ABSTRACT
Rab2 is a conserved Rab GTPase with a well-established role in secretory pathway function and phagocytosis. Here we demonstrate that Drosophila Rab2 is recruited to late endosomal membranes, where it controls the fusion of LAMP-containing biosynthetic carriers and lysosomes to late endosomes. In contrast, the lysosomal GTPase Gie/Arl8 is only required for late endosome-lysosome fusion, but not for the delivery of LAMP to the endocytic pathway. We also find that Rab2 is required for the fusion of autophagosomes to the endolysosomal pathway, but not for the biogenesis of lysosome-related organelles. Surprisingly, Rab2 does not rely on HOPS-mediated vesicular fusion for recruitment to late endosomal membranes. Our work suggests that Drosophila Rab2 is a central regulator of the endolysosomal and macroautophagic/autophagic pathways by controlling the major heterotypic fusion processes at the late endosome.
Introduction
The endolysosomal pathway plays a critical role in the turnover of cellular components and pathogens, downregulation of endocytosed signaling proteins, mobilization of energy and nutrients, and more specialized functions such as antigen presentation, plasma membrane repair and neurite outgrowth [Citation1–Citation7]. The upstream part of the pathway is characterized by endosomal maturation, whereby Rab5-containing early endosomal vesicles gradually mature into Rab7-containing late endosomes (LEs) through Rab conversion [Citation8], mediated by the Rab7 guanine nucleotide exchange factor (GEF) complex Mon1-Ccz1 [Citation9,Citation10]. In contrast, the downstream delivery of material from LEs to lysosomes is thought to occur by transient and permanent fusion between LEs and lysosomes [Citation11]. Simultaneously, autophagosomes containing cellular components destined for recycling fuse both to LEs and directly to lysosomes [Citation12]. These processes generate hybrid endo- and autolysosomal compartments, in which degradation catalyzed by lysosomal enzymes takes place. Following the completion of digestion, endolysosomes condense to reform small lysosomes that function as storage organelles for resident hydrolases [Citation13].
Table 1. Oligonucleotides used in the study
The current understanding of the mechanism underlying LE-lysosome fusion originates from work in baker’s yeast (Saccharomyces cerevisiae) and relies on the highly conserved heterohexameric homotypic fusion and vacuole protein sorting (HOPS) tethering complex, along with a set of endosomal and lysosomal soluble NSF attachment protein receptors (SNAREs) [Citation14–Citation19]. In yeast, the HOPS complex bridges the membranes of vacuoles (the yeast equivalent of lysosomes) and LEs by binding to the yeast Rab7 ortholog Ypt7 present on both membranes [Citation15,Citation16,Citation20]. The interaction with Ypt7/Rab7 occurs through the accessory subunits Vps39 and Lt/Vps41 that attach to the class C core subunits Vps11, Vps16, Vps18, and Vps33 comprising the central part of the HOPS complex [Citation20]. The HOPS complex then induces membrane fusion by the assembly of appropriate SNAREs into a trans-SNARE complex [Citation15,Citation21]. This model is largely believed to hold true also in metazoan systems, albeit with the RAB7A effector RILP together with the small lysosomal GTPase ARL8B and its effector PLEKHM2/SKIP playing the role of HOPS receptors in mammals [Citation22–Citation25]. Loss of HOPS subunits causes failure of lysosomal delivery of endocytosed and autophagic material in Drosophila and mammals [Citation26–Citation31].
The lysosome contains a large number of hydrolases that catalyze the degradation of endocytosed material, as well as proteins that mediate supporting functions, ranging from pH-regulating components in the form of the V-ATPase and CLC-7 H+/Cl− exchanger, to metabolite transporters and proteins that control chaperone-mediated autophagy and lysosomal fusion to other organelles [Citation4]. Biosynthetic delivery of lysosomal proteins to the lysosomal system broadly follows two routes. Most soluble lysosomal proteins such as acid hydrolases are transported in coated vesicles from the trans-Golgi network (TGN) to an early stage in the endosomal system, in a pathway that in vertebrates depends on the target protein being labeled with mannose-6-phosphate [Citation32]. In contrast, lysosomal membrane proteins (LMP) such as LAMPs are transported in non-clathrin coated carriers from the TGN directly to a late-stage compartment in the endolysosomal pathway. In mammals, the fusion of these carriers to multivesicular LEs requires VPS41 and the SNARE VAMP7, but not VPS39 or the core HOPS component VPS18 [Citation33]. In Drosophila, transport intermediates containing LMPs fuse to late endosomal compartments in an Lt/Vps41-dependent manner [Citation29]. In addition, a not insubstantial proportion of LMPs traffic to lysosomes via early endosomes and the plasma membrane [Citation4,Citation29,Citation32–Citation34].
Small GTPases belonging to the Rab family are molecular switches that control organelle identity and properties in all eukaryotic cells [Citation35]. In their active GTP-bound form, Rabs bind to membranes via a lipid anchor and recruit effector proteins such as lipid-modifying enzymes, molecular motors and motor adaptors, and tethering and fusion factors. Transition between the active, membrane-anchored, state of the Rabs and the soluble inactive GDP-bound state is controlled by the GEFs and specific GTPase activating proteins (GAPs).
RAB2 is a highly conserved Rab protein previously linked to Golgi function in both Drosophila and mammals and retrograde trafficking to the endoplasmic reticulum (ER) in mammalian cells [Citation36–Citation38]. More recent data from C. elegans have demonstrated a role in the regulated secretory pathway, in the maturation of dense core vesicles [Citation39–Citation41]. In addition, Rab2 has a role in phagosome maturation [Citation42–Citation45]. In C. elegans, mutations in Rab2/UNC-108 impair progression through the conventional endolysosomal pathway at both early and late stages [Citation46,Citation47]. However, the mechanism behind this function remains poorly understood.
Here we demonstrate that Drosophila Rab2 localizes to membranes of late endosomal and lysosomal compartments and is required for the delivery of LAMP-positive LMP carriers to the endosomal pathway. We also show that Rab2 is indispensable for HOPS-dependent fusion of LEs to lysosomes. In contrast to Rab2, we find that Drosophila Gie/Arl8 is required for LE-lysosome fusion, but not for LMP carrier delivery. While Vps39 is a Rab2 effector [Citation48] and is also involved in LMP carrier delivery, our data suggest that Rab2 does not rely on Vps39 for its role in this process. Surprisingly, we find that recruitment of Rab2 to LE membranes does not require HOPS-mediated vesicular fusion, suggesting that Rab2 is directly recruited from the cytosol during endosomal maturation.
Results
Drosophila Rab2 is essential and ubiquitously expressed
To examine the function of Rab2 in flies, we generated a P-element imprecise excision allele, Rab2Δ1, in which the entire coding region of Rab2 was deleted, except for the first exon containing the initial 15 codons (). Rab2Δ1 homozygous flies survived until the third instar larval stage (L3) and progressively died off during the pupal stages, with ~ 20% reaching the pharate adult stage, and none eclosing ()). Hemizygotes carrying Rab2Δ1 over the deficiency Df(2R)BSC326 that covers the Rab2 locus showed an essentially identical developmental survival profile, demonstrating that Rab2Δ1 is a null allele ()). Viability of Rab2Δ1/Df(2R)BSC326 hemizygotes was rescued to the wild type and Rab2Δ1/+ heterozygote levels by ubiquitous expression of mCherry-Rab2 under a αTub84B-Gal4 driver ()).
Figure 1. Rab2 genetics, expression pattern and loss-of-function phenotype. (A) Map of the Rab2 genomic region, with neighboring genes and 2 large deficiencies covering Rab2. The region covered by the HA-Rab2 transgene and the structure of the Rab2 locus is indicated, as well as the location of the HA and GFP tags inserted in the genomic transgenes, the P-element used in excision mutagenesis, and the extent of the deletion in Rab2Δ1. Coding region in red. (B) Developmental survival profile. The initial numbers of flies were: w1118, 90; Rab2Δ1/+, 92; Rab2Δ1, 92; Rab2Δ1/Df(2R)BSC326, 89; Rab2Δ1/Df(2R)BSC326;tub-Gal4,mCherry-Rab2/+, 32.(C) Confocal sum intensity projections showing the expression of HA-Rab2 in filleted L3 larvae carrying two copies of the HA-Rab2 allele in a Rab2Δ1 background, and w1118 controls without HA-Rab2. Anti-HA staining was supplemented by anti-HRP staining to mark the nervous system. Br, brain hemispheres; VNC, ventral nerve cord; PN, peripheral nerves, M, muscles; RG, ring gland; SG, salivary gland. (D and E) Accumulation of GFP-LAMP in the CNS of Rab2Δ1 mutant larvae. (D) Maximum projections of fixed CNS from control, Rab2Δ1 and rescued L3 larvae ubiquitously expressing GFP-LAMP. VNC, ventral nerve cord; LH/RH, left/right hemisphere. (E) Amount of LAMP signal in GFP-LAMP granules in the VNC and hemispheres. Each genotype represented by 5 CNS preparations. ANOVAs followed by the Tukey honest significant difference (HSD) test. (F and G) Localization of GFP-LAMP-containing granules accumulating in Rab2Δ1 L3 brains, in relation to the Golgi and late endosomes. High-magnification single optical sections through the surface of a Rab2Δ1 brain hemisphere prepared as in (D) and stained for GM130 to mark Golgi bodies (F) or Rab7 to mark late endosomes (G). Arrows indicate accumulations of GFP-LAMP coinciding with Rab7-positive LEs. Arrowheads indicate Rab7-negative GFP-LAMP granules. The detailed genotypes included in this and the following figures, supplemental figures, and videos are listed in Table S1
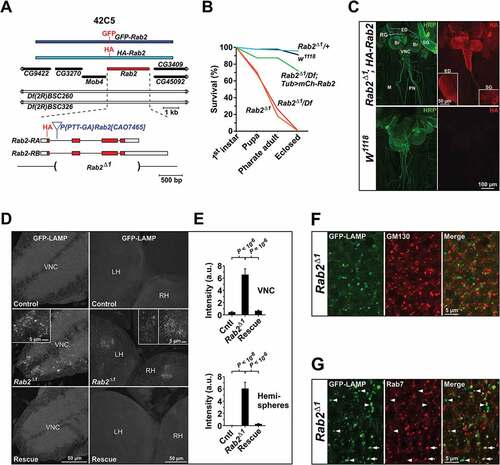
Because we were unsuccessful in raising a polyclonal antibody against the Rab2 protein, we generated HA-Rab2 and GFP-Rab2 transgenes controlled by endogenous Rab2 regulatory elements ()) to determine the Rab2 expression pattern and subcellular localization. The HA-Rab2 allele was fully capable of rescuing the viability and fecundity of Rab2Δ1 homozygotes, confirming its functionality (data not shown). When we immunostained rescued L3 larvae for the HA epitope, HA-Rab2 appeared to be ubiquitously expressed, but with strong enrichment in the nervous system and imaginal discs ()). No significant immunosignal was detected in w1118 controls devoid of HA-Rab2 ()). Likewise, the GFP-Rab2 transgene showed strong enrichment in larval VNC, and also in the testes (Fig. S1).
Lysosomal trafficking is perturbed in Rab2 mutants
Transmembrane lysosomal components such as LAMP and the V-ATPase originating in the ER and Golgi are carried to LEs through an AP-3 complex and Lt/Vps41-dependent pathway [Citation29], with a minor fraction trafficking through the plasma membrane, and are delivered from LEs to lysosomes by way of complete or transient fusion events. Perturbation of this delivery can be monitored using a weakly expressing chimeric reporter consisting of lumenal GFP fused to the combined transmembrane and cytosolic domain of human LAMP1 [Citation28]. Upon transfer to lysosomes, hydrolases quickly degrade the lumenal GFP, thereby eliminating the GFP signal. In case of a block in the delivery process, however, the GFP signal accumulates [Citation30,Citation49]. When introducing ubiquitously expressed GFP-LAMP into the Rab2Δ1 null mutants, we observed a massive accumulation of GFP in granular structures in the L3 CNS ()). This phenotype was completely rescued by a single copy of the HA-Rab2 allele ()).
We next sought to determine where the block in LAMP transport occurs in the endomembrane system. The granular GFP-LAMP structures that accumulated in the Rab2Δ1 L3 CNS did not overlap with Golgi bodies visualized by immunostaining for GM130 (ortholog of mammalian GOLGA2/Golgin A2), suggesting that loss of Rab2 does not block LAMP export from the Golgi ()). Furthermore, a subpopulation of GFP-LAMP granules corresponded to Rab7-positive LEs (), arrows), while another subpopulation did not overlap with Rab7 (), arrowheads). In mammalian cells transgenic Rab7 localizes to Golgi-derived LMP carriers as well as to LEs, implying that this protein may be an unreliable marker in this context [Citation33]. With this caveat, these findings are compatible with the notion that transport of LAMP to the lysosomal compartment in Rab2 mutants is partially blocked both at the late endosomal stage and also along the route between the Golgi-apparatus and LEs.
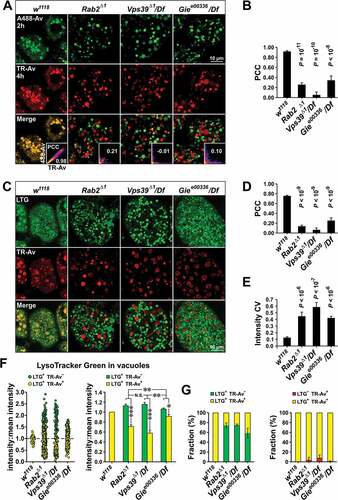
LAMP trafficking defects were also observed in the salivary glands and fat body of Rab2Δ1 mutants (). Unlike in neurons, in wild-type salivary gland cells the weakly expressed GFP-LAMP did not degrade below detectability and was almost entirely contained in a tubulo-vesicular lysosomal network staining positively for the acidophilic dye LysoTracker Red (LTR) ()). In Rab2Δ1 salivary glands, some GFP-LAMP still reached LTR-labeled lysosomes, but a substantial fraction of the GFP-LAMP signal accumulated in LTR-negative vesicles and vesicular clusters in the cytoplasm ()). In addition, while LTR-positive lysosomal tubules still formed in Rab2Δ1 mutants, the density of LTR-positive structures was reduced and the network appeared more fragmented compared to the wild type ()).
