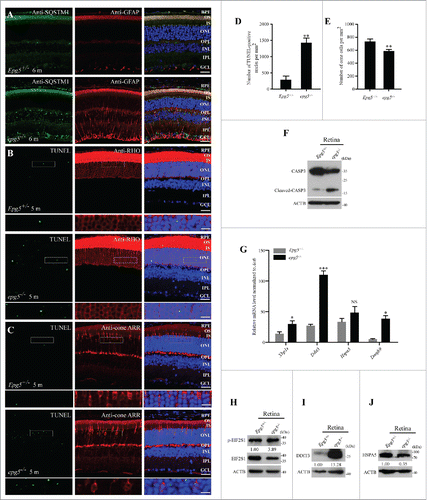
The expression of GFAP (glial fibrillary acidic protein), which is indicative of reactive astrogliosis, was increased throughout the retinas of epg5−/− mice, including the ONL, INL, OPL, IPL and GCL (). Therefore, although only the ONL exhibited degeneration, other neurons may be also damaged. To determine whether the photoreceptor loss was apoptotic, we performed the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay. The number of TUNEL-positive nuclei localized in ONL was dramatically increased in 5-mo-old epg5−/− retinas compared to controls (). The TUNEL-positive nuclei were mainly located in anti-RHO/Rhodopsin-stained rod photoreceptors, and also in anti-Cone ARR/Arrestin-stained cone photoreceptors (). Most of the ONL cells are rods (97% of photoreceptors are rods in the mouse retina);Citation27 therefore, the much thinner ONL in epg5−/− retinas indicates massive degeneration of rods. The number of cones was reduced by 15% in epg5−/− retinas (). Caspase activity was also assessed. Levels of cleaved CASP3 were dramatically increased, while levels of full-length CASP3 were reduced in epg5−/− retinas compared with controls (). The TUNEL-positive nuclei and increased levels of cleaved CASP3 indicate that apoptotic cell death leads to loss of photoreceptors in epg5−/− mice.
Figure 3. Photoreceptors undergo apoptosis and ER stress is activated in epg5−/− retinas. (A) Immunostaining analysis of cytoplasmic SQSTM1 and GFAP in the retinas of epg5−/− and Epg5+/− mice aged 6 mo. Few GFAP-stained structures are detected in Epg5+/− retinas, while many GFAP-stained tubular structures are observed throughout the retinas of epg5−/− mice. Scale bars: 20 µm. (B and C) TUNEL staining of 5-mo-old epg5−/− and Epg5+/− retinas that are costained with anti-RHO (rhodopsin) or anti-cone ARR/Arrestin antibodies. Scale bars: 20 µm. Enlargements of the boxed areas were shown on the bottom for each panel. Scale bars in enlargements: 60 µm. (D) Quantification of TUNEL-positive cells in retinas of epg5−/− and Epg5+/− mice. Means ± SEM of 6 paired mice at the age of 5 mo are shown. **P < 0.01. Statistical significance was determined by a 2-tailed, unpaired Student t test. (E) Quantification of anti-cone ARR-positive cones in epg5−/− and Epg5+/− mice at the age of 10 mo is shown in (E). Means ± SEM of 6 paired mice are shown. **P < 0.01. Statistical significance was determined by a 2-tailed, unpaired Student t test. Scale bars: 20 µm. (F) Immunoblotting analysis of full-length CASP3 and cleaved-CASP3 in retinas of epg5−/− and Epg5+/− mice at the age of 6 mo. Levels of cleaved CASP3 are dramatically increased in epg5−/− retinas compared with Epg5+/− retinas. (G) Transcriptional levels of genes involved in the UPR, including spliced Xbp1 (Xbp1s), Ddit3 and Eidj4, are significantly increased in epg5−/− retinas compared to controls at the age of 6 mo. *P < 0.05, ***P < 0.001. Data were compared with 2-tailed, unpaired Student t tests. (H, (I)and J) In epg5−/− retinas, DDIT3 and p-EIF2S1 protein levels are elevated, while the protein level of HSPA5, a downstream marker of the ATF6 pathway, is reduced.
