ABSTRACT
The master regulator of lysosome biogenesis, TFEB, is regulated by MTORC1 through phosphorylation at S211, and a S211A mutation increases nuclear localization. However, TFEBS211A localizes diffusely in both cytoplasm and nucleus and, as we show, retains regulation by MTORC1. Here, we report that endogenous TFEB is phosphorylated at S122 in an MTORC1-dependent manner, that S122 is phosphorylated in vitro by recombinant MTOR, and that S122 is important for TFEB regulation by MTORC1. Specifically, nuclear localization following MTORC1 inhibition is blocked by a S122D mutation (despite S211 dephosphorylation). Furthermore, such a mutation inhibits lysosomal biogenesis induced by Torin1. These data reveal a novel mechanism of TFEB regulation by MTORC1 essential for lysosomal biogenesis.
Introduction
TFEB (transcription factor EB) functions as an oncogene in a subset of RCCs (renal cell carcinomas).Citation1-3 In these tumors, the TFEB gene is translocated and overexpressed. TFEB is a bHLH (basic helix-loop-helix) leucine zipper transcription factor of the MITF family.Citation4 TFEB is a master transcriptional regulator of lysosomal biogenesisCitation5 and translocation carcinomas overexpress lysosomal proteases.Citation6 TFEB is also implicated in other functions like autophagy,Citation7 endocytosis,Citation8 exocytosis,Citation9 lipid metabolism,Citation10 the antiviral response,Citation11 and lysosomal calcium signaling.Citation12 Mice deficient for Tfeb/Tcfeb die at midgestation due to deficient placental vascularizationCitation13 and may exhibit defects in endoderm development.Citation14
MTORC1 (mechanistic target of rapamycin complex 1), an atypical Ser/Thr kinase, controls the balance between anabolism and catabolism and responds to a variety of signals including nutrients. Nutrient signals are relayed to MTORC1 through RRAG/Rag GTPases.Citation15,16 In the absence of amino acids, RRAGs stay in an inactive state in which RRAGA or RRAGB is bound to GDP and RRAGC or RRAGD is bound to GTP. Amino acids induce a switch in the GDP/GTP binding state of the RRAGs, activating the complex. When the Rag complex is active, MTORC1 translocates to the surface of the lysosome, where it signals to activate anabolic functions such as protein synthesis. The RRAGs are controlled by multiple factors, including the Ragulator complex,Citation17,18 which is a GEF (guanine nucleotide exchange factor) for RRAGA and RRAGB.Citation17 RRAGA/B GTP binding is also controlled by GATOR1, a complex with GAP (GTPase activating protein) activity.Citation19 The nucleotide bound state of RRAGC/D is regulated by FLCN (folliculin), which also functions as a GAP.Citation20
We reported previously that TFEB is regulated by MTORC1,Citation8 but TFEB regulation by MTORC1 is complex and cell context-dependent.Citation8,21 Nutrient deprivation leads to MTORC1 inhibition, reduces TFEB phosphorylation, and promotes TFEB nuclear translocation and expression of lysosomal genes.Citation22,23 While other mechanisms have been proposed,Citation7 MTORC1 is thought to phosphorylate TFEB on S211Citation24,25 and also on S142.Citation23 S211 phosphorylation by MTORC1 creates a high affinity binding site for YWHA proteins, leading to TFEB cytosolic retention. When MTORC1 is inhibited, S211 is dephosphorylated, and TFEB enters the nucleus. S142 is also dephosphorylated following conditions resulting in MTORC1 inhibition, but this has unclear functional significance.
Interestingly, several large-scale phosphoproteomic studies have shown that TFEB is phosphorylated at more than 20 sites,Citation26-30 including multiple sites that are regulated by MTORC1,Citation31 such as S122. We show here that TFEB dephosphorylation at S211 is not sufficient to exclusively localize TFEB into the nucleus and that a TFEB mutant (S211A) retains regulation by MTORC1. We identified S122 as a site directly phosphorylated by MTOR whose phosphomimetic substitution largely blocks the effects of MTORC1 inhibition on TFEB. Our results support a multistep mechanism of TFEB regulation by MTORC1.
Results
Torin1 induces TFEB dephosphorylation and nuclear localization
In the presence of excess nutrients and growth factors, TFEB is predominantly cytosolic. However, following treatment with the ATP-competitor MTOR inhibitor Torin1Citation32 TFEB is dephosphorylated (resulting in a faster migrating form on SDS-PAGE) and predominantly localizes to the nucleus. This phenomenon has been observed in multiple cell types.Citation23-25,33 Consistent with these results, treatment with Torin1 of both HeLa cells and mouse embryonic fibroblasts (MEFs) shifted TFEB to a fast migrating, hypophosphorylated, form that was predominantly localized in the nucleus (Fig. S1A to D). This increased nuclear localization is also found after depletion of MTOR (Fig. S1E), or RPTOR, a specific component of MTORC1 (Fig. S1F).