Figure 2. Lysosomal traffic and autophagy are perturbed by loss of Rab2. (A) Localization of ubiquitously expressed GFP-LAMP in relation to LysoTracker Red staining in L3 salivary glands. Single optical sections of gland cortex near the basolateral surface (live imaging). Arrowheads indicate GFP-LAMP-positive vesicular structures not labeled with LysoTracker Red. Arrows indicate structures labeled with both GFP-LAMP and LysoTracker Red. Brackets indicate coclustering of GFP-LAMP-positive structures with structures labeled with LysoTracker Red. (B) Quantification of data in (E), from 9 control, 14 Rab2Δ1 and 10 lt11/Df larvae. The fraction of GFP-LAMP signal located outside of LysoTracker Red-positive structures is shown. (C) Localization of GFP-LAMP relative to LysoTracker Red in the fat body. Left, late L3 fat bodies from control, Rab2Δ1 and lt11/Df larvae ubiquitously expressing GFP-LAMP. Single optical sections (live imaging). For better visibility, the intensity of the LysoTracker Red signal in the Rab2Δ1 and lt11/Df fat body was digitally increased relative to control. Right, intensity scatter plots for GFP-LAMP versus LysoTracker Red. Warm colors mark a high point density in the plot. The Pearson correlation coefficient (PCC) is indicated. (D and E) Quantification of (C). Control and Rab2Δ1 each represented by 5 larvae, lt11/Df by 6 larvae. (D) Correlation between the GFP-LAMP and LysoTracker Red signals. (E) Mean LysoTracker Red signal density (left) and cross-sectional area of LysoTracker Red-positive structures (right). (F) Eye imaginal discs of control and Rab2Δ1 late L3 larvae expressing the GFP-mCherry-Atg8a autophagy reporter. Arrows indicate accumulation of GFP and mCherry dual-positive autophagosomes in the ommatidia of Rab2Δ1 imaginal discs. Live confocal imaging of eye disc area posterior to the morphogenetic furrow. (G) Quantification of (F). The GFP:mCherry signal ratio in mCherry-positive autophagosomal structures in 8 w1118 and 7 Rab2Δ1 imaginal discs is shown. (H) Eye pigmentation is not perturbed by loss of Rab2. Micrographs of the eyes of control and Rab2Δ1 pharate adult flies. (B, D, and E) ANOVA followed by Tukey HSD test, (G) Unpaired Student’s t test
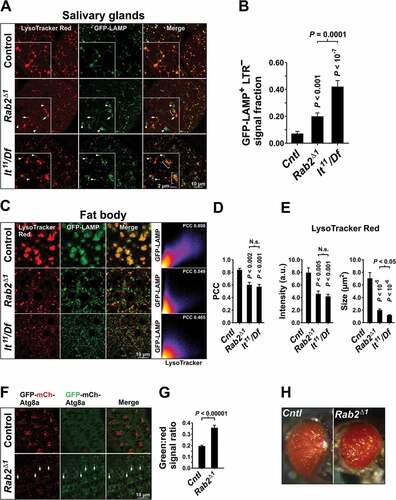
Lt/Vps41 is a HOPS complex subunit required for both LE-lysosome fusion [Citation26,Citation30] and delivery of LMPs to late endosomal and lysosomal compartments [Citation29,Citation33]. Salivary glands from animals hemizygous for light11 (lt11/Df), a strong hypomorphic allele of Drosophila lt/Vps41 [Citation50], showed a similar, but more severe phenotype compared to that of Rab2Δ1 ()). Notably, distinct GFP-LAMP and LTR-positive structures often appeared to cluster with each other in Rab2Δ1 and lt11 mutants, suggesting that loss of Rab2 and Lt/Vps41 impairs fusion rather than clustering (), brackets).
Autolysosomes accumulate in the fat body tissue of late L3 larvae due to developmentally programmed autophagy [Citation51]. In wild-type late L3 fat bodies, almost all GFP-LAMP localized to large autolysosomes strongly stained with LTR ()). As in salivary glands, there was a strong decrease in the association between LTR staining and the GFP-LAMP signal in fat bodies of both Rab2Δ1 and lt11/Df animals ()). In Rab2Δ1 and lt11/Df fat bodies, LAMP-positive structures were highly variable in size and GFP intensity, and many showed only weak or no staining with LTR ()). In addition, the average LTR labeling intensity and size of LTR-positive structures were strongly reduced in lt11/Df compared to controls ()). This is consistent with previous observations that HOPS is necessary for fusion of autophagosomes to endosomes and lysosomes [Citation30,Citation31]. A similar, but somewhat weaker phenotype was observed in the Rab2Δ1 fat body, suggesting that Rab2 is also required for autophagosome delivery to the endolysosomal pathway ()).
Taken together, these results suggest that transport of the transmembrane lysosomal protein LAMP to lysosomes is blocked in Rab2-null mutants.
Rab2 is required for autophagy but not for biogenesis of lysosome-related organelles
During autophagy, autophagosomes fuse to LEs and lysosomes to initiate digestion of their contents. This step can be monitored using a transgene encoding Atg8a fused to tandem GFP-mCherry (GFP-mCherry-Atg8a). Fusion of the relatively neutral autophagosomes to acidic degradative LEs and lysosomes selectively quenches the more pH-sensitive and proteolytically labile GFP fluorescence, shifting the emitted fluorescence ratio from green/yellow towards red [Citation52]. Autophagy is active in Drosophila photoreceptor cells, as evidenced by massive accumulation of autophagosomes in the eyes of flies with loss-of-function mutations in the HOPS complex components car/Vps33A and dor/Vps18, due to a failure of HOPS-dependent autophagosome-lysosome fusion [Citation28,Citation49]. Consistent with this observation, expression of a GFP-mCherry-Atg8a transgene under an endogenous promoter produced strong red fluorescence in the developing ommatidia in wild-type L3 eye imaginal discs ()). When expressed in Rab2Δ1 larvae, the same GFP-mCherry-Atg8a transgene produced a massive accumulation of puncta dispersed along the ommatidial periphery, which labeled strongly for green but weakly for red fluorescence ()). Together with our previous observation that Rab2Δ1 perturbs the formation of autolysosomes in the late L3 fat body, this strongly suggests that Rab2 is required for autophagosome fusion to organelles of the endosomal and lysosomal pathways in Drosophila.
Formation of lysosome-related organelles (LROs) has many mechanistic commonalities with that of conventional lysosomes, although with some molecular differences [Citation53–Citation55], and also requires the HOPS complex [Citation18,Citation53,Citation56,Citation57]. Mutations in Drosophila HOPS components disrupt the formation of the pigment granule, an insect LRO, leading to a loss of eye coloration [Citation30,Citation58–Citation63]. In view of Rab2’s function in lysosomal trafficking, we investigated if Rab2 is required for pigment granule biogenesis. Surprisingly, eyes of Rab2Δ1 pharate adults did not show any obvious defect in coloration compared to controls ()). This suggests that Rab2 is not involved in the formation of LROs, but only in the biogenesis of conventional lysosomes.
Rab2 is necessary for LE-lysosome fusion
To further elucidate the function of Rab2 in the endolysosomal system, we examined the effect of Rab2 loss on postendocytic trafficking in garland nephrocytes (garland cells), where the endocytic pathway can be readily visualized using an endocytic tracer. These cells form a wreath attached to the junction between the esophagus and proventriculus. Together with the pericardial nephrocytes, they belong to a class of large, highly endocytically active cells that filter and remove toxic materials from the hemolymph [Citation64].
Garland cells take up and deliver endocytic tracer to spherical Rab7-positive LEs within 5 min [Citation65]. Within 40 min of endocytosis, the tracer is delivered to an acidic, LTG-positive lysosomal compartment [Citation66]. We pulsed garland cells from wild-type control and Rab2Δ1 animals for 3 min with Texas Red-avidin (TR-avidin), followed by a 2-h chase. After 2 h, endocytosed TR-avidin resided in a dense LTG-stained tubulo-vesicular network interspersed with larger vacuoles in the cytoplasm of wild-type garland cells (). These vacuoles likely correspond to β-vacuoles previously described by other investigators [Citation67–Citation69]. A 7 h chase yielded a similar tracer localization, indicating that this morphology represents the terminal lysosomal compartment in this cell type (Fig. S2). Time-lapse imaging revealed the lysosomal network to be highly dynamic, with tubules constantly extending and retracting and small vesicles shifting position (Video S1). In striking contrast, after a 2 h chase in Rab2Δ1 garland cells all endocytosed avidin was found in large, highly spherical organelles with a complete absence of tubules (), see also , and ). This aberrant morphology could be fully rescued by the expression of mCherry-Rab2 in the mutant Rab2Δ1 background ()).
Figure 3. Perturbation of postendocytic trafficking in Rab2Δ1 garland cells. Garland cells from L3 larvae pulsed for 3 min with TR-avidin (TR-Av) or A488-avidin (A488-Av), followed by a 2 h chase. Single optical sections through the cortical cytoplasm of living garland cells. (A) Wild-type cells loaded with TR-avidin and stained with LysoTracker Green (LTG). The lysosomal network is detailed in the high-magnification inset, with three tubular extensions marked by arrowheads and a vacuolar structure by an asterisk. (B) Garland cells from control (left) and Rab2Δ1 (right) animals expressing YFP-Rab7 under direct control of the αTub84BcathD promoter and loaded with TR-avidin. (C) Rab2Δ1 garland cells expressing mCherry-Rab2 and loaded with A488-avidin (live imaging). The inset highlights the tubular network both labeled by endocytic tracer and positive for mCherry-Rab2
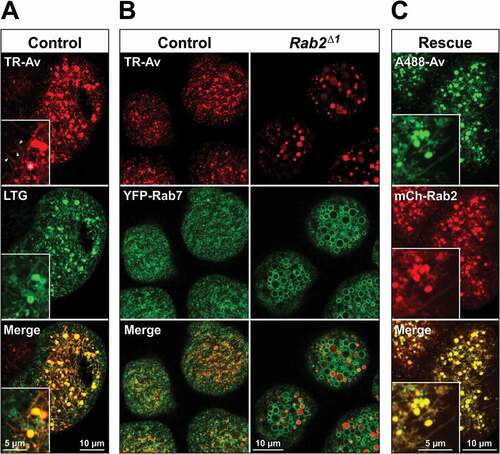
The lysosomal network in wild-type garland cells also labeled weakly with YFP-Rab7 expressed at a low level (), middle row, left). In contrast, spherical organelles showing strong YFP-Rab7 labeling of their limiting membrane dominated the cytoplasm of Rab2Δ1 cells (), middle row, right). The previously described aberrant 2-h tracer-positive compartments corresponded to a subpopulation of these organelles (), right), suggesting that the endocytic pathway is stalled at the late endosomal stage in Rab2Δ1 garland cells. Enlarged Rab7-positive LEs have also recently been reported in mutant garland cells devoid of Vps11, Vps39 and Lt/Vps41 where LE-lysosome fusion is blocked [Citation69]. An accumulation of enlarged YFP-Rab7-positive LEs was also observed in the larval CNS of Rab2Δ1 animals (Fig. S3A, B) showing that this phenotype is not exclusive to garland cells. Furthermore, GFP-Rab7 overexpressed in somata of large peptidergic neurons in the larval CNS dramatically redistributed from the cytosol to vesicular structures in Rab2Δ1 mutants, compared to the wild type (Fig. S3C, D).
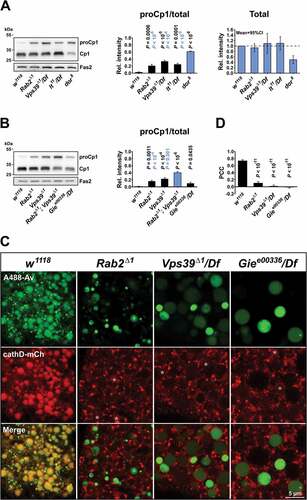
Soluble lysosomal enzymes such as the cathepsins cathD and Cp1/Cath L are delivered to the endosomal pathway from the Golgi apparatus through a Lt/Vps41-independent, but Dor/Vps18-dependent, pathway [Citation29,Citation33] and are concentrated in lysosomal compartments to undergo proteolytic processing into their mature forms. We performed western blotting of the CNS with attached imaginal disks, pro-ventriculi and garland cells from wild-type, Rab2Δ1 and lt11/Df L3 larvae, as well as L3 larvae hemizygous for a CRISPR/Cas9-generated deletion mutant of the HOPS subunit Vps39, Vps39Δ1 (see Materials and Methods). We found that although in Rab2Δ1, Vps39Δ1/Df and lt11/Df there was a noticeable increase in the amount of immature unprocessed endogenous pro-Cp1/Cath L compared to the wild type, most of the Cp1/Cath L in these mutants existed in the mature, processed form (), left and middle). This suggests that most soluble lysosomal enzyme still resided in late endosomal or lysosomal compartments. In contrast, in dor8 mutants, in which delivery of lysosomal enzymes to the endolysosomal pathway is blocked [Citation29], most Cp1/Cath L was in the immature pro-form (), left and middle), and the total amount of Cp1/Cath L was dramatically decreased (), left and right). This is consistent with soluble lysosomal cargo being secreted to the extracellular space when it cannot be delivered to endosomal compartments [Citation70].
