 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Mitochondrial dysfunction is causatively linked to organismal aging and the development of degenerative diseases. Here we describe stress-dependent opposing roles of mitophagy, the selective autophagic degradation of mitochondria, in aging and life-span control. We report that the ablation of the mitochondrial superoxide dismutase which is involved in reactive oxygen species (ROS) balancing, does not affect life span of the fungal aging model Podospora anserina, although superoxide levels are strongly increased and complex I-dependent respiration is impaired. This unexpected phenotype depends on functional autophagy, particularly mitophagy, which is upregulated during aging of this mutant. It identifies mitophagy as a prosurvival response involved in the control of mitohormesis, the well-known beneficial effect of mild mitochondrial oxidative stress. In contrast, excessive superoxide stress turns mitophagy to a prodeath pathway and leads to accelerated aging. Overall our data uncover mitophagy as a dynamic pathway that specifically responds to different levels of mitochondrial oxidative stress and thereby affects organismal aging.
Introduction
Maintenance of cellular homeostasis in response to changing endogenous and environmental conditions is a key for proper organismal development. Various pathways including those involved in preventing the accumulation of stressors like ROS, in repair, degradation, resynthesis, and remodeling of damaged molecules, larger functional molecular complexes, organelles, or whole cells are effective and keep biological systems functional over time. The coordinated induction of the appropriate pathways requires sensing of stressors and damage, and the induction of subsequent signaling processes. The molecular basis of these regulatory circuits is currently insufficiently elucidated.
ROS are molecules for which a potential damaging capacity has been emphasized and intensively studied for more than half a century. The accumulation of ROS has been suggested as the cause of cellular and organismal degeneration.Citation1-2 However, at low concentrations, ROS play a key role in cellular signaling and are important for proper development.Citation3-4 Consequently, balancing of cellular ROS is important to prevent molecular damage but still allows ROS signaling to occur.
Mitochondria are eukaryotic organelles with an important role in the regulation of ROS homeostasis. They are involved in many essential functions including energy transduction, iron/sulfur cluster synthesis, amino acid biosynthesis and lipid metabolism, generate most of the cellular superoxide free anion, hereafter termed superoxide, and emerge as an integral sensor for cellular ROS measurement and ROS balancing. Mitochondrial dysfunction is long known to be involved in aging and the development of a larger number of degenerative diseases (e.g., Alzheimer and Parkinson diseases).Citation5
The filamentous ascomycete Podospora anserina is an established aging model with a strong mitochondrial etiology of aging. In particular, pathways like mitochondrial DNA stability/instability, proteostasis and dynamics, which are involved in the control of mitochondrial integrity have been demonstrated to be of key relevance.Citation6-8 More recent investigations aimed at manipulating the mitochondrial ROS scavenging machinery revealed unexpected and counterintuitive results. Overexpression of PaSod3 coding for mitochondrial superoxide dismutase (in P. anserina SOD3, UniProtKB: B2AER6; in mammals SOD2, HGNC: 11180) active in the degradation of superoxide, leads to a decreased life span. This effect is due to insufficient hydrogen peroxide scavenging.Citation9 In contrast, the life span of a Pasod3 deletion strain (Pasod3Δ) does not significantly differ from that of the wild type.Citation10 Recently, we have identified autophagy, another cellular quality-control pathway that leads to vacuolar degradation of cytoplasmatic material and organelles, as a longevity-assurance mechanism.Citation11-12 Since ROS scavenging is affected in the Pasod3 deletion strain and since ROS can act as a signal to induce autophagy,Citation13-17 we set out to investigate autophagy in the Pasod3 deletion mutant in more detail.
Here we report several pronounced changes in the Pasod3Δ strain, which are consistent with a signaling function of mitochondrial ROS to regulate age-dependent mitophagy. Furthermore, functional autophagy is required for the unexpected healthy phenotype of the mutant. Most strikingly, challenging of the mutant with exogenous oxidative stress does not lead to the mitohormetic increase in life span that is seen in the wild type, but results in a strong life-span decrease. Overall our data identify the induction of mitophagy as a ‘backup’ pathway of ROS scavenging and a stress-dependent switch from prosurvival to prodeath. This dual role of this type of selective autophagy provides organisms with increased flexibility to respond to changing environmental and cellular conditions.
Results
Deletion of PaSod3 leads to the accumulation of superoxide and to mitochondrial impairments
To elucidate the mechanistic basis of the unexpected healthy phenotype of a mutant in which the gene coding for the mitochondrial manganese superoxide dismutase (PaSOD3) was deleted, we compared superoxide levels in the wild type and the Pasod3 deletion mutant (Pasod3Δ). We found that, in comparison to the wild type, superoxide levels were strongly increased in the mutant (). In contrast, hydrogen peroxide release in the 2 strains of the same chronological age was basically identical and, like previously reported for the wild type,Citation18 increased during aging.
Figure 1. Increased superoxide level of Pasod3Δ leads to mitochondrial impairments. (A) Determination of superoxide and hydrogen peroxide in juvenile and senescent wild-type and Pasod3Δ strains (n = 8) by NBT and DAB staining. (B) BN-PAGE analysis of mitochondrial protein extracts from wild type and Pasod3Δ (n = 3). (C) Oxygen consumption rate (OCR) of Pasod3Δ and wild-type mitochondria (for each strain 4 mitochondrial preparations with 10 to 22 technical replicates were analyzed). (i) State 4 CI: addition of pyruvate and malate to assess complex I-dependent state 4 respiration; (ii) state 3 CI: same substrates as in (i) plus ADP to measure complex I-dependent state 3 respiration; (iii) state 3 CI/II: same as (ii) plus succinate to assess complex I/II-dependent state 3 respiration; (iv) state 3 CII: same as (iii) plus complex I inhibitor rotenone to determine complex II-dependent respiration. (D) Residual OCR after complex I inhibition with the specific inhibitor rotenone of Pasod3Δ compared with wild type (for each strain 3 mitochondrial preparations with 10 measurements). (E) Mitochondrial membrane potential (mtMP) determined by the mtMP-dependent accumulation of TMRM in the mitochondria (for each strain 2 biologic replicates with 6 technical replicates). (F)-(H) GFP-fluorescence microscopy of 4- and 20-d-old PaSod3H26L-Gfp vs. Pasod3Δ PaSod3H26L-Gfp. Black arrowhead indicates vacuoles, white arrowhead demonstrates ‘dot-like’ GFP aggregates in the proximity of vacuoles. Scale bar: 10 µm. (C) to (E) Error bars correspond to the standard deviation. DIC, differential interference contrast.
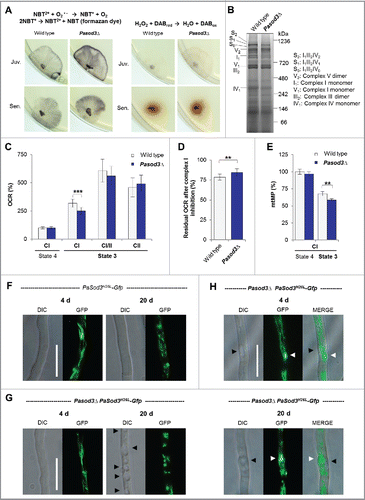
Next, we investigated mitochondrial function and morphology. In contrast to what has been reported for a mitochondrial sodΔ mutant of Caenorhabditis elegans,Citation19 no obvious differences of mitochondrial respiratory chain supercomplexes were visible in blue-native polyacrylamide gels (). However, the analysis of the mitochondrial oxygen consumption rate (OCR) revealed striking differences. While state 4 respiration via complex I measured in isolated mitochondria of the wild type and Pasod3Δ did not differ, complex I-dependent state 3 respiration of the mutant () was significantly decreased. Strikingly, this lower respiration could be compensated by the addition of the complex II substrate succinate, suggesting an increased complex II capacity of mutant mitochondria. To validate this possibility, we inhibited complex I by rotenone and found that, compared with the wild type, the residual OCR () was significantly higher in mitochondria of Pasod3Δ. Since a decrease in complex I activity is known to lower the mitochondrial membrane potential (mtMP), we compared the mtMP of both strains and found it significantly lowered in mitochondria of the mutant (). These data suggest that the mutant respires via a standard cytochrome-c-dependent electron transport chain in which respiration is slightly shifted from complex I toward complex II. This switch likely results from the partial inhibition of complex I due to a superoxide-induced loss of iron from the 4Fe-4S clusters of complex I proteins as it has been demonstrated in other systems like human lung carcinoma cells and mice with modulated mitochondrial SOD activity.Citation20–21
Interestingly, despite the increased superoxide load, no differences in mitochondrial morphotypes were observed when mitochondria of the wild type and Pasod3Δ were compared ( and ). Mitochondria from young mutant cultures were of the same ‘healthy’ filamentous morphotype as those from the wild type. As first described for the wild type of P. anserina,Citation18 mitochondrial morphology changes during aging of the mutant from filamentous to punctate. Interestingly, in Pasod3Δ, but not in the wild type, large vacuoles ( black arrows) and numerous dot-like GFP-aggregates of high density were observed in the vicinity of vacuoles ( white arrows). These cellular characteristics are the first evidence for a stress-induced increase in mitophagy in Pasod3Δ and were supported by previous work that identified autophagy as a longevity-assurance mechanism in P. anserina.Citation11–12
The healthy phenotype of Pasod3Δ depends on functional autophagy
Since it has been shown that ROS can act as a signal to induce autophagy,Citation13–17 we studied autophagy in Pasod3Δ in more detail. First, we investigated whether the unexpected healthy phenotype of the mutant depends on functional autophagy. We generated a Pasod3Δ Paatg1Δ (in mammals: Ulk1) double mutant () in which both superoxide scavenging and autophagy were affected. Strikingly, in comparison to the wild-type and the single deletion strains, the life span and growth rate of Pasod3Δ Paatg1Δ were decreased ( to) demonstrating that the wild-type-like phenotype of Pasod3Δ depends on functional autophagy.