TFEB regulation by Torin1 has been attributed to changes in S211 phosphorylation, and mutations of serine 211 to alanine increase TFEB nuclear localization.Citation24,25 MTORC1 is thought to phosphorylate S211 (although no evidence of direct phosphorylation of S211 by MTOR in vitro has been reported to date) creating a docking site for YWHA proteins and leading to cytoplasmic sequestration. Conversely, MTORC1 inhibition by Torin1 induces S211 dephosphorylation (indirectly measured using an antibody against a consensus YWHA binding siteCitation24,33 and more recently using a phospho-S211 antibodyCitation34). We observed similar results with a novel phospho-S211 antibody we developed in collaboration with Bethyl that specifically recognizes phosphorylated S211 (). Specifically, Torin1 inhibited phosphorylation of S211 in HeLa cells () reducing the amount of TFEB bound to YWHA proteins (). A TFEBS211A mutant failed to interact with YWHA proteins () and was no longer excluded from the nucleus (). Specifically, TFEBS211A-GFP, unlike wild-type TFEB-GFP, diffusely localized throughout the cytosol and nucleus ().
Figure 1. Torin1 enhances TFEBS211A nuclear localization. (A) Validation of the newly generated p-TFEB (S211) antibody. HeLa cells were transfected with the indicated MYC-tagged plasmids (WT, wild-type TFEB; S211A TFEB mutant or EV, empty vector), lysed, and treated or not with lambda phosphatase (“λ”) before MYC immunoprecipitation (IP) and western blot analysis. (B) p-TFEB (S211) analysis of Torin1-treated HeLa cells (250 nM). Endogenous TFEB was immunoprecipitated and analyzed by western blot. (C) HeLa cells transfected with the indicated MYC-tagged TFEB constructs were subjected to immunoprecipitation with YWHA or MYC antibodies and analyzed by western blot. Torin1 (250 nM) for 6 h. Where indicated, immunoprecipitates were treated with lambda phosphatase (λ). (D) Quantification of TFEB-GFP or TFEBS211A-GFP subcellular localization in HeLa cells transfected as indicated (C, exclusively cytoplasmic; C and N, cytoplasmic and nuclear; N, exclusively nuclear). Graph illustrates average (n = 2; ≥100 cells per experiment) ± SD. *, P < 0.05 for comparison between TFEB-WT and TFEBS211A for each of the fractions. (E) HeLa cells were transfected with the indicated plasmids and biochemical fractionation was performed after Torin1 treatment (250 nM for 3 h). (F) HeLa cells stably expressing TFEB-GFP (WT vs. S211A) were treated with vehicle (DMSO) or Torin1 (250 nM for 6 h) and analyzed by immunofluorescence (left panel, representative images; middle panel, schematic representation; right panel, quantification). Two-factor ANOVA was used to compare the global effect of the TFEBS211A mutation. ***, P < 0.001; ns, nonsignificant. Scale bar: 20 μm.
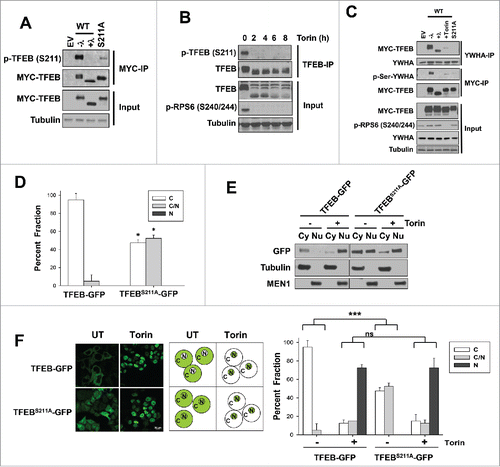
Importantly, however, we observed that TFEBS211A-GFP was still regulated by Torin1. Torin1 treatment, in fact, changed the distribution of TFEB from a diffuse pattern throughout the cell to almost exclusively nuclear (). Thus, while S211 regulation was important for cytoplasmic retention of TFEB, other mechanisms exist that enrich TFEB in the nucleus in response to Torin1.