UPR is activated in epg5−/−retinas
Disruption of protein homeostasis in photoreceptors activates UPR, which induces apoptotic cell death.Citation11-13 We next examined the expression of genes involved in ER-stress in the retinas of 6-mo-old epg5−/− mice. Expression of genes downstream of the ER stress pathway, including spliced Xbp1 (Xbp1s), Ddit3 and Dnajb9/Erdj4, were significantly higher in epg5−/− retinas than in controls (). Ddit3, a key mediator of ER stress-induced apoptosis,Citation28 was most significantly elevated. Immunoblotting assays also revealed that p-EIF2S1/p-eIF2α and DDIT3 protein levels were increased in the epg5−/− retina (). Interestingly, HSPA5/BIP protein levels were reduced (), which is probably due to attenuation of cytoprotective UPR by prolonged ER-stress.Citation11,29 Together, these results indicate that elevated production of the apoptotic transcriptional regulator DDIT3 may contribute to apoptotic photoreceptor cell death in the epg5−/− retina.
Discussion
We showed here that Epg5-deficient mice exhibit characteristic features of retinitis pigmentosa. In the retina of epg5 knockout mice, the thickness of the outer nuclear layer, consisting of rod and cone photoreceptor nuclei, is greatly decreased, and the amplitudes of the rod and cone ERG responses are reduced, whereas the morphology of other retinal layers, including RPE, the inner nuclear layer (composed of amacrine cells, bipolar cells and horizontal cells) and the ganglion cell layer, are largely preserved. Autophagy flux, however, is impaired throughout the epg5 knockout retina and SQSTM1 aggregates accumulate in various cell types, suggesting that photoreceptors are more sensitive to Epg5 loss.
Accumulation of SQSTM1 and ubiquitin aggregates, an indicator of impaired autophagy, shows no correlation with neuronal loss. In epg5 knockout mice, autophagy activity is systemically impaired and SQSTM1 aggregates accumulate in many regions of the brain and spinal cord, while specific subsets of neurons, such as spinal cord motor neurons, are lost.Citation25 Loss of motor neurons in epg5 KO mice appears not to result from impaired autophagic flux.Citation25 Impairment of the ubiquitin-proteasome system results in motor neuron loss in mice, while motor neuron-specific knockout of Atg7 causes no motor dysfunction despite accumulation of ubiquitin and SQSTM1.Citation30 Mice deficient in Wdr45/Wipi4 have abnormal accumulation of SQSTM1 aggregates in various brain regions, but fail to show evident neuronal loss.Citation31 Thus, different cell types may exhibit differential sensitivity to autophagy impairment. Autophagy genes also function in autophagy-independent processes, such as Atg5 and Atg7 in unconventional secretion, Epg5 in endocytic trafficking and Ulk1/ATG1 in axon structure and guidance and in coatomer protein complex-II (COPII) vesicle transport.Citation32,33 Neurodegeneration previously attributed to dysfunction of autophagy may instead be linked to autophagy-independent functions of autophagy proteins.
Photoreceptor apoptotic cell death in RP results from the sustained UPR due to disruption of protein homeostasis.Citation11 In transgenic animals, the P23H and T17M rhodopsin mutations activate the UPR, which is followed by elevated expression of the proapoptotic protein DDIT3.Citation12 In epg5−/− retinas, the photoreceptor loss is also apoptotic and levels of cleaved CASP3 are dramatically increased. UPR genes including Ddit3 are also upregulated in epg5−/− retina. Therefore, apoptotic photoreceptor cell death in epg5−/− retinas may result from the elevated UPR. In Epg5-deficient mice, accumulation of nondegradative autophagic vacuoles, together with defective endocytic trafficking,Citation25 may activate UPR and lead to degeneration of certain neuronal populations.
Human genetic studies showed that recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder characterized by agenesis of the corpus callosum, cataracts, hypopigmentation, cardiomyopathy and combined immunodeficiency.Citation34-37 Epg5-deficient mice display some phenotypic similarities with Vici syndrome patients, including autophagy defects, corpus callosum changes and myopathy.Citation38 Nearly all Vici syndrome patients suffer recurrent infections and show combined immunodeficiency.Citation35-37 Epg5-deficient mice, however, exhibit elevated basal lung inflammation and influenza resistance.Citation39 Cataract is another common trait of Vici syndrome that is not obviously detected in Epg5-deficient mice. Vici syndrome patients also show retinal changes and a defect in neurophysiological visual function.Citation40 However, many Vici syndrome patients may not survive to the age when RP develops. Approximately 20% to 30% of RP patients suffer from one of more than 30 different syndromes in which retinal degeneration is associated with nonocular phenotypes.Citation9 Nevertheless, Epg5-deficient mice provide a model of retinitis pigmentosa, which may help to unravel the mechanism of photoreceptor degeneration and to evaluate the efficacy of therapeutic interventions.