Figure 2. Functional autophagy is required for the healthy phenotype of Pasod3Δ. (A) Southern blot analysis of HindIII digested genomic DNA from wild type, Pasod3Δ, Paatg1Δ and Pasod3Δ Paatg1Δ by using a Ble- (phleomycin resistance), PaSod3- and PaAtg1-hybridization probe. (B) Survival curves of the wild type (n = 27), Paatg1Δ (n = 27; P < 0.001), Pasod3Δ (n = 25) and Pasod3Δ Paatg1Δ (n = 26; P < 0.001). (C) Relative mean life span of Paatg1Δ (n = 27), Pasod3Δ (n = 25) and Pasod3Δ Paatg1Δ (n = 26) resulting from the comparison of the mean life span of each strain with the mean life span of the wild type (n = 27, set to 100%). (D) Relative mean growth rates of Paatg1Δ (n = 27), Pasod3Δ (n = 25) and Pasod3Δ Paatg1Δ (n = 26) derived from the comparison of the mean growth rate of each strain with the mean growth rate of the wild type (n = 27, set to 100%). (E) Southern blot analysis of HindIII digested genomic DNA from wild type, Pasod3Δ, Gfp-PaAtg8 and Pasod3Δ Gfp-PaAtg8 using a PaAtg8 and PaSod3 hybridization probe, respectively. (F) and (G) LSFM of hyphae from 4- and 20-d-old wild-type and Pasod3Δ strains expressing Gfp-PaAtg8. The number of distinctive dots in the shown hyphal sections is indicated. (H) Quantification of autophagosomes of 4- and 20-d-old wild-type and Pasod3Δ strains expressing Gfp-PaAtg8 (n = 10). P values were determined between 4- and 20-d-old strains and between wild type and mutant of the same age. (C) and (D), (H) Error bars correspond to the standard error.
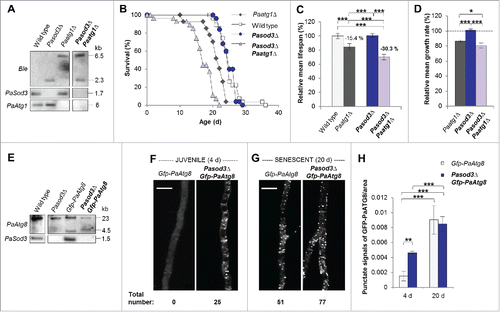
Next, we investigated whether there are signs of increased autophagy in Pasod3Δ and analyzed strains expressing the Gfp-PaAtg8 (in mammals: LC3 and GABARAP ortholog families of Atg8) fusion gene encoding the GFP-labeled autophagosomal marker PaATG8 (). Significantly, fluorescence microscopy analysis revealed a strong increase of autophagosomes in Pasod3Δ already at a young age. As in the wild type,Citation12 autophagosome abundance increased during aging also in Pasod3Δ ( to ).
Construction and validation of marker proteins for the analysis of autophagy
Since the microscopy analysis does only provide evidence for the initiation of autophagy but can neither prove delivery of autophagosomes to the vacuole and their subsequent degradation, nor discriminate between nonselective and selective autophagy, we aimed to perform a biochemical analysis.Citation22–24 We included the previously introduced PaSOD1-GFP fusion proteinCitation12 for the analysis of general autophagy as a control (). In addition, we generated 2 strains expressing mitochondrial PaSod3-Gfp fusion genes (). To avoid an interference of a potential PaSOD3-GFP fusion protein activity with the endogenous active PaSOD3, we used a PaSOD3-GFP construct with a mutation of histidine 26 (H26) to leucine in the active center of PaSOD3 (PaSOD3H26L-GFP). Inactivity of the protein was verified by the analysis of mitochondrial protein extracts using an ‘in gel’ SOD activity assay (). As assumed, in the PaSod3H26L-Gfp mutant there was no SOD3 activity detectable beside the activity of the endogenous PaSOD3. In the mutant expressing both, the endogenous PaSod3 gene copy and the introduced PaSod3-Gfp fusion gene, 3 protein bands with SOD3 activity were observed (). The 2 slower migrating bands, which were present in increased abundance, most likely correspond to different conformations or to the monomer and dimer of the PaSOD3-GFP fusion protein.
Figure 3. Generation and verification of autophagy- and mitophagy-reporter strains. (A) Southern blot analysis of HindIII digested genomic DNA validating the genetic constitution of wild type, PaSod1-Gfp, Pasod3Δ PaSod1-Gfp, PaSod3H26L-Gfp and Pasod3Δ PaSod3H26L-Gfp, as well as Paatg1Δ PaSod1-Gfp, Paatg1Δ PaSod3H26L-Gfp. In addition, Southern blot analysis of EcoRV-digested genomic DNA demonstrates the genetic constitution of Paatg1Δ PaSod3-Gfp, Paatg1Δ and PaSod3-Gfp, PaSod3H26L-Gfp and Paatg1Δ PaSod3H26L-Gfp. Ble-, Hyg- (hygromycin resistance) and PaSod3- hybridization probe were used as indicated. (B) ‘In-gel’ SOD3 enzyme activity assay from mitochondrial extracts of the wild type, PaSod3-Gfp and PaSod3H26L-Gfp. (C) Life span of monokaryotic wild type (n = 19) and PaSod1-Gfp (n = 15), PaSod3-Gfp (n = 15) and PaSod3H26L-Gfp (n = 15). P values were determined between the wild type and PaSod1-Gfp (P > 0.05), between the wild type and PaSod3-Gfp (P< 0.001) and between wild type and the PaSod3H26L-Gfp mutant (P > 0.05). (D) Growth rates of wild type (n = 19) and PaSod1-Gfp (n = 15), PaSod3-Gfp (n = 15) and PaSod3H26L-Gfp (n = 15). Error bars correspond to the standard deviation. (E) Western blot analysis of total protein extracts of PaSod1-Gfp vs. Paatg1Δ PaSod1-Gfp, PaSod3-Gfp vs. Paatg1Δ PaSod3-Gfp and PaSod3H26L-Gfp vs. Paatg1Δ PaSod3H26L-Gfp using a GFP-antibody. The protein amount of SOD1 served as a loading control. (F) Western blot analysis from mitochondrial extracts of the wild type, PaSod3-Gfp and PaSod3H26L-Gfp using an GFP-antibody. The Coomassie Blue-stained gel served as a loading control.
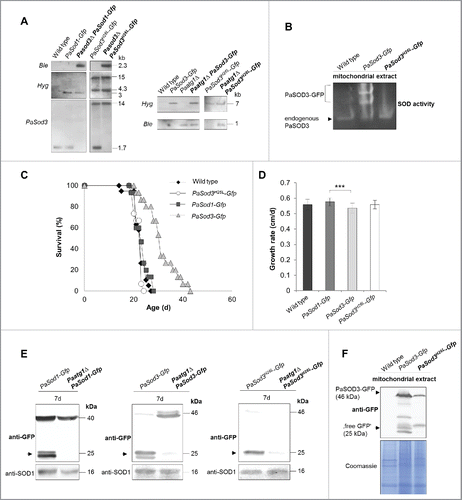
To see whether or not the different SOD reporter proteins affect physiology of the mutant strains, we performed life-span and growth-rate experiments and found that, in comparison to the wild type, both were unchanged in PaSod1-Gfp and PaSod3H26L-Gfp strains ( and ). In contrast, life span and growth rate of the PaSod3-Gfp mutant significantly varied from that of the other strains ( and ), which most likely is due to the increase in PaSOD3 activity.