Endogenous S122 is phosphorylated and its phosphorylation is regulated by MTORC1
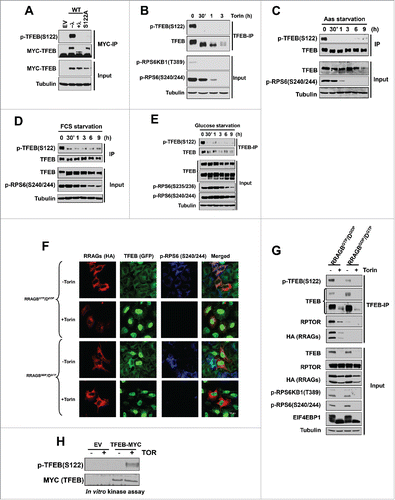
According to large-scale phosphoproteomic studies,Citation26-30 there are multiple serine residues in TFEB that are potentially phosphorylated including S122. S122 is conserved across species (Fig. S2), represents a putative MTORC1 site,Citation23,35 and was affected by MTORC1 inhibitors.Citation31 In collaboration with Bethyl Laboratories, we developed a second antibody that specifically recognized p-TFEB (S122) and whose signal was eliminated after lambda-phosphatase treatment or when TFEB (S122) was mutated (). Using this antibody, we found that S122 was rapidly dephosphorylated by multiple conditions inhibiting MTORC1, including Torin1 (), amino acid starvation (), serum starvation (), glucose starvation () as well as in response to expression of dominant negative RRAG proteins ( and ). These data suggest that S122 is regulated in an MTORC1-dependent manner. To determine whether MTORC1 directly phosphorylates S122, we performed in vitro kinase assays. As shown in , recombinant MTOR directly phosphorylated TFEB immunoprecipitates.
Figure 2. TFEB serine 122 is regulated by MTORC1. (A) Validation of the p-TFEB (S122) antibody. HeLa cells were transfected with the indicated MYC-tagged TFEB constructs followed by MYC-IP and western blot. λ, lambda-phosphatase treatment of immunoprecipitates. HeLa cells were treated with Torin1 (250 nM) (B), or starved for amino acids (C), serum (D), or glucose (E). (F, G) HeLa cells were transfected with active (RRAGBGTP/DGDP) or inactive (RRAGBGDP/DGTP) RRAG-GTPases and its effects on TFEB localization and phosphorylation were assessed. (H) MTOR in vitro kinase assay of epitope tag immunoprecipitates from cells transfected with empty vector (EV) vs. MYC-TFEB and incubated with or without recombinant MTOR. Scale bar: 10 μm.
Serine 122 dephosphorylation is essential for TFEB nuclear localization and TFEB (S122D) fails to induce lysosomal biogenesis
We tested whether S122 dephosphorylation was essential for TFEB nuclear localization in response to Torin1 treatment. We transfected HeLa cells with full-length wild-type or TFEBS122D constructs and analyzed their localization by confocal and biochemical fractionation experiments. We observed that Torin1 induced nuclear localization of ectopically expressed TFEB, but this was significantly blunted by S122D mutation ( and ).
Figure 3. Serine 122 regulation is essential for TFEB-mediated lysosome biogenesis. (A, B) Subcellular localization of transfected full-length-TFEB or S122D mutant in HeLa cells analyzed by confocal microscopy or biochemical subcellular fractionation in cells treated or not with Torin1 (250 nM, 3 h). Graph represents average ± SD (n = 6; ≥ 100 cells per experiment; *, P < 0.05). (C, D) HeLa cells depleted of TFEB were transfected with the indicated constructs and treated with Torin1 for 36 h. Cells were stained with LysoTracker Red (C) or an antibody against LAMP1 (D) and analyzed by FACS. Graphs indicate mean ± SE (n = 3). Asterisks illustrate statistically significant differences in LysoTracker Red or LAMP1 intensity. **, P < 0.01. (E) qPCR analysis of TFEB-target genes in HeLa cells depleted of TFEB and transfected with the indicated constructs (Torin1 250 nM, 36 h). ***, P < 0.001.
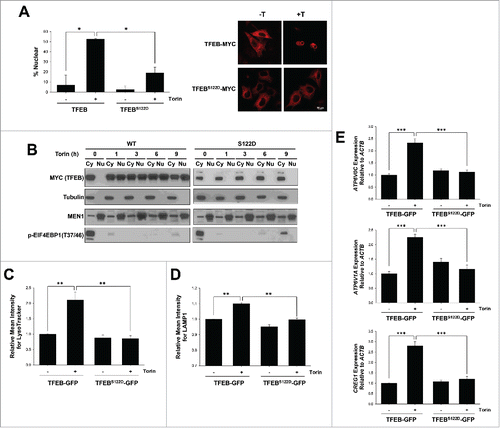
To ascertain the functional consequences of a S122D mutation, we evaluated lysosomal biogenesis. TFEB is a master regulator of lysosomal biogenesis, and this process is induced following MTORC1 inhibition by Torin1. For these experiments, we reconstituted HeLa cells depleted of endogenous TFEB by shRNACitation8 with shRNA-resistant plasmids encoding wild-type TFEB fused to GFP (TFEB-GFP) or a TFEB mutant (TFEBS122D-GFP). To assess the effects of MTORC1 inhibition on lysosomal biogenesis, we evaluated cells by FACS using LysoTracker Red or a LAMP1 antibody. As expected, cells expressing wild-type TFEB induced lysosomal biogenesis following MTORC1 inhibition ( and ), and increased the expression of its target genes (). However, the S122D phosphomimetic mutation largely blocked the effects of Torin1 on lysosome biogenesis and target gene expression ( to ). These data suggest that TFEB S122 dephosphorylation is essential for Torin1-mediated regulation of TFEB and lysosome biogenesis.