Figure 4. Processing of Cp1/Cath L and spatial dissociation between cathD and endolysosomal tracer in mutant garland cells. (A) Left, western blot of the CNS and attached tissues from L3 larvae of the indicated genotypes probed for endogenous Cp1/Cath L. Middle, Intensity of the pro-Cp1/Cath L immunosignal relative to the total Cp1/Cath L signal (i.e., both pro- and mature forms). P values in black and blue represent comparisons to w1118 and dor8, respectively. Right, Intensity of the total Cp1/Cath L signal relative to that of w1118. The mean and 95% confidence interval is shown. (B) Left, western blot of the CNS and attached tissues from L3 larvae of the indicated genotypes, probed for endogenous Cp1/Cath L. Right, Ratio of pro-Cp1/Cath L to total Cp1/Cath L as in (A), middle. P values in black and blue represent comparisons to w1118 and Rab2Δ1; Vps39Δ1, respectively. Quantifications in (A and B) are each based on 4 western blots and analyzed by ANOVA followed by Tukey HSD test. Fas2/Fasciclin II was used as loading control in (A and B). (C) Garland cells from larvae of the indicated genotypes expressing cathD-mCherry and pulsed with A488-avidin (3 min), then chased for 2 h. Confocal sections through the cortical cytoplasm of living garland cells are shown. Asterisks indicate low-level cathD-mCherry signal in the lumen of late endosomal vacuoles in Rab2, Vps39 and Gie/Arl8 mutants. (D) Quantification of the colocalization in (C), showing the Pearson correlation coefficient (PCC) for A488-avidin versus cathD-mCherry. Number of garland cell clusters analyzed: w1118, 5; Rab2Δ1, 5; Vps39Δ1/Df, 7; Gie e00336/Df, 10. ANOVA followed by Tukey HSD test
We next examined the localization of soluble lysosomal proteases relative to endocytic cargo, using a cathD-mCherry chimeric reporter [Citation30] in garland cells in combination with live imaging. Due to the high resistance of the mCherry tag to low pH and lysosomal proteolysis, it is highly suitable as a marker for resident lumenal lysosomal proteins [Citation52]. As expected, in wild-type garland cells almost all cathD-mCherry resided in the 2-h tracer-positive lysosomal compartments (Fig. (c,d)). Conversely, in Rab2Δ1 garland cells most of the cathD-mCherry signal resided in small granular structures separate from the spherical tracer-filled late endosomal vacuoles ()). Garland cells hemizygous for Vps39Δ1 displayed a phenotype essentially identical to that of Rab2Δ1 garland cells, both with respect to the localization of cathD-mCherry and the morphological defect of the 2-h tracer-positive compartment (), see also )). In addition, we observed low levels of cathD-mCherry signal in the lumen of the late endosomal vacuoles in both mutants (), asterisks). Since Vps39 is required for LE-lysosome fusion [Citation26,Citation30,Citation69] and in view of the observation that most of the Cp1/Cath L residing in Rab2Δ1 and Vps39Δ1/Df tissues was in the processed, mature form ()), we conclude that the small strongly cathD-positive vesicular compartments in Rab2Δ1 and Vps39Δ1 mutants are storage lysosomes that fail to fuse to tracer-accessible late endosomal vacuoles (see Discussion).
Figure 5. Deficient lumenal exchange in the terminal endocytic compartment of Rab2, Vps39 and Gie/Arl8 mutant garland cells. (A and B) Temporally spaced pulses of endocytic tracer equilibrate in terminal endocytic compartments in the wild type but not in Rab2Δ1, Vps39Δ1/Df and Giee00336/Df garland cells. (A) L3 garland cells pulsed with TR-avidin (3 min) and chased for 2 h, then pulsed with A488-avidin (3 min) and chased again for 2 h. Insets, intensity scatter plots for a cell representative of each genotype. The Pearson correlation coefficient (PCC) is indicated. (B) Quantification of the colocalization in (A). The PCC for TR-avidin versus A488-avidin is shown. Number of garland cell clusters analyzed: w1118, 12; Rab2Δ1, 11; Vps39Δ1/Df, 3; Giee00336/Df, 6. (C to G) Endocytic tracer fails to reach most acidic vacuoles stained with LysoTracker Green (LTG) in Rab2Δ1, Vps39Δ1/Df and Giee00336/Df garland cells. (C) L3 garland cells pulsed with TR-avidin (3 min), then chased for 2 h and stained with LTG. (D) Quantification of the colocalization in (C), showing Pearson correlation coefficient (PCC) for TR-avidin versus LTG. Number of garland cell clusters analyzed: w1118, 10; Rab2Δ1, 16; Vps39Δ1/Df, 6; Giee00336/Df, 8. (E) The within-cell coefficient of variance (CV) of the LTG signal density in LTG-positive vacuoles. Number of garland cell clusters analyzed: w1118, 5; Rab2Δ1, 6; Vps39Δ1/Df, 6; Giee00336/Df, 8. (F) Tracer-positive endolysosomal compartments are less acidic than tracer-negative compartments in Rab2Δ1 and Vps39Δ1/Df garland cells. Left, scatter plot of the LTG signal density in LTG-positive vacuoles. Data were normalized to the mean vacuolar LTG signal in each cell. Green circles represent vacuoles that are LTG-positive but TR-avidin-negative. Yellow circles represent vacuoles that are both LTG-positive and TR-avidin-positive. Plot constructed from pooled vacuolar measurements from 3 cells of each genotype. Right, quantification of data on the left. Mean LTG signal densities of vacuoles negative (green) or positive (yellow) for TR-avidin are shown. (G) Left, fraction of LTG-positive large vacuoles positive (yellow) or negative (green) for TR-avidin. Means and 95% confidence intervals are shown. Right, fraction of TR-avidin-positive vacuoles either positive (yellow) or negative (red) for LTG. Geometric means and 95% confidence intervals are shown. Data in (F and G) are from 7 w1118, 15 Rab2Δ1, 6 Vps39Δ1/Df, and 8 Giee00336/Df larvae. All images in (A and C) are confocal sections through the cortical cytoplasm of living garland cells. (B, D and E) ANOVA followed by Tukey HSD. (F) Right, ANOVA followed by Scheffé contrasts. *P < 0.05, **P < 0.001, ****P < 10−8, N.S., not significant
The small GTPase ARL8B is required for LE-lysosome fusion as the lysosomal HOPS receptor in mammals [Citation25]. Gie/Arl8, the single Drosophila ortholog for mammalian ARL8A/B, also localizes to lysosomes [Citation71], but a phenotypic description of Gie/Arl8 loss-of-function in flies is lacking. We found that Giee00336, caused by the insertion of a ~ 6 kb transposable element into the first intron of the Drosophila Gie/Arl8 gene [Citation72], is pupal lethal in combination with the overlapping deficiency Df(3R)BSC222, suggesting that it represents a strong loss-of-function allele. We therefore examined post-endocytic trafficking in garland cells from these hemizygotes, denoted Giee00336/Df. Strikingly, Giee00336/Df garland cells displayed a phenotype identical to that of Rab2Δ1 and Vps39Δ1/Df, with many small cathD-mCherry-positive lysosomes located separately from the enlarged spherical 2-h tracer-accessible vacuoles ()). Western blots revealed that, as in Rab2Δ1 and Vps39Δ1, most of the endogenous Cp1/Cath L was present in the processed mature form in Giee00336/Df mutants, with only a very small increase in the immature form compared to the wild type ()). These results demonstrate that Drosophila Gie/Arl8 is required for LE-lysosome fusion, similar to its mammalian ortholog ARL8B.
We hypothesized that the rate of exchange within the lumen of the organelles of the garland cell terminal lysosomal network is high to ensure efficient mixing of endocytic cargo, resident lysosomal degradative enzymes and accessory proteins. To test this idea, we examined the fate of 2 temporally separated pulses of endocytic tracer. Garland cells were first pulsed with TR-Avidin followed by a 2 h chase to allow it to reach the terminal endocytic compartment, and then pulsed with Alexa Fluor 488-Avidin followed by an additional 2 h chase. In wild-type cells, this protocol resulted in tracer from the 2 pulses being highly equilibrated in the terminal lysosomal compartment ()). In contrast, in cells from both Rab2Δ1, Vps39Δ1/Df and Giee00336/Df larvae the tracers from the 2 pulses were poorly mixed, with many of the spherical terminal compartments containing a large amount of tracer from one pulse, but little or nothing from the other ()). Similar results were obtained in lt11/Df cells (Fig. S4A). These data demonstrate that the blocked access to the terminal lysosomal compartment in HOPS, Rab2 and Gie/Arl8 mutant garland cells leads to poor content mixing in the endolysosomal pathway.
We then examined the location of tracer in relation to acidic compartments, labeled by LTG. As described above, and consistent with previous work [Citation66], TR-avidin showed a very high degree of colocalization with LTG in the terminal lysosomal network after a 2 h chase in wild-type garland cells ()). In Rab2Δ1, Vps39Δ1/Df, lt11/Df and Giee00336/Df cells, however, no lysosomal network was apparent, and spherical compartments strongly labeled with LTG filled the cytoplasm (); Fig. S4B). The overall correlation between LTG and 2-h tracer was also dramatically reduced in these mutants (); Fig. S4B). A quantitative analysis revealed that in large compartments of wild-type cells, LTG and tracer always colocalized ()). In contrast, although a fraction of LTG-positive vacuoles had received some tracer after 2 h, most (~ 70%) remained tracer-negative in Rab2Δ1, Vps39Δ1/Df and Giee00336/Df cells ()). Also, vacuoles that were tracer-positive but LTG-negative occurred in Rab2Δ1 and Vps39Δ1/Df, but not in wild-type cells, and were fewer in Giee00336/Df cells ()). We also observed that tracer-positive LTG compartments in Rab2Δ1 and Vps39Δ1/Df exhibited a LTG signal substantially lower than average, indicating that they were less acidic than the tracer-negative compartments ()). This reduction in acidity was much less pronounced in Giee00336/Df ()). Surprisingly, the overall LTG labeling intensity of vacuoles in all mutants was much higher than in the wild type (Fig. S5), but also exhibited a much higher within-cell variation, indicating a lack of equilibration between acidic compartments ()). Taken together, these results suggest that in Rab2, HOPS and Gie/Arl8 mutants LEs are still capable of acidification, but do not fuse with the terminal lysosomal network.
In summary, these findings demonstrate that garland cells lacking Rab2, HOPS components or Gie/Arl8 are deficient in fusing LEs to cathD-positive lysosomes and cannot form the rapidly exchanging terminal lysosomal network that characterizes wild-type garland cells.
Rab2, but not Gie/Arl8, is required for lysosomal membrane protein delivery to LEs
Next, we examined the localization of GFP-LAMP in relation to 2-h tracer-positive endocytic compartments. LAMP and other LMPs are transported from the Golgi in specialized carriers that require Lt/Vps41 to fuse with LEs [Citation29,Citation33]. Due to the high sensitivity of the GFP signal to pH and proteolytic degradation, our live imaging approach favors the visualization of GFP-LAMP that has not reached endolysosomal compartments. In control garland cells, GFP-LAMP was nearly undetectable, but residual signal largely overlapped with the TR-Avidin-labeled lysosomal network ()). In Rab2Δ1, lt11/Df, and Vps39Δ1/Df garland cells, the GFP-LAMP signal was elevated compared to the wild type and accumulated in vesicles and ring-like conformations encircling 2-h tracer-positive and negative enlarged late endosomal vacuoles ()). While some of the ring-shaped GFP-LAMP accumulations in lt11/Df and Vps39Δ1/Df cells may suggest the presence of GFP-LAMP in the vacuolar membrane, the location and fragmented nature of most of them indicate that they represent vesicular structures docked at the surface of the vacuoles (), inset in 3rd column). This notion is consistent with the observation that carriers accumulating in Lt/Vps41-depleted S2R+ cells are often docked to late endosomal and lysosomal compartments [Citation29]. Weak lumenal accumulation of the GFP signal was also observed in some vacuoles in all 3 mutant genotypes (blue arrow in ), 3rd column; and data not shown), likely due to liberation of the GFP-tag from the transmembrane domain by the action of residual cathepsins. This suggests that while the loss of Rab2, Vps39 and Lt/Vps41 strongly impedes the delivery of LAMP carriers to late endosomes in garland cells, it does not completely abolish it.
Figure 6. Perturbed biosynthetic delivery of LAMP to LEs in Rab2 and HOPS but not Gie/Arl8 mutant garland cells. (A) Garland cells from L3 larvae of the indicated genotypes expressing GFP-LAMP under direct control of the αTub84B promoter. Single optical sections through the cortical cytoplasm (live imaging). The cells were pulsed with TR-avidin (3 min), then chased for 2 h. Insets depict GFP-LAMP signal accumulating in tracer-negative vesicular structures (white arrows) and perivacuolar rings (white closed and open arrowheads) around tracer-positive vacuoles in Rab2Δ1, HOPS mutants and Rab2Δ1; Vps39Δ1 double mutants, or in the lumen (blue arrowheads) of tracer-positive and tracer-negative vacuoles in Giee00336 hemizygotes. Blue arrow indicates a vacuole with some degree of lumenal GFP accumulation in a Vps39Δ1/Df cell. (B and C) Quantification of data in (A). (B) Density of the GFP-LAMP signal inside tracer-filled vacuoles relative to the density in a 0.2 µm wide external band surrounding the vacuoles. (C) Density of high-intensity GFP-LAMP vesicles larger than 0.1 µm2 (equivalent to a diameter of ~ 360 nm) and located within a 2-µm band surrounding the tracer-filled vacuoles. In (B and C), the following numbers of garland cell clusters were analyzed: 10 Rab2Δ1, 16 Vps39Δ1/Df, 19 lt22/Df, 10 Rab2Δ1; Vps39Δ1, and 11 Giee00336/Df. (B) P values in black and blue represent comparisons to Rab2Δ1; Vps39Δ1 and Giee00336/Df, respectively. (C) Thick horizontal lines indicate pooling of genotypes before statistical testing. ANOVA followed by Tukey HSD test (B) and Scheffé contrasts (C)
In contrast to the largely perivacuolar accumulation of GFP-LAMP in Rab2Δ1, lt11/Df, and Vps39Δ1/Df garland cells, almost all the GFP-signal was contained in the lumen of the enlarged tracer-positive and negative late endosomal vacuoles in cells from Giee00336/Df animals ()). To quantify GFP-LAMP delivery to the vacuoles, we measured the ratio between the GFP fluorescence inside 2-h tracer-positive vacuoles and that in a 0.2 µm wide band around them ()). Although this analysis tends to underestimate the extent of GFP-LAMP delivery – for example because the GFP signal is quenched by intralumenal acidity of Les – it is suitable for revealing relative differences. We found that Giee00336/Df cells displayed a significantly higher inside:outside ratio of GFP fluorescence compared to Rab2Δ1, lt11/Df, and Vps39Δ1/Df cells ()). Furthermore, unlike in Rab2Δ1 and lt11/Df late L3 fat bodies, in which a large proportion of the GFP-LAMP signal did not overlap LTR-positive organelles ()), in Giee00336/Df most of the GFP-LAMP was contained in LTR positive autolysosomes or amphisomes, similar to the wild type (Fig. S6). The PCC for GFP-LAMP versus LTR in Giee00336/Df mutant fat bodies was 0.804 ± 0.019 (mean ± s.e.m; n = 22); this did not differ significantly from the wild-type PCC. These data demonstrate that Lt/Vps41, Vps39 and Rab2, but not Gie/Arl8, are required for efficient delivery of LMP carriers to LEs in Drosophila.