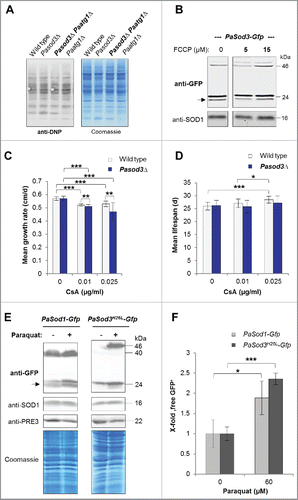
Next we investigated whether or not the different fusion proteins are suitable marker proteins for autophagy studies and analyzed their vacuolar degradation in the presence or absence of PaATG1. In western blots of total protein extracts ‘free GFP,’ as an indicator of vacuolar degradation, was only observed in strains containing a functional autophagic machinery (). Of note, signals identifying PaSOD3-GFP fusion proteins are less intense than of PaSOD1-GFP because these proteins are lower in abundance in total protein extracts due to their mitochondrial localization. In mitochondrial extracts they are clearly visible (). Under normal growth conditions, the fusion protein is not visible in Paatg1Δ PaSod3H26L-Gfp probably due to the increased proteasomal activity in this strain. To prove that the formation of ‘free GFP’ is indeed a measure for mitophagy, we attempted to at least partially inhibit proteasome activity of this strain by paraquat or MG132 (a known proteasome inhibitor), and by both drugs (Fig. S1), respectively. As expected, this treatment increased the amount of fusion protein with most protein detected in protein extracts of strains treated with both chemicals. In the Paatg1-deletion background, there was also a small proportion of ‘free GFP’ present, suggesting a minor ATG1-independent degradation of PaSOD3H26L-GFP. However, the amount was lower than in the wild-type background, demonstrating that most of the ‘free GFP’ was formed in an ATG1-dependent manner. Overall, in particular because of the unaltered phenotype of the corresponding strains, our data indicate that PaSOD1-GFP and PaSOD3H26L-GFP are suitable marker proteins to study general autophagy and mitophagy, respectively.
Mitophagy is a ‘backup’ protection pathway that is upregulated during aging of Pasod3Δ
Next we investigated autophagic and mitophagic flux in the Pasod3 deletion strain. In comparison to the wild type, we found no difference in the degradation of PaSOD1-GFP (). As has previously been shown for the wild type,Citation12 general autophagy increased significantly during aging (20 d) in Pasod3Δ and decreased in cultures of very old age (24 d) (). At this time, we cannot exclude that the increase in cytoplasmic PaSOD1 in the background of the PaSod1-Gfp strain may slightly decrease the ROS burden in the Pasod3Δ mutant and thus may also affect cellular homeostasis and autophagy. However, this effect may be rather negligible, since in previous experiments investigating the abundance and localization of PaSOD1 in the Pasod3Δ mutant we did not obtain any evidence for a compensatory response by PaSOD1 (unpublished).
Figure 4. Mitophagy is induced during aging of Pasod3Δ. (A) Monitoring autophagy by western blot analysis of 7-, 10-, 20- and 24-d-old PaSod1-Gfp compared with Pasod3Δ PaSod1-Gfp. (B) Monitoring mitophagy by western blot analysis of 7-, 10-, 20- and 24-d-old PaSod3H26L-Gfp compared with Pasod3Δ PaSod3H26L-Gfp. (C) GFP protein levels of 7- (n = 10), 10- (n = 3), 20- (n = 6) and 24- (n = 3) d-old PaSod3H26L-Gfp and Pasod3Δ PaSod3H26L-Gfp normalized to the level of PaSOD1. Protein abundance in 7-d-old PaSod3H26L-Gfp was set to 1. Error bars correspond to the standard deviation. P values were determined between cultures of the same strain or between wild type and mutant of the same age by the paired Student t test.
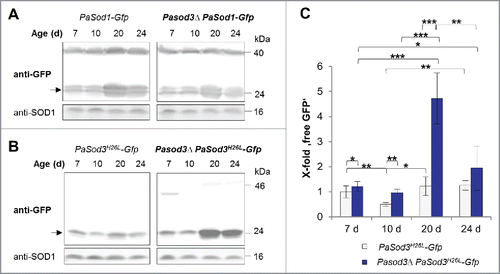
In contrast, a slight but significant increase in vacuolar degradation of mitochondrial PaSOD3H26L-GFP occurred already in Pasod3Δ of young age (7 d). At older age (20 d), mitophagy was strongly increased and slightly decreased again at very old age (24 d) ( and ). These data are consistent with those of the fluorescence microscopy analysis and demonstrate that autophagosomes are indeed delivered to the vacuole were they are degraded. Since such a mutant-specific increase is not observed for the cytosolic reporter, the induction of autophagic protein degradation specifically affects mitochondrial proteins. These data and the dependence of the wild-type-like phenotype on a functional autophagy machinery indicate that the observed induction of mitophagy compensates impairments in mitochondrial ROS scavenging in Pasod3Δ.
Impairments in mitochondrial homeostasis lead to a transcriptional adaptive response
In Pasod3Δ, mitophagy degrades mitochondria already at young age. Since this strain remains healthy, degraded mitochondria need to be replaced by new ones, a process requiring the expression of an appropriate gene set. To demonstrate such an effect of Pasod3Δ, we performed a comparative genome-wide transcriptome analysis of both, juvenile (5 d old) and aged (17 d old) Pasod3Δ vs wild type. For the identification of genes with robust differences in transcript abundance, we set the relative transcription threshold to ≥ 3 with a P value of P ≤ 0.05. We found that 23 transcripts were upregulated (Table S1) and 34 downregulated for 5 and 17 d old Pasod3Δ versus wild type (Table S2). represents an overview of these transcriptional changes in a graphical diagram (heatmap). To further obtain clues about the affected molecular pathways, we performed a Gene Ontology (GO) enrichment analysisCitation25 and found that in particular genes coding for components of mitochondrial processes involved in mitochondrial biogenesis, mitochondrial metabolism or mitochondrial respiration were upregulated, whereas extracellular processes were downregulated (). It appears that constitutive perturbation of mitochondrial homeostasis in Pasod3Δ leads to a transcriptional adaptive response.
Figure 5. Transcript levels of genes involved in the control of mitochondrial energy metabolism and biogenesis, are increased. (A) Transcriptome data of juvenile and senescent Pasod3Δ vs. wild type, demonstrated as a heatmap. Colors indicate relative expression of up- and downregulated transcripts of all genes, classified by Pa-number, with the relative transcription threshold to ≥ 3 and P-value of P ≤ 0.05. (B) GO enrichment analysis of transcriptome data of Pasod3Δ showing differential expression compared with the P. anserina wild type. All differentially expressed genes (Factor > 3; P < 0.05) were analyzed. GO terms with P < 1E-5 are shown. CC: Cellular Component; BP: Biological Process; C: Count (number of genes of respective GO term in the group [up- or downregulated]); S: Size (total number of P. anserina genes with the respective GO term).
![Figure 5. Transcript levels of genes involved in the control of mitochondrial energy metabolism and biogenesis, are increased. (A) Transcriptome data of juvenile and senescent Pasod3Δ vs. wild type, demonstrated as a heatmap. Colors indicate relative expression of up- and downregulated transcripts of all genes, classified by Pa-number, with the relative transcription threshold to ≥ 3 and P-value of P ≤ 0.05. (B) GO enrichment analysis of transcriptome data of Pasod3Δ showing differential expression compared with the P. anserina wild type. All differentially expressed genes (Factor > 3; P < 0.05) were analyzed. GO terms with P < 1E-5 are shown. CC: Cellular Component; BP: Biological Process; C: Count (number of genes of respective GO term in the group [up- or downregulated]); S: Size (total number of P. anserina genes with the respective GO term).](/cms/asset/6b8c687f-d17a-4236-86a9-36b9f7092ca1/kaup_a_1303021_f0005_c.gif)
Oxidative stress induces autophagy and mitophagy in Pasod3Δ
Next we addressed the question of how mitophagy is induced in Pasod3Δ. Protein carbonylation, lowered membrane potential or opening of the mitochondrial permeability transition pore (mPTP) have been reported to induce mitophagy in other systems.Citation26–28 However, none of these signals was found to be responsible for the mitophagy induction in Pasod3Δ ( to ). Instead, we identified superoxide as such a trigger. The analysis of total protein extracts of PaSod1-Gfp and PaSod3H26L-Gfp expressing strains grown in the presence of the superoxide inducer paraquat (60 µM) revealed more ‘free GFP’ than in strains grown in the absence of the drug ( and ). Moreover, mitophagy was more strongly induced than general autophagy, and the abundance of the GFP fusion proteins was higher in paraquat-treated samples which also may affect the amount of ‘free GFP’. At the transcript level we found only a slight, nonsignificant increase in PaSod1 and no increase in PaSod3 transcript levels of the corresponding genes linked to paraquat treatment (Fig. S2A and B). Moreover, compared with the protein extracts from untreated cultures, the amount of PaSOD1 and total Coomassie Blue-stained proteins were basically identical in paraquat-stressed samples while the abundance of the proteasome core particle subunit β1 (PRE3) was decreased. Previous studies demonstrated an age-related decline of proteasomal activity resulting from a decreased expression of genes coding for proteasomal proteins and their oxidative modification, respectively.Citation11,29–30 Such impairments in proteasome functionCitation31–32 may explain the accumulation of the 2 analyzed GFP fusion proteins in the paraquat-treated wild type. Thus, we conclude that normalization to the amount of fusion protein would be misleading and used a cytosolic protein that is not affected by paraquat treatment. Nevertheless, ‘free GFP’ that increases significantly after paraquat treatment strongly depended on the presence of ATG1 (Fig. S1A). According to our data, ATG1-independent autophagic processes, as they have been described recently,Citation33–34 may also contribute to the formation of ‘free GFP’, an issue that remains to be addressed in future investigations. Due to the high sensitivity of Pasod3Δ to oxidative stress,Citation10 we were unable to study the described effects of paraquat in this mutant.