Serine 122 and 211 cooperate to regulate TFEB nuclear localization
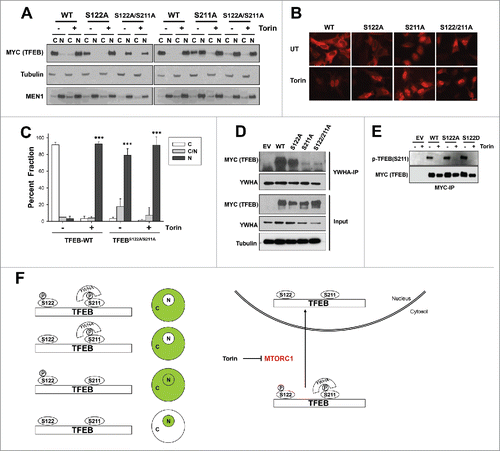
Next, we tested the S122A mutation. We evaluated the effects of a S122A single mutant and also in the context of a double mutation (S122A;S211A). The S122A single mutant behaved similarly to wild-type TFEB. The S122A mutant primarily localized to the cytosol, was enriched in the nucleus upon Torin1 treatment ( and Fig. S3A) and bound to YWHA proteins under basal conditions (). However, the S122A;S211A double mutant predominantly localized to the nucleus in basal conditions (to ). Indeed the subcellular localization of S122A;S211A was very similar to that of wild-type TFEB following Torin1 treatment. Parenthetically, both residues appeared to have similar dephosphorylation kinetics following Torin1 treatment (Fig. S3B), and mutation at S122 to either alanine or aspartate did not affect the S211 phosphorylation at baseline ().
Figure 4. Serine 122 contributes to TFEB nuclear enrichment in the context of S211A mutation (A-C) Subcellular localization of transfected full-length-TFEB, S122A, S211A or S122A;S211A mutants in HeLa cells by biochemical fractionation (A) or confocal microscopy (B, C) following (or not) treatment with Torin1 (250 nM, 3 h). Graph represents average ± SD (n = 3; ≥ 100 cells per experiment; ***, P < 0.01). (D) YWHA protein immunoprecipitation from HeLa cells transfected with the indicated TFEB constructs. (E) Western blot for S211 phosphorylation of wild-type or mutant MYC-TFEB immunoprecipitated from transfected HeLa cells. (F) Proposed model for TFEB regulation by MTORC1. Scale bar: 20 μm.
Discussion
Our data show that maximal nuclear enrichment is achieved by simultaneous mutation of S122 and S211 to nonphosphorylatable residues. However, whereas a S211A mutation causes diffuse localization through both the nucleus and cytosol, a S122A mutation, by itself, does not. These data suggest that YWHA binding, which is mediated by S211, is sufficient for TFEB cytosolic retention. Conversely, a S122D mutation is sufficient to block nuclear localization of an S211A mutant. Barring untoward effects from an aspartate substitution, these data suggest that dephosphorylation at S122 is necessary for TFEB nuclear localization following MTORC1 inhibition. Overall, our results support a model whereby TFEB is regulated by MTORC1 in a multi-step process that involves at least 2 different residues, S122 and S211. There is precedent for the phosphorylation of MTORC1 substrates at multiple residues, including EIF4EBP1,Citation36,37 ULK1Citation38-40 and GRB10Citation31,35 and for the regulation of a variety of transcription factors by MTORC1 such as HIF1A/HIF-1α (hypoxia inducible factor 1 α subunit),Citation41-43 SREBPs,Citation44 PPARGC1A/PGC1α (PPARG coactivator 1 α),Citation45 YY1 (YY1 transcription factor),Citation45 PPARG/PPARγ (peroxisome proliferator-activated receptor gamma)Citation46,47 and STAT3 (signal transducer and activator of transcription 3).Citation47,48
The molecular mechanism whereby changes in phosphorylation at S122 affect its distribution through the cell remains to be determined. However, multiple lines of evidence suggest that this mechanism is different from S211 and does not involve YWHA binding. Specifically, whereas an S211D mutation does not mimic phosphorylated S211 (possibly because this substitution is not sufficient to trigger YWHA binding), an S122D has functional consequences. Moreover, YWHA binding is largely abrogated by an S211A mutation showing that this is the primary site mediating the interaction. Notably, while the subcellular localization of several transcription factors is regulated through phosphorylation-mediated binding to 14–3–3 proteins, including the Forkhead transcription factor FOXO3/FOXO3a,Citation49 YY1AP1/YAP (YY1 associated protein 1),Citation50 and WWTR1/TAZ (WW domain containing transcription regulator 1),Citation51 to our knowledge, TFEB is the first transcription factor in which the same kinase regulates subcellular localization in a multistep process involving YWHA-dependent and -independent mechanisms.