Materials and methods
Animals
epg5−/− mice were generated as previously described.Citation25 Briefly, we constructed the Epg5 targeting vector by substituting exons 18 and 19 of Epg5 genomic DNA with a neomycin cassette, then the targeting vector was transfected into 129 R1 embryonic stem cells. And heterozygous Epg5+/− embryonic stem cell clones were microinjected into C57BL/6N blastocysts. Heterozygotes from this chimeric offspring mated with wild-type C57BL/6N mice were mated to obtain homozygous mutant mice. All mice were kept under specific pathogen-free conditions in the animal facility at the Institute of Biophysics, Chinese Academy of Sciences, Beijing. All animal experiments were approved by the institutional committee of the institute. epg5−/− mice start to die at 10 mo, but a few can survive longer. Three pairs of mice aged 15 mo and 3 pairs of mice aged 12 mo were used for histological analysis of retinal sections.
Antibodies
The following antibodies were used in this study: rabbit anti-SQSTM1 (MBL International, PM045), mouse anti-SQSTM1 (Abcam, ab56416), mouse anti-ubiquitin (Cell Signaling Technology, 3936S), rabbit anti-GFAP (Abcam, ab48050), rabbit anti-LC3 (Cell Signaling Technology, 2775S), rabbit anti-CASP3 (Cell Signaling Technology, 9662S), rabbit anti-Cone ARR (Millipore, AB15282), mouse anti-RHO (Millipore, clone 4D2, MABN15), mouse anti-DDIT3 (Cell Signaling Technology, 2895S), rabbit anti-HSPA5 (Cell Signaling Technology, 3177S), rabbit anti-EIF2S1/eIF2α (Cell Signaling Technology, 9722S), rabbit anti-p-EIF2S1/p-eIF2α (Cell Signaling Technology, 3597S) and mouse anti-ACTB (Protein Tech, 60008–1-Ig).
Histology and immunohistochemistry
epg5−/− mice and control mice were euthanized, then eyes were isolated, fixed and embedded in paraffin. For histological analysis, 5-µm sections were stained with hematoxylin-eosin and examined by light microscopy (Imager A1, Carl Zeiss, Jena, Germany) with a 40×/0.75 NA objective lens (Plan-Neofluar, Carl Zeiss, Jena, Germany) and a camera (AxioCamMRc 5, Carl Zeiss, Jena, Germany). Images were processed and viewed with AxioVision 40 (v4.6.3.0, Carl Zeiss, Jena, Germany) software. TUNEL staining was performed using the DeadEnd™ Fluorometric TUNEL System (Promega, G3250) according to the manufacturer's instructions. For immunostaining, sections were deparaffinized and rehydrated, and antigens were retrieved by microwaving in 0.01 M citrate buffer for 10 min. After blocking with 5% goat serum (ZSGB, ZLI-9056) for 60 min at room temperature, sections were incubated with primary antibodies overnight followed by 1-h incubation with FITC- or rhodamine-labeled secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) and counterstained with DAPI (Vector Laboratories). Slides were viewed under a confocal microscope (LSM 710 Meta plus Axiovert zoom, Carl Zeiss, Jena, Germany) with a 63×/1.40 NA oil immersion objective lens (PlanApochromat, Carl Zeiss) and a camera (AxioCamHRm, Carl Zeiss, Jena, Germany) at RT.
Protein extraction and western blotting
After removal of the eyes, neuro-retinas were dissected free of other tissues and lysed with radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% NP-40(Amresco, E109) supplemented with 1 mM PMSF and protease inhibitor cocktail (Roche, 04693116001). Thirty to 40 µg of protein was used for immunoblotting with the indicated antibodies. Results are representative of at least 3 experiments.
Electroretinograms (ERGs)
Mice were dark-adapted overnight, and all subsequent procedures were performed under dim red light. Before the recording, mice were anesthetized and the eyes were dilated with a combination of 1% cyclopentolate hydrochloride and 2.5% phenylephrine hydrochloride. Then mice were placed prostrate onto the ERG platform, which was kept at 37°C. ERGs were recorded simultaneously from the right eye of each animal using a Burian-Allen bipolar electrode (Hansen Laboratories, Coralville, IA) on the cornea of the right eye and a needle ground electrode placed in the tail. ERGs were recorded and analyzed with the Espion Visual Electrophysiology System from Diagnosis, LLC (Littleton, MA, USA). Full-field stimulation was produced with a Ganzfeld-stimulator (Roland Consult, Brandenburg, Germany). Dark-adapted (scotopic) ERGs were recorded first. Four to 64 consecutive stimuli were averaged with an interval between light flashes in scotopic conditions of 10 s for dim flashes and of up to 60 s for the highest intensity. Stimulus intensities of −2 cd·s·m−2 and 1.5 log cd·s·m−2 were used to record the rod response and mixed response, respectively. Light-adapted (photopic) ERGs were then recorded with light flashes of 2 log cd·s·m−2 on a rod-saturating background of 30 cd·m−2. Baseline removal and auto-zeroing were automatically performed with software provided by the manufacturer (Espion).