Figure 6. Influence of oxidized proteins, membrane potential, mPTP and oxidative stress on mitophagy induction in Pasod3Δ. (A) ‘Oxy blot’ analysis of total protein extracts from wild type, Pasod3Δ, Pasod3Δ Paatg1Δ and Paatg1Δ (n = 3) Carbonyl groups of oxidized proteins derivatized to 2,4-dinitrophenylhydrazone (DNP-hydrazone) were detected with a DNP-specific antibody. The Coomassie Blue-stained gel after blotting served as a loading control. (B) Monitoring mitophagy during FCCP treatment (0, 5 and 15 µM) via western blot of PaSod3-Gfp. (C) Growth rates of wild type (n = 10) and Pasod3Δ (n = 11) treated with 0, 0.01 and 0.025 µg/ml cyclosporine A (CsA). Error bars correspond to the standard deviation. P values were determined between wild type and Pasod3Δ (2-tailed Mann-Whitney-Wilcoxon U test). (D) Life span of monokaryotic wild type (n = 10) and Pasod3Δ (n = 11) treated with 0, 0.01 and 0.025 µg/ml CsA. (E) Monitoring autophagy and mitophagy during paraquat treatment (60 µM) via western blot analysis of PaSod1-Gfp and PaSod3H26L-Gfp. (F) GFP protein levels of PaSod1-Gfp and PaSod3H26L-Gfp (n = 4) untreated (0 µM, set to 1) and treated (60 µM) with paraquat were normalized to the amount of proteins on the Coomassie Blue-stained gel. P values were determined between paraquat-untreated and treated cultures of each strain.
The role of mitophagy as a prosurvival or prodeath pathway depends on the strength of oxidative stress
We have described previously a life-span-prolonging effect for the P. anserina wild type by mild paraquat-induced oxidative stress.Citation35 Such a beneficial effect of mild stress is termed hormesis, a process whereby exposure to a low dose of a potentially harmful stressor promotes adaptive changes leading to an increased stress tolerance.Citation36–37 In recent years, this concept has been applied specifically to mitochondria (mitohormesis) and was reported to occur in different species (for more detail see refs. Citation38, 39). To test whether a mitohormetic effect is linked to autophagy we investigated the life span of the wild type and the Paatg1Δ mutant on medium containing 20 and 80 µM paraquat ( to ). Strikingly, the mitohormetic increase in life span of the wild type on 20 µM paraquat was not autophagy-dependent, indicating that there were also other quality-control pathways responding to cellular stress ( and ). However, challenging strains with 80 µM paraquat stress led to a significant decrease of the Paatg1Δ life span and to an increased life span of the wild type ( and ), demonstrating that autophagy participates in mitohormesis.
Figure 7. Mitohormetic response to oxidative stress depends on mitophagy. (A) Mean life span of 20 µM paraquat-treated wild type (n = 25) and Paatg1Δ (n = 12). (B) Relative mean life span of paraquat-treated (20 µM) wild type (n = 25) and Paatg1Δ (n = 12). (C) Mean life span of 80 µM paraquat-treated wild type (n = 25) and Paatg1Δ (n = 12). (D) Relative mean life span of 80 µM paraquat-treated wild type (n = 25) and Paatg1Δ (n = 12). (E) Relative mean growth rates of wild type (n = 25), Paatg1Δ (n = 12), Pasod3Δ (n = 20) and Pasod3Δ Paatg1Δ (n = 10) treated with 20 µM paraquat. The mean growth rate of the untreated wild type (n = 25) was set to 100% (baseline). (F) Relative mean life span of paraquat-treated (20 µM) wild type (n = 25), Paatg1Δ (n = 12) and Pasod3Δ compared with Pasod3Δ Paatg1Δ (n = 10). The mean life span of the untreated wild type (n = 25) was set to 100% (baseline). (G) Survival curves of the wild type (n = 25), Paatg1Δ (n = 12; P < 0.001), Pasod3Δ (n = 20) and Pasod3Δ Paatg1Δ (n = 10; P < 0.001) under 20 µM paraquat. (A) to (F) Error bars correspond to the standard deviation.
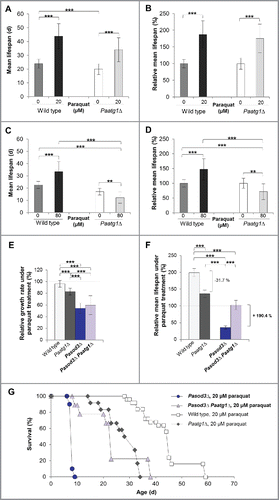
In a next series of experiments, we investigated the impact of oxidative stress and functional autophagy on growth and life span of Pasod3Δ. We compared wild-type, Paatg1Δ, Pasod3Δ and Pasod3Δ Paatg1Δ strains treated with 20 µM paraquat. Treatment with 80 µM was not possible since the Pasod3 deletion mutant, due to its high sensitivity to oxidative stress,Citation10 did not grow on paraquat concentrations above 20 µM. We found that the growth rate on 20 µM paraquat of Pasod3Δ Paatg1Δ was not significantly changed compared with that of the Pasod3Δ strain (). In contrast, in comparison to the untreated wild type (), the relative mean life span of paraquat-treated wild typeCitation35 and Paatg1Δ was significantly increased ( and ). In concordance with the data described above ( to ), the relative mean life span of Paatg1Δ was reduced by 31.7%, implying that the life-span-prolonging effect in the wild type under mild oxidative stress conditions depended at least partially on functional autophagy ( and ). The life span of paraquat-treated Pasod3Δ was strongly reduced when compared with the untreated wild type as well as to the paraquat-treated wild type and the Paatg1Δ control ( and ). Surprisingly, compared with the life span of Pasod3Δ, ablation of PaATG1 in the Pasod3Δ background led to a 190.4% increased life span ( and ) suggesting that excessive autophagy, as it occurs in paraquat-treated Pasod3Δ, leads to ‘autophagic cell death’Citation40–41 that cannot be executed in the absence of PaATG1. Thus, in Pasod3Δ, mitophagy acts as a prosurvival mechanism under normal growth conditions, and turns to a prodeath mechanism under excessive oxidative stress.
Discussion
Mitochondria are eukaryotic organelles involved in several essential functions including the synthesis of Fe/S clusters, ATP and ROS. Due to their dual role in signaling and molecular damaging, a tight control of cellular ROS is of paramount importance.
A key component of the molecular network involved in mitochondrial ROS balancing is the mitochondrial SOD isoenzyme. In the current study we investigated PaSOD3, the mitochondrial manganese SOD of P. anserina. Previously, life-span shortening due to a limited detoxification of hydrogen peroxide generated by PaSOD3 has been demonstrated in a PaSod3-overexpressing strain.Citation9–10 Surprisingly, the deletion of Pasod3 does not lead to accelerated aging and Pasod3 deletion strains are characterized by a wild-type-like life span. Also in other systems, including the nematode C. elegans and mammals,Citation42–44 unexpected results and conflicting data about the role of SODs in aging and life span control are repeatedly reported.