The simplest explanation for how phosphorylation at S122 affects TFEB subcellular localization is that phosphorylation at this site regulates protein-protein interactions or nuclear transport. However, other possibilities such as modulation of compartment-specific degradation exist. Independently of these considerations, our data show that MTORC1 coordinately regulates the phosphorylation of S211 and S122, and that substitution to nonphosphorylatable residues at both positions, but not either one, is sufficient to reproduce the predominantly nuclear localization of TFEB observed following MTORC1 inhibition by Torin1. While phosphorylation at other sites on TFEB is similarly regulated by MTORC1 (Citationref. 23 and data not shown), our data suggest that both S122 and S211 play a critical role.
Materials and methods
Cell culture, antibodies and reagents
HeLa cells and MEFs were grown in DMEM (Sigma-Aldrich, D5671) containing 10% fetal bovine serum (GE Healthcare, Hyclone SH30088.03) and 1% penicillin/streptomycin (P/S) (Gibco, 15140122). Stable HeLa cells depleted of TFEB were reported elsewhere.Citation8 For starvation experiments, cells were washed with phosphate-buffered saline (PBS; Sigma, D8662) and amino acid-free medium was added (USBiological, R9010–03), containing or not 10% dialyzed fetal bovine serum (Invitrogen, 26400036). Torin1 was from R&D Systems (4247).
Antibodies were from the following sources (all used at a 1:1000 dilution for western blot unless otherwise specified): Bethyl Laboratories: TFEB (A303–673A), MEN1/Menin (A300–105A), p-TFEB (S122), p-TFEB (S211) (these antibodies will be released for commercial use after publication); Cell Signaling Technology: p-RPS6KB1/S6K1 (T389) (9205), p-RPS6 (S240/244) (2215), p-Ser-YWHA binding motif (9601), p-EIF4EBP1 (T37/46) (9459), EIF4EBP1 (9452), MTOR (2983); Sigma-Aldrich: Tubulin (1:5000; T5168), LAMP1 (1:500 for FACS analysis, L1418); Santa Cruz Biotechnology, MYC (1:500; sc-40); Invitrogen: GFP (A11122); Thermo-Scientific: pan-YWHA (MS-1504-P0), HRP-conjugated secondary antibodies for western blot (1:5000; 31430, 31460); Covance: HA.11 (MMS-101P); Millipore: RPTOR (90217); Jackson Immunolabs: fluorescence-labeled secondary antibodies for immunofluorescence experiments (111– 175–144, 715–165–150).
Plasmids
Deletions and mutations of the full-length human TFEB cDNACitation8,52 were generated by conventional cloning and site-directed mutagenesis techniques and validated by sequencing. Plasmids (Brugarolas Lab Database ID): pcDNA3.1-TFEB-WT-MYC (#647), pcDNA3.1-TFEBS122D-MYC (#777), pcDNA3.1-TFEBS211A-MYC (#805), pEGFP-N1-TFEB-WT-GFP (#845), pcDNA3.1-TFEBS122A-MYC (#776), pcDNA3.1-TFEBS122,S211A-MYC (#929). TFEB-shRNA-resistant constructs were pEGFP-N1-TFEB-WT-(MMsh111)-GFP (#889) and pEGFP-N1-TFEB-S122D-(MMsh111)-GFP (#890). pRK5-HA-GST-RRAG plasmids [RRAGBT54L (#831, mutant constitutively bound to GDP), RRAGBQ99L (#828, mutant constitutively bound to GTP), RRAGDS77L (#829 mutant constitutively bound to GDP), RRAGDQ121L (#830, mutant constitutively bound to GTP)] were obtained from Addgene (David Sabatini's laboratory; plasmid numbers: 19302, 19303, 19308, 19309).
Biochemical fractionation
Subcellular fractionation of nuclear and cytosolic fractions was performed as reported.Citation8 Cells were rinsed in ice-cold PBS, scraped and collected after centrifugation. Cell pellets were resuspended in 2 cell volumes of hypotonic lysis buffer (HLB; 10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2) containing protease [0.1 μM aprotinin (USBiological, 162669), 0.02 mM leupeptin (USBiological, L2050), 0.01 mM pepstatin (USBiological, P3280), 0.5 mM benzamidine (Sigma, 434760), 0.5 mM PMSF (Sigma, 78830), 0.01 M NaF (Sigma, 201154)] and phosphatase [2 mM imidazole (Sigma, I5513), 1.15 mM sodium molybdate (Sigma, 243655), 1 mM sodium orthovanadate (Sigma, 450243), 5 nM microcystin (Calbiochem, 475815)] inhibitors for 10 min on ice. NP40 (Sigma, I3021) was added to 0.1% and incubated for 10 min on ice. After centrifugation at 1000 x g, for 5 min, the cytosolic fractions were transferred to fresh tubes and nuclei were washed with HLB-NP40 (0.1%). Both nuclear and cytosolic fractions were lysed in 5× lysis buffer and equivalent amounts of the indicated fractions were analyzed by western blot.