Rab2 does not strongly rely on Vps39 for LAMP delivery to the endolysosomal system
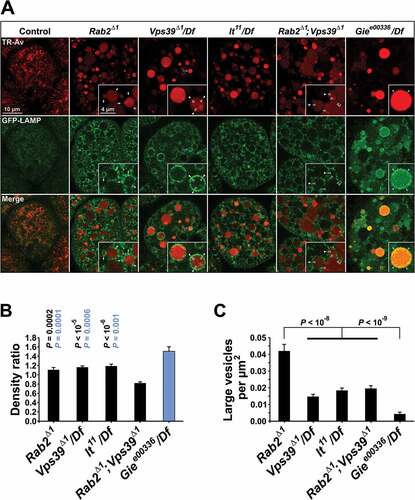
A striking feature of the Rab2Δ1 phenotype with respect to the GFP-LAMP distribution in garland cells is that much of the GFP-LAMP signal accumulated in larger (up to 500 nm in diameter) brightly fluorescent vesicles ()). This was not the case in cells from lt11/Df and Vps39Δ1/Df mutants, where GFP-LAMP was predominantly found in smaller vesicular structures often forming tight ring-like clusters around the late endosomal vacuoles ()). As described above, few GFP-LAMP vesicles of any kind were observed outside of the endosomal vacuoles in Giee00336/Df cells ()). As shown in ), these differences in the GFP-LAMP signal distribution were confirmed using an image processing algorithm counting large particles (diameter >355 nm) of high circularity surrounding the vacuoles (see Materials and Methods).
Drosophila Vps39 has recently been reported to be a Rab2 effector [Citation48]. We therefore investigated to what extent the function of Rab2 is mediated by Vps39, by comparing the phenotype of Rab2Δ1; Vps39Δ1 double mutants to that of the Rab2Δ1 and Vps39Δ1/Df single mutants. Surprisingly, the GFP-LAMP delivery phenotype in Rab2Δ1; Vps39Δ1 garland cells was stronger than in Rab2Δ1 and Vps39Δ1/Df cells, with a significantly lower inside:outside ratio of GFP fluorescence for the 2-h tracer-filled vacuoles ()). In addition, the late endosomal vacuoles tended to be somewhat larger and more irregularly shaped in double-mutant garland cells compared to Rab2Δ1 and Vps39Δ1/Df cells ()). Interestingly, the large GFP-LAMP vesicles that characterized Rab2Δ1 mutants were replaced by small vesicles in Rab2Δ1; Vps39Δ1 double mutants ()), strongly suggesting that the large vesicles in Rab2Δ1 cells represent either HOPS-dependent homotypic aggregation or fusion of LMP carriers, or HOPS-dependent heterotypic docking or fusion of LMP carriers to a different organelle.
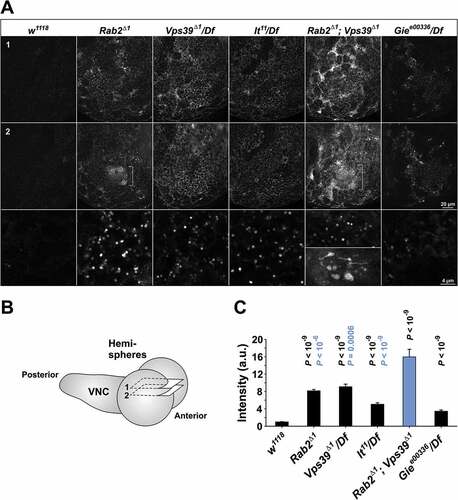
To further establish the nature of the interaction between the Rab2Δ1 and Vps39Δ1 alleles, we examined the GFP-LAMP granule accumulation initially observed in the fixed Rab2Δ1 CNS ()). As expected, Giee00336/Df showed the least amount of granular GFP-LAMP accumulation among all the mutants tested, consistent with GFP-LAMP still being delivered to degradative LEs in the absence of Gie/Arl8A (). Rab2Δ1 and Vps39Δ1/Df showed similar amounts of GFP-LAMP signal accumulating in granular structures in the dorsal aspect of L3 brain hemispheres, whereas lt11/Df displayed a slightly lower granular GFP-LAMP signal ()). However, while the shape and distribution of the GFP-LAMP granules were largely identical in Vps39Δ1/Df and lt11/Df brains, the GFP-LAMP distribution in Rab2Δ1 brains showed several unique features. Most prominent was a massive accumulation of clustered GFP-LAMP granules in a paired structure corresponding to the mushroom body calyx ()). Strikingly, Rab2Δ1; Vps39Δ1 double-mutant brains showed a strong increase in granular GFP-LAMP signal compared to Rab2Δ1 and Vps39Δ1/Df single mutants ()). Furthermore, the Rab2Δ1; Vps39Δ1 brains also exhibited synthetic effects in the form of a marked accumulation of diffuse GFP in the brain surface and the appearance of large GFP-LAMP-positive vacuoles up to 2 µm in diameter ()). In addition, western blotting revealed that the relative amount of unprocessed pro-Cp1/Cath L was elevated even further in Rab2Δ1; Vps39Δ1 tissues compared to Rab2Δ1 and Vps39Δ1 ()), again illustrating that the endolysosomal defect is more severe upon loss of both Rab2 and Vps39 compared to the loss of either protein.
Figure 7. Accumulation of GFP-LAMP in the CNS of Rab2, HOPS and Gie/Arl8 mutant larvae. (A) Micrographs of right brain hemispheres from fixed L3 larvae ubiquitously expressing GFP-LAMP. The plane of section for the top row (labeled ‘1’) is near the dorsal surface of the hemisphere, while that for the middle row (labeled ‘2’) is about 2 µm deeper in the preparation. The schematic in (B) shows the approximate position of the two optical sections in a live brain, with labels ‘1’ and ‘2’ corresponding to those in (A). (A, middle) A prominent structure (brackets) with clusters of GFP-LAMP granules appeared exclusively in Rab2Δ1 mutants and Rab2Δ1; Vps39Δ1 double mutants. (A, bottom) Areas representative of GFP-LAMP signal accumulation at high resolution. Large GFP-LAMP-positive vacuoles, only occurring in Rab2Δ1; Vps39Δ1 double mutants, are also shown (bottom inset). Single optical slices; medial is right, anterior down. (C) The amount of GFP signal contained in GFP-LAMP granules in the dorsal aspect of the brain hemispheres. The following numbers of larvae (hemispheres) were analyzed (ANOVA followed by Tukey HSD test): 16 w1118, 15 Rab2Δ1, 7 Vps39Δ1/Df, 10 lt22/Df, 9 Rab2Δ1; Vps39Δ1, and 11 Giee00336/Df. P-values in black and blue represent comparisons to w1118 and Rab2Δ1; Vps39Δ1, respectively
In summary, these data show that Rab2 and Vps39 to a large extent function independently in LMP carrier delivery to LEs.
Rab2 is required for proteolytic degradation of endocytosed material
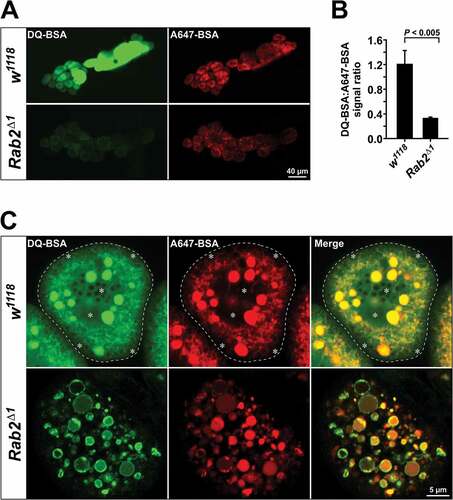
To test if the impaired function of the endolysosomal system in Rab2 mutants impedes proteolytic degradation of endocytosed material, we used DQ-BSA, an endocytic substrate that becomes fluorescent upon proteolytic cleavage [Citation73]. We pulsed garland cells with a 1:1 mix of DQ-Green BSA and Alexa Fluor 647-BSA (A647-BSA) and determined the ratio of DQ-BSA and Alexa Fluor 647 fluorescence. A647-BSA served as a marker of endocytic uptake. After 2 h, wild-type garland cells exhibited a strong DQ-BSA fluorescence, indicating proteolysis of the endocytosed tracer (). Strikingly, Rab2Δ1 cells exhibited a 3.6-fold reduction in the DQ-BSA:A647-BSA ratio ()). Furthermore, high-magnification imaging of wild-type cells revealed that, while A647-BSA was contained in the tubulo-vacuolar lysosomal system, much of the DQ-BSA signal resided in the cytosol ()). This most likely reflects that the highly fluorescent BODIPY-labeled free amino acids and oligopeptides generated by DQ-BSA proteolysis are exported from the lysosomal lumen into the cytosol by lysosomal transporters. In contrast, in Rab2Δ1 cells, the DQ-BSA signal was generally contained in the endocytic vacuoles, with very little reaching the cytosol ()). This could indicate that the proteolytic degradation in Rab2Δ1 is only partial and does not generate a substantial amount of small degradation products, or that export of catabolites from the aberrant late endocytic compartments does not occur. These results indicate that proteolytic degradation of endocytosed material is markedly impaired in cells lacking Rab2, presumably as a combined effect of an uneven acidification of LEs ()) and abolished fusion between LEs and lysosomes that function as storage compartments for degradative enzymes ()).
Figure 8. Reduced proteolysis of endocytosed material in Rab2Δ1 garland cells. (A) Garland cells from wild-type and Rab2Δ1 mutant larvae pulsed for 3 min with a 1:1 mix of DQ-Green-BSA and Alexa Fluor 647-BSA, followed by a 2 h chase. Sum projections of z-stacks through garland cell clusters using a low-power objective (live imaging). (B) Quantification of data in (A). Ratio of DQ-Green-BSA and Alexa Fluor 647-BSA fluorescence intensity from 14 w1118 and 18 Rab2Δ1 garland cell clusters. P value from unpaired Student’s t test. (C) DQ-BSA signal in the cytosol of wild-type but not Rab2Δ1 garland cells. At the top, the dashed line marks the border of a wild-type garland cell. The asterisks indicate areas with strong diffuse DQ-BSA signal in the cytosol, distinct from the tubulo-vacuolar network labeled by A647-BSA. In the Rab2Δ1 cell (bottom), the DQ-BSA and A647-BSA signals are both contained in vacuoles, with no apparent cytoplasmic DQ-BSA labeling
Rab2 localizes to LEs and lysosomes in a HOPS-independent manner
In light of Rab2’s function in LMP carrier delivery to LEs and LE-lysosome fusion, we sought to determine the localization of the Rab2 protein within the endolysosomal system. Wild-type mCherry-Rab2 accumulated to very high levels in the lumen of the terminal lysosomal compartment in garland cells (Fig. S7A, see also )). Curiously, the lumenal lysosomal accumulation of mCherry-Rab2 was at least partially activity-dependent, as it was much lower in non-active GDP-locked mCherry-Rab2S20N (Fig. S7A), and was also displayed by HA-Rab2 expressed under the endogenous Rab2 promoter (Fig. S7B), suggesting that it is part of the normal life cycle of the Rab2 protein. The lumenal level of mCherry-Rab2 was strongly reduced in a Atg7d77 null mutant background, in which macroautophagy is abolished [Citation74] (Fig. S7C). Therefore, macroautophagy appears to be a major mechanism contributing to the intra-lysosomal accumulation of Rab2. However, residual accumulation of mCherry-Rab2 in Atg7d77 garland cell lysosomal compartments revealed that other pathways, such as endosomal microautophagy [Citation75], must be involved as well.
Though intriguing, the lumenal accumulation of Rab2 in lysosomes confounds visualization of active, membrane-bound Rab2. To overcome this problem, we used Rab2 transgenes fused to the strongly pH-sensitive super-ecliptic pHluorin (pHl). In agreement with the work of others in Drosophila S2 cells [Citation48], the gain-of-function Rab2 proteins pHluorin-RabQ65L and mCherry-RabQ65L overexpressed in garland cells were strongly recruited to the limiting membranes of enlarged spherical endolysosomal compartments that were positive for 2-h tracer and GFP-LAMP ( and Fig. S7A). Moreover, in L3 salivary glands pHluorin-RabQ65L strongly labeled the limiting membrane of both the tubulovesicular lysosomal network described previously and giant LTR-positive vacuoles (3 to 5 µm in diameter) that generally did not appear in controls ()).
Figure 9. Rab2 is present on the limiting membranes of LEs and lysosomes. (A to E) High-magnification single optical sections (live imaging) through cortical garland cells (A to C) or cortical salivary gland cells from L3 larvae (D and E). (A) Garland cells from larva overexpressing wild-type pHluorin-Rab2 (top) or gain-of-function pHluorin-Rab2Q65L (bottom) pulsed with TR-avidin (3 min) and chased for 2 h. (B) Garland cells from larva expressing GFP-Rab2 under endogenous promoter regulation, incubated for 7ʹ with TR-avidin (top), or pulsed for 3 min with TR-avidin and chased for 2 h (bottom). (C) Garland cell from larva overexpressing gain-of-function mCherry-Rab2Q65L and expressing GFP-LAMP under control of the αTub84B promoter, pulsed with A647-BSA (3 min) and chased for 2 h. (D) Salivary gland cells from larva expressing GFP-Rab2 under endogenous promotor regulation, stained with LysoTracker Red (LTR). GFP-Rab2 labels LTR-positive lysosomal tubules (top, open arrowheads) and membranes of LTR-positive lysosomal vacuoles (bottom, closed arrowheads). Inset shows a pair of adjacent LTR and GFP-Rab2-positive vacuoles at higher magnification. The strongly GFP-Rab2-labeled, LTR-negative structures are mostly Golgi bodies (arrows). (E) Salivary gland cells from larva overexpressing gain-of-function pHluorin-Rab2Q65L, stained with LTR. Top, Tangential section of the basolateral gland surface. The limiting membranes of smaller LTR-positive lysosomal vacuoles (arrows) and lysosomal tubules extending from them (open arrowheads) are labeled with pHluorin-Rab2Q65L. Bottom, Giant lysosomal vacuole containing strongly LTR-labeled internal vesicles. The limiting membrane is labeled with pHluorin-Rab2Q65L (arrowheads). Plane of section deeper within the gland cytoplasm compared to top.