Here we demonstrated that the healthy phenotype of the Pasod3 mutant depends on a functional autophagy machinery and identified mitophagy as a molecular pathway that can compensate for impairments in mitochondrial ROS scavenging. Similar effects have also been observed in yeast, where the role of mitophagy in aging is indicated by an increase of mitophagy in cells harboring dysfunctional mitochondria.Citation45–47 Mitophagy thus represents a ‘backup’ protection pathway in cases when components of other pathways of the mitochondrial quality-control network are impaired. Such a function can explain the counterintuitive results of specific, well-defined experiments that otherwise lead to false conclusions. We have previously obtained similar counterintuitive results when we ablated the mitochondrial AAA protease PaIAP, a protein of the inner mitochondrial membrane of P. anserina that is involved in mitochondrial.Citation48 Under standard laboratory growth conditions at 27°C the life span of the mutant is strongly increased. However, at 37°C, a temperature that under natural conditions is often approached during a day, life span in ΔPaIap is shortened.Citation48 Finally, it turns out that PaIAP is active in heat stress response and as such is an important component for survival in the changing natural habitat of P. anserina.Citation48
Most importantly, our current experiments identified a stress-dependent dual and opposing role for mitophagy. Under low oxidative stress, as occurs in wild-type cultures grown under standard conditions, selected autophagy of mitochondria and nonselective autophagy as prosurvival or longevity-assurance pathways. This is also the case for the Pasod3 deletion strain, which is characterized by increased endogenous superoxide stress. Challenging the wild type with exogenous paraquat leads to a hormetic increase in relative mean life-span. In contrast, growth of the Pasod3 deletion strain on medium supplemented with paraquat leads to a strong life-span reduction. Strikingly, in the Pasod3Δ Paatg1Δ double knockout the life span is reverted to the life span of the wild type grown on medium without paraquat. Under these conditions superoxide stress, resulting from the ablation of PaSOD3 and from paraquat in the growth medium, has passed critical thresholds and turns mitophagy to an ‘autophagic cell death’ pathway. This is a type of programmed cell death (PCD) accompanied by autophagy that has also been reported for other systems including mammalian cell cultures.Citation49–50
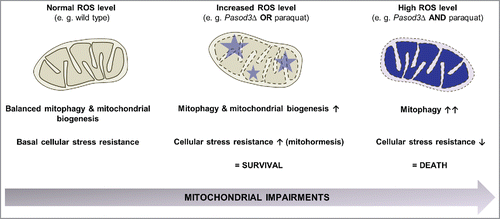
Overall, from our data we conclude that accumulated superoxide resulting from the PaSOD3 ablation provides the stimulus for the mitophagy induction of this mutant. After challenging the mutant with exogenous paraquat stress the level of stimulation passes a hormetic threshold and turns the signal from a survival into a death signal.Citation51 In contrast, mitohormesis by mild mitochondrial stress can initiate and transduce signals to the nucleus to coordinate a transcriptional response resulting in both mitochondrial and nonmitochondrial adaptations involved in maintenance of cellular homeostasis. Such responses are not limited to acute cytoprotective mechanisms but can induce long-term metabolic alterations and stress resistance.Citation38 In Pasod3Δ, this regulation depends on adaptive changes in mitophagic activity and expression of genes coding for mitochondrial proteins in response to cellular metabolic state, stress and other intracellular or environmental signals. Overall it appears that a balanced interplay between mitophagy and mitochondrial biogenesis is a prerequisite for mitochondrial homeostasis linked to cellular adaptation and stress resistance (). The elucidation of the precise regulatory circuits coordinating the involved molecular pathways, including those active in the regulation of crosstalk between autophagy with other pathways, is of far-reaching relevance. This knowledge also provides keys for the development of effective strategies to intervene into the various kinds of degenerative processes that occur during the lifetime of biological systems.
Figure 8. Stress-dependent role of mitophagy. In the wild type, coordination between mitophagy and mitochondrial biogenesis leads to a basal stress resistance under ‘normal’ ROS levels. When ROS is slightly increased as is demonstrated for example in the Pasod3 deletion mutant, mitochondrial function is partially impaired, leading to an induction of mitophagy and mitochondrial biogenesis to compensate impairments in ROS scavenging via a mitohormetic response. In contrast, when oxidative stress levels pass critical thresholds (e.g., Pasod3Δ treated with paraquat), mitophagy is further increased in response to enhanced mitochondrial impairments. In this situation mitophagy can no longer rescue the healthy phenotype and this leads to autophagic cell death.
Materials and methods
P. anserina strains and cultivation
P. anserina wild-type strain ‘s’,Citation52 the PaSod1-Gfp, PaSod3-Gfp and Pasod3Δ,Citation10 as well as the Paatg1Δ, Gfp-PaAtg8 strainsCitation12 and the newly generated mutants Pasod3Δ Paatg1Δ, Pasod3Δ PaSod1-Gfp, Pasod3Δ PaSod3H26L-Gfp, PaSod3H26L-Gfp and Pasod3Δ Gfp-PaAtg8 were used. Strains were grown on standard cornmeal agar/biomalt maize medium at 27°C under constant light.Citation53 For germination of spores, standard cornmeal agar/biomalt maize medium with 60 mM ammonium acetate was used and incubated at 27°C in dark for 2 d. All strains used in this study were derived from monokaryotic ascospores.Citation54
Cloning procedures and generation of P. anserina mutants
All transgenic strains are in the genetic background of wild-type strain ‘s’. For the generation of a strain with mutated PaSod3, a plasmid has been constructed in which a mutated gene version is encoded under control of the endogenous PaSod3 promoter region. Two PCR fragments were amplified (promoter and 5′ part of PaSod3 ORF; 3′ part of PaSod3 ORF with point mutation and without stop codon), introducing the necessary point mutation via the oligonucleotide Sod3H26L2 (5′-P-CTCCACAAGAAGCACCACC-3′, mutation underlined, Biomers, Ulm, Germany). The 2 fragments were cloned in the EcoRI and NcoI (Thermo scientific, ER0271 and ER0571) site of plasmid pSM4,Citation10 derived from pSM2.Citation55 The mutation results in the replacement of histidine 26 (H26) to leucine. H26 is part of the active site of this manganese superoxide dismutase and replacement by leucine in the human MnSOD results in an inactive protein version.Citation56 The plasmid was transformed into the wild-type strain. A transformant with single ectopic integration (PaSod3H26L-Gfp) was selected for subsequent experiments. Lack of enzymatic activity was demonstrated by an ‘in-gel’ SOD activity assay ().
Double mutants were obtained after crossing the single mutant strains with each other and the selection of strains from the progeny containing both mutations. In the case of the Pasod3Δ PaSod3H26L-Gfp double mutant, crossing of both single mutant strains was not possible. Therefore, we transformed the PaSod3H26L-Gfp plasmid into the Pasod3Δ strain. A transformant with a single ectopic integration of the plasmid and hygromycin (PaSod3H26L-Gfp) and phleomycin resistance (Pasod3Δ) was used for the subsequent experiments.
Transformation of P. anserina spheroplasts
For preparation of spheroplasts, strains were grown on standard cornmeal agar/biomalt maize medium at 27°C under constant light for 3 d and subsequently under the same conditions in liquid complete medium (CM) for 2 d. 20 g of the resulting mycelium was washed with TPS buffer (5 mM Na2HPO4, 45 mM KH2PO4, 0.8 M sucrose [Roth, 4621.2], pH 5.5) and TPS buffer containing 20 mg/ml ‘Glucanex’ (Novozymes) was added to a final volume of 100 ml. After chopping the mixture in a Waring blender the resulting suspension was incubated for 1.5 hat 35°C. Following filtration through gauze and glass wool, the suspension was centrifuged for 10 min at 4.000 rpm to pelletize the spheroplasts and the pellet was washed 3 times with TPS buffer.
To regenerate spheroplasts, the pellet was recovered in TPS buffer and the spheroplasts were plated on regeneration agar (3.7 g/l NH4Cl, 2 g/l tryptone, 1 g/l casamino acids, 1 g/l yeast extract, 10 g/l glucose, 342.3 g/l sucrose, 1.5 g/l KH2PO4, 0.5 g/l KCl, 0.54 g/l MgSO4 and 1 mg/l MnSO4 × 1 H2O, FeSO4 × 7 H2O, CuSO4 × 5 H2O and ZnSO4 × 7 H2O) containing 100 μ g/ml hygromycin B (Calbiochem, 400051). After 7–10 d of growth, mycelia of developing cultures were transferred to standard medium agar plates. Integrative transformation of P. anserina spheroplasts was performed as described previously.Citation57–58
Southern blot analysis
Isolation of total DNA of P. anserina was performed according to the protocol developed by Lecellier and Silar.Citation59 DNA digestion, gel electrophoresis and Southern blotting were performed according to standard protocols. For Southern blot hybridization analysis, digoxigenin-labeled hybridization probes (DIG DNA Labeling and Detection Kit; Roche Applied Science, 11175033910) were used according to the manufacturer's protocol. The PaSod3-specific probe (290 bp) was generated by SacI digestion of the plasmid PaSod3 overexpression plasmid pPaSod3_OEx.Citation10 The PaAtg1- and PaAtg8-specific probes were generated according to Knuppertz and colleagues.Citation12 As a hybridization probe specific for the phleomycin resistance gene (Ble), the 1293 bp BamHI-fragment of the plasmid pKO460 was used. The 736 bp XhoI-fragment of the plasmid pSM410 was used as a hygromycin resistance gene (Hyg) specific hybridization probe.