Quantitative RT-PCR
Total RNA was extracted from neuro-retinas using TRIzol (Invitrogen, 15596018) and cDNA was synthesized using a Super Script® III First-Strand kit (Invitrogen, 11752050). Quantitative PCR was carried out on a Step One Plus™ Real-Time PCR system (Applied Biosystems, Foster city, USA) using SYBR® Premix Ex TaqTM (TaKaRa, RR820A). Data were normalized to the actin level. Results are representative of at least 3 experiments. The following primers were used:
F-Ddit3, 5′-CTGCCTTTCACCTTGGAGAC-3′;
R-Ddit3, 5′-CGTTTCCTGGGGATGAGATA-3′;
F-Hspa5, 5′-GGTGCAGCAGGACATCAAGTT-3′;
R-Hspa5, 5′-CCCACCTCCAATATCAACTTGA-3′;
F-Xbp1s (spliced Xbp1), 5′-GAGTCCGCAGCAGGTG-3′;
R-Xbp1s (spliced Xbp1), 5′-GTGTCAGAGTCCATGGGA-3′;
F-Dnajb9, 5′-TGAATTTGCAGAGGTTTCACTG-3′;
R-Dnajb9, 5′-CAAACTCAGCCCGACACATA-3′;
F-Actb/β-actin, 5′-TGGCTCCTAGCACCATGAAGAT-3;
R-Actb, 5′-GGTGGACAGTGAGGCCAGGAT-3.
Statistical analysis
Results are expressed as mean ± SEM. Data from at least 3 pairs of samples were used for statistical analysis. Statistical -significance was determined by a 2-tailed, unpaired Student t test. *P < 0.05, **P < 0.01, ***P < 0.001.
Abbreviations
| ACTB/β-actin | = | actin, β |
| Atg | = | autophagy-related |
| CNS | = | central nervous system |
| epg | = | ectopic P granule |
| ERG | = | electroretinogram |
| GCL | = | ganglion cell layer |
| GFAP | = | glial fibrillary acidic protein |
| H&E | = | hematoxylin and eosin |
| INL | = | inner nuclear layer |
| IPL | = | inner plexiform layer |
| IS | = | inner segment |
| KO | = | knockout |
| MAP1LC3B/LC3B | = | microtubule-associated protein 1 light chain 3 |
| ONL | = | outer nuclear layer |
| OPL | = | outer plexiform layer |
| OS | = | outer segment |
| RP | = | retinitis pigmentosa |
| RPE | = | retinal pigment epithelium |
| TUNEL | = | terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling |
| UPR | = | unfolded protein response |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KAUP_S_1238554.docx
Download MS Word (1.2 MB)Acknowledgments
We are grateful to Dr. Isabel Hanson for editing work. We also thank the animal facility at the Institute of Biophysics, Chinese Academy of Sciences for maintaining the mice.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (31421002, 31561143001, 31225018 to H.Z., 31401184 to Y.G.Z. and 31401175 to H.Y.Z.), and a grant from the National Basic Research Program of China (2013CB910100) to H.Z. The research of Hong Zhang was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute.