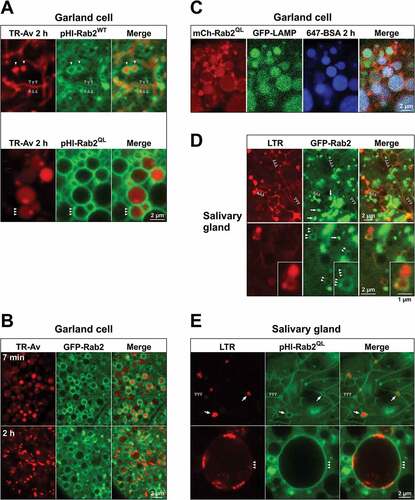
Importantly, overexpressed wild-type pHluorin-Rab2 also labeled the limiting membranes of many of the tubules and vacuoles of the 2-h tracer-positive lysosomal compartment in garland cells (), open arrowheads). Labeling of endolysosomal membranes could also be observed with wild-type mCherry-Rab2 overexpressed in the Atg7d77 background, where the lumenal mCherry signal was reduced (Fig. S7C). Similar results were obtained using the moderately pH-sensitive GFP-Rab2 expressed under the endogenous Rab2 promoter ()), showing that endolysosomal membrane labeling is not caused by Gal4-driven overexpression. Weak labeling of some the tubules and vesicles of the LTR-positive lysosomal network in salivary glands with GFP-Rab2 could also be demonstrated ()), although this signal was orders of magnitude lower than that from adjacent Rab2-positive Golgi bodies ()). In agreement with previous work [Citation48,Citation76–Citation78] GFP-Rab2 strongly concentrated in structures that capped or encircled Golgi bodies in salivary glands (Fig. S8).
To determine at what stage in the endolysosomal pathway Rab2 becomes associated with endosomal and lysosomal organelles, we exposed GFP-Rab2 garland cells to TR-avidin-containing medium and imaged them during endocytosis and further downstream processing of the tracer. We found that, in many cells, GFP-Rab2 was present in the membranes of spherical LEs that were filled by tracer as early as approximately 6 min after the start of the experiment (the earliest time point allowed to be evaluated by our method) (), top).
We then analyzed the circumstances of Rab2’s recruitment to late endosomes in garland cells, by examining the localization of GFP-Rab2 in different mutant backgrounds. In Giee00336/Df cells, GFP-Rab2 was strongly recruited to the limiting membranes of many of the enlarged late endosomal vacuoles (), demonstrating that it does not reach LEs through fusion with lysosomal compartments. It has been suggested that Rab2 arrives to LEs on Golgi-derived vesicles in a HOPS-mediated fusion event and is subsequently quickly released from the membrane [Citation48,Citation79]. Surprisingly, we found that in both Vps39Δ1/Df and lt11/Df garland cells GFP-Rab2 was still present on the membranes of many of the 2-h tracer-positive and negative enlarged late endosomal vacuoles ()).
Figure 10. Location of Rab2 in HOPS component and Gie/Arl8 mutants and schematic representation of the function of Rab2 in the endolysosomal system. (A) Recruitment of Rab2 to the limiting membrane of LEs is not blocked by the absence of Gie/Arl8 or HOPS complex activity. Garland cells from Vps39Δ1/Df, lt22/Df, and Giee00336/Df animals expressing GFP-Rab2 under the endogenous Rab2 promoter, pulsed (3 min) and chased (2 h) with TR-avidin. Insets show GFP-Rab2 labeling the limiting membranes of enlarged late endosomal vacuoles in high magnification. (B) Simplified diagram of the endolysosomal trafficking pathways investigated in this study. Material is passed from early endosomes to LEs, which fuse to lysosomes sometimes exhibiting a tubular morphology. Rab2, Gie/Arl8 and the HOPS complex are necessary for LE-lysosome fusion. Lysosomal membrane proteins are transported directly to LEs from the Golgi apparatus in AP-3 dependent carriers [Citation29]. In Drosophila, Rab2 and both Lt/Vps41 [Citation29] and Vps39, but not Gie/Arl8, are necessary for efficient delivery of these carriers. In addition, Rab2 and the HOPS complex are required for the fusion of autophagosomes to LEs and lysosomes [Citation30,Citation31,Citation79,Citation89]. In contrast, the AP-3 and HOPS complexes, but not Rab2, are necessary for trafficking to LROs such as pigment granules [Citation30,Citation58–Citation63]
![Figure 10. Location of Rab2 in HOPS component and Gie/Arl8 mutants and schematic representation of the function of Rab2 in the endolysosomal system. (A) Recruitment of Rab2 to the limiting membrane of LEs is not blocked by the absence of Gie/Arl8 or HOPS complex activity. Garland cells from Vps39Δ1/Df, lt22/Df, and Giee00336/Df animals expressing GFP-Rab2 under the endogenous Rab2 promoter, pulsed (3 min) and chased (2 h) with TR-avidin. Insets show GFP-Rab2 labeling the limiting membranes of enlarged late endosomal vacuoles in high magnification. (B) Simplified diagram of the endolysosomal trafficking pathways investigated in this study. Material is passed from early endosomes to LEs, which fuse to lysosomes sometimes exhibiting a tubular morphology. Rab2, Gie/Arl8 and the HOPS complex are necessary for LE-lysosome fusion. Lysosomal membrane proteins are transported directly to LEs from the Golgi apparatus in AP-3 dependent carriers [Citation29]. In Drosophila, Rab2 and both Lt/Vps41 [Citation29] and Vps39, but not Gie/Arl8, are necessary for efficient delivery of these carriers. In addition, Rab2 and the HOPS complex are required for the fusion of autophagosomes to LEs and lysosomes [Citation30,Citation31,Citation79,Citation89]. In contrast, the AP-3 and HOPS complexes, but not Rab2, are necessary for trafficking to LROs such as pigment granules [Citation30,Citation58–Citation63]](/cms/asset/e0b3f99b-6ff2-4685-a59c-963c604a3073/kaup_a_1458170_f0010_c.jpg)
Collectively, these data show that Rab2 becomes associated with late endosomal membranes independently of HOPS-mediated vesicular fusion and remains membrane-bound into the lysosomal stage.
Discussion
The endolysosomal pathway is critical for cellular functions ranging from energy metabolism and pathogen neutralization to protein and organellar quality control. Here we demonstrate that Rab2 is required at two distinct steps in the endolysosomal pathway in Drosophila ()). First, Rab2 mediates delivery of LAMP-containing lysosomal membrane protein transport intermediates to LEs. Second, Rab2 is necessary for the fusion of LEs to pre-existing lysosomes. In addition, we observe a strong perturbation of macroautophagy in tissues lacking Rab2, consistent with a requirement of Rab2 in fusion of autophagosomes with LEs and lysosomes.
Many LMPs are carried directly from the Golgi apparatus to the endolysosomal pathway in specialized biosynthetic transport intermediates bypassing early endosomal compartments [Citation29,Citation33,Citation80]. While the structure of these transport carriers is very similar across species (they are non-coated and contain internal membranes), the detailed mechanistic requirements of this pathway vary. In yeast, the carriers fuse directly to the vacuole in a process that requires the entire HOPS complex and Ypt7/Rab7 [Citation81,Citation82], whereas in mammals they fuse to LEs depending only on VPS41 [Citation33]. In fly hemocytes, the pathway is similarly targeted to LEs, and depends on Lt/Vps41, but not Dor/Vps18 [Citation29].
We find that Drosophila Rab2 and Vps39 are also required for delivery of LAMP-containing LMP carriers to LEs. Loss of Rab2 elicits a LAMP trafficking phenotype in multiple tissues that in many ways is similar to that caused by loss of Lt/Vps41 and Vps39. Some differences were, however, observed. In particular, in the absence of Rab2 in garland cells, accumulating LAMP carriers appeared to aggregate or fuse together (or possibly to non-degradative storage lysosomes) in a Vps39-dependent manner, thus forming larger granules. In contrast, without Lt/Vps41 or Vps39 small LAMP carriers tended to dock to the surface of LE vacuoles, often forming tightly packed ring-like conformations. We also observed that Rab2 is associated with LE membranes, and this association was not visibly perturbed by absence of Vps39 or Lt/Vps41 ()). Collectively, these data suggest that Rab2 functions as the late endosomal receptor for LMP carrier docking and fusion.
In contrast to Rab2, we found that the small GTPase Gie/Arl8, while being required for LE-lysosome fusion, is not required for LAMP carrier delivery to LEs. In addition to showing that these processes clearly differ mechanistically, this fits well with the predominantly lysosomal localization of Drosophila Gie/Arl8 and its mammalian orthologs [Citation25,Citation71].
Transmembrane components of the vacuolar protein pump are among the proteins transported to LEs in LMP carriers, together with LAMP [Citation29]. Interestingly, perturbation of LMP carrier delivery in Rab2, Vps39 or lt/Vps41 mutants did not prevent acidification of late endosomal vacuoles in garland cells, possibly reflecting that the block in LMP delivery is incomplete or that substantial amounts of V-ATPase are already present in early endosomes [Citation83]. However, we did observe that vacuoles accessed by endocytic tracer after a 2-h chase were less acidic in Rab2 and Vps39 mutants, both compared to tracer-negative vacuoles, and also to tracer-positive vacuoles in Gie/Arl8 mutants where LMP carriers are still efficiently delivered ()). Assuming that the tracer-accessible vacuoles are on average younger than the tracer-inaccessible ones, this suggests that delivery of LMP carriers accelerates the acidification of LEs.
Even though Vps39 is a Rab2 effector [Citation48], combining the loss of Rab2 with a loss of Vps39 almost doubled the strength of the LAMP-trafficking defect in the CNS and considerably enhanced it in garland cells ( and ). This suggests that Rab2 and Vps39 function independently in LMP carrier delivery. Rab2 must therefore recruit other effectors such as non-Vps39 HOPS components or tethering factors unrelated to HOPS to mediate tethering and fusion of LMP carriers to LEs.
In addition to Rab2’s role in delivery of LAMP carriers to LEs, we also find that Drosophila Rab2 is required for LE-lysosome fusion. It is possible that this requirement is indirect and stems from Rab2-dependent LMP carriers supplying LEs with crucial factors needed for LE-lysosome fusion, such as SNAREs and the vacuolar acidification machinery. However, several arguments can be made against this hypothesis. First, the Rab2Δ1; Vps39Δ1 double-mutant experiments demonstrate that the block in LMP carrier delivery to LEs in Rab2 mutants is not complete. In addition, moderate amounts of LMPs traffic to LEs via the plasma membrane [Citation29]. Therefore, moderate delivery of transmembrane fusion factors would be expected to occur and allow for some degree of LE-lysosome fusion. However, the LE-lysosome fusion defect in Rab2 null garland cells is essentially as severe as in cells lacking Gie/Arl8 and Vps39 ( and ), which are presumably directly required for this process. Moreover, while acidification has indeed been proposed to be important for fusion [Citation84], the enlarged LEs in Rab2 mutants still become highly acidic () and Fig. S5), even if this acidification is possibly delayed. Finally, recent work in Drosophila has shown that V-ATPase-dependent acidification is not required for fusion of autophagosomes to LEs and lysosomes and for biosynthetic delivery of LMPs to lysosomes, and therefore presumably also not LE-lysosome fusion, but only for lysosomal degradation [Citation85].
Recent work has shown that after ended hydrolytic digestion, endolysosomes form into small non-acidic storage lysosomes containing inactive lysosomal hydrolases [Citation13]. These storage lysosomes can then fuse to LEs and existing endolysosomes to recycle their content of lysosomal constituents and regenerate lysosomal compartments (see model in Fig. S9A). This provides a likely explanation for the presence of small cathepsin-positive, tracer-inaccessible lysosomes when LE-lysosome fusion is abrogated in Rab2, Vps39 and Gie/Arl8 mutant garland cells ()). We speculate that as the maternal gene products for these factors are depleted during development, the cathepsins that have already been delivered to the lysosomal system become trapped in small storage lysosomes that can no longer fuse with LEs (Fig. S9B). After this time point, cathepsins delivered to endosomes can no longer reach the lysosomal compartment. Consistent with this, we observe low but discernable levels of lumenal cathD-mCherry in many of the enlarged LE vacuoles in Rab2, Vps39 and Gie/Arl8 mutants.
The current model for LE-lysosome fusion in mammals entails the ~ 30-nm long, seahorse-shaped HOPS complex bridging lysosomal and endosomal membranes by binding the RAB7A effector RILP in the LE membrane and ARL8B and its effector SKIP on the lysosome [Citation22–Citation25]. The binding is accomplished by the Vps39 and Vps41 subunits that are positioned at each end of the HOPS complex, based on electron tomography of the complex in yeast [Citation20]. However, Gillingham et al. detected no strong interaction between Drosophila Rab7 and Vps39 or the sole fly RILP ortholog [Citation48]. Similarly, activated Drosophila Rab7 does not precipitate Lt/Vps41 [Citation48]. In contrast, activated Rab2 precipitates the entire HOPS complex and binds Vps39 directly [Citation48]. We find that both Drosophila Rab2 and Gie/Arl8 are required for LE-lysosome fusion. We also observe that Rab2 resides on LE membranes, both in the wild type and in the absence of LE-lysosome fusion in Gie/Arl8 and HOPS mutants ()). These results suggest that, in Drosophila, the HOPS tethering complex predominantly interacts with Rab2 rather than Rab7 on the LE membrane, while Gie/Arl8 (or its conserved effector SKIP) likely functions as the lysosomal HOPS receptor. However, the observation that the combined loss of Rab2 and Vps39 leads to a slight increase in the severity of the morphological phenotype of the stalled late endosomal compartment in garland cells could imply that Rab2 also interacts with other factors involved in the LE-lysosome transition. In addition, Rab7 is still required for LE-lysosome fusion in Drosophila [Citation79,Citation86]. This suggests that Rab7 may assist in the tethering process, perhaps by recruiting the conserved LE/lysosomal adaptor PLEKHM1 (FlyBase Annotation Symbol CG6613) which is strongly precipitated by activated Drosophila Rab7 [Citation48] and in mammals facilitates LE-lysosome fusion [Citation87,Citation88].