Western blot analysis
For extraction of total protein extracts, mycelia from different P. anserina strains were allowed to overgrow a cellophane foil covered M2 agarCitation54 plate for 2 d at 27°C and constant light. Subsequently, the pieces of mycelia were transferred into CM-liquid mediumCitation54 grown for 3 d at 27°C under constant light and shaking. In contrast, paraquat-untreated and treated cultures were transferred for 2 d in CM-liquid medium containing no or 60 µM paraquat (Sigma-Aldrich, 856177). For proteasome inhibition, cultures were transferred for 2 d in CM-liquid medium supplemented with 25 µM MG132 (Sigma-Aldrich, C2211), dissolved in DMSO (Carl Roth, 4720.1; final DMSO concentration 0.25%).
Harvested mycelia were pulverized in liquid nitrogen and the protein was isolated from the powder as describedCitation54 using a protein extraction buffer containing 50 mM HEPES, 100 mM NaCl, 5 mM DTT, pH 7.4 (NaOH). 100 to 200 μg total protein extracts of P. anserina strains, were fractionated by 2-phase SDS-PAGE (12% separating gels) according to a standard protocol.Citation61 After electrophoresis, proteins were transferred to PVDF membranes (Immobilon Transfer Membranes; Merck Millipore, IPFL00010). Blocking, antibody incubation and washing steps were performed according to the Odyssey western blot analysis handbook (LI-COR Biosciences, Bad Homburg, Germany). The antibodies used are described in Knuppertz et al.Citation12 Western blot analysis of total protein samples derivatized with di-nitrophenylhydrazine from an ‘Oxy blot’ kit (Merck Millipore, S7150) were performed using an anti-DNPH antibody as previously shown.Citation62
Growth rate and life-span determination
The determination of the life span and growth rate of cultures derived from monokaryotic ascospores was performed on M2 medium as described previously.Citation54 The determination of growth rates and life spans under stress conditions was performed on M2 medium with 20 µM paraquat (Sigma-Aldrich, 856177) at 27°C and constant light or with 0.01–0.025 µg/ml cyclosporine A (CsA) (Sigma-Aldrich, 30024) at 27°C in the dark.
Superoxide release measurements
Qualitative determination of superoxide release from mycelia was performed by monitoring reduction of nitroblue tetrazolium (NBT, Sigma-Aldrich, N6876) according to Munkres.Citation63
Hydrogen peroxide release measurements
Qualitative determination of hydrogen peroxide release from mycelia was performed by monitoring the oxidation of diaminobenzidine (DAB, Sigma-Aldrich, D-8001) according to Munkres.Citation63
Fluorescence microscopy
P. anserina mycelia were grown on glass slides with a central depression filled with CM agar and incubated for 1 d at 27°C and constant light. Hyphae were visualized and documented using a fluorescence microscope (DM LB/11888011, Leica, Wetzlar, Germany) equipped with the appropriate excitation and emission filters. Light sheet-based fluorescence microscopy (LSFM) and quantification of autophagosomes was performed as described previously.Citation12
Isolation of mitochondria
P. anserina strains were grown on cellophane foil covered solid M2 agar for 2 d at 27°C and constant light. After transferring pieces of grown mycelia into CM-liquid medium, 3 d of incubation at 27°C under constant light and shaking followed. Mitochondria of P. anserina cultures were isolated as described previously by differential centrifugation for the measurement of mitochondrial oxygen consumption and by sucrose gradient centrifugation for BN-PAGE, respectively.Citation54
‘In-gel’ SOD enzyme activity assay
Mitochondrial proteins (100 μg) were fractionated on native polyacrylamide gels (10% separating gels). For staining of gels, nitroblue tetrazolium, riboflavin and TEMED were used as described.Citation64
Blue-native polyacrylamide gel electrophoresis (BN-PAGE)
BN-PAGE was performed according to Wittig et al.Citation61 For preparation of each sample, 100 μg of mitochondrial protein extracts were solubilized using a digitonin (Sigma-Aldrich, D141): protein ratio of 3:1 (w:w). Linear gradient gels (4–13%) overlaid with 3.5% stacking gels were used for separation of the solubilized samples. Respiratory chain components were visualized by Coomassie Blue-staining and assigned as described previously.Citation65
Mitochondrial oxygen consumption
Determination of mitochondrial oxygen consumption was performed at 27°C using a high-resolution respirometer (Oxygraph-2k series G, Oroboros Instruments, Innsbruck, Austria). 200 µg freshly prepared mitochondria were injected into 2 ml air saturated oxygen buffer (0.3 M sucrose, 10 mM KH2PO4, 5 mM MgCl2, 1 mM EDTA, 10 mM KCl, 0.1% BSA, pH 7.2). The following substances were added serially: 20 mM pyruvate (Sigma-Aldrich, P2256) and 5 mM malate (Sigma-Aldrich, M1000), to promote complex I-dependent state 4 respiration (state 4 CI); 1.5 mM ADP (Sigma-Aldrich, A5285) was added to determine complex I-dependent state 3 respiration (state 3 CI); 10 mM succinate (Sigma-Aldrich, S2378) was used to stimulate complex I/II-dependent state 3 respiration (state 3 CI/II) and finally 1 µM rotenone (Sigma-Aldrich R8875) was added for inhibition of complex I to detect complex II-dependent state 3 respiration (state 3 CII). Data were analyzed using the Oroboros Software DatLab 6.
Mitochondrial membrane potential measurement
Oxygen consumption measurements were performed with complex I-dependent substrates as described above in the presence of tetramethylrhodamine-methyl-ester-perchlorate (TMRM; Sigma-Aldrich, T5428). Freshly prepared mitochondria were incubated in air-saturated oxygen buffer to which 1.5 µM TMRM was injected.
Isolation of total RNA
Total RNA for qRT-PCR analysis was isolated from mycelia of different P. anserina strains grown for 2 d at 27°C and constant light on M2 agar covered with cellophane foil. Subsequently, mycelia were transferred into CM-liquid medium without or with 60 µm paraquat for 2 further d at 27°C under constant light and shaking conditions. The grown mycelium was harvested and ground in liquid nitrogen and RNA was isolated using the Nucleo Spin® RNAClean-Up kit (Macherey-Nagel, 740948.50). Total RNA for transcriptome analysis was isolated using a CsCl density gradient as described previously.Citation66
Mapping of the transcriptomic reads of wild-type and Pasod3Δ strains against available genes of P. anserina
Transcriptome analysis was performed for each of the 5- and 17-d-old wild-type and Pasod3Δ samples, respectively. RNA from 3 individual cultures (biological replicates) of each strain was processed by LGC Genomics Berlin, Germany. Paired end reads of the transcriptomes were generated using Illumia HiSeq2000 sequencing platform (LGC Genomics Berlin). TruSeq™ adapters were trimmed in all reads using Illumina's CASAVA software. High quality trimmed reads from strains were used for mapping against the available 10,639 protein coding genes of P. anserinaCitation67 which were downloaded from P. anserina Genome Project database release version 6.32 (downloaded from http://podospora.igmors.u-psud.fr/) on a local Mascot server (Matrix Science, UK). Mapping was done using Bowtie268 with default parameters. SAMCitation69 output files of Bowtie2 were converted into sorted BAMCitation69 format. Relative expression of each gene was calculated from BAM files by using the formula
where Cov(X) = coverage per base of the gene under consideration X, n = numbers of bases in the gene under consideration X, M = number of bases mapped to all genes, m = number of bases mapped to gene X, N = number of bases in all genes.
Gene Ontology enrichment analysis
The assignment of proteins was deduced by Gene Ontology (GO)-term analysis using a P. anserina specified database.Citation11 For this database a “BLAST” search using UniProt with each P. anserina protein has been applied. The GO-terms of each hit with an e-value of ≤ 1e-20 of any other species protein had been adopted for the regarding P. anserina protein.Citation11 Briefly, a list of differentially expressed genes of Pasod3Δ versus wild type (Factor > 3; P < 0.05) was imported into the GO annotation library that is provided in a previous study.Citation11 The assignment of transcripts to the 3 GO categories ‘biological process’, ‘cellular component’ and ‘molecular function’ and the enrichment of transcripts was performed using the programming language R by means of the R package GOstatsCitation70 and a one-sided hyper geometric test (test for overrepresentation). GO terms were assumed to be statistically significantly enriched if the probability P value was ≤ 0.01.