siRNA and plasmid transfections
For siRNA experiments, HeLa cells were transfected using the DharmaFECT3 reagent according to manufacturer's instructions (ThermoScientific, T-2003–01). siRNA oligos were from Dharmacon: Scrambled siRNACitation8 (5′-CAAUGAGUAACAAUCCAUUGAUU-3′), siGENOME human MTOR (A) siRNA (5′- CCAAAGUGCUGCAGUACUA-3′), and MTOR (B) siRNA (5′- GAGAAGAAAUGGAAGAAAU-3′), siON-TARGET-PLUS for human RPTOR and siGENOME SMART pool for human RICTOR (RPTOR independent companion of MTOR complex 2). siRNAs were transfected using TransIT-LT1 reagent (MirusBio. MIR 2300), or Lipofectamine 2000 LTL reagent (Invitrogen, 11668027), according to the manufacturer's instructions.
In vitro kinase assays
MTOR in vitro kinase assays were performed as in Yu and coworkers.Citation31 Briefly, HeLa cells were transfected with plasmids containing MYC-TFEB. 24 h later, cells were amino acid starved for 2 h, lysed with IP buffer and immunoprecipitation was performed according to conventional protocols (see IP section). MYC immunoprecipitates were washed twice with IP buffer, twice with FRAP kinase buffer (Invitrogen, PV4794), and resuspended in FRAP kinase buffer containing DTT (2 mM), ATP (10 μM; Invitrogen, PV3227) and MTOR kinase (250 ng; Calbiochem, 475987). Assays were performed for 30 min at 37°C and reactions were stopped by the addition of 3× protein loading buffer and boiled for 5 min. Analyses were performed by western blot.
Cell lysates, immunoprecipitation and western blot
Cells were rinsed with ice-cold PBS and lysed with lysis buffer containing protease and phosphatase inhibitors for 10 min at 4°C as previously reported.Citation53 Lysates were cleared by centrifugation at 16,000 g for 10 min at 4°C. 3× loading buffer was added and samples were boiled for 10 min. Lysate analysis was done by western blot. For immunoprecipitation experiments, cells were rinsed with ice-cold PBS and lysed using IP buffer (with protease and phosphatase inhibitors) for 10 min at 4°C as previously reported.Citation53 Cleared lysates were incubated with proteinG-Sepharose beads (Invitrogen, 101242) for 1 h at 4°C and transferred to a new tube for further antibody incubation. Immunoprecipitation was performed for 6 h in the cold and protein G-Sepharose beads were added for an additional hour. IPs were washed 3 times with IP buffer and boiled for 5 min in 1× loading buffer.
Immunofluorescence
Cells were grown in coverslips and transfected with the indicated plasmids. Cells were rinsed in PBS and fixed with 10% formalin-buffered phosphate for 5 min at room temperature. Cells were washed 3 times with PBS, then blocked and permeabilized with 3% BSA (Sigma, A7906)-PBS containing 0.5% Tween-20 (RPI, P20370) for 30 min at room temperature. Blocking solution was rinsed with PBS and the corresponding antibodies were added diluted in 1% BSA-PBS. Samples were analyzed in a blinded manner in a Zeiss LSM510 confocal microscope (Etters, PA, USA), and counting was performed blinded.
FACS analysis
HeLa cells depleted of TFEB were transfected with the indicated plasmids and treated with Torin1 (250 nM) for 36 h. LysoTracker Red (Molecular Probes, L7528) incubation was performed for 20 min and cells were harvested for FACS analysis using a MoFlo flow cytometer (Beckman, Indianapolis, IN, USA). LAMP1 staining on permeabilized samples was performed for 1 h at room temperature. GFP-positive cells were analyzed for lysosomal content with FlowJo software (Tree Star Inc., OR, USA). Median LysoTracker Red or LAMP1 intensities are shown normalized to the samples not incubated with LysoTracker Red or LAMP1 antibody.
qPCR
HeLa cells depleted of TFEB were transfected with the indicated plasmids and treated with Torin1 (250 nM) for 36 h. RNA was extracted with RNeasy (Qiagen, 74104) and reverse transcribed with M-MLV reverse transcriptase (Invitrogen, 28025013) with random hexamer primers from 1 μg RNA according to the manufacturer instructions. qPCR was performed with a Bio-Rad CFX96 Real-Time PCR Detection System (Hercules, CA, USA) as described previouslyCitation8 using the following primers: ATP6V0C Fwd 5′-GTATGCTTCGTTTTTCGCCG-3′, ATP6V0C Rev 5′- CGATGATGCCAGCCATGACCAC-3′, ATP6V1A Fwd 5′-GAAACTTCTGGTGTGTCTGT-3′, ATP6V1A Rev 5′- CCATAATGCCAGGACCAAG-3′, CREG1 Fwd 5′-CAAAAATCGTGACACCAGAAG-3′, CREG1 Rev 5′-CTAAATTCACCACAGTCTGCTTC-3′, ACTB Fwd 5′-CACCCGCCGCCAGCTCACCATG-3′, ACTB Rev 5′-CCATGCCCACCATCACGCCCTG-3′.