While this work was in revision, two papers have been published describing the role of Rab2 in LE-lysosome fusion and autophagosome-lysosome fusion, respectively [Citation79,Citation89]. Lőrincz et al. use Drosophila garland cells and fat bodies and human cells to essentially show the same requirement for Rab2 in LE-lysosome fusion and autophagy described here [Citation79]. Fujita et al. describe the dependence of autophagic clearance and autophagosome delivery to endosomal and lysosomal compartments on Rab2 in Drosophila and mouse embryonic fibroblasts [Citation89]. The picture that emerges from our work and the findings of Lőrincz et al. and Fujita et al. is that Rab2 controls all major heterotypic fusion processes at the LE, i.e. biosynthetic LMP carrier delivery, LE-lysosome fusion and autophagosome-LE fusion ()). Interestingly, mammalian Rab2A has been found to promote LE exocytosis in cancer cells, suggesting that LE-plasma membrane fusion is also controlled by Rab2 [Citation90]. The role of Rab2 in phagocytosis [Citation42,Citation43] likely parallels its function in the endolysosomal pathway, as the two pathways are mechanistically related. A LE-centric role of Rab2 in the endolysosomal pathway also explains why we do not detect any obvious perturbation of LRO formation in Rab2 null flies. Although the LRO biogenesis pathway in many aspects resembles that of conventional lysosomes, there is evidence suggesting that, at least for some types of LROs, it bypasses the LE compartment [Citation53].
An important question is on which membranes the presence of Rab2 is required in LE-related fusion events, and also how Rab2 arrives on endosomal and lysosomal organelles. Lőrincz et al. argue that Rab2 traffics to LEs on the membranes of Golgi-derived vesicles that fuse with Rab7-positive LEs in a HOPS-dependent manner [Citation79]. Likewise, Fujita et al. suggest that Rab2 is present on the outer membrane of autophagosomes that fuse to Rab7-positive LEs or lysosomes [Citation89]. However, as mentioned, we observe that in garland cells Rab2 is bound to the limiting membrane of LE vacuoles even when HOPS-mediated vesicular fusion was blocked by the absence of Vps39 or Lt/Vps41, and when macroautophagy was abrogated by the absence of Atg7. In fact, in Vps39, lt/Vps41 and Gie/Arl8 mutants the association of Rab2 with LE vacuoles appeared more pronounced than in wild-type cells. While this does not exclude that Rab2 is present in the membranes of Golgi-derived vesicles or autophagosomes, it suggests that Rab2 is – at least in some cell types – recruited to LE membranes irrespective of vesicular fusion processes and is likely required on the LE membrane to accept incoming traffic of lysosomes, autophagosomes and biosynthetic Golgi-derived LMP carriers. The failure to easily detect wild-type Rab2 on LE membranes in many cell types [48, 79, 89; and data not shown] may be a consequence of very tight regulation of active membrane-bound Rab2 due to its high fusogenicity illustrated by the enlarged aberrant endolysosomal compartments formed by gain-of-function Rab2 ()(a,e) and Fig. S7A). Autophagic turnover may assist in keeping Rab2 activity down, forming a feed-back loop that controls formation of endolysosomes and autolysosomes.
Materials and methods
Molecular biology
To generate the wild-type UAS-mCherry-Rab2, the gain-of-function UAS-mCherry-Rab2Q65L, and the dominant-negative UAS-mCherry-Rab2S20N transgenes, a pUAST-mCherry plasmid was produced by amplifying the mCherry coding region from pV2-mCherry [Citation91] with primers mCh-3 and mCh-4(primer oligonucleotides specified in ), and subcloning it into pUAST via the EcoRI-NotI restriction sites. Using oligos Rab2-3 and Rab2-4, the Rab2 coding region was then amplified from pUAST-YFP-Rab2, pUASp-YFP-Rab2Q65L and pUAST-YFP-Rab2S20N plasmids (a gift from Dr. Hugo J. Bellen, Baylor College of Medicine). The resulting fragments were cloned into NotI-XbaI sites of the pUAST-mCherry plasmid.
The genomic HA-Rab2 transgene was generated by amplifying a 4.1 kb fragment from genomic DNA using oligos Rab2-23 and Rab2-24 and cloning it into the SpeI-NotI restriction sites of the pCasper4 vector. The genomic fragment extended from the second codon of the Rab2 gene to approximately 1.0 kb downstream of the Rab2 transcriptional termination site. A second genomic fragment was then amplified that corresponded to the region stretching from 4.7 kb upstream of the Rab2 start ATG to the start ATG, using oligos Rab2-21 and Rab2-22. This fragment was inserted via XhoI-SpeI restriction sites into the pCasper4 plasmid carrying the first fragment to complete the genomic Rab2 transgene. The Rab2-22 primer includes a sequence coding for YPYDVPDYA, resulting in insertion of a hemagglutinin tag downstream of the Rab2 start methionine and an intervening glycine, and followed by a short TSG linker before resumption of the wild-type Rab2 amino acid sequence.
To generate the UAS-pHluorin-Rab2 and UAS-pHluorin-Rab2Q65L transgenes the mCherry cassette in the UAS-mCherry-Rab2 and UAS-mCherry-Rab2Q65L plasmids, respectively, was exchanged for a super-ecliptic pHluorin cassette using the GenScript gene editing service. Likewise, to generate the genomic GFP-Rab2 transgene the HA coding sequence in the pCasper4-HA-Rab2 construct was exchanged for a EGFP coding sequence using the GenScript gene editing service.
For CRISPR/Cas9-mediated mutagenesis, gRNA target sequences GGAGTCCAACGACTATTCGC and GAAGTCCACAACCGGACGTG were inserted into the pBFv-U6.2 vector to obtain the two gRNA constructs targeting the Vps39 gene. The donor DNA construct for homology-directed repair was generated by inserting ~ 1kb homology arms flanking the Cas9 cleavage sites into the pHD-DsRed-attP vector. All DNA work for CRISPR/Cas9 mediated mutagenesis was performed by GenScript.
Fly strains and genetics
Flies were reared on Nutri-Fly™ Bloomington Formulation medium (Genesee Scientific) at 26°C. For experiments involving Rab2 mutants, larvae were reared on apple juice plates supplemented with yeast paste.
DNA injections and selection of P-element transformants in a w1118 background was performed by BestGene Inc.. The Rab2Δ1 allele was generated by mobilizing the P-element P{PTT-GA}Rab2CA07465 inserted into the first intron of the Rab2 gene using the P{ry[+ t7.2] = Delta2-3}99B transposase. The UAS-GFP-Rab7 (Bloomington stock no. 42706), 386Y-Gal4 (25410), CyO/Df(2R)BSC326 (24351), UAS-mito-HA-GFP (8443), UASp-RFP.Golgi (30908), tubP-Gal4/TM3Sb (5138), Df(3R)C7, ry506/TM3, ryRK Sb1 Ser1 P{ry[+ t7.2] = Delta2-3}99B (1808), Df(3R)Exel6178, P{w[+ mC] = XP-U}Exel6178/TM6B, Tb1 (7657), lt11/In(2LR)Gla, wgGla−1 PPO1Bc (26262), Df(2L)lt45, PPO1Bc/SM1, lt16 (26187), PBac[7]Giee00336/TM6B, Tb1 (17846), Df(3R)BSC222/TM6B, Tb+ (9699), dor8/FM6 (28), and P{PTT-GA}Rab2CA07465 (50821) lines were obtained from the Bloomington Stock Collection. Gliotactin-Gal4 was a gift from M. Stern (Rice University). prospero-Gal4 was a gift from Dr. Fumio Matsuzaki, RIKEN Center for Developmental Biology, Kobe, Japan. αTub84B-GFP-LAMP [Citation28] was a gift from Dr. Helmut Krämer (University of Texas Southwestern Medical Center). The αTub84B-Rab7.YFP strain [Citation92] was a gift from Dr. Suzanne Eaton (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany). The line carrying the cathD-mCherry transgene controlled by the endogenous cathD promoter [Citation30] and the Atg7d77 line [Citation74] were a gift from Dr. Gábor Juhász (Eötvös Loránd University, Budapest, Hungary). pm-Cherry-GFP-Atg8a was a gift from Dr. Tor Erik Rusten (University of Oslo, Oslo, Norway). c929-Gal4 [Citation93] was kindly provided by Dr. Paul Taghert (Washington University, St. Louis).
The Vps39Δ1 knockout line was generated using CRISPR/Cas9-catalyzed homology-directed repair according to the method outlined in [Citation94]. In brief, two gRNA-expressing DNA constructs targeted near each end of the coding sequence of the Vps39 gene were injected into yw; nos-Cas9(II-attP40) embryos expressing Cas9, together with a donor DNA construct based on the pHD-DsRed-attP plasmid. As a result, most of the coding sequence of Vps39 (3R:18273942–18272027) was replaced with a DsRed transgene, allowing for the expression of only the first 120 amino acids of the Vps39 protein. This covers only a third of the N-terminal β-propeller domain, most likely resulting in a completely non-functional protein. The DNA injections and selection of transformants were performed by BestGene Inc. The Vps39Δ1 allele is homozygous lethal, and it is hemizygous lethal when placed over the Df(3R)Exel6178 deficiency covering Vps39. Both Vps39Δ1 homozygotes and Vps39Δ1/Df(3R)Exel6178 hemizygotes survive until pupation, but die in pharate adult stage, indicating that Vps39Δ1 is a null allele.
To establish the lethal stage of Rab2 mutants, first instar larvae from overnight lays were deposited on apple juice plates supplemented with yeast (10 animals per plate) and allowed to develop at 26°C. The number of pupae, the most advanced developmental stage reached, and the number of eclosed animals was subsequently scored.
Immunohistochemistry and preparation for fluorescent protein imaging of fixed tissue
For immunohistochemistry and fluorescent protein visualization in fixed tissues, third instar (L3) larval fillets were prepared or isolated L3 brains were dissected out in phosphate-buffered saline (PBS) containing 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 6.5 mM Na2HPO4, pH 7.4. Isolated brains were briefly stored in Schneider insect cell medium (Life Technologies, A820) prior to fixation. The tissues were fixed in 3.7% formaldehyde in PBS on ice for 2 h or at room temperature (RT) for 45 min, followed by 6 × 10 min washes in PBX (PBS with 0.3% Triton-X 100 [Sigma-Aldrich, T8787]) at RT. After a 2-h blocking step in blocking buffer (PBX with 5% normal goat serum [Sigma-Aldrich, G9023]) at RT, the tissues were incubated for 24 to 48 h with primary antibodies in blocking buffer at 4°C under gentle agitation. They were then washed 6 × 10 min in PBX, incubated with secondary antibodies in blocking buffer for 2 h at RT, and again washed 6 × 10 min in PBX. Finally, the tissues were washed 2 × 5 min in PBS and mounted in ProLong® Gold antifade reagent (Life Technologies, P36934).
Rabbit anti-HRP (1:250) (P-7899, Lot: 083K4840) and monoclonal rat anti-HA (100 ng/ml) (clone 3F10, REF: 11867423001, Lot: 14553800) were from Sigma. Rabbit anti-GM130 (D. melanogaster) (1:500) (ab30637, Lot:GR199711-1) was from Abcam. Mouse anti-Rab7 (D. melanogaster) [Citation95] (1:200) was from Developmental Studies Hybridoma Bank, University of Iowa.
Secondary antibodies, Alexa Fluor 488 goat anti-rabbit IgG (H + L) (A11034, lot: 1423009), Alexa Fluor 647 goat anti-rabbit IgG (H + L) (A21245, lot: 1558736), Alexa Fluor 647 goat anti-mouse IgG (H + L) (A21236, LOT: 1360398) and Alexa Fluor 647 goat anti-rat IgG (H + L) (A21247, lot: 1524910) were from Life Technologies and were all used in a concentration of 1:500.
For imaging of YFP-Rab7 and αtub-GFP-LAMP in fixed third instar (L3) brains, the tissue was dissected out in PBS or Schneider insect cell medium (Life Technologies, A820) and fixed in 3.7% formaldehyde dissolved in PBS for 40ʹ at RT or 2 h on ice. The brains were then washed 6 × 10 min in PBX, followed by 2 × 5 min in PBS and mounted in antifade reagent.
Live imaging
Salivary glands and eye imaginal disks were dissected out of late L3 larvae and imaged in PBS in an imaging chamber constructed from vacuum grease and covered with a coverslip. For live imaging of peptidergic somata in the dorsal ventral nerve cord (VNC) of 386Y > GFP-Rab7 animals, fillet dissections of L3 larvae were performed in modified HL3 solution (70 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 20 mM MgCl2, 5 mM trehalose [Merck KGaA, 108216], 115 mM sucrose [Sigma-Aldrich, S1888], 0.5 mM EGTA, 5 mM HEPES, pH 7.2). The VNC was imaged directly in the dissection dish using a LSM710 confocal microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) equipped with a 40x/1.0 dipping objective.
Salivary gland and fat body lysotracker red staining
Salivary glands and fat body tissue was stained in 1 µM LysoTracker Red DND-99 (Invitrogen, L7528) in PBS for 10 min at RT. After 2 rinses in PBS, the tissues were mounted for imaging as described above.
Garland cell fluorescent tracer uptake and imaging
Proventriculi with attached garland cells were dissected out of L3 larvae in PBS. The tissue was pulsed with 80 µg/ml Texas Red or Alexa Fluor 488-conjugated Avidin (Life Technologies, A820 and A21370) in Schneider medium with 5% heat-inactivated fetal bovine serum (FBS) for 3 min at RT. This was followed by 4 rinses in PBS and a chase of 2 h (or a different duration depending on the experiment) in Schneider medium with 5% FBS at RT. The tissue was then mounted for imaging in PBS as described for salivary glands. For TR-Avidin uptake without chase (0 min chase time), garland cells were mounted for imaging directly in 80 µg/ml TR-Avidin dissolved in Schneider medium with 5% FBS. Garland cells that had taken up TR-Avidin were stained in 1 µM LysoTracker Green DND-26 (Invitrogen, L7526) after the 2 h chase as described above. For sequential tracer uptake, garland cells were first pulsed with TR-Avidin and chased for 2 h, then pulsed with A488-Avidin and chased for an additional 2 h before being mounted for imaging. Garland cells were imaged live except for HA staining experiment shown in ).
DQ-BSA proteolysis assay
Garland cells were pulsed for 3 min at RT with a mix of 80 µg/ml Alexa Fluor 647 BSA (Life Technologies, A34785) and 80 µg/ml DQ Green BSA (Life Technologies, D12050) in Schneider medium without fetal bovine serum and chased for 2 h at RT, before being mounted for imaging in PBS as described. Confocal stacks of garland cell clusters were acquired using a 10x/0.3 objective.