Quantitative real-time PCR (qRT-PCR)
Total RNA samples of the following strains and conditions were analyzed: PaSod1-Gfp and PaSod3H26L-Gfp grown in CM- medium without or with 60 µM paraquat, respectively. The cDNA was synthesized from 480 ng total RNA using the iScript kit (Bio-Rad, 170–8891) and was used for qRT-PCR reaction (iQ SybrGreen SuperMix; Bio-Rad, 170–8880). Three biological and 3 technical replicates were analyzed for each strain and each condition. Oligonucleotides (Biomers, Ulm, Germany) used were: Sod1–1 (5′ CAT CAC TGG CCA TGA TGC 3′), Sod1–2 (5′ GCT TGA TGA GGT TGT CGG 3′), Sod3H26L-1 (5′ GAG CCT AAG GGA GAC CTC 3′), Sod3H26L-2 (5′ CTT GCT CAC CAT GGC GTC 3′) Porin-rt-for (5′ TCTCCTCCGGCAGCCTTG 3′) and Porin-rt-rev (5′ GAGGGTGTCGGCAAGTTC 3′). For each gene, the PCR efficiency was determined according to Pfaffl et al.Citation71 The relative expression level (normalized to the level of the PaPorin transcript) was calculated according to Servos et al.Citation72
Statistical analysis
For statistical analyses of life span, growth rate, oxygen consumption and mitochondrial membrane potential measurements, a 2-tailed Mann-Whitney-Wilcoxon U test was used. For the statistical analysis of protein amounts during oxy- or western blot analyses we used the paired Student t test. The respective samples were compared with the appropriate wild-type sample. For all analyses the minimum level of statistical significance was set at P < 0.05 (not significant different means P > 0.05; significant different (*) means P < 0.05; highly significant different (**) means P < 0.01; very highly significant different (***) means P < 0.001).
Abbreviations
| AAA | = | ATPases associated with diverse cellular activities |
| Atg | = | autophagy-related gene |
| ATP | = | adenosine triphosphate |
| Ble | = | phleomycin resistance gene |
| BN-PAGE | = | blue-native polyacrylamide gel electrophoresis |
| bp | = | base pairs |
| CI | = | complex I |
| CII | = | complex II |
| CM | = | complete medium |
| CsA | = | cyclosporine A |
| DAB | = | diaminobenzidine |
| DIC | = | differential interference contrast |
| DIG | = | digoxigenin |
| DMSO | = | dimethyl sulfoxide |
| DNPH | = | 2,4-dinitrophenylhydrazone |
| Fe/S | = | iron/sulfur |
| GFP | = | green fluorescent protein |
| GO | = | gene ontology |
| Hyg | = | hygromycin-resistance gene |
| kDa | = | kilodalton |
| LSFM | = | light sheet-based fluorescence microscopy |
| mtMP | = | mitochondrial membrane potential |
| mPTP | = | mitochondrial permeability transition pore |
| NBT | = | nitroblue tetrazolium |
| OCR | = | oxygen consumption rate |
| PaIAP | = | P. anserina i-AAA protease |
| PaPRE3 | = | P. anserina proteasome core particle subunit β1 |
| PaSOD | = | P. anserina superoxide dismutase |
| PCD | = | programmed cell death |
| PCR | = | polymerase chain reaction |
| qRT-PCR | = | quantitative real-time PCR |
| ROS | = | reactive oxygen species |
| TMRM | = | tetramethylrhodamine-methyl-ester-perchlorate. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1303021_Supplemental_Material.docx
Download MS Word (7.1 MB)Acknowledgements
We thank A. Werner for skillful preparation of total RNA, D. K. Gupta and Prof. Dr. M. Thines (Senckenberg Institute, Frankfurt) for handling and annotation of sequencing raw data of the transcriptome analysis, and O. Philipp for transcriptome data preparation and sorting. We are grateful to Prof. E. Stelzer and Dr. F. Pampaloni for their support in light sheet-based fluorescence microscopy.
Funding
This work was supported by grants of the Deutsche Forschungsgemeinschaft (Os75/15–1; SFB1177) to HDO and by the LOEWE excellence initiative (project: Integrated Fungal Research) of the state of Hesse (Germany).
References
- Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol 1956; 11:298-300; PMID:13332224; https://doi.org/10.1093/geronj/11.3.298
- Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc 1972; 20:145-7; PMID:5046729; https://doi.org/10.1111/j.1532-5415.1972.tb00787.x
- Werner E, Werb Z. Integrins engage mitochondrial function for signal transduction by a mechanism dependent on Rho GTPases. J Cell Biol 2002; 158:357-68; PMID:12119354; https://doi.org/10.1083/jcb.200111028
- Severin FF, Hyman AA. Pheromone induces programmed cell death in S. cerevisiae. Curr Biol 2002; 12:R233-5; PMID:11937036; https://doi.org/10.1016/S0960-9822(02)00776-5
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006; 443:787-95; PMID:17051205; https://doi.org/10.1038/nature05292
- Bernhardt D, Hamann A, Osiewacz HD. The role of mitochondria in fungal aging. Curr Opin Microbiol 2014; 22:1-7; PMID:25299751; https://doi.org/10.1016/j.mib.2014.09.007
- Osiewacz HD, Brust D, Hamann A, Kunstmann B, Luce K, Müller-Ohldach M, Scheckhuber CQ, Servos J, Strobel I. Mitochondrial pathways governing stress resistance, life, and death in the fungal aging model Podospora anserina. Ann N Y Acad Sci 2010; 1197:54-66; PMID:20536834; https://doi.org/10.1111/j.1749-6632.2010.05190.x
- Osiewacz HD, Kimpel E. Mitochondrial-nuclear interactions and life span control in fungi. Exp Gerontol 1999; 34:901-9; PMID:10673144; https://doi.org/10.1016/S0531-5565(99)00063-7
- Grimm C, Osiewacz HD. Manganese rescues adverse effects on life span and development in Podospora anserina challenged by excess hydrogen peroxide. Exp Gerontol 2015; 63:8-17; PMID:25616172; https://doi.org/10.1016/j.exger.2015.01.042
- Zintel S, Schwitalla D, Luce K, Hamann A, Osiewacz HD. Increasing mitochondrial superoxide dismutase abundance leads to impairments in protein quality control and ROS scavenging systems and to life span shortening. Exp Gerontol 2010; 45:525-32; PMID:20080171; https://doi.org/10.1016/j.exger.2010.01.006
- Philipp O, Hamann A, Servos J, Werner A, Koch I, Osiewacz HD. A genome-wide longitudinal transcriptome analysis of the aging model Podospora anserina. PLoS One 2013; 8:e83109; PMID:24376646; https://doi.org/10.1371/journal.pone.0083109
- Knuppertz L, Hamann A, Pampaloni F, Stelzer E, Osiewacz HD. Identification of autophagy as a longevity-assurance mechanism in the aging model Podospora anserina. Autophagy 2014; 10:822-34; PMID:24584154; https://doi.org/10.4161/auto.28148
- Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ 2009; 16:1040-52; PMID:19407826; https://doi.org/10.1038/cdd.2009.49
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007; 26:1749-60; PMID:17347651; https://doi.org/10.1038/sj.emboj.7601623
- Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol 2007; 17:422-7; PMID:17804237; https://doi.org/10.1016/j.tcb.2007.07.009
- Scherz-Shouval R, Shvets E, Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy 2007; 3:371-3; PMID:17438362; https://doi.org/10.4161/auto.4214
- Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy 2012; 8:1462-76; PMID:22889933; https://doi.org/10.4161/auto.21211
- Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nyström T. Osiewacz HD Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol 2007; 9:99-105; PMID:17173038; https://doi.org/10.1038/ncb1524
- Suthammarak W, Somerlot BH, Opheim E, Sedensky M, Morgan PG. Novel interactions between mitochondrial superoxide dismutases and the electron transport chain. Aging Cell 2013; 12:1132-40; PMID:23895727; https://doi.org/10.1111/acel.12144
- Powell CS, Jackson RM. Mitochondrial complex I, aconitase, and succinate dehydrogenase during hypoxia-reoxygenation: modulation of enzyme activities by MnSOD. Am J Physiol Lung Cell Mol Physiol 2003; 285:L189-198; PMID:12665464; https://doi.org/10.1152/ajplung.00253.2002
- Williams MD, Van Remmen H, Conrad CC, Huang TT, Epstein CJ, Richardson A. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J Biol Chem 1998; 273:28510-5; PMID:9774481; https://doi.org/10.1074/jbc.273.43.28510
- Meiling-Wesse K, Barth H, Thumm M. Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett 2002; 526:71-6; PMID:12208507; https://doi.org/10.1016/S0014-5793(02)03119-8
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016; 12:1-222; PMID:26799652; https://doi.org/10.1080/15548627.2015.1100356
- Kanki T, Kang D, Klionsky DJ. Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy 2009; 5:1186-9; PMID:19806021; https://doi.org/10.4161/auto.5.8.9854
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-9; PMID:10802651; https://doi.org/10.1038/75556