References
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458-67; PMID:19491929; http://dx.doi.org/10.1038/nrm2708
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/10.1038/cr.2013.168
- Lu Q, Wu F, Zhang H. Aggrephagy: lessons from C. elegans. Biochemical J 2013; 452:381-90; http://dx.doi.org/10.1042/BJ20121721
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27-42; PMID:18191218; http://dx.doi.org/10.1016/j.cell.2007.12.018
- Zhang H, Baehrecke EH. Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol 2015; 25:376-87; PMID:25862458; http://dx.doi.org/10.1016/j.tcb.2015.03.001
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885-9; PMID:16625204; http://dx.doi.org/10.1038/nature04724
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880-4; PMID:16625205; http://dx.doi.org/10.1038/nature04723
- Zhao YG, Zhao HY, Miao L, Wang L, Sun F, Zhang H. The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. J Biol Chem 2012; 287:42053-63; PMID:23074225; http://dx.doi.org/10.1074/jbc.M112.415968
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet 2006; 368:1795-809; PMID:17113430; http://dx.doi.org/10.1016/S0140-6736(06)69740-7
- Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet 2010; 11:273-84; PMID:20212494; http://dx.doi.org/10.1038/nrg2717
- Tzekov R, Stein L, Kaushal S. Protein misfolding and retinal degeneration. Cold Spring Harb Perspect Biol 2011; 3:a007492; PMID:21825021; http://dx.doi.org/10.1101/cshperspect.a007492
- Kunte MM, Choudhury S, Manheim JF, Shinde VM, Miura M, Chiodo VA, Hauswirth WW, Gorbatyuk OS, Gorbatyuk MS. ER Stress Is Involved in T17M Rhodopsin-Induced Retinal Degeneration. Invest Ophthalmol Visual Sci 2012; 53:3792-800; PMID:22589437; http://dx.doi.org/10.1167/iovs.11-9235
- Kosmaoglou M, Schwarz N, Bett JS, Cheetham ME. Molecular chaperones and photoreceptor function. Prog Retin Eye Res 2008; 27:434-49; PMID:18490186; http://dx.doi.org/10.1016/j.preteyeres.2008.03.001
- Yao J, Jia L, Khan N, Lin C, Mitter SK, Boulton ME, Dunaief JL, Klionsky DJ, Guan JL, Thompson DA, et al. Deletion of autophagy inducer RB1CC1 results in degeneration of the retinal pigment epithelium. Autophagy 2015; 11:939-53; PMID:26075877; http://dx.doi.org/10.1080/15548627.2015.1041699
- Reme CE. Autophagy in visual cells and pigment epethelium. Invest Ophthalmol Vis Sci 1977; 16:807-14; PMID:302253
- Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, et al. Noncanonical Autophagy Promotes the Visual Cycle. Cell 2013; 154:365-76; PMID:23870125; http://dx.doi.org/10.1016/j.cell.2013.06.012
- Rodriguez-Muela N, Germain F, Marino G, Fitze PS, Boya P. Autophagy promotes survival of retinal ganglion cells after optic nerve axotomy in mice. Cell Death Differ 2012; 19:162-9; PMID:21701497; http://dx.doi.org/10.1038/cdd.2011.88
- Sternberg C, Benchimol M, Linden R. Caspase dependence of the death of neonatal retinal ganglion cells induced by axon damage and induction of autophagy as a survival mechanism. Braz J Med Biol Res 2010; 43:950-6; PMID:20802972; http://dx.doi.org/10.1590/S0100-879X2010007500082
- Knoferle J, Koch JC, Ostendorf T, Michel U, Planchamp V, Vutova P, Tönges L, Stadelmann C, Brück W, Bähr M, et al. Mechanisms of acute axonal degeneration in the optic nerve in vivo. Proc Natl Acad Sci U S A 2010; 107:6064-9; PMID:20231460; http://dx.doi.org/10.1073/pnas.0909794107
- Rodriguez-Muela N, Koga H, Garcia-Ledo L, de la Villa P, de la Rosa EJ, Cuervo AM, Boya P. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell 2013; 12:478-88; PMID:23521856; http://dx.doi.org/10.1111/acel.12072
- Zhou Z, Doggett TA, Sene A, Apte RS, Ferguson TA. Autophagy supports survival and phototransduction protein levels in rod photoreceptors. Cell Death Differ 2015; 22:488-98; PMID:25571975; http://dx.doi.org/10.1038/cdd.2014.229
- Chen Y, Sawada O, Kohno H, Le YZ, Subauste C, Maeda T, Maeda A. Autophagy protects the retina from light-induced degeneration. J Biol Chem 2013; 288:7506-18; PMID:23341467; http://dx.doi.org/10.1074/jbc.M112.439935