Microscopy
All microscopy was carried out at the Core Facility for Integrated Microscopy (Department of Biomedical Sciences, University of Copenhagen). Confocal imaging was performed on LSM 710 and LSM 780 confocal microscopes (Zeiss), using the following objectives: Plan-Apochromat 63x/1.4 Oil DICIII, W Apochromat 421462–9900 40x/1.0 Water dipping, EC Plan-NEOFLUAR 10x/0.3.
Western blotting
For western blotting, late L3 larval CNS with imaginal discs, mouthparts, esophagus, pro-ventriculus and garland cells attached were dissected out in PBS and stored in Schneider medium with 5% FBS prior to freezing. Upon thawing, samples were immediately boiled in loading buffer, run on SDS-PAGE gels and electroblotted to PVDF membranes. Each sample consisted of pooled tissues from 5 individuals. Membranes were probed with rabbit anti-Cp1/CathL/Cathepsin L (1:500; Abcam, ab58991) and mouse anti-Fas2/Fasciclin II (1:25; Developmental Studies Hybridoma Bank, 1D4, cell culture supernatant), and then with HRP-conjugated donkey anti-rabbit (1:5000; Thermo Fisher scientific, 31458) and HRP-conjugated goat anti-mouse (1:5000) (Thermo Fisher scientific, 31430). Blots were developed using Amersham ECL Prime Western Blotting Detection Reagent (RPN2232) and the resulting chemiluminescence imaged with an Alpha Innotech MultiImage Light Cabinet equipped with a CCD camera (Proteinsimple, San Jose, CA, USA).
Image processing
Analysis of confocal images was performed using pre-existing plugins or custom-designed macros in the Fiji distribution of the ImageJ software [Citation96], supplemented by calculations in Excel (Microsoft). Details of the macros are available upon request. Background subtraction was performed by employing the Fiji rolling ball plugin or after measuring the signal intensity in an empty area located outside the specimen. To the largest extent possible, thresholding was done objectively using a fixed percentile (specified below) of the signal intensity distribution.
GFP-LAMP in the CNS
To quantify the GFP-LAMP signal in the larval CNS of controls, Rab2Δ1 mutants, and rescuants, sum projections of image stacks were background-subtracted and smoothed (Gaussian blur, 0.1 µm). The 99th percentile of the pixel intensity distribution was set as threshold, before obtaining the total intensity of suprathreshold GFP-LAMP-positive areas (lower size cutoff 0.25 µm2) using the particle analysis feature in Fiji. Essentially the same procedure was used for the experimental series involving controls, Rab2Δ1, Vps39Δ1/Df, lt11/Df, Rab2Δ1; Vps39Δ1, and Giee00336/Df.
YFP-Rab7 and GFP-Rab7 in the larval CNS
To obtain size distributions of YFP-Rab7-positive granules in the CNS of Rab2Δ1 mutants and controls, maximum projections of 3 optical slices from the middle of stacks of third instar VNCs were background-subtracted and smoothed. An intensity threshold was set to vary from 200 to 11,800 with an increment of 200. Each threshold setting was followed by a particle analysis (lower size cutoff 0.1 µm2; circularity 0.8–1.0). The particle size distribution corresponding to the threshold resulting in the largest number of particles was evaluated. To assess the distribution of GFP-Rab7 in peptidergic neurons in the larval VNC, a blinded analysis (genotype unknown to the evaluator) was carried out. GFP-Rab7-positive granules were outlined by placing the Fiji wand tool at local intensity maxima in average projections of substacks (5 slices) close to the ventral surface of the VNC.
GFP-LAMP in garland cells
To quantify the internalization of GFP-LAMP in the enlarged endosomal vacuoles of Rab2Δ1, Vps39Δ1/Df, lt11/Df, Rab2Δ1; Vps39Δ1, and Giee00336 garland cells, ROIs corresponding to individual vacuoles were generated using particle analysis (lower size cutoff 1 µm2) on the thresholded red (tracer) channel. After switching to the green channel, the mean GFP-LAMP signal intensity in each vacuolar ROI was divided by the mean intensity in a 0.2 µm wide band surrounding the individual ROIs, yielding the density ratio. After automatic thresholding (95th percentile), particle analysis restricted to perivacuolar bands (witdth 1 µm) was employed to detect GFP-LAMP-positive vesicles (circularity 0.8 to 1.0, area > 0.1 µm2) surrounding the vacuoles.
LAMP and lysotracker red colocalization in salivary glands and fat body
To quantify the fraction of GFP-LAMP signal located outside LysoTracker Red-positive organelles in salivary glands, ROIs with similar signal intensities across preparations were selected. Following smoothing of the GFP-LAMP channel, both channels were thresholded at the 97.5th percentile. This generally led to inclusion of the vesicular aspect of the tubulo-vacuolar network, while leaving out the tubular system. The total intensity of pixel values exceeding the threshold in the GFP-LAMP channel but not in the LysoTracker Red channel, divided by the total intensity of all pixels with intensities above the GFP-LAMP threshold, was used as readout. Quantification of the intensity and/or size of LysoTracker Red-positive and GFP-LAMP-positive structures was done with particle analysis (lower size cutoff 0.5 µm2). In fat bodies, the Pearson correlation coefficient (PCC) for LAMP versus LysoTracker Red was obtained from a square ROI (225*225 µm2).
Lysotracker green and tr-avidin in garland cells
To quantify the LysoTracker Green signal in garland cell vacuoles, individual vacuoles were mapped for intensity measurement using a semi-automated approach (lower size cutoff 0.35 µm2). For both LysoTracker Green and TR-avidin, a threshold equal to 7.5% of the maximal vacuole labeling intensity in the analyzed cell was used to classify each vacuole as either labeled or not.
PCC calculation in garland cells
Using a macro capable of processing irregularly shaped ROIs the PCC was calculated for each cell, before averaging over all cells to obtain a single PCC value representing the whole preparation.
Eye disc autophagy
To determine the ratio between the GFP and mCherry signals emitted by the GFP-mCherry-Atg8a reporter in eye discs, both channels were background-subtracted before applying particle analysis on the red (mCherry) channel to map areas with pixel intensities exceeding the 95th percentile. For each area, the ratio between the mean intensity in the green (GFP) and red channels was calculated, with the average of this ratio representing one disc.
Garland cell proteolysis
Proventriculi with garland cells attached were imaged as stacks in toto at low magnification, and the ratio of the total intensities of the DQ-BSA and A647-BSA channels in background-subtracted sum projections was evaluated.
Statistics
Analysis of variance (ANOVA), followed by the Tukey HSD (honest significant difference) test or the Scheffé test, was performed using JMP (SAS Institute, Inc.). Student’s t tests and the Kolmogorov-Smirnov test were performed in Excel. When appropriate, data were transformed to a normal distribution by taking the logarithm or the square root before performing statistical tests [Citation97], or nonparametric tests were used (http://www.vassarstats.net/). The level of statistical significance was set to 5%. Data are presented as mean ± s.e.m unless specified otherwise in the legend.
Abbreviations
| ANOVA | = | analysis of variance |
| AP-3 complex | = | adaptor protein 3 complex |
| Atg | = | autophagy-related |
| BSA | = | bovine serum albumin |
| CNS | = | central nervous system |
| Cp1 | = | cysteine proteinase-1 |
| CRISPR | = | clustered regularly interspaced short palindromic repeats |
| CV | = | coefficient of variation |
| Df | = | deficiency |
| ER | = | endoplasmic reticulum |
| FBS | = | fetal bovine serum |
| GAP | = | GTPase-activating protein |
| GDP | = | guanosine-5ʹ-diphosphate |
| GEF | = | guanine nucleotide exchange factor |
| GFP | = | green fluorescent protein |
| Gie | = | GTPase indispensable for equal segregation of chromosomes |
| GTP | = | guanosine-5ʹ-triphosphate |
| HA | = | hemagglutinin |
| HOPS | = | homotypic fusion and vacuole protein sorting |
| HSD | = | honest significant difference |
| LAMP | = | lysosomal-associated membrane protein |
| LE | = | late endosomes |
| LMP | = | lysosomal membrane proteins |
| LRO | = | lysosome-related organelle |
| LTG | = | LysoTracker Green |
| LTR | = | LysoTracker Red |
| PBS | = | phosphate-buffered saline |
| PBX | = | PBS with Triton X-100 |
| PLEKHM | = | pleckstrin homology domain-containing family M member |
| PCC | = | Pearson correlation coefficient |
| Rab | = | ras-related in brain |
| RILP | = | Rab-interacting lysosomal protein |
| SKIP | = | SifA and kinesin-interacting |
| SNARE | = | endosomal and lysosomal soluble NSF attachment protein receptor |
| TGN | = | trans-Golgi network |
| TR | = | Texas Red |
| V-ATPase | = | vacuolar-type proton-translocating ATPase |
| VAMP | = | vesicle-associated membrane protein |
| VNC: | = | ventral nerve cord |
| Vps | = | vacuolar protein sorting |
| YFP | = | yellow fluorescent protein. |
Supplemental Material
Download Zip (13.8 MB)Acknowledgments
We thank the Bloomington Stock Center, the Vienna Drosophila Resource Center, and the following colleagues for fly lines and antibodies: Julie Brill, Suzanne Eaton, Gábor Juhász, Helmut Krämer, Tor Erik Rusten, Paul Taghert, and Benjamin White. We are grateful to Ulrik Gether for helpful discussions and Jacob Balslev Sørensen and Stefan Kins for critical reading of the manuscript. Anders Bohl Pedersen and the staff at the Core Facility for Integrated Microscopy provided excellent technical assistance. We thank the developers of the ImageJ/Fiji image processing package, and Richard Lowry for maintaining the Vassarstats website for statistical computation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemenatry material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009 Sep;10(9):623–635. PubMed PMID: 19672277.
- Settembre C, Fraldi A, Medina DL, et al. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013 May;14(5):283–296. PubMed PMID: 23609508; PubMed Central PMCID: PMC4387238.
- Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007 Aug;8(8):622–632. PubMed PMID: 17637737.
- Schwake M, Schroder B, Saftig P. Lysosomal membrane proteins and their central role in physiology. Traffic. 2013 Jul;14(7):739–748. PubMed PMID: 23387372.
- Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001 Jul 27;106(2):157–169. PubMed PMID: 11511344.
- Repnik U, Stoka V, Turk V, et al. Lysosomes and lysosomal cathepsins in cell death. Biochim Biophys Acta. 2012 Jan;1824(1):22–33. PubMed PMID: s21914490.
- Arantes RM, Andrews NW. A role for synaptotagmin VII-regulated exocytosis of lysosomes in neurite outgrowth from primary sympathetic neurons. J Neurosci. 2006 Apr 26;26(17):4630–4637. PubMed PMID: 16641243.
- Rink J, Ghigo E, Kalaidzidis Y, et al. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005 Sep 9;122(5):735–749. PubMed PMID: 16143105.
- Poteryaev D, Datta S, Ackema K, et al. Identification of the switch in early-to-late endosome transition. Cell. 2010 Apr 30;141(3):497–508. PubMed PMID: 20434987.
- Nordmann M, Cabrera M, Perz A, et al. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010 Sep 28;20(18):1654–1659. PubMed PMID: 20797862.
- Bright NA, Gratian MJ, Luzio JP. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol. 2005 Feb 22;15(4):360–365. PubMed PMID: 15723798.
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013 Dec;14(12):759–774. PubMed PMID: 24201109.
- Bright NA, Davis LJ, Luzio JP. Endolysosomes are the principal intracellular sites of acid hydrolase activity. Curr Biol. 2016 Sep 12;26(17):2233–2245. PubMed PMID: 27498570; PubMed Central PMCID: PMCPMC5026700.
- Nakamura N, Hirata A, Ohsumi Y, et al. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997 Apr 25;272(17):11344–11349. PubMed PMID: 9111041.
- Hickey CM, Stroupe C, Wickner W. The major role of the Rab Ypt7p in vacuole fusion is supporting HOPS membrane association. J Biol Chem. 2009 Jun 12;284(24):16118–16125. PubMed PMID: 19386605; PubMed Central PMCID: PMC2713515.
- Wickner W. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. PubMed PMID: 20521906.
- Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J Cell Sci. 2013 Mar 15;126(Pt 6):1307–1316. PubMed PMID: 23645161.
- Solinger JA, Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013 Jun;280(12):2743–2757. PubMed PMID: 23351085.
- Spang A. Membrane tethering complexes in the endosomal system. Front Cell Dev Biol. 2016;4:35. PubMed PMID: 27243003; PubMed Central PMCID: PMCPMC4860415.
- Brocker C, Kuhlee A, Gatsogiannis C, et al. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci U S A. 2012 Feb 7;109(6):1991–1996. PubMed PMID: 22308417; PubMed Central PMCID: PMCPMC3277535.
- Baker RW, Jeffrey PD, Zick M, et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015 Sep 4;349(6252):1111–1114. PubMed PMID: 26339030; PubMed Central PMCID: PMC4727825.
- van der Kant R, Jonker CT, Wijdeven RH, et al. Characterization of the Mammalian CORVET and HOPS complexes and their modular restructuring for endosome specificity. J Biol Chem. 2015 Dec 18;290(51):30280–30290. PubMed PMID: 26463206; PubMed Central PMCID: PMCPMC4683254.
- Lin X, Yang T, Wang S, et al. RILP interacts with HOPS complex via VPS41 subunit to regulate endocytic trafficking. Sci Rep. 2014;4:7282. PubMed PMID: 25445562; PubMed Central PMCID: PMCPMC4250914.
- van der Kant R, Fish A, Janssen L, et al. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci. 2013 Aug 1;126(Pt 15):3462–3474. PubMed PMID: 23729732.
- Khatter D, Raina VB, Dwivedi D, et al. The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J Cell Sci. 2015 May 1;128(9):1746–1761. PubMed PMID: 25908847; PubMed Central PMCID: PMCPMC4432227.
- Pols MS, Ten Brink C, Gosavi P, et al. The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic. 2013 Feb;14(2):219–232. PubMed PMID: 23167963.
- Sriram V, Krishnan KS, Mayor S. deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J Cell Biol. 2003 May 12;161(3):593–607. PubMed PMID: 12743107; PubMed Central PMCID: PMCPMC2172926.
- Akbar MA, Ray S, Kramer H. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol Biol Cell. 2009 Mar;20(6):1705–1714. PubMed PMID: 19158398; PubMed Central PMCID: PMCPMC2655250.