- Carreira RS, Lee Y, Ghochani M, Gustafsson ÅB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy 2010; 6:462-72; PMID:20364102; https://doi.org/10.4161/auto.6.4.11553
- Gomes LC, Scorrano L. Mitochondrial morphology in mitophagy and macroautophagy. Biochim Biophys Acta 2012; 1833:205-12; PMID:22406072; https://doi.org/10.1016/j.bbamcr.2012.02.012
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 2007; 143:291-9; PMID:17098847; http//dx.doi.org/10.1104/pp.106.092106
- Carrard G, Bulteau AL, Petropoulos I, Friguet B. Impairment of proteasome structure and function in aging. Int J Biochem Cell Biol 2002; 34:1461-74; PMID:12200039
- Chondrogianni N, Stratford FL, Trougakos IP, Friguet B, Rivett AJ, Gonos ES. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J Biol Chem 2003; 278:28026-37; PMID:12736271; http//dx.doi.org/10.1074/jbc.M301048200
- Dröge W. Free radicals in the physiological control of cell function. Physiol Rev 2002; 82:47-95; PMID:11773609; https://doi.org/10.1152/physrev.00018.2001
- Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol 2002; 192:1-15; PMID:12115731; https://doi.org/10.1002/jcp.10119
- Feng Y, Backues SK, Baba M, Heo JM, Harper JW, Klionsky DJ. Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy 2016; 12:648-58; PMID:27050455; https://doi.org/10.1080/15548627.2016.1157237
- Braden CR, Neufeld TP. Atg1-independent induction of autophagy by the Drosophila Ulk3 homolog, ADUK. FEBS J 2016; 283:3889-97; PMID:27717182; https://doi.org/10.1111/febs.13906
- Wiemer M, Osiewacz HD. Effect of paraquat-induced oxidative stress on gene expression and aging of the filamentous ascomycete Podospora anserina. Microbial Cell 2014; 1:225-40; https://doi.org/10.15698/mic2014.07.155
- Calabrese EJ. Hormesis: a revolution in toxicology, risk assessment and medicine. EMBO Rep 2004; 5:37-40; PMID:15459733; https://doi.org/10.1038/sj.embor.7400222
- Zimmermann A, Bauer MA, Kroemer G, Madeo F, Carmona-Gutierrez D. When less is more: hormesis against stress and disease. Microbial Cell 2014; 1:150-3; https://doi.org/10.15698/mic2014.05.148
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis). Exp Gerontol 2010; 45:410-8; PMID:20350594; https://doi.org/10.1016/j.exger.2010.03.014
- Merry TL, Ristow M. Mitohormesis in exercise training. Free Radic Biol Med 2016; 34:768-80; PMID:26654757; https://doi.org/10.1016/j.freeradbiomed.2015.11.032
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. Classification of cell death: recommendations of the Nomenclature committee on cell death. Cell Death Differ 2009; 16:3-11; PMID:18846107; https://doi.org/10.1038/cdd.2008.150
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Selfeating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8:741-52; PMID:17717517; https://doi.org/10.1038/nrm2239
- Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev 2008; 22:3236-41; PMID:19056880; https://doi.org/10.1101/gad.504808
- Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends life span in Caenorhabditis elegans. PLoS Genet 2009; 5:e1000361; PMID:19197346; https://doi.org/10.1371/journal.pgen.1000361
- Pèrez VI, Van Remmen H, Bokov A, Epstein CJ, Vijg J, Richardson A. The overexpression of major antioxidant enzymes does not extend the life span of mice. Aging cell 2009; 8:73-5; PMID:19077044; https://doi.org/10.1111/j.1474-9726.2008.00449.x
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 2005; 8:3-5; PMID:15798367; https://doi.org/10.1089/rej.2005.8.3
- Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ 2007; 14:1647-56; PMID:17541427; https://doi.org/10.1038/sj.cdd.4402167
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ 2005; 12:1613-21; PMID:15947785; https://doi.org/10.1038/sj.cdd.4401697
- Weil A, Luce K, Dröse S, Wittig I, Brandt U, Osiewacz HD. Unmasking a temperature-dependent effect of the P. anserina i-AAA protease on aging and development. Cell Cycle 2011; 10:4280-90; PMID:22134244; http:dx.doi.org/10.4161/cc.10.24.18560
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase 8. Science 2004; 304:1500-2; PMID:15131264; https://doi.org/10.1126/science.1096645
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 2004; 6:1221-8; PMID:15558033; https://doi.org/10.1038/ncb1192
- Cerella C, Teiten MH, Radogna F, Dicato M, Diederich M. From nature to bedside: pro-survival and cell death mechanisms as therapeutic targets in cancer treatment. Biotechnol Adv 2014; 32:1111-22; PMID:24681093; https://doi.org/10.1016/j.biotechadv.2014.03.006
- Rizet G. Impossibility of obtaining uninterrupted and unlimited multiplication of the ascomycete Podospora anserina (in English). C R Hebd Seances Acad Sci 1953; 237:838-40; PMID:13107134
- Esser K. Podospora anserina. In: King RC, ed. Handbook of Genetics. New York: Plenum Press, 1974; 531-551
- Osiewacz HD, Hamann A, Zintel S. Assessing organismal aging in the filamentous fungus Podospora anserina. Methods Mol Biol 2013; 965:439-62; PMID:23296676; https://doi.org/10.1007/978-1-62703-239-1_29
- Pöggeler S, Masloff S, Hoff B, Mayrhofer S, Kück U. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr Genet 2003; 43:54-61; PMID:12684845
- Zhang Y, Smith BJ, Oberley LW. Enzymatic activity is necessary for the tumor-suppressive effects of MnSOD. Antioxid Redox Signal 2006; 8:1283-93; PMID:16910776; https://doi.org/10.1089/ars.2006.8.1283
- Osiewacz HD, Skaletz A, Esser K. Integrative transformation of the ascomycete Podospora anserina: identification of the mating-type locus on chromosome VII of electrophoretically separated chromosomes. Appl Microbiol Biotechnol 1991; 35:38-45; PMID:1367277; https://doi.org/10.1007/BF00180633
- Stumpferl SW, Stephan O, Osiewacz HD. Impact of a disruption of a pathway delivering copper to mitochondria on Podospora anserina metabolism and life span. Eukaryot Cell 2004; 3:200-11; PMID:14871950; https://doi.org/10.1128/EC.3.1.200-211.2004
- Lecellier G, Silar P. Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Curr Genet 1994; 25:122-3; PMID:8087879; https://doi.org/10.1007/BF00309536
- Luce K, Osiewacz HD. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat Cell Biol 2009; 11:852-8; PMID:19543272; https://doi.org/10.1038/ncb1893
- Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc 2006; 1:418-28; PMID:17406264; https://doi.org/10.1038/nprot.2006.62
- Fischer F, Filippis C, Osiewacz HD. RCF1-dependent respiratory supercomplexes are integral for life span-maintenance in a fungal ageing model. Sci Rep 2015; 5:12697; PMID:26220011; https://doi.org/10.1038/srep12697
- Munkres KD. Histochemical detection of superoxide radicals and hydrogen peroxide by Age-1 mutants of Neurospora. Fungal Genet Newsl 1990; 37:24-5
- Flohé L, Ötting F. Superoxide dismutase assays. Methods Enzymol 1984; 105:93-104; PMID:6328209; https://doi.org/10.1016/S0076-6879(84)05013-8
- Krause F, Scheckhuber CQ, Werner A, Rexroth S, Reifschneider NH, Dencher NA, Osiewacz HD. Supramolecular organization of cytochrome c oxidase- and alternative oxidase-dependent respiratory chains in the filamentous fungus Podospora anserina. J Biol Chem 2004; 279:26453-61; PMID:15044453; https://doi.org/10.1074/jbc.M402756200
- Borghouts C, Werner A, Elthon T, Osiewacz HD. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol Cell Biol 2002; 21:390-9; PMID:11134328; https://doi.org/10.1128/MCB.21.2.390-399.2001
- Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, Porcel BM, Couloux A, Aury JM, Ségurens B, Poulain J, et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol 2008; 9:R77; PMID:18460219; https://doi.org/10.1186/gb-2008-9-5-r77
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25; PMID:19261174; https://doi.org/10.1186/gb-2009-10-3-r25
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25:2078-9; PMID:19505943; https://doi.org/10.1093/bioinformatics/btp352
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007; 23:257-8; PMID:17098774; https://doi.org/10.1093/bioinformatics/btl567
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; https://doi.org/10.1093/nar/29.9.e45
- Servos J, Hamann A, Grimm C, Osiewacz HD. A differential genome-wide transcriptome analysis: impact of cellular copper on complex biological processes like aging and development. PLoS One 2012; 7:e49292; PMID:23152891; https://doi.org/10.1371/journal.pone.0049292