Statistics
Means were compared with t tests unless otherwise indicated. Two-tailed Student t tests were performed for samples with equal variances and 2-tailed Welch t tests were performed for samples with unequal variances. Two-factor analysis of variance (ANOVA) was performed using SPSS Statistics 17.
Protein sequence alignment
Alignments of selected sequences were done using the Vector NTI® software (Life Technologies).
Abbreviations
| EIF4EBP1 | = | eukaryotic translation initiation factor 4E binding protein 1 |
| GFP | = | green fluorescent protein |
| LAMP1 | = | lysosomal associated membrane protein 1 |
| MEF | = | mouse embryo fibroblast |
| MTOR | = | mechanistic target of rapamycin |
| MTORC1 | = | mechanistic target of rapamycin complex 1 |
| MYC | = | v-myc avian myelocytomatosis viral oncogene homolog |
| RRAG | = | Ras related GTP binding |
| RPS6 | = | ribosomal protein S6 |
| RPS6K1B | = | ribosomal protein S6 kinase B1 |
| RPTOR | = | regulatory associated protein of MTOR complex 1 |
| siRNA | = | small interfering RNA |
| TFEB | = | transcription factor EB |
| YWHA | = | 14–3–3 phospho-serine/phospho-threonine binding proteins |
Disclosure of potential conflicts of interest
Dr. Brugarolas is a member of the scientific advisory board at Bethyl Laboratories.
KAUP_A_supplementaryfigures.pdf
Download PDF (304.9 KB)Acknowledgments
We thank Dr. Beth Levine for critically reviewing the manuscript and members of the Brugarolas lab for helpful discussions.
Funding
J.B is supported by R01CA175754, CPRIT RP130603 and P50CA196516.
References
- Srigley JR, Delahunt B. Uncommon and recently described renal carcinomas. Mod Pathol 2009; 22(Suppl 2):S2-S23; PMID:19494850; http://dx.doi.org/10.1038/modpathol.2009.70
- Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A 2003; 100:6051-6; PMID:12719541; http://dx.doi.org/10.1073/pnas.0931430100
- Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, Schuuring E, Schoenmakers EF, van Kessel AG. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Human Mol Genet 2003; 12:1661-9; PMID:12837690; http://dx.doi.org/10.1093/hmg/ddg178
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Ann Rev Genet 2004; 38:365-411; PMID:15568981; http://dx.doi.org/10.1146/annurev.genet.38.072902.092717
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science (New York, NY 2009; 325:473-7.
- Zheng G, Martignoni G, Antonescu C, Montgomery E, Eberhart C, Netto G, Taube J, Westra W, Epstein JI, Lotan T, et al. A broad survey of cathepsin K immunoreactivity in human neoplasms. Am J Clin Pathol 2013; 139:151-9; PMID:23355199; http://dx.doi.org/10.1309/AJCPDTRTO2Z4UEXD
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science (New York, NY 2011; 332:1429-33; http://dx.doi.org/10.1126/science.1204592
- Peña-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR, Brugarolas J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J 2011; 30:3242-58; http://dx.doi.org/10.1038/emboj.2011.257
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 2011; 21:421-30; PMID:21889421; http://dx.doi.org/10.1016/j.devcel.2011.07.016
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 2013; 15:647-58; PMID:23604321; http://dx.doi.org/10.1038/ncb2718
- Hasan M, Koch J, Rakheja D, Pattnaik AK, Brugarolas J, Dozmorov I, Levine B, Wakeland EK, Lee-Kirsch MA, Yan N. Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes. Nat Immunol 2013; 14:61-71; PMID:23160154; http://dx.doi.org/10.1038/ni.2475
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 2015; 17:288-99; PMID:25720963; http://dx.doi.org/10.1038/ncb3114
- Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development (Cambridge, England) 1998; 125:4607-16; PMID:9806910
- Young NP, Kamireddy A, Van Nostrand JL, Eichner LJ, Shokhirev MN, Dayn Y, Shaw RJ. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev 2016; 30:535-52; PMID:26944679; http://dx.doi.org/10.1101/gad.274142.115
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science (New York, NY 2008; 320:1496-501; http://dx.doi.org/10.1126/science.1157535
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008; 10:935-45; PMID:18604198; http://dx.doi.org/10.1038/ncb1753
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012; 150:1196-208; PMID:22980980; http://dx.doi.org/10.1016/j.cell.2012.07.032
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010; 141:290-303; PMID:20381137; http://dx.doi.org/10.1016/j.cell.2010.02.024
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science (New York, NY 2013; 340:1100-6; http://dx.doi.org/10.1126/science.1232044
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The Folliculin Tumor Suppressor Is a GAP for the RagC/D GTPases That Signal Amino Acid Levels to mTORC1. Mol Cell 2013; PMID:24095279.