- Swetha MG, Sriram V, Krishnan KS, et al. Lysosomal membrane protein composition, acidic pH and sterol content are regulated via a light-dependent pathway in metazoan cells. Traffic. 2011 Aug;12(8):1037–1055. PubMed PMID: 21535339.
- Takats S, Pircs K, Nagy P, et al. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014 Apr;25(8):1338–1354. PubMed PMID: 24554766; PubMed Central PMCID: PMCPMC3982998.
- Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014 Apr;25(8):1327–1337. PubMed PMID: 24554770; PubMed Central PMCID: PMCPMC3982997.
- Braulke T, Bonifacino JS. Sorting of lysosomal proteins. Biochim Biophys Acta. 2009 Apr;1793(4):605–614. PubMed PMID: 19046998.
- Pols MS, van Meel E, Oorschot V, et al. hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nat Commun. 2013;4:1361. PubMed PMID: 23322049.
- Ihrke G, Kyttala A, Russell MR, et al. Differential use of two AP-3-mediated pathways by lysosomal membrane proteins. Traffic. 2004 Dec;5(12):946–962. PubMed PMID: 15522097.
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009 Aug;10(8):513–525. PubMed PMID: 19603039.
- Tisdale EJ, Jackson MR. Rab2 protein enhances coatomer recruitment to pre-Golgi intermediates. J Biol Chem. 1998 Jul 3;273(27):17269–17277. PubMed PMID: 9642298.
- Aizawa M, Fukuda M. Small GTPase Rab2B and its specific binding protein golgi-associated Rab2B Interactor-like 4 (GARI-L4) regulate golgi morphology. J Biol Chem. 2015 Sep 4;290(36):22250–22261. PubMed PMID: 26209634; PubMed Central PMCID: PMCPMC4571976.
- Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011 Jun;3(6). PubMed PMID: 21436057; PubMed Central PMCID: PMC3098672.
- Ailion M, Hannemann M, Dalton S, et al. Two Rab2 interactors regulate dense-core vesicle maturation. Neuron. 2014 Apr 2;82(1):167–180. PubMed PMID: 24698274; PubMed Central PMCID: PMCPMC3997996.
- Sumakovic M, Hegermann J, Luo L, et al. UNC-108/RAB-2 and its effector RIC-19 are involved in dense core vesicle maturation in Caenorhabditis elegans. J Cell Biol. 2009 Sep 21;186(6):897–914. PubMed PMID: 19797081; PubMed Central PMCID: PMCPMC2753160.
- Edwards SL, Charlie NK, Richmond JE, et al. Impaired dense core vesicle maturation in Caenorhabditis elegans mutants lacking Rab2. J Cell Biol. 2009 Sep 21;186(6):881–895. PubMed PMID: 19797080; PubMed Central PMCID: PMCPMC2753164.
- Mangahas PM, Yu X, Miller KG, et al. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol. 2008 Jan 28;180(2):357–373. PubMed PMID: 18227280; PubMed Central PMCID: PMCPMC2213587.
- Guo P, Hu T, Zhang J, et al. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc Natl Acad Sci U S A. 2010 Oct 19;107(42):18016–18021. PubMed PMID: 20921409; PubMed Central PMCID: PMC2964220.
- Garg A, Wu LP. Drosophila Rab14 mediates phagocytosis in the immune response to Staphylococcus aureus. Cell Microbiol. 2014 Feb;16(2):296–310. PubMed PMID: 24119134; PubMed Central PMCID: PMC4120862.
- Maringer K, Saheb E, Bush J. Vesicular trafficking defects, developmental abnormalities, and alterations in the cellular death process occur in cell lines that over-express dictyostelium GTPase, Rab2, and Rab2 Mutants. PubMed PMID: 25157910; PubMed Central PMCID: PMC4192625 Biology (Basel). 2014;33:514–535.
- Lu Q, Zhang Y, Hu T, et al. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 2008 Mar;135(6):1069–1080. PubMed PMID: 18256195.
- Chun DK, McEwen JM, Burbea M, et al. UNC-108/Rab2 regulates postendocytic trafficking in Caenorhabditis elegans. Mol Biol Cell. 2008 Jul;19(7):2682–2695. PubMed PMID: 18434599; PubMed Central PMCID: PMC2441656.
- Gillingham AK, Sinka R, Torres IL, et al. Toward a comprehensive map of the effectors of rab GTPases. Dev Cell. 2014 Nov 10;31(3):358–373. PubMed PMID: 25453831; PubMed Central PMCID: PMCPMC4232348.
- Pulipparacharuvil S, Akbar MA, Ray S, et al. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci. 2005 Aug 15;118(Pt 16):3663–3673. PubMed PMID: 16046475.
- Wakimoto BT, Hearn MG. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics. 1990 May;125(1):141–154. PubMed PMID: 2111264; PubMed Central PMCID: PMC1203996.
- Rusten TE, Lindmo K, Juhasz G, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004 Aug;7(2):179–192. PubMed PMID: 15296715.
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007 Sep-Oct;3(5):452–460. PubMed PMID: 17534139.
- Delahaye JL, Foster OK, Vine A, et al. Caenorhabditis elegans HOPS and CCZ-1 mediate trafficking to lysosome-related organelles independently of RAB-7 and SAND-1. Mol Biol Cell. 2014 Apr;25(7):1073–1096. PubMed PMID: 24501423; PubMed Central PMCID: PMCPMC3967972.
- Wasmeier C, Romao M, Plowright L, et al. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006 Oct 23;175(2):271–281. PubMed PMID: 17043139; PubMed Central PMCID: PMC2064568.
- Ma J, Plesken H, Treisman JE, et al. Lightoid and Claret: a rab GTPase and its putative guanine nucleotide exchange factor in biogenesis of Drosophila eye pigment granules. Proc Natl Acad Sci U S A. 2004 Aug 10;101(32):11652–11657. PubMed PMID: 15289618; PubMed Central PMCID: PMC511034.
- Feng L, Seymour AB, Jiang S, et al. The beta3A subunit gene (Ap3b1) of the AP-3 adaptor complex is altered in the mouse hypopigmentation mutant pearl, a model for Hermansky-Pudlak syndrome and night blindness. Hum Mol Genet. 1999 Feb;8(2):323–330. PubMed PMID: 9931340.
- Dell’Angelica EC, Shotelersuk V, Aguilar RC, et al. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999 Jan;3(1):11–21. PubMed PMID: 10024875.
- Warner TS, Sinclair DA, Fitzpatrick KA, et al. The light gene of Drosophila melanogaster encodes a homologue of VPS41, a yeast gene involved in cellular-protein trafficking. Genome. 1998 Apr;41(2):236–243. PubMed PMID: 9644832.
- Shestopal SA, Makunin IV, Belyaeva ES, et al. Molecular characterization of the deep orange (dor) gene of Drosophila melanogaster. Mol Gen Genet. 1997 Feb 20;253(5):642–648. PubMed PMID: 9065698.
- Lloyd V, Ramaswami M, Kramer H. Not just pretty eyes: drosophila eye-colour mutations and lysosomal delivery. Trends Cell Biol. 1998 Jul;8(7):257–259. PubMed PMID: 9714595.
- Sevrioukov EA, He JP, Moghrabi N, et al. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999 Oct;4(4):479–486. PubMed PMID: 10549280.
- Mullins C, Hartnell LM, Wassarman DA, et al. Defective expression of the mu3 subunit of the AP-3 adaptor complex in the Drosophila pigmentation mutant carmine. Mol Gen Genet. 1999 Oct;262(3):401–412. PubMed PMID: 10589826.
- Mullins C, Hartnell LM, Bonifacino JS. Distinct requirements for the AP-3 adaptor complex in pigment granule and synaptic vesicle biogenesis in Drosophila melanogaster. Mol Gen Genet. 2000 Jul;263(6):1003–1014. PubMed PMID: 10954086.
- Weavers H, Prieto-Sanchez S, Grawe F, et al. The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature. 2009 Jan 15;457(7227):322–326. PubMed PMID: 18971929; PubMed Central PMCID: PMCPMC2687078.
- Chang HC, Hull M, Mellman I. The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J Cell Biol. 2004 Mar 29;164(7):1055–1064. PubMed PMID: 15051737; PubMed Central PMCID: PMCPMC2172058.
- Lloyd TE, Atkinson R, Wu MN, et al. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002 Jan 25;108(2):261–269. PubMed PMID: 11832215.
- Koenig JH, Ikeda K. Transformational process of the endosomal compartment in nephrocytes of Drosophila melanogaster. Cell Tissue Res. 1990 Nov;262(2):233–244. PubMed PMID: 2127554.
- Kosaka T, Ikeda K. Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J Cell Biol. 1983 Aug;97(2):499–507. PubMed PMID: 6411734; PubMed Central PMCID: PMC2112522.
- Lorincz P, Lakatos Z, Varga A, et al. MiniCORVET is a Vps8-containing early endosomal tether in Drosophila. Elife. 2016;5. PubMed PMID: 27253064; PubMed Central PMCID: PMC4935465. DOI:10.7554/eLife.14226
- Piper RC, Whitters EA, Stevens TH. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post-Golgi transport vesicles. Eur J Cell Biol. 1994 Dec;65(2):305–318. PubMed PMID: 7720726.
- Hofmann I, Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J Cell Sci. 2006 Apr 15;119(Pt 8):1494–1503. PubMed PMID: 16537643.
- Thibault ST, Singer MA, Miyazaki WY, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004 Mar;36(3):283–287. PubMed PMID: 14981521.
- Vazquez CL, Colombo MI. Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol. 2009;452:85–95. PubMed PMID: 19200877.
- Juhasz G, Erdi B, Sass M, et al. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007 Dec 1;21(23):3061–3066. PubMed PMID: 18056421; PubMed Central PMCID: PMCPMC2081972.
- Mukherjee A, Patel B, Koga H, et al. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 2016 Nov;12(11):1984–1999. PubMed PMID: 27487474; PubMed Central PMCID: PMCPMC5103356.
- Sinka R, Gillingham AK, Kondylis V, et al. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol. 2008 Nov 17;183(4):607–615. PubMed PMID: 19001129; PubMed Central PMCID: PMCPMC2582897.
- Diao A, Rahman D, Pappin DJ, et al. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J Cell Biol. 2003 Jan 20;160(2):201–212. PubMed PMID: 12538640; PubMed Central PMCID: PMCPMC2172652.
- Short B, Preisinger C, Korner R, et al. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001 Dec 10;155(6):877–883. PubMed PMID: 11739401; PubMed Central PMCID: PMCPMC2150909.
- Lorincz P, Toth S, Benko P, et al. Rab2 promotes autophagic and endocytic lysosomal degradation. J Cell Biol. 2017 Jul 03;216(7):1937–1947. PubMed PMID: 28483915; PubMed Central PMCID: PMCPMC5496615.
- Cowles CR, Snyder WB, Burd CG, et al. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997 May 15;16(10):2769–2782. PubMed PMID: 9184222; PubMed Central PMCID: PMCPMC1169886.
- Angers CG, Merz AJ. HOPS interacts with Apl5 at the vacuole membrane and is required for consumption of AP-3 transport vesicles. Mol Biol Cell. 2009 Nov;20(21):4563–4574. PubMed PMID: 19741093; PubMed Central PMCID: PMCPMC2770944.
- Cabrera M, Langemeyer L, Mari M, et al. Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J Cell Biol. 2010 Nov 15;191(4):845–859. PubMed PMID: 21079247; PubMed Central PMCID: PMCPMC2983053.
- Lafourcade C, Sobo K, Kieffer-Jaquinod S, et al. Regulation of the V-ATPase along the endocytic pathway occurs through reversible subunit association and membrane localization. PLoS One. 2008 Jul 23;3(7):e2758. PubMed PMID: 18648502; PubMed Central PMCID: PMCPMC2447177.
- Klionsky DJ, Elazar Z, Seglen PO, et al. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008 Oct;4(7):849–850. PubMed PMID: 18758232.
- Mauvezin C, Nagy P, Juhasz G, et al. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. 2015 May 11;6:7007. PubMed PMID: 25959678; PubMed Central PMCID: PMCPMC4428688.
- Yousefian J, Troost T, Grawe F, et al. Dmon1 controls recruitment of Rab7 to maturing endosomes in Drosophila. J Cell Sci. 2013 Apr 1;126(Pt 7):1583–1594. PubMed PMID: 23418349; PubMed Central PMCID: PMC3647437.
- Marwaha R, Arya SB, Jagga D, et al. The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes. J Cell Biol. 2017 Apr 03;216(4):1051–1070. PubMed PMID: 28325809; PubMed Central PMCID: PMCPMC5379943.
- McEwan DG, Popovic D, Gubas A, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015 Jan 08;57(1):39–54. PubMed PMID: 25498145.
- Fujita N, Huang W, Lin TH, et al. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. Elife. 2017 Jan 07:6. PubMed PMID: 28063257; PubMed Central PMCID: PMCPMC5249261. DOI: 10.7554/eLife.23367
- Kajiho H, Kajiho Y, Frittoli E, et al. RAB2A controls MT1-MMP endocytic and E-cadherin polarized Golgi trafficking to promote invasive breast cancer programs. EMBO Rep. 2016 Jul;17(7):1061–1080. PubMed PMID: 27255086; PubMed Central PMCID: PMC4931572.
- Lund VK, DeLotto Y, DeLotto R. A set of P-element transformation vectors permitting the simplified generation of fluorescent fusion proteins in Drosophila melanogaster. Fly (Austin). 2011 Jul-Sep;5(3):255–260. PubMed PMID: 21406969.
- Marois E, Mahmoud A, Eaton S. The endocytic pathway and formation of the Wingless morphogen gradient. Development. 2006 Jan;133(2):307–317. PubMed PMID: 16354714.
- Hewes RS, Park D, Gauthier SA, et al. The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development. 2003 May;130(9):1771–1781. PubMed PMID: 12642483.
- Gratz SJ, Ukken FP, Rubinstein CD, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014 Apr;196(4):961–971. PubMed PMID: 24478335; PubMed Central PMCID: PMC3982687.
- Riedel F, Gillingham AK, Rosa-Ferreira C, et al. An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol Open. 2016 Jul 15;5(7):987–992. PubMed PMID: 27256406; PubMed Central PMCID: PMCPMC4958275.
- Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012 Jul;9(7):676–682. PubMed PMID: 22743772; PubMed Central PMCID: PMC3855844.
- Altman DG. Practical statistics for medical research. 1st ed. London; New York: Chapman and Hall; 1991.