- Rodriguez-Muela N, Hernandez-Pinto AM, Serrano-Puebla A, Garcia-Ledo L, Latorre SH, de la Rosa EJ, Boya P. Lysosomal membrane permeabilization and autophagy blockade contribute to photoreceptor cell death in a mouse model of retinitis pigmentosa. Cell Death Differ 2015; 22:476-87; PMID:25501597; http://dx.doi.org/10.1038/cdd.2014.203
- Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 2010; 141:1042-55; PMID:20550938; http://dx.doi.org/10.1016/j.cell.2010.04.034
- Zhao H, Zhao YG, Wang X, Xu L, Miao L, Feng D, Chen Q, Kovács AL, Fan D, Zhang H. Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J Cell Biol 2013; 200:731-41; PMID:23479740; http://dx.doi.org/10.1083/jcb.201211014
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313-26; PMID:20144757; http://dx.doi.org/10.1016/j.cell.2010.01.028
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci 1998; 18:8936-46; PMID:9786999
- Cao SS, Kaufman RJ. Unfolded protein response. Curr Biol 2012; 22:R622-6; PMID:22917505; http://dx.doi.org/10.1016/j.cub.2012.07.004
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science 2007; 318:944-9; PMID:17991856; http://dx.doi.org/10.1126/science.1146361
- Tashiro Y, Urushitani M, Inoue H, Koike M, Uchiyama Y, Komatsu M, Tanaka K, Yamazaki M, Abe M, Misawa H, et al. Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J Biol Chem 2012; 287:42984-94; PMID:23095749; http://dx.doi.org/10.1074/jbc.M112.417600
- Zhao YG, Sun L, Miao G, Ji C, Zhao H, Sun H, Miao L, Yoshii SR, Mizushima N, Wang X, et al. The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy 2015; 11:881-90; PMID:26000824; http://dx.doi.org/10.1080/15548627.2015.1047127
- Joo JH, Wang B, Frankel E, Ge L, Xu L, Iyengar R, Li-Harms X, Wright C, Shaw TI, Lindsten T, et al. The Noncanonical Role of ULK/ATG1 in ER-to-Golgi Trafficking Is Essential for Cellular Homeostasis. Mol Cell 2016; 62:491-506; http://dx.doi.org/10.1016/j.molcel.2016.04.020
- Zhao YG, Zhang H. The Incredible ULKs: Autophagy and Beyond. Mol Cell 2016; 62:475-6; PMID:27203174; http://dx.doi.org/10.1016/j.molcel.2016.05.005
- Cullup T, Kho AL, Dionisi-Vici C, Brandmeier B, Smith F, Urry Z, Simpson MA, Yau S, Bertini E, McClelland V, et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet 2012; 45:83-7; PMID:23222957; http://dx.doi.org/10.1038/ng.2497
- Vici CD, Sabetta G, Gambarara M, Vigevano F, Bertini E, Boldrini R, Parisi SG, Quinti I, Aiuti F, Fiorilli M. Agenesis of the corpus callosum, combined immunodeficiency, bilateral cataract, and hypopigmentation in two brothers. Am J Med Genet 1998; 29:1-8; http://dx.doi.org/10.1002/ajmg.1320290102
- Chiyonobu T, Yoshihara T, Fukushima Y, Yamamoto Y, Tsunamoto K, Nishimura Y, Ishida H, Toda T, Kasubuchi Y. Sister and brother with Vici syndrome: agenesis of the corpus callosum, albinism, and recurrent infections. Am J Med Genet 2002; 109:61-6; PMID:11932994; http://dx.doi.org/10.1002/ajmg.10298
- Al-Owain M, Al-Hashem A, Al-Muhaizea M, Humaidan H, Al-Hindi H, Al-Homoud I, Al-Mogarri I. Vici syndrome associated with unilateral lung hypoplasia and myopathy. Am J Med Genet 2010; 152A:1849-53; PMID:20583151; http://dx.doi.org/10.1002/ajmg.a.33421
- Zhao YG, Zhao H, Sun H, Zhang H. Role of Epg5 in selective neurodegeneration and Vici syndrome. Autophagy 2013; 9:1258-62; PMID:23674064; http://dx.doi.org/10.4161/auto.24856
- Lu Q, Yokoyama CC, Williams JW, Baldridge MT, Jin X, DesRochers B, Bricker T, Wilen CB, Bagaitkar J, Loginicheva E, et al. Homeostatic Control of Innate Lung Inflammation by Vici Syndrome Gene Epg5 and Additional Autophagy Genes Promotes Influenza Pathogenesis. Cell Host Microbe 2016; 19:102-13; PMID:26764600; http://dx.doi.org/10.1016/j.chom.2015.12.011
- Byrne S, Jansen L, U-King-Im JM, Siddiqui A, Lidov HG, Bodi I, Smith L, Mein R, Cullup T, Dionisi-Vici C, et al. EPG5-related Vici syndrome: a paradigm of neurodevelopmental disorders with defective autophagy. Brain 2016; 139:765-81.