- Peña-Llopis S, Brugarolas J. TFEB, a novel mTORC1 effector implicated in lysosome biogenesis, endocytosis and autophagy. Cell cycle (Georgetown, Tex 2011; 10:3987-8; http://dx.doi.org/10.4161/cc.10.23.18251
- Settembre C, Ballabio A. TFEB regulates autophagy: an integrated coordination of cellular degradation and recycling processes. Autophagy 7:1379-81; PMID:21785263; http://dx.doi.org/10.4161/auto.7.11.17166
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095-108; PMID:22343943; http://dx.doi.org/10.1038/emboj.2012.32
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423; http://dx.doi.org/10.1126/scisignal.2002790
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012; 8:903-14; PMID:22576015; http://dx.doi.org/10.4161/auto.19653
- Chen RQ, Yang QK, Lu BW, Yi W, Cantin G, Chen YL, Fearns C, Yates JR 3rd, Lee JD. CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res 2009; 69:2663-8; PMID:19276368; http://dx.doi.org/10.1158/0008-5472.CAN-08-3222
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A 2008; 105:10762-7; PMID:18669648; http://dx.doi.org/10.1073/pnas.0805139105
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010; 143:1174-89; PMID:21183079; http://dx.doi.org/10.1016/j.cell.2010.12.001
- Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal 2009; 2:ra46; PMID:19690332; http://dx.doi.org/10.1126/scisignal.2000007
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 2010; 3:ra3; PMID:20068231; http://dx.doi.org/10.1126/scisignal.2000475
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science (New York, NY 2011; 332:1322-6; http://dx.doi.org/10.1126/science.1199484
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009; 284:8023-32; PMID:19150980; http://dx.doi.org/10.1074/jbc.M900301200
- Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol 2013; 200:475-91; PMID:23401004; http://dx.doi.org/10.1083/jcb.201209135
- Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 2013; 202:1107-22; PMID:24081491; http://dx.doi.org/10.1083/jcb.201307084
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science (New York, NY 2011; 332:1317-22; http://dx.doi.org/10.1126/science.1199498
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A 1998; 95:1432-7; PMID:9465032; http://dx.doi.org/10.1073/pnas.95.4.1432
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel 2-step mechanism. Genes Dev 1999; 13:1422-37; PMID:10364159; http://dx.doi.org/10.1101/gad.13.11.1422
- Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A 2011; 108:4788-93; PMID:21383122; http://dx.doi.org/10.1073/pnas.1100844108
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http://dx.doi.org/10.1038/ncb2152
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science (New York, NY 2011; 331:456-61; http://dx.doi.org/10.1126/science.1196371
- Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG, Jr. TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell 2003; 4:147-58; PMID:12957289; http://dx.doi.org/10.1016/S1535-6108(03)00187-9
- Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, Simons JW, Semenza GL. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res 2000; 60:1541-5; PMID:10749120.
- Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol 2002; 22:7004-14; PMID:12242281; http://dx.doi.org/10.1128/MCB.22.20.7004-7014.2002
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metabolism 2008; 8:224-36; PMID:18762023; http://dx.doi.org/10.1016/j.cmet.2008.07.007
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007; 450:736-40; PMID:18046414; http://dx.doi.org/10.1038/nature06322
- Kim JE, Chen J. regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes 2004; 53:2748-56; PMID:15504954; http://dx.doi.org/10.2337/diabetes.53.11.2748
- Zhang HH, Huang J, Duvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PloS One 2009; 4:e6189; PMID:19593385; http://dx.doi.org/10.1371/journal.pone.0006189
- Kim JH, Yoon MS, Chen J. Signal transducer and activator of transcription 3 (STAT3) mediates amino acid inhibition of insulin signaling through serine 727 phosphorylation. J Biol Chem 2009; 284:35425-32; PMID:19875458; http://dx.doi.org/10.1074/jbc.M109.051516
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96:857-68; PMID:10102273; http://dx.doi.org/10.1016/S0092-8674(00)80595-4
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell 2003; 11:11-23; PMID:12535517; http://dx.doi.org/10.1016/S1097-2765(02)00776-1
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J 2000; 19:6778-91; PMID:11118213; http://dx.doi.org/10.1093/emboj/19.24.6778
- Esumi N, Kachi S, Campochiaro PA, Zack DJ. VMD2 promoter requires 2 proximal E-box sites for its activity in vivo and is regulated by the MITF-TFE family. J Biol Chem 2007; 282:1838-50; PMID:17085443; http://dx.doi.org/10.1074/jbc.M609517200
- Vega-Rubin-de-Celis S, Abdallah Z, Kinch L, Grishin NV, Brugarolas J, Zhang X. Structural analysis and functional implications of the negative mTORC1 regulator REDD1. Biochemistry 2010; 49:2491-501; PMID:20166753; http://dx.doi.org/10.1021/bi902135e
